Abstract
We recently described a mouse model that reproduces important pathophysiological features of mitochondrial DNA (mtDNA) mutation diseases. The gene for mouse mitochondrial transcription factor A, Tfam (also called mtTFA), a nucleus-encoded key regulator of mtDNA expression, was targeted with loxP sites (TfamloxP) and disrupted in vivo by transgenic expression of cre-recombinase from the muscle creatinine kinase (Ckmm) promoter. This promoter is active from embryonic day 13, and the knockouts had normal respiratory chain function in the heart at birth and developed mitochondrial cardiomyopathy postnatally. In this paper, we describe a heart-knockout strain obtained by mating TfamloxP mice to animals expressing cre-recombinase from the α-myosin heavy chain (Myhca) promoter. This promoter is active from embryonic day 8, and the knockouts had onset of mitochondrial cardiomyopathy during embryogenesis. The age of onset of cardiac respiratory chain dysfunction can thus be controlled by temporal regulation of cre-recombinase expression. Further characterization demonstrated that ≈75% of the knockouts died in the neonatal period, whereas, surprisingly, ≈25% survived for several months before dying from dilated cardiomyopathy with atrioventricular heart conduction blocks. Modifying gene(s) affect the life span of the knockouts, because ≈95% of the knockout offspring from an intercross of the longer-living knockouts survived the neonatal period. Thus, the tissue-specific knockouts we describe here not only reproduce important pathophysiological features of mitochondrial cardiomyopathy but also provide a powerful system by which to identify modifying genes of potential therapeutic value.
The biogenesis of the respiratory chain is unique in its bipartite dependence on nuclear and mtDNA genes, and mitochondrial disorders may be caused by mutations of both genomes (1, 2). Mutations of mtDNA cause several genetic syndromes with deficient oxidative phosphorylation and have also been implicated in common age-associated disorders, e.g., heart failure, diabetes mellitus, and neurodegeneration (3–5). The mtDNA mutation disorders are extremely pleiotropic, and affected patients may have symptoms from almost any organ with varying ages of onset (6). One important explanation for this clinical variability is heteroplasmy with random segregation of normal and mutated mtDNA (1, 2, 7). This condition will result in widely varying levels of mutated mtDNA in different organs, and levels exceeding a certain threshold will impair the respiratory chain function (8). Clinical and experimental studies have suggested that nuclear genes also may influence the phenotypic expression of mtDNA mutations (9, 10).
We have generated an animal model for mtDNA mutation disorders by using the cre-loxP conditional knockout strategy to disrupt Tfam, which regulates replication and transcription of mtDNA (11). Animals expressing cre-recombinase in heart and skeletal muscle from embryonic day (E)13, under the control of the Ckmm promoter, were mated to TfamloxP animals to obtain tissue-specific knockouts (TfamloxP/TfamloxP, +/Ckmm-cre) (12). The knockouts had a normal respiratory chain function in the heart at birth, but developed a dilated mitochondrial cardiomyopathy with a severe heart-specific respiratory chain deficiency in the postnatal period (12). These tissue-specific Tfam knockouts reproduce important pathophysiological features of mtDNA mutation disorders (12), i.e., (i) respiratory chain deficiency caused by impaired mtDNA expression, (ii) tissue-specific patterns of respiratory chain deficiency, (iii) mosaicism of respiratory chain-deficient cells in affected organs, (iv) progressive deterioration of respiratory chain function with time, and (v) accumulation of morphologically abnormal mitochondria in affected organs.
In this paper, we describe a heart-knockout strain obtained by mating TfamloxP mice to animals expressing cre-recombinase from the α-myosin heavy chain (Myhca) promoter, which is active from E8. The resulting knockouts (TfamloxP/TfamloxP, +/Myhca-cre) had onset of cardiomyopathy during embryogenesis. It is thus possible to control the age of onset of cardiac respiratory chain dysfunction by temporal regulation of cre-recombinase expression. Further studies showed that ≈75% of the knockouts died in the neonatal period, whereas, unexpectedly, ≈25% survived for several months before dying from dilated cardiomyopathy with atrioventricular heart-conduction blocks. Our data show that modifying gene(s) affect the lifespan of the knockouts, because ≈95% of the knockout offspring from an intercross of the longer-living knockouts survived the neonatal period. The nature of the modifying gene(s) is unknown, and our results suggest that they do not affect the cre-loxP recombination efficiency, but rather act to stabilize mitochondrial protein levels. Determination of the exact nature of the modifying gene(s) will require mapping, positional cloning, and probably also functional tests.
Materials and Methods
Construction of Myhca-cre Transgenic Mice.
To create a construct for heart-specific expression of cre-recombinase, a 6-kb fragment, containing the Myhca promoter (13), exon 1, intron 1, exon 2, and part of intron 2 of the Myhca gene, was inserted into the SalI/BamHI sites of the plasmid pIBI131 to obtain pBSαMHC. A 2.1-kb SalI/KpnI fragment, containing the cre gene and β-actin 3′ untranslated region, was isolated from the plasmid pML78 and inserted into the SalI/KpnI sites of pBSαMHC to obtain pBSαMHCCre, where the cre gene is inserted into exon 2. The insert with the Myhca promoter and the cre gene was released by NotI/KpnI digestion and used for pronuclear injections to obtain Myhca-cre transgenic FVB mice.
PCR, Southern Blot, and Western Blot Analyses.
Determination of the Tfam genotype and detection of the cre transgene were performed by PCR and Southern blot analyses as described (11). Southern blot analyses were used to determine mtDNA copy number in DNA samples from heart, liver, kidney, and skeletal muscle as described (11). Western blot analyses of Tfam, ATP8 (ATP synthase subunit 8, a respiratory chain subunit encoded by mtDNA), and actin protein levels in heart, liver, kidney, and skeletal muscle protein extracts were performed as described (11).
Histological and Biochemical Evaluation of Respiratory Chain Function.
Enzyme histochemical staining of succinate dehydrogenase (SDH) and cytochrome c oxidase (COX) activities was performed on cryostat sections of different tissues from staged embryos and adult animals as described (11). The activities of the respiratory chain enzyme complexes NADH dehydrogenase (complex I), SDH (complex II), cytochrome c reductase (complex III), and COX (complex IV) were determined as described (14). The activities of the citric acid cycle enzymes citrate synthase, isocitrate dehydrogenase, and aconitase were determined as described (14).
Statistical Analyses.
The life-table method was used to analyze survival of mutant animals (15). The log-rank test was used to statistically compare lifespans of different knockout strains. The one-sided t test was used to statistically analyze other data. The symbol “n” refers to the number of analyzed animals.
Electrocardiographic (ECG) Recordings and Echocardiography.
ECG recordings during isoflurane anesthesia and telemetric ECG recordings were performed as described (12).
Results
Most Mice with Heart-Specific Disruption of Tfam Die in the Neonatal Period.
The Tfam gene was selectively disrupted in the heart by mating TfamloxP/TfamloxP mice of mixed genetic background to Myhca-cre transgenic FVB mice. Double-heterozygous offspring (+/TfamloxP, +/Myhca-cre) from this cross were identified and mated with TfamloxP/TfamloxP mice to obtain the heart-specific Tfam knockouts with the TfamloxP/TfamloxP, +/Myhca-cre genotype. This latter mating, referred to as the “standard mating,” produced four genotypes (Table 1). Analysis of staged embryos obtained by the standard mating demonstrated that the knockouts (TfamloxP/TfamloxP, +/Myhca-cre) and the three other genotypes were recovered at the expected frequencies of ≈25% each (Table 1). The mean litter size of staged embryos was normal (Table 1).
Table 1.
Genotypes of embryos and weaned pups produced by standard mating
| Stage | Total offspring analyzed, n | Mean litter size | Genotypes, n (% of total offspring analyzed)
|
|||
|---|---|---|---|---|---|---|
| TfamloxP/TfamloxP, +/Myhca-cre | +/TfamloxP, +/Myhca-cre | TfamloxP/TfamloxP | +/TfamloxP | |||
| E10.5–E16.5 | 130 | 8.7 | 29 | 27 | 35 | 39 |
| E18.5 | 153 | 8.5 | 51 | 37 | 36 | 29 |
| All staged embryos | 283 | 8.6 | 80 (28%) | 64 (23%) | 71 (25%) | 68 (24%) |
| Postnatal age 18–20 days (weaning) | 483 | 7.8 | 28 (6%) | 144 (30%) | 144 (30%) | 167 (34%) |
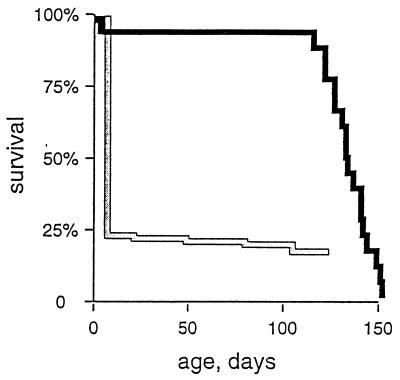
At birth, we observed an increased frequency of neonatal death and reduced litter sizes (Table 1). We recovered and genotyped 15 newborn pups that died in the neonatal period, and 11 of these were knockouts (TfamloxP/TfamloxP, +/Myhca-cre). We observed no increased lethality between the age of 1 week and weaning at age 3 weeks. Genotyping at weaning demonstrated that the knockouts constituted only ≈6% of the remaining pups (Table 1). In summary, these data show that the proportion of knockouts is reduced from 25% to 6% during the neonatal period, and thus ≈75% of them die during the first week of life. Surprisingly, the remaining ≈25% of the knockouts, which survive the neonatal period, have a mean lifespan of >3 months (Fig. 1). Heart-specific knockouts produced by the standard mating can thus be divided into two groups with dramatically different lifespans (Table 1; Fig. 1).
Figure 1.
Survival of animals with heart-specific knockout of Tfam. The life-table method was used to analyze lifespan of heart-knockout animals obtained by the standard mating (gray line) or the mutant mating (black line). The neonatal survival of the standard-mating knockouts was estimated by comparing genotype frequencies in staged embryos and weaned pups. The survival curve from day 7 of the standard-mating knockouts is based on analysis of 17 animals that survived the neonatal period. About 75% of the mutant animals obtained by the standard mating died within the first week of life, whereas the remaining ≈25% survived the neonatal period and had a mean lifespan of >3 months. The survival curve of the mutant-mating knockouts is based on analysis of 26 animals. Two of the mutant-mating knockouts died in the neonatal period, whereas the remaining animals had a mean lifespan of ≈4 months. The lifespan of the adult animals obtained by the standard and mutant matings is not significantly different (P = 0.086) on statistical analysis.
Heart-Specific Knockout Embryos Exhibit Mitochondrial Cardiomyopathy.
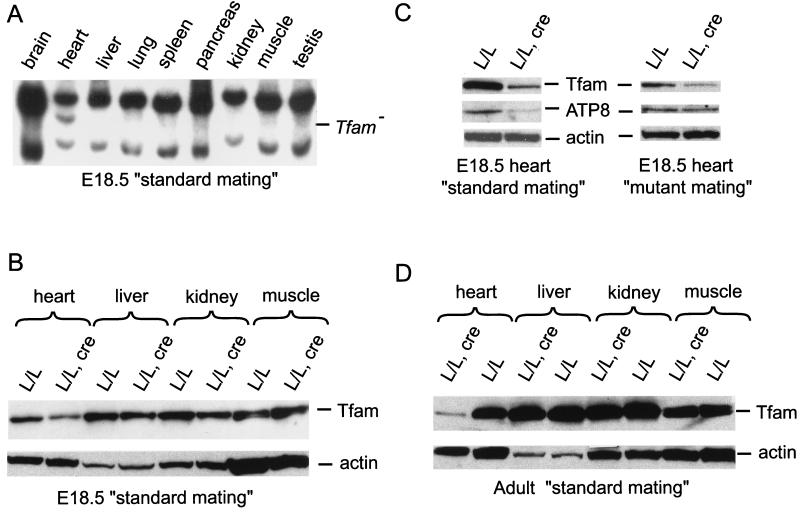
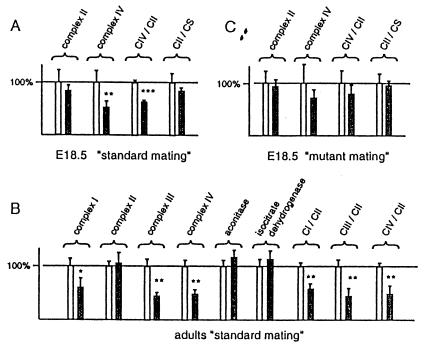
The E18.5 knockout embryos obtained by the standard mating had a highly tissue-specific cre-loxP recombination pattern with presence of the knockout allele (Tfam−) only in the heart, as determined by Southern blot (Fig. 2A) and PCR analyses (data not shown). Western blot analyses confirmed the tissue-specific nature of the knockout and demonstrated reduced Tfam protein levels in the heart, but not in other organs, of the knockout embryos (Fig. 2 B and C). Southern blot analyses showed decreased mtDNA copy number in the heart of the E18.5 knockout embryos (31 ± 2%; mean ± 1 SEM; n = 4) in comparison with TfamloxP/TfamloxP littermates. The levels of the mtDNA-encoded ATP8 protein were reduced in the heart of the E18.5 knockout embryos in comparison with the TfamloxP/TfamloxP littermates (Fig 2C, Left). Biochemical measurements of respiratory chain function demonstrated a deficient activity of complex IV, which contains mtDNA-encoded catalytic subunits, whereas the activity of complex II, which is exclusively nucleus-encoded, was normal in the heart of E18.5 knockout embryos (Fig. 3A). Enzyme histochemical double staining for SDH and COX activities disclosed a highly tissue-specific pattern with presence of COX-deficient cells only in hearts of the E18.5 knockout embryos (data not shown). By further analysis of staged embryos, we determined that COX-deficient cells were present in a mosaic fashion in the hearts of knockouts from E10.5 and onwards (data not shown). This finding indicates that the Myhca-cre transgene is expressed before E10.5, which is consistent with studies by others reporting an even distribution of Myhca transcripts in E8 mouse hearts (16). The Myhca promoter is highly expressed in atria between E10.5 and birth, and there is expression in both atria and ventricles in the postnatal period (16, 17). Determination of absolute heart weight (hw) and the ratio of heart weight to body weight (hw/bw) showed no difference between knockouts (hw = 5.9 ± 0.25 mg; hw/bw = 0.44 ± 0.02%; mean ± 1 SEM; n = 26) and their TfamloxP/TfamloxP littermates (hw = 5.7 ± 0.3 mg; hw/bw = 0.41 ± 0.02%; n = 13) at E18.5. Histological studies showed no gross structural malformations in hearts from knockout E18.5 embryos.
Figure 2.
Patterns of cre-loxP recombination and Western blot analysis of Tfam, ATP8, and actin protein levels. (A) Southern blot analysis of Tfam genotypes in different tissues of E18.5 knockouts (TfamloxP/TfamloxP, +/Myhca-cre) obtained by the standard mating. The knockout allele (Tfam−) is present only in the heart of the knockout embryos. (B) Western blot analyses of Tfam and actin protein levels in different organs of a standard-mating E18.5 knockout (TfamloxP/TfamloxP, +/Myhca-cre; L/L, cre) and a littermate control (TfamloxP/TfamloxP; L/L). (C) Western blot analyses of Tfam, ATP8, and actin protein levels in hearts of standard- or mutant-mating E18.5 knockouts (TfamloxP/TfamloxP, +/Myhca-cre; L/L, cre) and littermate controls (TfamloxP/TfamloxP; L/L). (D) Western blot analyses of Tfam and actin protein levels in different organs of a standard-mating adult knockout (TfamloxP/TfamloxP, +/Myhca-cre; L/L, cre) and a littermate control (TfamloxP/TfamloxP; L/L).
Figure 3.
Biochemical measurements of the respiratory chain function in hearts from knockout and control E18.5 embryos and adults. The bars indicate the relative enzyme activities (mean ± 1 SEM) of the knockouts (TfamloxP/TfamloxP, +/Myhca-cre; gray bars) and their littermate controls (TfamloxP/TfamloxP; white bars). The relative enzyme activities of respiratory chain enzyme complexes (complexes I–IV) and citric acid cycle enzymes (citrate synthase, aconitase, and isocitrate dehydrogenase) are shown. The relative ratios of the activities of complexes IV and II (CIV/CII), complex II and citrate synthase (CII/CS), complexes I and II (CI/CII), and complexes III and II (CIII/CII) also are shown. The results from statistical analyses are indicated by asterisks (*, P <0.05; **, P <0.01; ***, P <0.001). (A) Standard-mating E18.5 knockouts (TfamloxP/TfamloxP, +/Myhca-cre; n = 6) and littermate controls (TfamloxP/TfamloxP; n = 3). (B) Standard-mating adult knockouts (TfamloxP/TfamloxP, +/Myhca-cre; n = 3) and littermate controls (TfamloxP/TfamloxP; n = 3). (C) Mutant-mating E18.5 knockouts (TfamloxP/TfamloxP, +/Myhca-cre; n = 6) and littermate controls (TfamloxP/TfamloxP; n = 3).
Adult Heart-Specific Knockouts Develop a Dilated Cardiomyopathy with Atrioventricular Blocks.
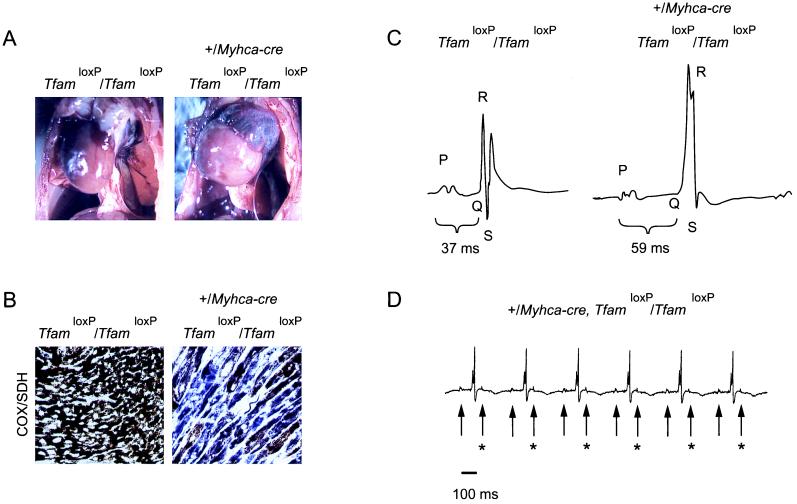
We also analyzed the adult knockouts obtained by the standard mating. At autopsy, at the ages of 2–4 months, all knockouts had significantly enlarged hearts (Fig. 4A) with dilation of ventricles and atria. Determination of heart weights showed a marked difference between knockouts (hw = 280 ± 30 mg; hw/bw = 1.15 ± 0.14%; mean ± 1 SEM; n = 13) and their TfamloxP/TfamloxP littermates (hw = 140 ± 10 mg; hw/bw = 0.50 ± 0.02%; n = 11). Western blot analyses showed a tissue-specific reduction of Tfam protein levels in hearts of the knockouts in comparison with their TfamloxP/TfamloxP littermates (Fig. 2D). Southern blot analyses demonstrated lower mtDNA copy number in hearts of the knockouts (31 ± 7%; mean ± 1 SEM; n = 4) in comparison with their TfamloxP/TfamloxP littermates. Biochemical measurement of respiratory chain function in hearts from adult knockouts demonstrated a reduced activity of complexes I, III, and IV, whereas the activities of the nucleus-encoded complex II, aconitase, and isocitrate dehydrogenase were normal (Fig. 3B). Enzyme histochemical double staining for SDH and COX activities demonstrated a highly tissue-specific mosaic pattern with presence of abundant COX-deficient cells in hearts of the knockouts (n = 12; Fig. 4B). ECG studies of knockout animals (Fig. 4 C and D) demonstrated the presence of atrioventricular blocks during anesthesia (n = 3) and during telemetric recordings in awake animals (n = 3).
Figure 4.
Characterization of standard-mating adult knockouts and controls. (A) Autopsy photographs of a standard-mating adult knockout animal (TfamloxP/TfamloxP, +/Myhca-cre; Right) and a littermate control (TfamloxP/TfamloxP; Left) at the age of 3 months. The knockout animal (Right) has a strikingly enlarged heart with dilation of the left ventricle and atria. (B) Enzyme histochemical double staining for COX and SDH activities in the myocardium of a standard-mating adult knockout animal (TfamloxP/TfamloxP, +/Myhca-cre; Right) and a littermate control (TfamloxP/TfamloxP; Left). A mosaic pattern with severe COX deficiency is seen in the myocardium of the mutant animal (Right). Cells appearing blue lack COX activity, whereas cells appearing brown have COX activity. (C) ECG recording during sustained isoflurane anesthesia of a standard-mating adult knockout (TfamloxP/TfamloxP, +/Myhca-cre; Right) and a littermate control (TfamloxP/TfamloxP; Left) at the age of ≈2 months. The computer-generated average ECG traces are depicted. The P, Q, R, and S waves are indicated by letters, and the PQ intervals are indicated by brackets. The control animal (Left) has a normal PQ interval of 37 ms, whereas the mutant animal (Right) exhibits atrioventricular block with a prolonged PQ interval of 59 ms. (D) Telemetric ECG recording in a standard-mating adult knockout (TfamloxP/TfamloxP, +/Myhca-cre) animal at the age of 2 months. An ECG recording 14 days after implantation of the telemetric transmitter is depicted. Arrows indicate the P waves. The bar indicates the time scale. The knockout animal exhibits atrioventricular block, where every second P wave (arrows with asterisks) is not followed by a QRS complex.
Lifespan of Heart-Specific Knockouts Is Affected by Modifying Gene(s).
We mated adult knockout animals obtained from the standard mating to each other to investigate whether the longer survival was determined genetically. This mating, referred to as “mutant mating,” is expected to produce ≈75% of animals that are knockouts (genotypes TfamloxP/TfamloxP, +/Myhca-cre and TfamloxP/TfamloxP, Myhca-cre/Myhca-cre) and ≈25% of TfamloxP/TfamloxP animals. The mutant mating generated knockouts at about the expected frequency (71% in E18.5 embryos and 68% in weaned pups), and there was no evidence for embryonic lethality (mean litter size = 10 at E18.5). The knockouts obtained by the mutant mating thus survive embryogenesis, and, surprisingly, nearly all of them (≈95%) also survive the neonatal period and have a mean lifespan of >3 months (Fig. 1). The knockouts obtained by the standard mating have widely varying lifespans, and our data demonstrate that the ability to survive the neonatal period is inherited. Thus, the lifespan of the knockouts is affected by the presence of modifying gene(s).
Differences in the Severity of Respiratory Chain Dysfunction Correlate with Lifespan.
We further investigated possible mechanisms that may prevent neonatal death by comparing cre-loxP recombination and mtDNA expression in E18.5 knockout embryos. There was no difference (P = 0.43) in the relative cre-loxP recombination efficiency (defined as the proportion of TfamloxP alleles that had undergone conversion to Tfam− alleles) in hearts from E18.5 knockouts of the standard (41 ± 4%; mean ± 1 SEM; n = 4) and mutant (36 ± 9%; n = 4) matings. This finding indicates a highly efficient recombination, because only a proportion of all cells in the heart is cardiomyocytes. In the adult heart, the cardiomyocytes constitute ≈25% of all cell types and contain ≈75% of the total cytoplasmic volume (18, 19). The cell-type composition of the fetal heart has not been determined with the same accuracy and is likely to vary with the developmental stage.
Western blot analyses demonstrated reduced Tfam protein levels in hearts of standard-mating E18.5 knockouts (n = 9; Fig. 2C, Left), whereas the levels were either reduced (n = 2) or normal (n = 2) in hearts of mutant-mating E18.5 knockouts (Fig. 2C, Right). Determination of mtDNA copy number in hearts showed low levels in standard-mating (31 ± 2%; mean ± 1 SEM; n = 4) but not in mutant-mating E18.5 knockout embryos (87 ± 18%; n = 3) in comparison with TfamloxP/TfamloxP littermates. We also noted a consistent difference in the levels of ATP8 protein; the levels were reduced in hearts of standard-mating E18.5 knockouts (n = 9; Fig. 2C, Left) and normal in hearts of mutant-mating E18.5 knockouts (n = 4; Fig. 2C, Right). Consistent with this difference in mtDNA expression, we observed a reduced complex IV activity in hearts of standard-mating E18.5 knockouts (Fig. 3A). The small difference in complex IV activity did not reach statistical significance in mutant-mating E18.5 knockouts (Fig. 3C). Enzyme histochemical double staining for COX/SDH activity demonstrated a profound COX deficiency in hearts of standard-mating E18.5 knockouts and only scattered COX-deficient cells in hearts of mutant-mating E18.5 knockouts (data not shown).
The adult heart knockouts generated by the mutant mating died from cardiomyopathy at the ages of ≈3–4 months (Fig. 1) and, thus, had a similar lifespan as the standard-mating knockouts that survived the neonatal period (Fig. 1). At autopsy, the mutant-mating adult knockouts had dilation of ventricles and atria with markedly increased heart weight (hw = 290 ± 20 mg; hw/bw = 1.61 ± 0.1%; mean ± 1 SEM; n = 3) in comparison with littermate controls. Double staining for COX/SDH demonstrated a highly tissue-specific mosaic pattern with abundant COX-deficient cells in the knockout hearts (n = 3) at the ages of 3–4 months.
Discussion
In this study, we show that the phenotype of tissue-specific mouse knockouts can be influenced strongly by modifying genes. It previously has been reported that the phenotype in germ-line mouse knockouts can differ dramatically depending on the genetic background. The epidermal growth factor receptor-knockout mice on the CF-1 background die around implantation, whereas knockouts on the CD-1 background die postnatally at the age of 3 weeks (20). The multiple intestinal neoplasia (MIN) mouse strain is heterozygous for a mutation (+/ApcMin) that causes intestinal neoplasms. A strong modifying locus that dramatically reduces tumor formation was mapped, and the responsible gene (Pla2 g2a) subsequently was identified by positional cloning and functional tests (21). A locus modifying the cystic fibrosis phenotype of knockout mice initially was mapped to mouse chromosome 7, and in a recent study of human patients, a corresponding modifying locus was identified in a region of conserved synteny on chromosome 19q13 (22). The presence of modifying genes may be an important explanation for the clinical heterogeneity often encountered in patients with monogenic disorders.
We recently have described animals obtained by mating TfamloxP mice to transgenic animals expressing cre-recombinase, regulated by the Ckmm promoter, with disruptions of Tfam in heart and muscle (12). These knockout animals (TfamloxP/TfamloxP, +/Ckmm-cre) develop a severe heart-specific mosaic respiratory chain deficiency, dilated cardiomyopathy, and atrioventricular heart conduction blocks, very similar to the phenotype in patients with mtDNA deletions (12). The Ckmm promoter is activated late (E13) in embryogenesis (23), which may explain why the knockouts have a normal respiratory chain function in the heart at birth and develop a dilated cardiomyopathy in the postnatal period (12). In the present study, we have used the Myhca promoter, which is active early in embryogenesis (E8), to express cre-recombinase in the heart (16, 23) and, thus, have generated a knockout strain (TfamloxP/TfamloxP, +/Myhca-cre) that develops cardiac respiratory chain dysfunction during late embryogenesis. These findings demonstrate that the age of onset of cardiac respiratory chain dysfunction in the knockouts can be manipulated by temporal regulation of cre expression in the heart. Future use of inducible cre-expression systems will allow the creation of heart-specific Tfam knockouts at virtually any age in the mouse. We observed a time lag between the onset of cre expression and the occurrence of a cardiac respiratory chain dysfunction in both the TfamloxP/TfamloxP, +/Ckmm-cre (12) and the TfamloxP/TfamloxP, +/Myhca-cre knockout strains. This delay likely is determined by turnover rates for Tfam transcripts, Tfam protein, mtDNA, mitochondrial transcripts, and mtDNA-encoded respiratory chain subunits.
Most of the TfamloxP/TfamloxP, +/Myhca-cre knockouts died during the first week of life, but we observed that ≈25% of the knockouts, surprisingly, survived the neonatal period and lived for several months. When these longer-living knockouts were mated to each other, they produced knockout offspring that almost always survived (≈95%) the neonatal period. The life-span of the knockouts may thus be influenced strongly by modifying gene(s). Our findings suggest that the modifying gene(s) do not affect the efficiency of the cre-loxP recombination, because hearts from standard- and mutant-mating E18.5 knockouts had similar levels of Tfam recombination. The presented molecular, biochemical, and morphological data suggest that the standard-mating E18.5 knockouts, on average, had a more pronounced respiratory chain deficiency in their hearts than did the mutant-mating E18.5 knockouts. The data are compatible with the speculation that the longer-living mutant-mating knockouts carry modifying gene(s) that compensate for the consequences of the knockout. These genes may act to stabilize mtDNA, mitochondrial transcripts, or mitochondrial proteins, for example. However, it should be emphasized that, at present, other modes of action cannot be excluded, and positional cloning will be necessary to determine the exact nature of the modifying gene(s). Studies that breed the tissue-specific Tfam knockouts on several different genetic backgrounds will allow researchers to search systematically for genes that modify the phenotype. Such genes may provide novel insights into the pathogenesis of respiratory chain disorders and may also be of potential therapeutic value.
Acknowledgments
We thank Gail Martin and Mark Lewandoski for the generous gift of the plasmid pML78. N.-G.L. is supported by grants from the Swedish Heart and Lung Foundation, the Swedish Medical Research Council (K98–13X-12197–02B and K98–13P-12204–02B), Pharmacia-Upjohn, and Ronald McDonald Barnfond. P.T. is supported by a grant from the Swedish Medical Research Council (K99-14X-4764-24A). H.W. is supported by stipends from Drottning Silvias Barnfond and Samariten. A.H. is supported by a fellowship from the Swedish Foundation for Strategic Research.
Abbreviations
- Ckmm
muscle creatinine kinase
- COX
cytochrome c oxidase
- En
embryonic day n
- Myhca
α-myosin heavy chain
- SDH
succinate dehydrogenase
- Tfam
mitochondrial transcription factor A
- hw
heart weight
- bw
body weight
References
- 1.Larsson N-G, Clayton D A. Annu Rev Genet. 1995;29:151–178. doi: 10.1146/annurev.ge.29.120195.001055. [DOI] [PubMed] [Google Scholar]
- 2.Wallace D C. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 3.Graff C, Clayton D A, Larsson N-G. J Int Med. 1999;246:11–23. doi: 10.1046/j.1365-2796.1999.00514.x. [DOI] [PubMed] [Google Scholar]
- 4.Larsson N-G, Luft R. FEBS Lett. 1999;455:199–202. doi: 10.1016/s0014-5793(99)00854-6. [DOI] [PubMed] [Google Scholar]
- 5.Schon E A, Hirano M, DiMauro S. J Bioenerg Biomembr. 1995;26:291–299. doi: 10.1007/BF00763100. [DOI] [PubMed] [Google Scholar]
- 6.Munnich A, Rotig A, Chretien D, Saudubray J M, Cormier V, Rustin P. Eur J Pediatr. 1996;155:262–274. doi: 10.1007/BF02002711. [DOI] [PubMed] [Google Scholar]
- 7.Lightowlers R N, Chinnery P F, Turnbull D M, Howell N. Trends Genet. 1997;13:450–455. doi: 10.1016/s0168-9525(97)01266-3. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi J-I, Ohta S, Kikuchi A, Takemitsu M, Goto Y-I, Nonaka I. Proc Natl Acad Sci USA. 1991;88:10614–10618. doi: 10.1073/pnas.88.23.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moraes C T, Ciacci F, Silvestri G, Shanske S, Sciacco M, Hirano M, Schon E A, Bonilla E, DiMauro S. Neuromuscul Disord. 1993;3:43–50. doi: 10.1016/0960-8966(93)90040-q. [DOI] [PubMed] [Google Scholar]
- 10.Hao H, Morrison L E, Moraes C T. Hum Mol Genet. 1999;8:1117–1124. doi: 10.1093/hmg/8.6.1117. [DOI] [PubMed] [Google Scholar]
- 11.Larsson N-G, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh G S, Clayton D A. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Wilhelmsson H, Graff C, Li H, Oldfors A, Rustin P, Brüning J C, Kahn C R, Clayton D A, Barsh G S, et al. Nat Genet. 1999;21:133–137. doi: 10.1038/5089. [DOI] [PubMed] [Google Scholar]
- 13.Gulick J, Subramaniam A, Neumann J, Robbins J. J Biol Chem. 1991;266:9180–9185. [PubMed] [Google Scholar]
- 14.Rustin P, Chretien D, Bourgeron T, Gerard B, Rotig A, Saudubray J M, Munnich A. Clin Chim Acta. 1994;228:35–51. doi: 10.1016/0009-8981(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 15.Peto R, Pike M C, Armitage P, Breslow N E, Cox D R, Howard S V, Mantel N, McPherson K, Peto J, Smith P G. Br J Cancer. 1977;35:1–38. doi: 10.1038/bjc.1977.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyons G E, Schiaffino S, Sassoon D, Barton P, Buckingham M. J Cell Biol. 1990;111:2427–2436. doi: 10.1083/jcb.111.6.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palermo J, Gulick J, Colbert M, Fewell J, Robbins J. Circ Res. 1996;78:504–509. doi: 10.1161/01.res.78.3.504. [DOI] [PubMed] [Google Scholar]
- 18.Grove D, Zak R, Nair K G, Aschenbrenner V. Circ Res. 1969;25:473–485. doi: 10.1161/01.res.25.4.473. [DOI] [PubMed] [Google Scholar]
- 19.Moore G W, Hutchins G M, Bulkley B H, Tseng J S, Ki P F. Am Heart J. 1980;100:610–616. doi: 10.1016/0002-8703(80)90224-0. [DOI] [PubMed] [Google Scholar]
- 20.Threadgill D W, Dlugosz A A, Hansen L A, Tennenbaum T, Lichti U, Yee D, LaMantia C, Mourton T, Herrup K, Harris R C, et al. Science. 1995;269:230–234. doi: 10.1126/science.7618084. [DOI] [PubMed] [Google Scholar]
- 21.Cormier R T, Hong K H, Halberg R B, Hawkins T L, Richardson P, Mulherkar R, Dove W F, Lander E S. Nat Genet. 1997;17:88–91. doi: 10.1038/ng0997-88. [DOI] [PubMed] [Google Scholar]
- 22.Zielenski J, Corey M, Rozmahel R, Markiewicz D, Aznarez I, Casals T, Larriba S, Mercier B, Cutting G R, Krebsova A, et al. Nat Genet. 1999;22:128–129. doi: 10.1038/9635. [DOI] [PubMed] [Google Scholar]
- 23.Lyons G E, Mühlebach S, Moser A, Masood R, Paterson B M, Buckingham M E, Perriard J-C. Development (Cambridge, UK) 1991;113:1017–1029. doi: 10.1242/dev.113.3.1017. [DOI] [PubMed] [Google Scholar]