Abstract
Conjugative plasmids are extra-chromosomal DNA elements that are capable of horizontal transmission and are found in many natural isolated bacteria. Although plasmids may carry beneficial genes to their bacterial host, they may also cause a fitness cost. In this work, we studied the evolution of the R1 plasmid and we found that, in spite of the R1 plasmid conferring an initial cost to its host, after 420 generations the cost disappeared in all five independent evolution experiments. In fact, in two of these five experiments evolved conjugative plasmids actually conferred a fitness advantage to their hosts. Furthermore, the relative fitness of the ancestral clone bearing one of the evolved plasmids is significantly higher than both the plasmid-free ancestral cells and the evolved cells carrying the evolved plasmid. Given that the R1 plasmid may spread among different species of enterobacteria, we wondered what the effect of the evolved plasmid would be inside Salmonella enterica cells. We found that the evolved plasmid is also able to dramatically increase the relative fitness of these cells. Our results suggest that even if general usage of antibiotics is halted, conjugative plasmids that have been selected with antibiotics in previous years can still persist among bacterial populations or even invade new strains.
Keywords: plasmid evolution, host–parasite, antibiotic resistance
1. Introduction
The fundamental importance of conjugative plasmids as mediators of DNA transfer arises from the following characteristics (Summers 1996): (i) many conjugative plasmids carry antibiotic resistance genes or virulence factors that can spread between commensal and pathogenic bacterial cells (Martinez & Baquero 2002); (ii) some of them have a broad host range; donor and recipient cells may belong to different species, genera, or even to different kingdoms (Amabile-Cuevas & Chicurel 1992; Courvalin et al. 1995), which is of major concern since bacteria often share the habitat with hundreds of other bacterial species (e.g. in human gut or skin; Berg 1996). Given these properties, some conjugative plasmids that evolved in a given strain can spread to other strains or species, including pathogenic ones. Therefore, studying the evolution of associations of bacterial cells with conjugative plasmids is of great importance.
We assessed the evolution of the R1 conjugative plasmid in Escherichia coli cells and found that, after about 420 generations of evolution, plasmids can strongly increase the fitness of other bacterial cells, including Salmonella.
2. Methods
(a) Bacterial strains, plasmid used and plasmid transfers
For the long-term evolution experiment, we used two spontaneous derivatives of the E. coli K12 MG1655 strain: one strain resistant to nalidixic acid and mecillinam (NalR, MecR) and the other resistant to rifampicin and fosfomycin (RifR, FosR). The R1 plasmid was introduced in both strains, conferring resistance to six antibiotics (chloramphenicol, kanamycin, ampicillin, streptomycin, spectinomycin and sulphonamides). R1 is a member of a group of plasmids (IncF II group) very well represented in nature (Sherley et al. 2003). The F group bears many of the virulence determinants of enteric species (Falkow 1996). We measured fitness (see §2b) using two reference strains: E. coli K12 MG1655 StrR, srl∷ Tn10 and E. coli K12 MG1655 Δara ValR, both strains bearing the R1 plasmid (StrR and ValR stand for resistance to streptomycin and valine, Δara stands for the deletion of the arabinose operon). We also used Salmonella enterica serovar typhimurium LT2 resistant to nalidixic acid and mecillinam.
Plasmid transfers were performed as in Dionisio et al. (2002). Transfer rates are calculated as the number of transconjugants divided by the square root of the product between the number of donor and recipient cells (see details in Dionisio et al. 2002).
(b) Fitness measurements
We performed fitness assays using head to head competitions against a reference strain. To measure, for instance, the fitness of a strain ‘A’ relative to its ancestral plasmid-free cells, our main procedure consisted in competing A against X and the ancestral against X, where X is the reference strain. To perform each of these competitions, we preconditioned the two strains in LB for 1 day, then 10 μl of each of these cultures was inoculated in 10 ml of LB with no antibiotics for 24 h at 37 °C with agitation. Competition experiments to measure fitness were performed at least three times. The actual number of measurements performed is indicated as n (minimum n=3, maximum n=15, depending on the strain or plasmid). Fitness could then be calculated according to formula 3a of Lenski et al. (1991) to infer the fitness of A relative to its ancestral plasmid-free strain, the latter being normalized to one. Then, all experiments were repeated with another reference strain, Y. We chose two different reference strains in order to measure the fitness advantage or disadvantage of the bacteria of interest when competing with different bacteria (eventually after migrating to other hosts containing a different bacterial community).
(c) Antibiotics and media
The concentrations of antibiotics were: 40 μg ml−1 of nalidixic acid, 10 μg ml−1 of mecillinam, 100 μg ml−1 of rifampicin, 30 μg ml−1 of fosfomycin, 100 μg ml−1 of kanamycin, 30 μg ml−1 of chloramphenicol, 40 μg ml−1 of tetracycline. In most experiments, we used Luria–Bertrani broth medium (LB), supplemented (or not) with agar and antibiotics. We also used minimal medium M9 supplemented with MgSO4 and 4 g l−1 of arabinose or 4 g l−1 of glucose with 40 μg ml−1 of valine.
(d) Evolution experiments
The evolution experiments were done as follows. (i) For the growth of strains, we cultured E. coli ‘RifR FosR’ and E. coli ‘NalR MecR’ (both containing the R1 plasmid) overnight in a liquid LB medium supplemented with antibiotics selecting for the chromosome and the R1 plasmid. (ii) For the competition, 25 μl of each overnight culture (about 25×106 bacterial cells) was mixed and spread on LB agar plates with ampicillin (ampicillin was used to prevent plasmid loss). Each overnight culture was previously ‘washed’ with MgSO4 10−2 M to prevent cell death owing to different antibiotics. After 24 h, a layer of bacteria had formed over the plate. (iii) For the selection of one of the strains: the layer was removed to 5 ml of MgSO4 10−2 M, and the ‘RifR FosR’ strain was selected by transferring 10 μl of this mixture to LB supplemented with rifampicin and fosfomycin as well as with chloramphenicol and kanamycin to select for the R1 plasmid. We performed these 2-day steps 21 times. In each competition step, the `RifR FosR' strain evolved in competition with the original ‘NalR MecR’ strain (grown from frozen stock). Therefore, the ‘NalR MecR’ strain was not allowed to evolve. Each cycle comprised about 20 bacterial generations, with 10 generations in the plate and 10 generations in the liquid LB for the selection process, calculated as Log2 of (final colony forming units)/(initial colony forming units).
The evolution experiment was carried out with both strains containing the R1 plasmid to prevent plasmid transfer between both strains (owing to surface exclusion, the transfer of R1 between these cells is extremely reduced (Achtman et al. 1977)).
(e) Curing bacteria
To obtain the E′(−) strain evolved bacterial cells containing the evolved plasmid were cured with acridine orange as described in Dionisio et al. (2002).
3. Results
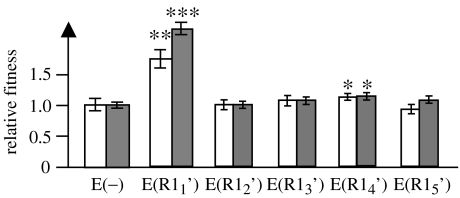
We performed five independent evolution experiments involving bacterial cells with the R1 plasmid. Because of its ability to transfer, we looked for possible effects in the relative fitness of evolved plasmids inside new cells (that had not evolved with the plasmid). None of the evolved plasmids confer fitness disadvantage to new cells (figure 1). Surprisingly, two of the evolved plasmids conferred a significant fitness advantage to new cells, which, to our knowlege, had never been observed before (figure 1).
Figure 1.
Relative fitness of E. coli bearing evolved R1 plasmids, R11′ to R15′ from five independent experiments with the fitness of E(−) normalized to one. E(R1) and E(−) stand for E. coli K12 cells bearing and not bearing the R1 plasmid, respectively. White bars show relative fitness measured by competition against E. coli srl∷Tn10, StrR (R1); grey bars show competition against E. coli K12 Δara, ValR (R1). Fitness values (±twofold the standard error, both throughout the text and in error bars) are relative to E(−), which is 1±0.077 for the white bar (n=15, n being the number of experiments) and 1.000±0.033 for the grey bar (n=15). Relative fitnesses of E(R11′)–E(R15′) are, respectively, 1.726±0.192 (n=15), 1.026±0.074 (n=3), 1.101±0.072 (n=3), 1.162±0.021 (n=3) and 0.951±0.067 (n=3) for white bars and 2.051±0.087 (n=12), 1.013±0.071 (n=3), 1.088±0.050 (n=3), 1.128±0.042 (n=3) and 1.092±0.053 (n=3) for grey bars. Significance levels over the bars (two-tailed t-test assuming unequal variances: *if p<0.05, **if p<0.01 and ***if p<0.001) refer to significant differences of fitness relative to E(−)).
The evolved plasmid conferring the highest advantage was studied in detail. In figure 2, we show the relative fitness of ancestral cells, E(−); ancestral cells with ancestral plasmid, E(R1); evolved cells with evolved plasmid, E′(R1′); evolved cells cured of the R1′ plasmid, E′(−); evolved cells with ancestral plasmid, E′(R1); and the ancestral cells with evolved plasmid, E(R1′). As expected, the ancestral plasmid confers a fitness cost to its host. However, as evolution proceeds the plasmid becomes less costly (Bouma & Lenski 1988; Modi & Adams 1991; Dahlberg & Chao 2003). Also, the evolved cells bearing the evolved plasmid, E′(R1′), have a higher fitness than the original plasmid-free cells, E(−) (figure 2), despite the similarity of relative fitness between E(−) and E′(−). Moreover, the ancestral plasmid in the evolved cells, E′(R1), bears no cost. This suggests that plasmid and bacterial cell coevolved, as previously observed by others (Modi & Adams 1991; Dahlberg & Chao 2003).
Figure 2.
Relative fitness of E. coli cells with the fitness of E(−) normalized to one. The plasmid R1′ is the evolved R11′ plasmid of figure 1, hence the values are those of figure 1. E′ stands for evolved E. coli cells. Other symbols as explained in figure 1. Fitness values (±twofold the standard error) are relative to E(−). The relative fitness values of E(R1), E′(R1′), E′(−) and E′(R1) are, respectively, 0.683±0.042 (n=3), 1.136±0.196 (n=3), 1.083±0.025 (n=3) and 1.028±0.098 (n=3) for white bars and 0.360±0.093 (n=3), 1.192±0.042 (n=3), 0.982±0.018 (n=3) and 1.092±0.054 (n=3) for grey bars.
The increase in relative fitness can be partially accounted for by R1′ replicating at a lower copy-number than the ancestral R1 plasmid (28% lower; see the Electronic Appendix). However, the advantage cannot be attributed solely to this decrease because the fitness of E′(R1′) is not as high as that of E(R1′). No differences were found between evolved and ancestral plasmid by restriction enzyme analysis using four restriction enzymes (see the Electronic Appendix). We also measured the plasmid transfer rate of evolved and ancestral plasmid; the rate from E(R1′) to E(−), 4.58×10−4±1.36×10−4, n=5, is not significantly different from E(R1) to E(−), 10.1×10−4±3.05×10−4, n=5, (t-test, d.f.=8, p-value=0.19).
Since this plasmid can also move between species, we investigated the effect of this evolved plasmid on the fitness of a pathogenic species: Salmonella enterica serovar typhimurium. We observed that the Salmonella cells bearing the evolved plasmid have a significantly higher fitness (1.558±0.059, n=3) than Salmonella cells without the R1 plasmid (1.000±0.074, n=3; two-tailed t-test, p-value=0.007). Consequently, the plasmid can increase the fitness of the species where it evolved as well as the fitness of other species.
4. Discussion
Whenever a bacterial strain colonizes a new host, it has to compete with resident bacterial cells. This is what happens when, for example, pathogenic bacteria consecutively infect human beings. In our evolution experiments, the successive passage of cells containing a conjugative plasmid for about 420 generations yielding a plasmid with the striking property of increasing relative fitness to its host. Not only did the evolved plasmid increase the relative fitness of other cells, including pathogenic species, but the evolution process also occurred in a very short period. This may have occurred owing to the competition imposed in our experiments, an experimental condition not used in previous works (Bouma & Lenski 1988; Modi & Adams 1991; Dahlberg & Chao 2003).
It is clear that the cells also changed: the evolved plasmid-free cells have a fitness similar to that of ancestral plasmid-free cells, while the ancestral cells bearing the evolved plasmid, E(R1′), have a much higher relative fitness than the evolved dyad, E′(R1′). This fact suggests that changes in plasmid have pleiotropic effects with chromosomal changes. Therefore, the outcome of these experiments differ in two ways from previous reports. (i) In the experiments reported by Bouma & Lenski (1988), where a non-conjugative plasmid was used, there was no indication that the plasmid changed, only the bacterial cell. Their evolved plasmid inside ancestral bacterial cells had no significant effect, contrary to our results. Two out of five evolved plasmids in our experiments had a significant fitness-increasing effect in the ancestral E. coli cells as well as in Salmonella cells. (ii) Both in the experiments by Modi & Adams (1991), where a non-conjugative plasmid was used, and in the experiments by Dahlberg & Chao (2003), where conjugative plasmids were used (the R1 and the IncP RP4 plasmids), both the plasmid and the bacterial cell evolved, as in the present work. However, unlike them, we found that the effect of the evolved plasmid inside other cells was to increase their relative fitness. Most importantly, even in another species (S. enterica), the fitness-increasing effect of the evolved R1 plasmid was very strong. If this is common in nature as in experimental evolution, then it has important implications for the maintenance of plasmids in bacterial populations (Bouma & Lenski 1988; Modi & Adams 1991; Bergstrom et al. 2000; Dahlberg & Chao 2003). Our observation that conjugative plasmids can evolve to confer a fitness advantage to new cells has implications both for maintenance and spread of plasmids.
In the real world, there is antibiotic-pressure in some habitats (e.g. in people taking antibiotics) whereas others are antibiotic-free. A migration of bacteria between these two types of habitats and evolving plasmids transferring between bacteria with different histories is a possible scenario. Therefore, it is conceivable that bacteria containing these evolved plasmids could spread in the entire population, for instance, during hospital acquired infections. Unfortunately, this may be happening every day.
Acknowledgments
We thank G. Koraimann for the R1 plasmid, I. Matic for some of the strains used here and R. Gardner for help in editing the manuscript. We are also grateful for discussions with the ESF exploratory workshop The Evolution of Cooperation and Cheating in Nature, Montpellier, May 2003. This work was supported by project POCTI/BSE/46856/2002 (I.G. and F.D.) through Fundação Ciência Tecnologia and fellowships SFRH/BPD/8104/2002 (I.G.) and SFRH/BPD/14820/2003 (F.D.).
References
- Achtman M, Kennedy N, Skurray R. Cell–cell interactions in conjugating Escherichia coli: role of traT protein in surface exclusion. Proc. Natl Acad. Sci. USA. 1977;74:5104–5108. doi: 10.1073/pnas.74.11.5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amabile-Cuevas C.F, Chicurel M.E. Bacterial plasmids and gene flux. Cell. 1992;70:189–199. doi: 10.1016/0092-8674(92)90095-t. [DOI] [PubMed] [Google Scholar]
- Berg R.D. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4:430–435. doi: 10.1016/0966-842x(96)10057-3. [DOI] [PubMed] [Google Scholar]
- Bergstrom C.T, Lipsitch M, Levin B.R. Natural selection, infectious transfer and the existence conditions for bacterial plasmids. Genetics. 2000;155:1505–1519. doi: 10.1093/genetics/155.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouma J.E, Lenski R.E. Evolution of a bacteria/plasmid association. Nature. 1988;335:351–352. doi: 10.1038/335351a0. [DOI] [PubMed] [Google Scholar]
- Courvalin P, Goussard S, Grillot-Courvalin C. Gene transfer from bacteria to mammalian cells. C. R. Acad. Sci. III. 1995;318:1207–1212. [PubMed] [Google Scholar]
- Dahlberg C, Chao L. Amelioration of the cost of conjugative plasmid carriage in Eschericha coli K12. Genetics. 2003;165:1641–1649. doi: 10.1093/genetics/165.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionisio F, Matic I, Radman M, Rodrigues O.R, Taddei F. Plasmids spread very fast in heterogeneous bacterial communities. Genetics. 2002;162:1525–1532. doi: 10.1093/genetics/162.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkow S. The evolution of pathogenicity in Escherichia, Shigella, and Salmonella. In: Neidhardt F.C, Curtiss R III, Ingraham J.L, Lin E.C.C, Low K.B, Magasanik B, Reznikoff W.S, Riley M, Schaechter M, Umbarger H.E, editors. In Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 2. ASM Publications; Washington DC: 1996. pp. 2723–2729. [Google Scholar]
- Lenski R.E, Rose M.R, Simpson S.C, Tadler S.C. Long-term experimental evolution in Escherichia coli. 1. Adaptation and divergence during 2,000 generations. Am. Nat. 1991;138:1315–1341. [Google Scholar]
- Martinez J.L, Baquero F. Interactions among strategies associated with bacterial infection: pathogenicity, epidemicity, and antibiotic resistance. Clin. Microbiol. Rev. 2002;15:647–679. doi: 10.1128/CMR.15.4.647-679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi R.I, Adams J. Coevolution in bacterial–plasmid populations. Evolution. 1991;45:656–667. doi: 10.1111/j.1558-5646.1991.tb04336.x. [DOI] [PubMed] [Google Scholar]
- Sherley M, Gordon D.M, Collignon P.J. Species differences in plasmid carriage in the Enterobacteriaceae. Plasmid. 2003;49:79–85. doi: 10.1016/s0147-619x(02)00107-5. [DOI] [PubMed] [Google Scholar]
- Summers D.K. The biology of the plasmids. Blackwell Science; Oxford: 1996. [Google Scholar]