Identifying a natural reservoir for Ebola virus has eluded researchers for decades [1,2]. Recently, Leroy et al. presented the most compelling evidence to date that three species of fruit bats (Hypsignathus monstrosus, Epomops franqueti, and Myonycteris torquata) may constitute a long-missing wildlife reservoir for Ebola virus Zaire (EBOVZ) [3]. These bats, caught near affected villages at the Gabon–Congo border, appear to have been asymptomatically infected and, in seven cases, yielded virus sequences that closely matched those found in the human outbreaks happening about the same time. Leroy et al.'s phylogenetic analysis of the partial sequences of the viral polymerase (L) gene derived from humans and bats emphasized the interspecific relationships to related filoviruses. Here, we show that (1) despite their short length (265 bp), these sequences also provide critical information about the intraspecific history of EBOVZ, and (2) based on the genetic data available so far, the association of the virus with fruit bats in the sampled area can only be traced back a few years.
Consistent with previous analysis using glycoprotein (GP) gene sequences [4], results for the L gene show that viruses amplified from more recently collected samples appear to be direct descendents of viruses seen during previous outbreaks. This relationship is not only apparent for viruses found in 1976–1995 compared with those found in 2001–2003, but also within the latter group (Figure 1). In essence, this means that all genetic variation seen thus far in EBOVZ, including virus amplified from fruit bats, appears to be the product of mutations that have accumulated within the last 30 years. Finding such strong evidence for temporal structure by chance seems highly unlikely, especially given the concordance with our earlier results from the GP gene [4]. Although the lack of any mutational differences between the sequences Mayinga 1976 and Kikwit 1995 is perplexing in this context, it is most likely a stochastic artifact due to the short length of the sequence considered. Full-length sequences are available for both these isolates, which over the entire genome are 1.2% different. Over 19 years this yields an ad hoc evolutionary rate estimate of 6.2 × 10−4 substitutions per site per year, close to the rate we had previously estimated for the GP gene (~8.0 × 10−4) [4] and to the point estimate for all partial L sequences in the current analysis (1.1 × 10−3; 95% highest posterior density interval: 6.3 × 10−7 to 2.4 × 10−3). Thus, even though the L sequences are rather short, they yield evolutionary rate estimates similar to the longer GP sequences.
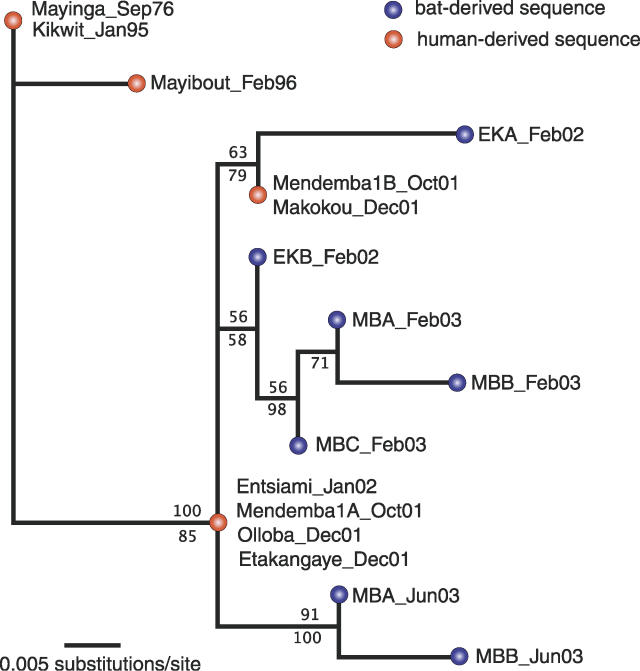
Figure 1. Maximum Likelihood Tree from Partial L Sequences of Ebola Virus Zaire.
Tree was obtained in PAUP* 4.0b10.8 [7] under a HKY + I model. Values above branches represent percent support based on 1,000 maximum likelihood bootstrap trees; values below branches represent posterior probabilities from a complementary Bayesian analysis in BEAST [8]. For details regarding methods (including tree rooting), see Walsh et al. [4]. Year and month of sampling is given for each sequence.
The temporal structure visible in the L gene genealogy implies that all viruses sampled from both humans and bats between 2001 and 2003 can be traced back to a very recent common ancestor, by which we mean a recent coalescence of genetic lineages, not an ancestral alternative reservoir species. In fact, according to our phylogenetic estimate, the partial L sequence of this genetic ancestor would have looked identical to that sampled from infected humans during outbreaks in late 2001 and early 2002 (Entsiami and Mendemba, Figure 1), suggesting that this ancestor could not be much older. This is in agreement with the previous analysis of the GP gene, which indicated that all viruses sampled from outbreaks since 2001 had a most recent common ancestor in 1999 (confidence region: 1998–2000) [4]. While these findings do not question whether fruit bats may represent a wild reservoir for EBOVZ, they do raise important issues.
If the three identified fruit bat hosts were the natural reservoir for EBOVZ, the recent common ancestry of all sequences derived from them so far is surprising because, at least at first sight, it seems to contradict the idea of a long-established association of bat and virus. The most reasonable explanation for this result is that the virus experienced a recent genetic bottleneck. We present three alternative scenarios of what could have caused such a bottleneck.
One possibility is that somewhere around 1999, the total number of infected bats within the sampled area became extremely small (likely much less than the peak 23% incidence determined by Leroy et al.). Under such a scenario, the most recent common ancestor could not be traced back further into the past because an extremely small, effective viral population size has caused the descendents of all but one of the previously existing viral lineages to be lost. Since no trapping study on bats was undertaken before 2001, we could not directly address this issue. However, some epidemiological and virological observations may account for this situation. The need to apply the very sensitive nested PCR to detect viral RNA suggests a very low viral load in organs of infected bats. Furthermore, the presence of a high prevalence rate of seropositive bats (16.7%, 4/24) compared with only 3.2% (2/63) that were PCR positive (but seronegative) just three months after the appearance of the first human cases in Mendemba, Gabon, indicates that viral replication within bats may be highly restricted and possibly only taking place prior to the onset of the host immune response. Especially if infections are synchronized, for example, by some environmental trigger, this may lead to periods with an extremely small number of productively infected bats, repeatedly forcing the virus population through a genetic bottleneck.
Alternatively, the recent common ancestor could be explained by infected bats introducing the virus into the EBOVZ-affected area of Gabon and Congo around 1999. Previous results for the GP gene actually support this hypothesis by revealing a consistent signature of geographic spread within the spatial, temporal, and genetic data for EBOVZ over the last 30 years [4]. Similar genetic patterns associated with local founder events followed by spatial spread have also been documented from rabies virus in wildlife host populations [5]. Some observed epidemiological changes in sampled bat populations over time may also support this hypothesis. Leroy et al. found that during the first visit to one of their sampling locations, 23% (7/31) of the bats were PCR positive, whereas 0% (0/10) were seropositive. At a second visit five months later, these numbers had changed to 2% (4/184) and 8% (12/160) [3]. Again, no bats were positive by both PCR and serology. Though other factors may also explain these opposing trends, the observed temporal pattern is consistent with an infection wave moving through the sampled population, resulting in a high proportion of infectious individuals at first, followed by an increased proportion of seropositive animals.
Given that the three implicated fruit bat species may not be the only reservoir for EBOVZ, as Leroy et al. were already careful to point out [3], another possible explanation for the existence of a common viral ancestor in the recent past is that the virus was introduced to these fruit bats around the same time it affected other wildlife populations and emerged in humans. It is important to note that this scenario does not rule out bats as reservoir species, a hypothesis for which there is additional independent support [6]. Instead, it would imply that the primary reservoir of EBOVZ, whether it involves additional bat species or representatives of other taxonomic groups, has yet to be found.
We expect that distinguishing between these possible scenarios will become increasingly easier as more temporal, spatial, and genetic data are generated. Additional viral sequences from infected fruit bats and large-scale serological prevalence in bat populations both within and outside the affected area should give some much needed answers regarding the dynamics of the virus in its wild reservoir. Together with other viral sequences in human cases and vulnerable animal species, and a better understanding of the factors associated with its emergence in human and wildlife populations, these combined approaches will hopefully lead to new and more successful strategies for preventing and controlling outbreaks of EBOVZ in the near future.
Supporting Information
Accession Numbers
The GenBank (http://www.ncbi.nlm.nih.gov/Genbank) accession numbers for the human-derived sequences discussed in this paper are Entsiami (DQ211657), Kikwit (AY354458), Mayibout (DQ205418), Mayinga (AY142960), Mendemba1A (DQ205416, identical to Olloba and Etakangaye), and Mendemba1B (DQ205417, identical to Makokou). Bat-derived sequences are available under DQ205409–DQ205415.
Acknowledgments
We thank an anonymous reviewer for helpful comments.
Abbreviations
- EBOVZ
Ebola virus Zaire
- GP
glycoprotein
- L
polymerase
Footnotes
Roman Biek and Leslie A. Real are at the Center for Disease Ecology, Department of Biology, Emory University, Atlanta, Georgia, United States of America. Peter D. Walsh is at the Max-Planck-Institute for Evolutionary Anthropology, Leipzig, Germany. Eric M. Leroy is at the Institut de Recherche pour le Développement de Franceville (CIRMF), Franceville, Gabon.
Competing interests. The authors have declared that no competing interests exist.
Author contributions. RB, PDW, and LAR conceived and designed the experiments. RB analyzed the data. EML contributed reagents/materials/analysis tools. RB, PDW, EML, and LAR wrote the paper.
Funding. The authors received no specific funding for this article.
References
- Pourrut X, Kumulungui B, Wittmann T, Moussavou G, Delicat A, et al. The natural history of Ebola virus in Africa. Microbes Infect. 2005;7:1005–1014. doi: 10.1016/j.micinf.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Feldmann H, Wahl-Jensen V, Jones SM, Stroher U. Ebola virus ecology: A continuing mystery. Trends Microbiol. 2004;12:433–437. doi: 10.1016/j.tim.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, et al. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- Walsh PD, Biek R, Real LA. Wave-like spread of Ebola Zaire. PLoS Biol. 2005. e371. DOI: 10.1371/journal.pbio.0030371. [DOI] [PMC free article] [PubMed]
- Real LA, Henderson JC, Biek R, Snaman J, Jack TL, et al. Unifying the spatial population dynamics and molecular evolution of epidemic rabies virus. Proc Natl Acad Sci U S A. 2005;102:12107–12111. doi: 10.1073/pnas.0500057102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanepoel R, Leman PA, Burt FJ, Zachariades NA, Braack LE, et al. Experimental inoculation of plants and animals with Ebola virus. Emerg Infect Dis. 1996;2:321–325. doi: 10.3201/eid0204.960407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP* 4.0b10. Phylogenetic analysis using parsimony (*and other methods). 4.0b8 edition. Sunderland (Massachusetts): Sinauer Associates; 2003. [Google Scholar]
- Drummond AJ, Rambaut A. BEAST version 1.3. 2003. Available: http://evolve.zoo.ox.ac.uk/beast. Accessed 24 September 2006.