Abstract
The interaction between mutations in the tumor-suppressor genes Apc and p53 was studied in congenic mouse strains to minimize the influence of polymorphic modifiers. The multiplicity and invasiveness of intestinal adenomas of ApcMin/+ (Min) mice was enhanced by deficiency for p53. In addition, the occurrence of desmoid fibromas was strongly enhanced by p53 deficiency. The genetic modifier Mom1 and the pharmacological agents piroxicam and difluoromethylornithine each reduced intestinal adenoma multiplicity in the absence of p53 function. Mom1 showed no influence on the development of desmoid fibromas, whereas the combination of piroxicam and difluoromethylornithine exerted a moderate effect. The ensemble of tumor suppressors and modifiers of a neoplastic process can be usefully analyzed in respect to tissue specificity and synergy.
Genes of several kinds regulate normal and neoplastic growth in the mammal. Positive and negative regulators can act either cell autonomously or nonautonomously to alter the proliferative potential of the stem cell that establishes the normal or neoplastic lineage. Individual genes in the ensemble can affect the transition between normal and neoplastic growth and/or the maintenance of one of the growth states. One important class of genes has been identified by loss-of-function alleles transmitted through the germline in heterozygous form, each predisposing a carrier to a particular spectrum of neoplasms. In these tumors, it is commonly found that the remaining wild-type allele has been lost or inactivated. The gene in question, commonly called a “tumor suppressor,” is formally a cell-autonomous negative regulator of the neoplastic state.
Humans and mice heterozygous for germline mutations that inactivate the Adenomatous polyposis coli (APC/Apc) gene develop only a limited range of neoplasms, including intestinal adenomas and desmoid fibromas (1). Because this gene is widely expressed (2), this restriction in neoplastic histotype is surprising. Does the limited range of neoplasia reflect cooperation between APC/Apc and other negative regulators?
The p53 gene is a negative regulator that is mutated in a broad range of human neoplasms, but its gene product is constitutively active in only a few cell types (3, 4). For example, the loss of p53 adenocarcinomas in the human intestine, but the protein becomes detectable in this tissue after stress such as ionizing radiation (5). Does an interaction between Apc and p53 affect intestinal neoplasia in experimental models of cancer (6)? Preliminary studies tested whether a lack of p53 activity affected neoplasia in the intestine of Min (multiple intestinal neoplasia) mice heterozygous for the Min nonsense allele of Apc, but no significant effect was observed (7–10). Those studies utilized populations of mice with heterogeneous genetic backgrounds. In this report, we reinvestigate whether a tissue-specific or stage-specific interaction between Apc and p53 affects the spectrum of neoplasms in mice, by using a homogeneous genetic background. A central issue addressed in these experiments is whether the major negative regulators each act singly in controlling neoplasia of a particular histotype, as implied by the “gatekeeper hypothesis” (11).
Beyond these major negative regulators, a number of other factors have been found to influence intestinal neoplasia in humans and mice. Such modifiers can act to either promote or retard tumor growth, thereby affecting tumor multiplicity in experimental models. The genetic modifier Mom1 encodes a secretory phospholipase, Pla2g2a, expressed throughout the intestinal tract. The active allele of Pla2g2a leads to a reduction in the growth rate and multiplicity of intestinal adenomas in the Min mouse. Similarly, pharmacological agents can affect tumorigenesis. Piroxicam, a nonsteroidal antiinflammatory agent that inhibits cyclooxygenase-1 and -2, and difluoromethylornithine (DFMO), a suicide substrate of ornithine decarboxylase, each reduce the multiplicity of intestinal adenomas. The strongest inhibition is seen with a combination of piroxicam and DFMO (12). In the present study, we have asked whether the action of either Mom1 or the piroxicam/DFMO combination is tissue specific. Further, we have asked whether either of these growth-inhibitory actions depends on p53 activity.
Materials and Methods
Mice.
The C57BL/6–p53 congenic mouse strain (B6-p53) was founded by a (129/Sv × B6) F2 female that carried a targeted disruption of the p53 gene (13). Designating the founder female as N1, an expanded N10 intercross population was produced by mating Apc+/+ Mom1R/S p53+/− females to ApcMin/+ Mom1R/S p53+/−, ApcMin/+ Mom1R/S p53−/−, and ApcMin/+ Mom1R/R p53−/− males. Mice were housed as described previously (14).
Drug Treatment.
After weaning at approximately 30 days of age, animals were housed in groups of one to five in microisolator cages under fluorescent lighting on a 12-hr cycle and weighed once per week. Tap water was available ad libitum for the duration of the experiment and was replaced weekly. The mice were treated with the appropriate drug or control vehicle mixed in the defined synthetic AIN-93G diet (Dyets, Bethlehem, PA), then killed after the specified duration of treatment.
The chemoprotective agents were stable for at least 7 days in a standard rodent diet at the concentrations used in these studies. New batches were prepared weekly by thorough mixing of the diet with the indicated doses of drug and were stored until use in sealed containers at 4°C. Fresh diet was added to protected feeders three times weekly and was completely changed after the feeders were emptied once weekly.
Piroxicam (CAS no. 36322–90-4) was purchased from Sigma, and DFMO (CAS no. 70052–12-9) was a gift from Ajit Verma (University of Wisconsin–Madison). Piroxicam at the intended concentration was mixed in the diet beginning at approximately 30 days of age; the mice were killed at age 90 days after 2 mo of treatment. Mice treated with DFMO were given water mixed with the intended concentration of that drug from age 30 to 90 days. When administered to mice after weaning, these cyclooxygenase and decarboxylase inhibitors were not overtly toxic at the doses used.
DNA Preparation.
Mice were anesthetized with ether, and 250 μl of blood was collected from the retroorbital sinus. DNA was isolated from blood as described previously (14).
Genotyping.
Mice were genotyped to identify carriers of the Min allele of Apc, the resistance allele of Mom1, and the knockout allele of p53 with PCR assays as described (14–16). In some cases, tail DNA was prepared from neonates to verify independently the p53 genotype by Southern blot analysis, as described previously (13).
Tumor Counts.
All mice were killed at 90 days by CO2 asphyxiation. The intestinal tract was removed, washed with PBS, opened longitudinally, and laid out as described previously (6). In this method, 4-cm sections from the proximal, medial, and distal regions of the small intestine and the entire colon were examined. The number of tumors was scored with a dissecting microscope by a single observer blind to the genotype of the mice or their treatment group. The samples were then fixed in 10% buffered formalin, washed in 70% ethanol, and stored in this solution. Tumor counts were verified by a second observer in a subset of the postfixed samples.
At sacrifice, the number of desmoid fibromas was scored by a single observer blind to the genotype of the mice or their treatment group. The abdominal body wall was then removed, stretched, fixed overnight in 10% formalin, washed in 70% ethanol, and stored in this solution. Tumor counts were verified by a second observer for a subset of the postfixed samples. To facilitate scoring, the tissue was stained with Fast green, a dye that detects collagen. Samples were incubated for 30 min each in the following series: distilled water; acetate buffer (1 M acetic acid/7.2 mM sodium acetate/0.25% formalin); 5% (wt/vol) phosphotungstic acid in distilled water; Fast green [1 M acetic acid/110 mM sodium acetate/0.05% (wt/vol) Fast green]; acetate buffer; 10 mM sulfuric acid; 50% ethanol; and 70% ethanol.
Histological Analysis of Intestinal Tumors.
The largest tumors were isolated from sections of the small intestine and the entire colon. After fixation in 10% buffered formalin, the tumors were embedded in paraffin, sectioned, and stained with hematoxylin and eosin. Sections were analyzed by light microscopy for signs of progression by H. C. Pitot (McArdle Lab), also blind to the genotype of the samples.
Histological Analysis of Desmoid Fibromas.
Desmoid fibromas were isolated from postfixed abdominal body walls. The tumors were embedded in paraffin, sectioned, and stained with either hematoxylin and eosin or Masson's trichrome.
Results
p53 Effect on Multiplicity of Intestinal Adenomas.
Does a lack of p53 activity affect the multiplicity of adenomas in the intestine of the Min mouse? Min mice homozygous for a null allele of p53 developed significantly more intestinal adenomas than those homozygous for the wild-type allele of p53, regardless of the Mom1 genotype (Table 1). Thus, p53 negatively regulates the development of adenomas in the intestine of the Min mouse.
Table 1.
Effect of p53 on intestinal tumor multiplicity in Min mice
|
Mom1S/S
|
Mom1R/S
|
Mom1R/R
|
Treated*
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Tumor count | P-value† | N | Tumor count | P-value† | N | Tumor count | P-value† | N | Tumor count | P-value‡ | |
| p53+/+ | 20 | 32 ± 11 | NA | 20 | 13 ± 5 | NA | 19 | 5 ± 4 | NA | 5 | 5 ± 2 | 0.0007 |
| p53+/− | 19 | 37 ± 9 | 0.072 | 21 | 15 ± 7 | 0.403 | 20 | 8 ± 4 | 0.053 | 10 | 6 ± 4 | 0.00001 |
| p53−/− | 21 | 45 ± 13 | 0.0014 | 14 | 26 ± 8 | 0.00002 | 16 | 13 ± 6 | 0.00008 | 4 | 12 ± 8 | 0.003 |
Mice were killed at 90 days of age. Tumor counts are shown as means ± SD.
*Mom1S/S mice were treated with piroxicam and DFMO (see Materials and Methods).
†P-values were calculated by using two-sided Wilcoxon rank sum tests compared with p53+/+ mice of the same Mom1 genotype.
‡P-values were calculated by using two-sided Wilcoxon rank sum tests compared with untreated mice of the same p53 genotype.
Min mice heterozygous for a null allele of p53 (ApcMin/+ p53+/−) also develop more tumors than those homozygous for the wild-type allele of p53 (Table 1). The difference is at the borderline of statistical significance for two of the Mom1 classes.
One interpretation for the effect of the p53 genotype is that a polymorphic modifier, linked to the p53 locus, affects the development of intestinal adenomas in the Min mouse (14). In deriving the B6-p53 congenic animals, a region from chromosome 11 of the 129 genome was moved onto the B6 genetic background. We have tested this hypothesis and found no evidence for such a modifier. The extent of the 129/SvEv genome carried by the p53 mutant line was assessed by genotyping Min mice heterozygous for a null allele of p53 with markers distributed along chromosome 11. This analysis indicated that the introgressed region of the 129/SvEv genome in the B6-p53 congenic strain extends at least from 19.7 cM to the p53 locus at 39 cM on chromosome 11. To test whether a polymorphic modifier was present in this region of the 129 genome, B6 females were mated to (129/SvPas × B6)F1 Min males. At least 20 resulting progeny were scored for the number of adenomas and genotyped with markers from chromosome 11. Tumor multiplicity was comparable between Min mice that were heterozygous for the 129/SvPas allele and those that are homozygous for the B6 allele at each locus (data not shown). Thus, the heterozygous effect of the p53 genotype cannot be explained by a dominant polymorphic modifier of intestinal tumorigenesis that is linked to the p53 locus.
p53 Effect on the Progression of Intestinal Adenomas.
Mutation of p53 has been correlated with the progression of tumors in the human colon (17). To investigate whether a lack of p53 function enhances tumor progression in Min mice, the largest tumors from three regions of the small intestine and the entire colon were analyzed for signs of morphological progression in Min mice of each p53 genotype. Two of the 42 tumors that were isolated from the small intestines of ApcMin/+ p53−/− mice were locally invasive with tumors invading the underlying muscularis mucosae (Fig. 1 B and C), whereas two of the nine tumors isolated from the colons of these mice exhibit features characteristic of in situ carcinomas (Fig. 1 E and F). By contrast, all 78 tumors isolated from ApcMin/+ p53+/+ and ApcMin/+ p53+/− mice were benign. This difference in the incidence of invasive tumors lies at the borderline of statistical significance [P = 0.05, Fisher's exact test, one sided]. Thus, the lack of p53 activity may affect the Min phenotype by permitting the progression of adenomas to adenocarcinomas. Alternatively, p53 deficiency may permit invasive tumors to arise from a stem cell population distinct from that giving rise to adenomas. Would a stronger effect be observed if the tumors could be analyzed in older mice? Note that mice were killed at 90 days of age in this study. Unfortunately, the average lifespan of ApcMin/+ p53−/− mice is 122 days, regardless of the Mom1 genotype. Furthermore, the loss-of-function p53 allele used in these studies may not fully simulate any gain-of-function p53 allele found in human tumors (18). Other experimental designs are necessary to explore more fully the extent to which alterations in p53 can lead to more advanced tumors in the Min mouse.
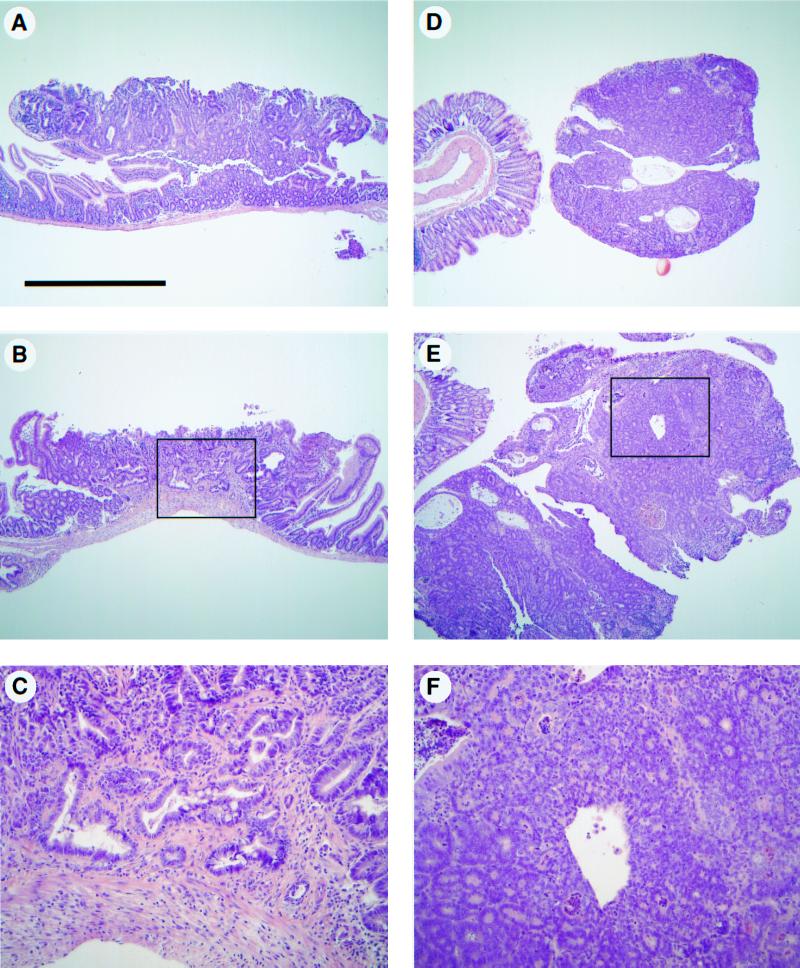
Figure 1.
Intestinal tumors exhibit signs of progression in ApcMin/+ p53−/− mice. Tumors were isolated from ApcMin/+ Mom1S/S mice carrying zero, one, or two copies of the p53 knockout allele and stained with hematoxylin and eosin. Most tumors from the small intestine and colon of p53-deficient mice were typical adenomas (A and D, respectively). However, signs of progression were exhibited by some of the tumors isolated from the proximal region of the small intestine and the colon (B and E, respectively). The regions that exhibited signs of progression are boxed in B and E and are shown at higher magnification in C and F. The magnification was the same for A, B, D, and E, with the scale bar in A representing 1 mm. C and F are ×4 of B and E, respectively.
Function of p53 on the Multiplicity of Desmoid Fibromas.
Min mice and humans carrying germline defects in APC develop desmoid fibromas. This type of tumor is characterized by nodules or penetrating masses of unusually dense connective tissue that result from the active proliferation of fibroblasts. Some of these features are evident in sections of tumors stained with dyes that detect collagen (Fig. 2).
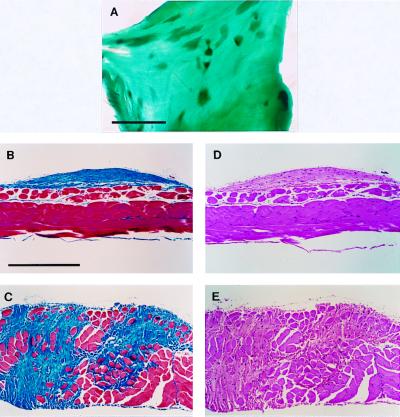
Figure 2.
Desmoid fibromas develop on the abdominal body wall of ApcMin/+ p53−/− mice. The abdominal body wall was isolated and fixed in 10% formalin. The postfixed tissue was either stained with Fast green (A) or further dissected to isolate individual tumors. Isolated tumors were sectioned and stained either with Masson's Trichrome (B and C), in which muscle stains red and collagen blue, or with hematoxylin and eosin (D and E). These lesions can form on the surface of the abdominal wall muscle (B and D) or infiltrate and disrupt the organization of this tissue (C and E). The scale bar in A represents 5 mm, whereas that in B represents 0.5 mm. The magnification in B–E was the same.
The multiplicity of these lesions in Min mice is dramatically affected by the lack of p53 activity (Table 2). These lesions formed on the musculature of the entire body but were scored only in the abdominal region. ApcMin/+ p53−/−mice develop on average 52 desmoid fibromas, whereas Apc+/+ p53+/−, ApcMin/+ p53+/+, and ApcMin/+ p53+/− mice develop on average less than a single tumor per animal. This difference indicates that both Apc and p53 must be altered for maximal transformation of normal fibroblasts into a neoplastic counterpart. A germline mutation in one of these genes is almost completely without effect in the presence of a wild-type form of the other. These results contrast with the observation by Smits et al. (19) of desmoid tumors in an Apc knockout strain wild type for p53. It is possible that the neomycin-resistance insertion in the knockout allele of Apc exerted a position effect on a gene adjacent to Apc (20).
Table 2.
Tests for effects of p53 and Mom1 on the development of desmoid fibromas in Min mice
|
Mom1S/S
|
Mom1R/S
|
Mom1R/R
|
Treated*
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Tumor count | N | Tumor count | P-value† | N | Tumor count | P-value† | N | Tumor count | P-value‡ | |
| p53+/+ | 27 | 0.5 ± 0.7 | 19 | 0.7 ± 1.0 | 0.69 | 19 | 0.6 ± 1.0 | 0.89 | 2 | 0.5 ± 0.7 | 0.84 |
| p53+/− | 40 | 0.8 ± 1.3 | 28 | 1.6 ± 2.2 | 0.11 | 26 | 1.1 ± 1.7 | 0.16 | 3 | 0 ± 0 | 0.25 |
| p53−/− | 23 | 53 ± 11 | 17 | 58 ± 10 | 0.12 | 14 | 51 ± 12 | 0.66 | 2 | 34 ± 9 | 0.04 |
Tumor counts are shown as means ± SD. Mice were killed at 90 days of age, and the number of desmoid fibromas in the abdominal region were scored. Using two-sided Wilcoxon rank sum tests, the results for Mom1S/S mice were compared with those for Mom1R/S (P = 0.122) and Mom1R/R mice (P = 0.661). Desmoid fibromas were not observed in Apc+/+ p53+/+ mice, regardless of Mom1 type.
*Mom1S/S mice were treated with piroxicam and DFMO (see Materials and Methods).
†P-values were calculated by using two-sided Wilcoxon rank sum tests compared with Mom1S/S mice of the same p53 genotype.
‡P-values were calculated by using two-sided Wilcoxon rank sum tests compared with untreated Mom1S/S mice of the same p53 genotype.
Effects of Mom1 and Pharmacological Agents on the Multiplicity of Intestinal Adenomas and Desmoid Fibromas.
The resistance allele of Mom1 (Mom1R) and the pharmacological agents piroxicam and DFMO slow the net growth rate of intestinal adenomas in the Min mouse and reduce multiplicity. Do these factors act by stimulating a p53-dependent activity, such as cell cycle arrest or apoptosis? The number of adenomas was scored in ApcMin/+ p53−/− mice carrying zero, one, or two copies of Mom1R. A single copy of this allele reduced the number of tumors by a factor of 1.7, two copies reduced the number by a factor of 3.5 (Table 1). The semidominant effect of Mom1 in p53-deficient animals was indistinguishable from its effect in mice heterozygous or wild type for p53. Similarly, the effect on tumor multiplicity of the piroxicam/DFMO combination was unaffected by the lack of p53 activity (Table 2). Thus, Mom1 and these pharmacological agents act independently of any p53-dependent processes. Furthermore, the growth inhibition exhibited by the piroxicam/DFMO combination does not depend on the active Mom1 resistance allele encoding the Pla2 g2a secretory phospholipase.
Do the resistance allele of Mom1 and the piroxicam/DFMO combination affect the development of desmoid fibromas? Tumors were scored on the abdominal body wall of ApcMin/+ p53−/− mice either carrying Mom1R or treated with both piroxicam and DFMO (Table 2). All Mom1 classes of mice were indistinguishable in desmoid multiplicity, indicating that the resistance allele of Mom1 does not suppress the development of desmoid fibromas. By contrast, mice treated with the piroxicam/DFMO combination showed a significant reduction in desmoid multiplicity. Additional mice need to be studied to confirm this effect. Thus, Mom1 action is tissue specific, affecting tumorigenesis only in the intestine of the ApcMin/+ mouse, whereas the action of the piroxicam/DFMO combination appears not to be restricted to the intestine.
Discussion
We are analyzing the genetic network controlling intestinal neoplasia in the laboratory mouse to further a fundamental understanding of human colon cancer. One of the mysteries in the development of human colon cancer is its nonlinear age dependence, fitted to a six-hit process (21). One strategy to identify the elements contributing to such a multihit process is to generate mice carrying germline mutations of candidate tumor-suppressor genes and to observe strong enhancement of neoplasia. The candidates currently available for such a test are the genes of the mouse homologous to those mutated in familial or sporadic colon cancer in the human. An impediment to a rigorous test of cooperation between negative regulators is the creation of heterogeneous genetic backgrounds when the distinct mutations are carried on different inbred backgrounds. This heterogeneity creates genetic variation for polymorphic modifiers of the phenotype (7, 15, 22) that can obscure any cooperation. This study has utilized congenic derivatives of a single inbred strain, B6, to detect cooperation between p53 and Apc in suppressing intestinal adenoma formation (Table 1) and progression (Fig. 1). More impressively, strong cooperation has been detected between these negative regulators in suppressing desmoid fibromas of the skeletal muscle wall (Fig. 2; Table 2).
Strong cooperation between distinct negative regulators of neoplasia (“tumor suppressors”) can provide a basis for multihit cancer development with a kinetic order higher than the two-hit process hypothesized by Knudson (23). Such a higher-order process may contain within it the canonical two-hit/one-locus process developed for retinoblastoma and the retinoblastoma gene in the human (24) and intestinal adenoma formation in the mouse (25). These simpler cases may fit the “Gatekeeper Hypothesis,” in which a single negative regulator controls the neoplastic potential of a particular cell type (11). However, a more general understanding of neoplasia in mammals must recognize instances of redundant regulation, such as that between p53 and Apc in controlling desmoid fibromas (Table 2) and that between p53 and Rb in controlling endocrine tumors (26).
The APC/Apc gene is expressed very broadly throughout mammals (27). The restricted spectrum of neoplasms found in humans and mice carrying germline mutations in APC/Apc could be explained by cooperation with other negative regulators. However, the p53 gene is not an obvious candidate for fulfilling such a role, because it appears to be constitutively expressed in only a few cell types (3, 4), becoming more broadly active under several conditions of stress (28, 29). Several possibilities can be considered for the involvement of p53 function in the development of invasive intestinal tumors and desmoid fibromas. The p53 gene may be constitutively active in the precursors of these neoplasms. Note that Li-Fraumeni patients carrying germline mutations in the p53 gene usually develop soft tissue sarcomas of mesenchymal origin (30). Alternatively, the p53 gene may be induced as the neoplasm is initiated. The loss of APC/Apc activity in the tumor might lead to the activation of the c-Myc protooncogene (31), which in turn might activate p53 via sequential steps of negative regulation from p19ARF to Mdm2 to p53 (32, 33). If this process activates p53-dependent apoptosis, then a deficiency of p53 would enhance tumor formation.
Other testable mechanisms for interaction between Apc and p53 can be proposed. The p53 transcription factor is required for Apc expression in vitro (34), so a deficiency of p53 may lead to silencing of Apc expression (35) and enhanced adenoma formation. A molecular analysis of the status of the wild-type Apc allele in the tumors that arise in Min mice lacking p53 may be informative. Indirect modes of interaction are also conceivable and testable, involving any of the polypeptides known to interact with APC/Apc (36) or any of the genes known to be regulated by p53 activity or polypeptides known to interact with p53 (28, 29, 37).
Beyond the negative regulatory genes of neoplasia, APC/Apc and p53, the modifier gene Mom1 encoding the secretory phospholipase Pla2g2a and the pharmacological agents piroxicam and DFMO must be placed in the network of interactions controlling neoplasia. The activity of each of these factors leads to a reduction in net growth rate of the adenomas of Min mice (12, 38). The modifying effect of Mom1 is tissue specific, affecting only the intestinal phenotype of the Min mouse. Early insight into the selective action of modifiers in pleiotropic cancer syndromes has been reported for familial neurofibromatosis type I (39).
Two questions are important in assessing these modifying agents for chemoprevention or therapy of tumors. Is the effect on growth rate lost in p53-deficient tumors? Clearly both Mom1 and the piroxicam/DFMO combination are active against intestinal adenomas under p53-deficient conditions (Tables 1 and 2). Is there synergistic action between modifiers? It has been found previously that the treatment of Min mice with the combination of piroxicam plus DFMO is much more effective than either agent alone, resulting in a significant number of tumor-free mice (12). These findings of successful combination chemoprevention pertain to Min mice on the B6 genetic background, deficient in the secretory phospholipase Pla2g2a. Finding that Mom1 resistance is also independent of p53 function points to the importance of investigating whether the secretory phospholipase or one of the products of its action will synergize with piroxicam/DFMO to give complete chemoprevention and/or therapy for adenomatous polyps. A parallel study has found p53 independence and strong synergy between the modifiers of intestinal tumors, Mom1 and Dnmt (R. T. Cormier and W.F.D., unpublished work). Single factors, each with a small effect on a disease process, can be combined usefully.
Acknowledgments
We thank Natalie Borenstein, Cheri Pasch, and Jen Triemstra for steady and highly capable technical assistance in mouse genetics. The development of the chemoprevention trials was performed by Dr. Anita Merritt and Mindy Grove. Finally, the histopathology was performed with the skilled technical assistance of Jane Weeks and Harlene Edwards and the histopathological assessments of Dr. Henry Pitot. Professors John Petrini and Stephen Gallinger have given scientific feedback, and Linda Clipson has filled crucial roles in the preparation of this report. This research has been supported by Core Grant CA 07175, Contract NO1 CN 65122, and research grants CA 50585 and CA 63677 from the National Cancer Institute. R.B.H. was supported by postdoctoral fellowships PF-4217 from the American Cancer Society and F32CA77946 from the National Institutes of Health (NIH). P.D.H. was supported by training grants to the Department of Biostatistics from NIH (T32EY07119-08 and T32CA09565-09). This is publication no. 3549 from the Laboratory of Genetics.
Abbreviation
- DFMO
difluoromethylornithine
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050585597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050585597
References
- 1.Bulow S. Dan Med Bull. 1987;34:1–15. [PubMed] [Google Scholar]
- 2.Smith M L, Chen I T, Zhan Q, Bae I, Chen C Y, Gilmer T M, Kastan M B, O'Connor P M, Fornace A J. Science. 1994;266:1376–1380. doi: 10.1126/science.7973727. [DOI] [PubMed] [Google Scholar]
- 3.MacCallum D E, Hupp T R, Midgley C A, Stuart D, Campbell S J, Harper A, Walsh F S, Wright E G, Balmain A, Lane D P, et al. Oncogene. 1996;13:2575–2587. [PubMed] [Google Scholar]
- 4.Komarova E A, Diatchenko L, Rokhlin O W, Hill J E, Wang Z J, Krivokrysenko V I, Feinstein E, Gudkov A V. Oncogene. 1998;17:1089–1096. doi: 10.1038/sj.onc.1202303. [DOI] [PubMed] [Google Scholar]
- 5.Merritt A J, Potten C S, Kemp C J, Hickman J A, Balmain A, Lane D P, Hall P A. Cancer Res. 1994;54:614–617. [PubMed] [Google Scholar]
- 6.Moser A R, Pitot H C, Dove W F. Science. 1990;247:322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 7.Dove W F, Luongo C, Connelly C S, Gould K A, Shoemaker A R, Moser A R, Gardner R L. Cold Spring Harbor Symp Quant Biol. 1994;59:501–508. doi: 10.1101/sqb.1994.059.01.055. [DOI] [PubMed] [Google Scholar]
- 8.Clarke A R, Cummings M C, Harrison D J. Oncogene. 1995;11:1913–1920. [PubMed] [Google Scholar]
- 9.Fazeli A, Steen R G, Dickinson S L, Bautista D, Dietrich W F, Bronson R T, Bresalier R S, Lander E S, Costa J, Weinberg R A. Proc Natl Acad Sci USA. 1997;94:10199–10204. doi: 10.1073/pnas.94.19.10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fodde R, Khan P M. Crit Rev Oncog. 1995;6:291–303. doi: 10.1615/critrevoncog.v6.i3-6.60. [DOI] [PubMed] [Google Scholar]
- 11.Kinzler K E, Vogelstein B. In: The Genetic Basis of Human Cancer. Vogelstein B, Kinzler K E, editors. New York: McGraw–Hill; 1998. pp. 241–242. [Google Scholar]
- 12.Jacoby, R. F., Cole, C. E., Tutsch, K., Newton, M. A., Kelloff, G., Hawk, E. T. & Lubet, R. A. (2000) Cancer Res., in press. [PubMed]
- 13.Donehower L A, Harvey M, Slagle B L, McArthur M J, Montgomery C A, Jr, Butel J S, Bradley A. Nature (London) 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 14.Gould K A, Dietrich W F, Borenstein N, Lander E S, Dove W F. Genetics. 1996;144:1769–1776. doi: 10.1093/genetics/144.4.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dietrich W F, Lander E S, Smith J S, Moser A R, Gould K A, Luongo C, Borenstein N, Dove W F. Cell. 1993;75:631–639. doi: 10.1016/0092-8674(93)90484-8. [DOI] [PubMed] [Google Scholar]
- 16.Timme T L, Thompson T C. BioTechniques. 1994;17:460–463. [PubMed] [Google Scholar]
- 17.Fearon E R, Vogelstein B. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 18.White A E, Livanos E M, Tlsty T D. Genes Dev. 1994;8:666–677. doi: 10.1101/gad.8.6.666. [DOI] [PubMed] [Google Scholar]
- 19.Smits R, Van Oordt W, Luz A, Zurcher C, Jagmohan-Changur S, Breukel C, Khan P M, Fodde R. Gastroenterology. 1998;114:275–283. doi: 10.1016/s0016-5085(98)70478-0. [DOI] [PubMed] [Google Scholar]
- 20.Gerard M, Chen J Y, Gronemeyer H, Chambon P, Duboule D, Zakany J. Genes Dev. 1996;10:2326–2334. doi: 10.1101/gad.10.18.2326. [DOI] [PubMed] [Google Scholar]
- 21.Armitage P, Doll R. Br J Cancer. 1954;8:1–12. doi: 10.1038/bjc.1954.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cormier R T, Hong K H, Halberg R B, Hawkins T L, Richardson P, Mulherkar R, Dove W F, Lander E S. Nat Genet. 1997;17:88–91. doi: 10.1038/ng0997-88. [DOI] [PubMed] [Google Scholar]
- 23.Knudson A G., Jr Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knudson A G., Jr Semin Oncol. 1978;5:57–60. [PubMed] [Google Scholar]
- 25.Luongo C, Dove W F. Genes Chromosomes Cancer. 1996;17:194–198. doi: 10.1002/1098-2264(199611)17:3<194::aid-gcc2870170302>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 26.Harvey M, Vogel H, Morris D, Bradley A, Bernstein A, Donehower L A. Nat Genet. 1995;9:305–311. doi: 10.1038/ng0395-305. [DOI] [PubMed] [Google Scholar]
- 27.Smith K J, Johnson K A, Bryan T M, Hill D E, Markowitz S, Willson J K V, Paraskeva C, Petersen G M, Hamilton S R, Vogelstein B, et al. Proc Natl Acad Sci USA. 1993;90:2846–2850. doi: 10.1073/pnas.90.7.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oren M. J Biol Chem. 1999;274:36031–36034. doi: 10.1074/jbc.274.51.36031. [DOI] [PubMed] [Google Scholar]
- 29.Prives C, Hall P A. J Pathol. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 30.Strong L C, Williams W R, Tainsky M A. Am J Epidemiol. 1992;135:190–199. doi: 10.1093/oxfordjournals.aje.a116271. [DOI] [PubMed] [Google Scholar]
- 31.He T C, Chan T A, Vogelstein B, Kinzler K W. Cell. 1999;99:335–345. doi: 10.1016/s0092-8674(00)81664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zindy F, Eischen C M, Randle D H, Kamijo T, Cleveland J L, Sherr C J, Roussel M F. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherr C J. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 34.Narayan S, Jaiswal A S. J Biol Chem. 1997;272:30619–30622. doi: 10.1074/jbc.272.49.30619. [DOI] [PubMed] [Google Scholar]
- 35.Shoemaker A R, Moser A R, Midgley C A, Clipson L, Newton M A, Dove W F. Proc Natl Acad Sci USA. 1998;95:10826–10831. doi: 10.1073/pnas.95.18.10826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polakis P. Biochim Biophys Acta. 1997;1332:F127–F148. doi: 10.1016/s0304-419x(97)00008-5. [DOI] [PubMed] [Google Scholar]
- 37.Yu J, Zhang L, Hwang P M, Rago C, Kinzler K W, Vogelstein B. Proc Natl Acad Sci USA. 1999;96:14517–14522. doi: 10.1073/pnas.96.25.14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gould K A, Luongo C, Moser A R, McNeley M K, Borenstein N, Shedlovsky A, Dove W F, Hong K, Dietrich W F, Lander E S. Genetics. 1996;144:1777–1785. doi: 10.1093/genetics/144.4.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Easton D F, Ponder M A, Huson S M, Ponder B A J. Am J Hum Genet. 1993;53:305–313. [PMC free article] [PubMed] [Google Scholar]