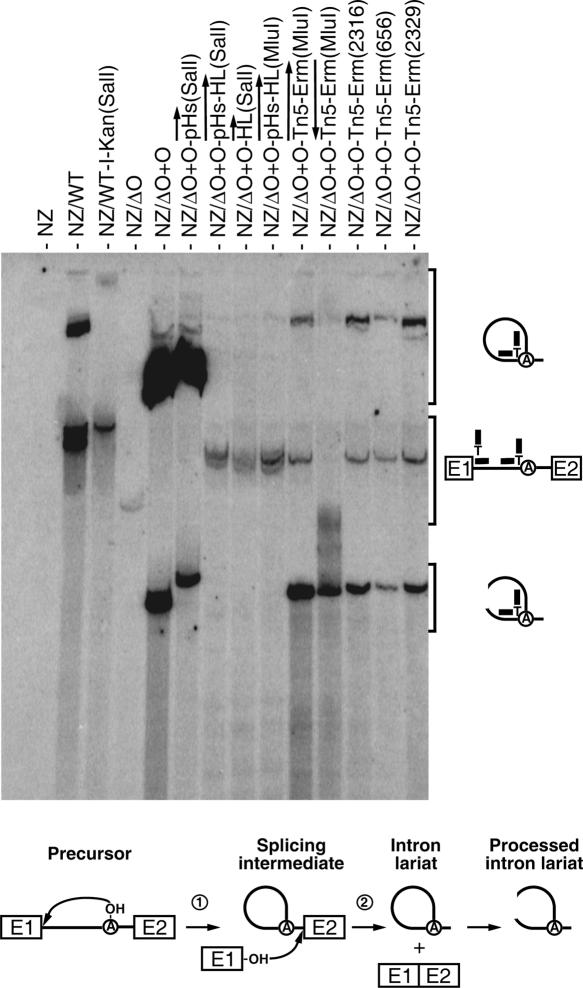
FIGURE 5.
Ll.LtrB lariat accumulation. Northern blot hybridizations were performed using total RNA extracted from NZ9800 L. lactis cells, which were induced with nisin for 3 h to express different variants of Ll.LtrB. The branch point probe used recognizes spliced Ll.LtrB lariats by base pairing with the 5′ extremity of the intron (12 nt) and the sequence upstream from the branch site (12 nt). The branch point probe (thick line), the intron (thin line), and exons (E1, E2) are depicted. The position of intron lariats, processed intron lariats, and precursor mRNA are denoted. A schematic of the Ll.LtrB splicing pathway is presented (bottom). The initial nucleophilic attack is made by the 2′ OH group of an internal adenosine residue (circled A, branch point) (step 1). The liberated 3′ OH group of exon I (E1) then attacks the phosphodiester bond located at the intron–exon II junction. This leads to the ligation of the two exons and liberation of the intron lariat (step 2). After splicing, the intron lariat accumulates and may be processed to create opened circular structures. (NZ) NZ9800, (WT) wild-type Ll.LtrB, (WT-I-Kan) wild-type Ll.LtrB harboring the td group I intron and a kanamycin resistance gene, (ΔO+O) Ll.LtrBΔORF+ORF construct. The position of the cargo within Ll.LtrB is denoted between parentheses.