Abstract
To evaluate whether alterations in the multidrug-resistance (MDR)-1 gene correlate with intestinal MDR-1 expression and uptake of orally administered P-glycoprotein (PGP) substrates, we analyzed the MDR-1 sequence in 21 volunteers whose PGP expression and function in the duodenum had been determined by Western blots and quantitative immunohistology (n = 21) or by plasma concentrations after orally administered digoxin (n = 8 + 14). We observed a significant correlation of a polymorphism in exon 26 (C3435T) of MDR-1 with expression levels and function of MDR-1. Individuals homozygous for this polymorphism had significantly lower duodenal MDR-1 expression and the highest digoxin plasma levels. Homozygosity for this variant was observed in 24% of our sample population (n = 188). This polymorphism is expected to affect the absorption and tissue concentrations of numerous other substrates of MDR-1.
Keywords: cancer therapy, drug resistance, molecular transport, pharmacogenetics
The human multidrug-resistance (MDR)-1 gene encodes an integral membrane protein, P-glycoprotein (PGP), whose function is the energy-dependent export of substances from the inside of cells and from membranes to the outside (1–4). Its physiological role is the protection of cells from toxic substances or metabolites. PGP may also play a role in steroid metabolism. Many drugs are substrates of MDR-1. Therefore, degree of expression and the functionality of the MDR-1 gene product can directly affect the therapeutic effectiveness of such agents. This is of particular importance in cancer therapy where high expression and activity of MDR-1 cause cancer cells to become refractory to treatment with many agents, all of which are PGP substrates (1, 2). MDR-1 is also expressed on different nonmalignant cells in various organs, e.g., in the intestine and at the blood–brain barrier. Modulation of MDR-1 expression in these normal cell types can also influence the activity and bioavailability of drugs (5, 6). In the intestine, modulation of MDR-1 may control the degree of drug uptake after drug ingestion (7–9). At the blood–brain barrier, PGP influences the uptake of substrates into the brain (10–13): high PGP levels may limit the uptake of sufficient amounts of desired drugs into the brain, and reduced PGP activity could lead to abnormally increased accumulation in the brain and undesired side effects of drugs.
The “overall MDR-1 activity” controlling PGP-dependent drug transport depends on two parameters: (i) the level of expression of the MDR-1 gene controls the amount of protein that is synthesized in the cells, and (ii) the functionality of the MDR-1-encoded PGP, which determines which substrates are recognized and transported with what effectiveness. The first parameter, level of expression of MDR-1, has been intensively analyzed, particularly because the sensitivity of tumor cells toward chemotherapy often correlates inversely with increased MDR-1 expression. MDR-1 overexpression can be attributed partially to gene amplifications (14–17), but it is known that other factors must also exist (18–20). For example, rifampine induces MDR-1 (9). Allelic differences in individual MDR-1 gene sequences may be associated with or even causative for different expression levels. Regions in the human MDR-1 gene where sequence differences could influence expression levels would be the promoter and/or enhancer, sequences that influence the mode or efficacy of processing of the pre-mRNAs, and sequences that influence mRNA stability (4, 18–20). In addition, expression levels can be influenced by structural differences in the genome, such as chromatin alterations, methylation, or acetylation, which may be relevant not only directly in the MDR-1 gene, but also in the genomic sequence that surrounds MDR-1. It is difficult to identify directly and characterize nonpromoter/enhancer-associated sequence variations that affect levels of gene expression. However, the linear structure of the human genome on defined chromosomes opens the possibility of utilizing polymorphisms that do not directly influence expression levels as marker for other so-far-unidentified changes in and around the MDR-1 gene. Because of this linkage effect, defined alleles can serve as a marker for an important phenotype (expression levels) even if the MDR-1 sequence variations by themselves are not causative for that phenotype (21).
The second parameter that influences the overall MDR-1 activity in individuals is the functionality of PGP. Which substrates are recognized by the protein and pumped out of the cell with which effectiveness? This parameter is defined by the amino acid sequence of the protein that is encoded by the MDR-1 allele. Amino acid changes may influence the function of PGP. So far, two naturally occurring MDR-1 polymorphisms have been described and correlated with potential clinical effects (22). In that study, no connection between these changes and PGP function could be made. On the other hand, experiments with artificially introduced MDR-1 mutations show clearly that PGP reacts quite sensitively to amino acid alterations. Mutations that change amino acids can alter the substrate spectrum of PGP, the effectiveness of transport, and also the sensitivity of PGP toward inhibition with specific inhibitory substances (2, 23–34). It is clear that some naturally occurring mutations and alleles—if they exist, and if we can discover them—may show similar effects. It is unknown, however, how many such variations exist, with what frequency and at what positions in the human MDR-1 gene, and whether any of these can be correlated in humans with altered activity and/or side effects of drugs that are substrates of PGP.
Here we describe the identification and distribution of 15 polymorphisms in the human MDR-1 gene in a Caucasian population, one of which shows a significant correlation with MDR-1 expression levels and PGP activity in vivo.
Methods
Volunteers and DNA Samples.
Samples were from Caucasian volunteers from the Institute of Clinical Pharmacology at the University Medical Center, Charité in Berlin and from the Dr. Margarete Fischer-Bosch Institute of Clinical Pharmacology in Stuttgart, obtained under consideration of all ethical and legal requirements. Twenty-one volunteers and patients whose MDR-1 expression and activity profile had been analyzed were from clinical studies at the Dr. Margarete Fischer-Bosch Institute of Clinical Pharmacology in Stuttgart [study 112, healthy volunteers, male, 29 ± 5 yr, 84 ± 9 kg, Greiner et al. (9) and a followup study (study 113, O. von Richter and M. Eichelbaum, personal communication). Fourteen samples whose digoxin uptake under steady-state conditions was evaluated were from healthy volunteers from the University Medical Center, Charité in Berlin (35). Genomic DNA was prepared from blood samples by using the Qiagen (Chatsworth, CA) (QiaAmp) blood kits on a Qiagen 9604 robot.
Analysis of MDR-1 Expression and PGP Activity in Human Volunteers.
Expression levels in the duodenum were analyzed by quantitative immunohistochemistry (eight volunteers) and Western blot analysis with antibody F4 (Sigma) and C219 (eight volunteers + thirteen patients) (9). In one study (8 volunteers, 112–1 to 112–8) duodenal biopsies and in the second study (13 patients, 113-A to 113-P) enterocyte preparations of surgical specimens were used. For volunteers 112–1 to 112–8, CYP3A4 expression was also determined before and after rifampin induction by Western blot. Blots were quantified by densitometric evaluation of the PGP signals. To assure that quantitative immunohistochemistry of PGP and CYP3A4 reflects expression in intestinal enterocytes, an additional marker protein that is expressed in enterocytes, villin, was analyzed before and after rifampin induction. For quantification, an image analysis workstation (histoanalyzer) was used. For 112–1 to 112–8 PGP activity before and after induction was determined. PGP activity was tested by measuring the plasma concentrations before and between 0.17 and 144 h after the oral administration of digoxin (9). The area under the plasma concentration time curve (AUC) of orally administered digoxin correlates well with the intestinal amount of PGP (9) and serves as indicator for PGP activity. To assay blood digoxin levels under steady-state conditions, another group of healthy volunteers from a clinical study at the University Medical Center, Charité in Berlin (35) was analyzed, 6 women and 8 men (mean age, 26 yr; range 22 to 33 yr; mean weight, 67 kg; range 50 to 90 kg), comprised of 7 individuals with TT genotype (4 male, 3 female) and 7 individuals with CC genotype (4 male, 3 female). The volunteers were treated with 0.25 mg digoxin per day orally (steady state); details of the study are described by Johne et al. (35). Maximum concentrations (Cmax) were taken for comparison. Values were standardized according to lean body weight of individuals.
Identification of MDR-1 Polymorphisms.
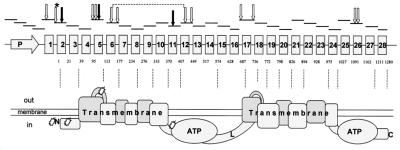
Specific oligonucleotide primers for amplification by PCR of MDR-1 gene fragments (28 exons and core promoter region) from genomic DNA were derived from known sequences [GenBank accession no: AC002457 for the promoter region and exon 1–7 and AC005068 for exons 8–28, (4)]; see Fig. 1. The primers were designed to bind to sequences upstream and downstream of the different exons to cover each exon, as well as sequences at the exon–intron boundaries that are important for mRNA splicing. Details regarding the primers, optimized PCR conditions, and subsequent purification and sequencing of the fragments are available at info@epidauros.com. Sequences of purified PCR fragments were obtained by automated DNA sequencing on ABI377 (gel) or ABI3700 (capillary) sequencers by using BigDye Terminator cycle sequencing reactions (Perkin–Elmer/Applied Biosystems). For allele-specific PCR, in a reaction volume of 20 μl, 50 ng genomic DNA template was added to buffer containing 1.5 mM MgCl2, 250 μM dNTPs,1 × Q-solution, 20 pmol of each primer, and 1 unit Taq polymerase. PCR was carried out in a GeneAmp PCR System 9700 (Perkin–Elmer) with an initial denaturation of 3 min at 95°C followed by 30 cycles of denaturation 94°C for 30 sec, primer annealing for 30 sec at temperatures between 50°C and 61°C depending on the individual primer combination, and 30 sec extension (8 min final) at 72°C. The sequences of the primer combinations for specific detection of MDR-1 alleles (exon 2), and the corresponding annealing temperature are listed in the legend to Fig. 2.
Figure 1.
Polymorphisms in the human MDR-1 gene. The positions of PCR fragments that we analyzed in our sequencing screen and the locations of the identified polymorphisms (arrows) are indicated in relation to the exon–intron structure of the human MDR-1 gene and the corresponding domain composition of PGP. *, SNP just before the translation start codon. White arrows depict noncoding polymorphisms, two of which are linked (broken line). Black arrows show the protein polymorphisms in exons 2, 5, and 11.
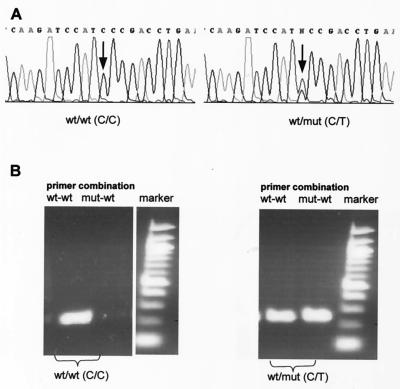
Figure 2.
Detection of MDR-1 polymorphisms. Examples for the identification of homozygous and heterozygous polymorphisms by sequence analysis (A) and by allele-specific PCR (B). Shown is the polymorphism in exon 2 that directly precedes the ATG start codon at cDNA pos.-1. MDR-1 sequence deviations can be detected directly in the DNA sequence profiles. A shows a wild-type sequence (Left) and (Right) the sequence of a fragment with a heterozygous polymorphism. (B) Allele-specific PCR (agarose gel of the PCR fragments) defines the MDR-1 genotype, i.e., the presence and/or absence of alleles. PCR was performed with human genomic DNA as with allele-specific primer combinations 5′-ggtttctcttcaggtcgg-G3′(wild type) or 5′-ggtttctcttcaggtcgg-A3′(mut), and 5′-ctcagccaacaaacttctgc-3′ (reverse) and annealing temperatures of 54°C for both reactions. The wild-type sample shows exclusively a fragment with the wild-type–primer combination; the heterozygous sample is positive for the wild-type primer combination as well as the primer combination that specifically detects the polymorphism.
Results
MDR-1 Polymorphisms.
All 28 exons and the core promoter region of MDR-1 were amplified by PCR from genomic DNA, covering the coding exons and sequences at the exon–intron boundaries that are important for mRNA splicing (Fig. 1). For the initial screen, sequence analyses were carried out for all gene fragments of 24 individuals. The screen was extended for selected MDR-1 gene fragments, some of which have been analyzed from a total of 188 individuals. The sequences were inspected for deviations from the original MDR-1 sequences (GenBank accession no.: AC002457; AC005068), which we define as “wild type.” Fifteen different polymorphisms were observed. The positions and sequences of the polymorphisms are listed in Table 1. Of the 15 polymorphisms, 12 do not alter the protein sequence. Seven are located in introns 4, 6, 12, 16, and 17, close to exon boundaries, and 3 at wobble positions that do not change amino acids, one in exon 12, and two in exon 26. One noncoding variation that occurs as heterozygous in 11.2% of our population was found at the nucleotide that directly precedes the ATG translation start codon. Three polymorphisms resulted in protein alterations in exons 2, 5, and 11. Exon 2 contains a polymorphism that changes Asn-21 to Asp. This variation occurs in 17.6% of the heterozygous volunteer population and in 0.5% homozygous. Exon 5 showed in 1.2% of the heterozygotes a polymorphism that changes Phe-103 to Leu and in exon 11, Ser-400 is changed to Asn in 12.9% (heterozygous) of the sample population. The sequences, positions, and frequencies of the polymorphisms are summarized in Table 1. A comparison of the experimental and predicted (Hardy–Weinberg equilibrium) genotype frequencies regarding different MDR-1 polymorphisms (Table 1) shows that the determined genotype distribution agrees with the predicted distribution. Sequencing of gene fragments is often not suitable to define linked SNPs, except for cases in which alleles occur homozygously. This is the case for a C/T SNP in intron 6 and the C/T SNP in exon 12 that appear linked. We found (with one exception; see Table 2) consistently at these positions the homozygous combinations T/T-T/T and C/C-C/C (indicated as broken line in Fig. 1). For other polymorphisms, linkage could not be predicted directly from our sequence data, but we used allele-specific PCR as alternative to analyze linkage. One example for specific detection of MDR-1 variations by allele-specific PCR is shown in Fig. 2. Exon 2 harbors two different polymorphisms that appear as singular SNPs in most samples. However, in seven samples, both polymorphisms were observed to be combined heterozygously. Fig. 2b shows that, by using allele-specific primers deduced from the SNP-containing DNA sequences in exon 2 and wild-type primers, PCR is suitable to distinguish defined MDR-1 alleles (shown for exon 2; not shown for other SNPs, e.g., in exon 5). However, our attempts to determine allele haplotypes in exon 2 by PCR did not give a clear answer. Therefore, the haplotypes of these samples were determined by sequencing of plasmids into which double-heterozygous PCR-fragment mixtures had been cloned. This proved unambiguously that the different polymorphisms in exon 2 were on separate alleles and not linked in our volunteers.
Table 1.
Positions, sequences, and frequencies of MDR-1 variations
| MDR-1 position, exon | Nucleotide sequence
|
Frequency, %
|
Region | Effect | ||||
|---|---|---|---|---|---|---|---|---|
| Wild type | mut | Analyzed individuals, N | Heterozygous | Homozygous | Homozygous calculated | |||
| 1b/12 | cgagTagcg | cgagCagcg | 85 | 11.8 | 0 | 0.4 | Exon 1 | Noncoding |
| 2/−1 | tcggGatgg | tcggAatgg | 188 | 11.2 | 0 | 0.4 | Exon 2 | TL initiation |
| 2/61 | gaacAataa | gaacGataa | 188 | 17.6 | 0.5 | 0.81 | Exon 2 | Asn-Asp |
| 5/−25 | aatgGtatg | aatgTtatg | 85 | 26 | 3.5 | 2.3 | Intron | |
| 5/−35 | aagaGacat | aagaCacat | 85 | 1.2 | 0 | 0.01 | Intron | |
| 5/307 | agggTtctt | agggCtctt | 85 | 1.2 | 0 | 0.01 | Exon 5 | Phe-Leu |
| 6/+139 | gcaaCaatg | gcaaTaatg | 85 | 48.2 | 16.5 | 16.8 | Intron | ## |
| 6/+145 | atgtCgtgt | atgtTgtgt | 85 | 2.4 | 0 | 0.01 | Intron | |
| 11/1199 | ttcaGttac | ttcaAttac | 85 | 12.9 | 0 | 0.4 | Exon 11 | Ser-Asn |
| 12/1236 | agggCctga | agggTctga | 188 | 48.9 | 13.3 | 14.4 | Exon 12 | Wobble ## |
| 12/+44 | cagtCacat | cagtTacat | 188 | 11.7 | 0 | 0.4 | Intron | |
| 17/−76 | ttacTaatt | ttacAaatt | 85 | 45.9 | 22.4 | 20.3 | Intron | |
| 17/+137 | gaagAtgta | gaagGtgta | 85 | 1.2 | 0 | 0.01 | Intron | |
| 26/3435 | agatCgtga | agatTgtga | 188 | 48.3 | 23.9 | 23 | Exon 26 | Wobble |
| 26/3396 | ttgcCtatg | ttgcTtatg | 188 | 0.53 | 0 | 0.01 | Exon 26 | Wobble |
The positions of the identified polymorphisms correspond to positions of the MDR-1 cDNA (GenBank accession no. M14758, codon TTC exon 10, F335, is missing in that sequence), with the first base of the ATG start codon set to 1. SNPs that are located in introns are presented as (exon+/−n), i.e., n nucleotides upstream (−) or downstream (+) of the exons that were defined by Chen et al. (4). Fifteen polymorphisms were identified. The number of samples that were included in the allele frequency determination is listed as (N). Homozygous calculated: the predicted ratios of the homozygous genotypes (q2) were calculated on the basis of the Hardy–Weinberg distribution, using the formulas p = (2 × AA + 1 × Aa)/2N and p + q = 1: AA = number of probands homozygous for the wild-type allele, Aa = number of heterozygotes, N = size of the sample test, p = frequency of the wild-type allele, q = frequency of the mut allele, q2 = frequency of the genotype homozygous for the mut allele. # indicates polymorphisms that are linked.
Table 2.
Polymorphisms in phenotyped individuals; correlation with MDR-1 expression
| MDR-1 position, exon | Type of polymorphism | Wild type (−) | 112 1 | 113 D | 113 F | 113 K | 113 P | 112 2 | 112 4 | 112 5 | 112 8 | 113 A | 113 C | 113 E | 113 G | 113 N | 113 O | 112 3 | 112 6 | 112 7 | 113 I | 113 L | 113 M |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1/12 | Exon, 5′ noncoding | T | — | — | — | — | — | — | — | T/C | — | — | T/C | T/C | — | — | — | — | T/C | — | — | — | — |
| 2/−1 | Exon, TL initiation | G | — | — | — | — | — | — | — | — | — | — | G/A | — | — | — | — | — | — | — | — | — | G/A |
| 2/61 | Exon, Asn-Asp | A | — | — | — | A/G | — | A/G | — | — | — | — | — | A/G | — | — | A/G | — | — | — | — | — | — |
| 5/−25 | Intron | G | — | — | — | — | — | G/T | — | — | — | — | — | G/T | G/T | — | — | G/T | G/T | — | — | T/T | — |
| 5/−35 | Intron | G | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | G/C | — | — |
| 5/307 | cDNA, Phe-Leu | T | — | — | — | — | — | — | — | T/C | T/C | — | — | — | — | — | — | — | — | — | — | — | — |
| 6/+139 | Intron | C | C/T | C/T | T/T | C/T | T/T | C/T | C/T | C/T | — | C/T | — | C/T | C/T | — | T/T | — | — | T/T | — | — | — |
| 6/+145 | Intron | C | — | — | — | C/T | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 11/1199 | cDNA, Ser-Asn | G | — | — | — | G/A | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 12/1236 | cDNA, wobble | C | C/T | C/T | T/T | C/T | T/T | C/T | C/T | C/T | — | C/T | — | C/T | C/T | — | — | — | — | T/T | — | — | — |
| 12/+44 | Intron | C | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | C/T | — |
| 17/−76 | Intron | T | T/A | — | — | T/A | — | — | T/A | T/A | T/A | T/A | A/A | — | — | A/A | — | T/A | T/A | — | A/A | T/A | A/A |
| 17/+137 | Intron | A | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | A/G | — | — | — |
| 26/3396 | cDNA, wobble | C | — | — | — | — | — | — | C/T | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 26/3435 | cDNA, wobble | C | T/T | C/T | C/C | ||||||||||||||||||
| PGP expression (OD) | 211 | 568 | 2142 | 176 | 40 | 529 | 1052 | 1430 | 828 | 981 | 246 | 1595 | 781 | 145 | 1972 | 365 | 1150 | 1108 | 554 | 1817 | 2658 | ||
| PGP expression, mean (N = 21) | 627 | 956 | 1275 | ||||||||||||||||||||
| PGP expression, % | 15 | 21 | 81 | 7 | 2 | 37 | 73 | 100 | 58 | 37 | 9 | 62 | 29 | 5 | 74 | 26 | 80 | 77 | 21 | 68 | 100 | ||
| PGP expression, %, mean (N = 21) | 25 | 48 | 62 | ||||||||||||||||||||
| Rifampin induction digoxin (AUC), mean (N = 8) | 57.3 | 39.9 | 28.6 | ||||||||||||||||||||
The MDR-1 allele distribution in volunteers and patients was determined by direct sequencing of MDR-1 gene fragments. − indicates homozygous wild-type sequences at the defined positions. The MDR-1 phenotype of the 21 samples was determined by Western blot analysis (9). The MDR-1 genotype in exon 26 was determined by DNA sequencing. Because the expression levels for the samples were determined in two distinct Western blot experiments, the absolute OD values for one set can differ from the values for the other set because of time differences during the exposition to the x-ray film. Therefore in each set the highest expression level was equated to 100% and, beside the densitometric OD values for the MDR-1 expression, the percentage of the expression is also listed. The correlation of the exon 26 SNP with MDR-1 expression and activity after rifampin induction was analyzed in eight volunteers of this group.
A Polymorphism in Exon 26 Correlates with Intestinal MDR-1 Expression in Vivo.
To analyze whether MDR-1 polymorphisms correlate with the levels of intestinal MDR-1 expression, we determined the MDR-1 genotype distribution in volunteers and patients of two experimental studies (n = 21) at the Dr. Margarete Fischer-Bosch Institute of Clinical Pharmacology in Stuttgart (9). The expression levels of MDR-1 in the intestine of these volunteers and patients had been determined by quantitative immunohistochemistry and Western blots of biopsies and enterocyte preparations of the duodenum. To assure that this analysis reflects the specific PGP expression in intestinal enterocytes, an additional marker protein that is expressed in enterocytes, villin, was simultaneously analyzed (see Methods and ref. 9). A substudy with eight volunteers also included the measurement of duodenal PGP after induction of MDR-1 expression with rifampin.
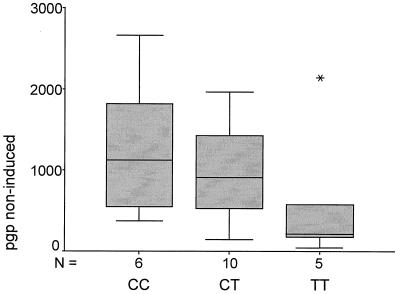
A comparison of the MDR-1 genotyping results and corresponding duodenal PGP levels of 21 volunteers and patients, listed in Table 2, shows a correlation of the levels of PGP expression and a polymorphism in exon 26 (C/T at wobble position 3435). The homozygous T-allele is associated with more than 2-fold lower MDR-1 expression levels compared with homozygous CC samples. Heterozygous individuals display an intermediate phenotype (Fig. 3). These differences have a significance of P = 0.056 (Jonckheere–Terpstra test). Consistent with this correlation, the individual with the lowest PGP levels displayed the homozygous T/T genotype and the individual with the highest expression levels the C/C phenotype (underlined in Table 2). The difference in PGP levels between these individuals was >65-fold (40 vs. 2,658 OD). A subpopulation of n = 8, for which MDR-1 induction experiments were performed (ref. 9; see below), also showed a correlation of the MDR-1 genotype at position 3435 with MDR-1 expression levels under rifampin-induced conditions. The mean of the MDR-1 protein levels of the C-allele-carrying volunteers is much higher compared with those carrying the T-allele (Fig. 4A). Consistent with that, we found that a volunteer homozygous for the T-allele had the lowest rifampin-induced PGP level, whereas a volunteer homozygous for the C-allele had the highest level of all probands tested.
Figure 3.
Correlation of the exon 26 SNP with MDR-1 expression. The MDR-phenotype (expression and activity) of 21 volunteers and patients was determined by Western blot analyses. The box plot shows the distribution of MDR-1 expression clustered according to the MDR-1 genotype at the relevant exon 26 SNP. The genotype–phenotype correlation has a significance of P = 0.056 (n = 21).
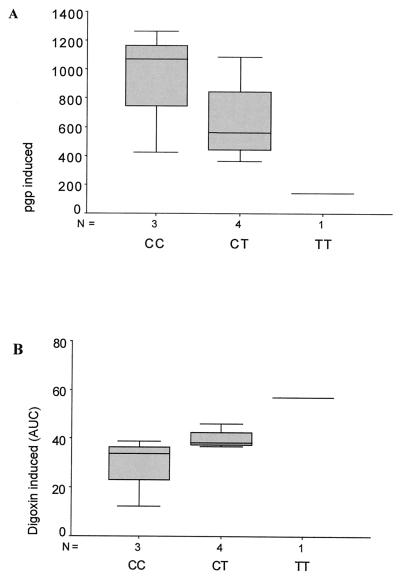
Figure 4.
MDR-1 expression and PGP in vivo activity after rifampin induction. MDR-1 genotype in exon 26 and distribution of rifampin-induced PGP protein expression (A) in the duodenum. (B) Distribution of plasma levels of digoxin after rifampin induction. The plasma levels of digoxin area under the curve, μg × h × L−1 (AUC) are inversely proportional to PGP activity in the duodenum (9). The correlation of MDR genotype and PGP activity has a significance of P = 0.053 (n = 8).
Correlation of a Polymorphism in Exon 26 with MDR-1 Activity in Vivo.
In eight volunteers, we analyzed not only expression levels but also the PGP activity in duodenum after rifampin induction, by measuring the blood concentrations of orally administered digoxin (1 mg), which is a PGP substrate (9). It is known by measurements of intestinal PGP protein levels and comparisons to plasma levels of digoxin after oral application that digoxin plasma concentrations show an inverse correlation with the MDR-1 expression in duodenum (9). In such experiments, measurements of digoxin uptake are performed shortly after oral intake, and therefore effects of digoxin metabolism are neglectable. Thus, digoxin plasma concentrations can be used to assay PGP activity.
We observe a statistically significant inverse correlation of the exon 26 polymorphism with digoxin plasma levels, which reflects PGP activity in vivo. Fig. 4B shows that volunteers that harbor the T-allele (weaker MDR-1 expression) contain higher plasma levels of digoxin compared with samples that have a C/C genotype. The mean of the rifampin-induced digoxin concentrations (see Table 2) of the C-allele population is consistently lower than those of the T-population (28.6 vs. 57.3/39.9 digoxin induced). In total agreement with that, a volunteer homozygous for the T-allele had the highest detectable digoxin concentration in the blood after rifampin induction (AUC 57.3), and a volunteer homozygous for the C-allele displayed the lowest level of all (AUC 12.3). The difference of the digoxin plasma levels of these individuals was more than 4-fold. Even in this small population of eight rifampin-induced volunteers (which could not be extended because the study has been closed), the correlation of the MDR-1 genotype and low/high PGP activity was statistically significant (P = 0.053, Jonckheere–Terpstra test).
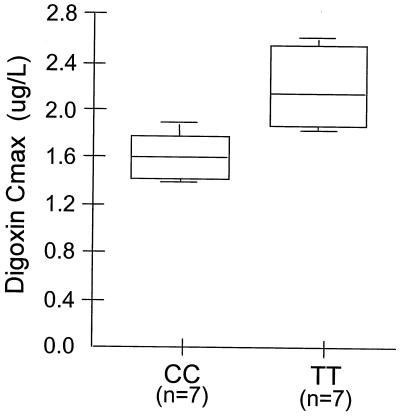
To validate further the correlation of MDR-1 genotype with intestinal digoxin uptake, additional volunteers of another clinical study that addresses blood levels of digoxin after oral application (without rifampin induction and PGP protein determination; ref. 35) were evaluated for their MDR-1 genotype in exon 26. In this study, maximum plasma concentrations (Cmax) were evaluated during steady-state conditions of digoxin. This pharmacokinetic parameter especially reveals differences in the absorption of digoxin between the different groups. Fig. 5 shows a comparison of digoxin Cmax of seven volunteers that carried homozygously the T/T-allele and seven volunteers with the homozygous C/C genotype in exon 26. The mean difference of 38% in digoxin Cmax between the groups is statistically significant (P = 0.006, Mann–Whitney U two-sample test) and reflects the impact of this polymorphism on digoxin pharmacokinetics.
Figure 5.
Correlation of MDR-1 genotype and digoxin uptake in vivo. The MDR-1 genotype in exon 26 was analyzed in 14 healthy volunteers who participated in a clinical study that addresses plasma levels of digoxin during steady-state conditions (35). A statistically significant difference (P = 0.006; Mann–Whitney U two-sample test) was found in the comparison of maximum concentrations (Cmax) of digoxin between two groups of seven healthy volunteers harboring either T/T or C/C genotype. The mean difference of 38% in Cmax may reflect the importance of genotype on the absorption of digoxin after oral application. A 0.25 mg dose was applied on steady-state of digoxin.
Discussion
We have identified polymorphisms in the human MDR-1 gene and described their distribution in a Caucasian population. Among 15 different polymorphisms that we found were 11 noncoding SNPs in introns and wobble positions and a noncoding SNP that directly preceded the ATG translation initiation codon. Three polymorphisms, located in exons 2, 5, and 11, changed the amino acid sequence. In the populations whose MDR-1 expression levels (n = 21) and activity in the duodenum (n = 8 + 14) were characterized, one wobble polymorphism in exon 26 correlates in a statistically significant manner with PGP expression and activity in vivo.
MDR-1 Polymorphisms.
The discovery and characterization of variations in the MDR-1 gene and diagnostic tests for the discrimination of different MDR-1 alleles in human individuals may provide a potent tool for improving the therapy of diseases with drugs that are substrates of the MDR-1 gene product and whose cellular uptake is therefore dependent on MDR-1. The identification of MDR-1 variations may also be useful to predict therapy outcome, an improved approach compared with determination of general MDR-expression. Furthermore, the characterization of novel MDR-1 variants may be useful for the development of novel inhibitors that specifically modulate the activity of the individual types of MDR-1. The feasibility of using inhibitors of individual (artificially created) MDR-1 variants and their potential therapeutic application has recently been demonstrated in a model system (30, 31). Mickley et al. have recently described two genetic polymorphisms in MDR-1 at positions 2677 and 2995 in exons 21 and 24 (22). These polymorphisms were identified in selected cell lines as well as in healthy volunteers and in refractory lymphomas. Although in that study the polymorphism at position 2677 was found heterozygously in 43% of the samples, we did not find this polymorphism, even though for this position 24 different individuals were screened. The positions on the MDR-1 gene of different polymorphisms that we found in our screen, in relation to the domain structure of the MDR-1-encoded PGP protein, are summarized in Fig. 1. Among the discovered polymorphisms, four appear to be particularly interesting. Three of these change the amino acid sequence of PGP: at PGP position 21 (exon 2), Asn is replaced by Asp, which results in a net charge change (basic to acidic) at this position. This polymorphism is located close to the N terminus of PGP. So far, mutational analyses of PGP have not revealed this region to be of major importance for the function of PGP (2). But we certainly cannot rule out that this particular mutation may still affect the function of PGP. Another protein alteration that we found changes phenylalanine 103 in exon 5 to leucine. This protein polymorphism is located on the extracellular side of PGP before the second transmembrane domain. The position of this polymorphism is close to glycosylation sites that are known in PGP. The polymorphism results in a change from a large aromatic residue to another large lipophilic residue. The impact on the structure of the exon-5-encoded region, e.g., whether the side-chain packing becomes disturbed and therefore the structure becomes altered, remains to be clarified. Because this polymorphism is located in an extracellular domain of the protein, it may be possible to use it for antibody-based diagnosis of this MDR-1 allele. In exon 11, a polymorphism replaces Ser-400 to asparagine, thus the side chain has a significantly different size and possibly a charge difference, dependent on pH and isoelectric environment of the residue within the protein structure. The polymorphism is located on the cytoplasmic side just preceding the first ATP-binding domain. Functional ATP domains are essential for the function of PGP (32–34). Therefore, further analyses are required to determine whether this polymorphism modulates the function of PGP.
Correlation with MDR-1 Phenotype.
The access to a population of 8 volunteers and 13 patients whose MDR-1 status, i.e., expression (n = 21) and activity in the duodenum (n = 8), had been thoroughly analyzed in experimental clinical trials led us to evaluate whether any of our identified polymorphisms correlated with a MDR-1 phenotype. Because of the limited size of the population, potential correlations with rare protein polymorphisms could not be tested; only one of the individuals carried heterozygously the exon 11 (Ser-Asn) polymorphism, four harbored heterozygously the exon 2 (Asn-Asp) polymorphism, and only two heterozygously the exon 5 (Phe-Leu) polymorphism. Also, effects of the SNP that directly precedes the translation initiation codon and thereby might influence translation efficacy and expression levels of MDR-1 could not be analyzed because only two individuals carried heterozygously the SNP at this position. None of the individuals that carried these four polymorphisms showed any unusually high or low PGP expression or activity that would indicate an effect of those polymorphisms.
Interestingly, among the noncoding SNPs, one at a wobble position in exon 26 correlated with expression levels and in vivo activity of PGP in this population (Table 2). This correlation regarding the PGP expression was observed with statistical significance in various approaches that determine MDR-1 expression as well as intestinal PGP activity: the levels of MDR expression in the duodenum correlate with the exon 26 SNP (P = 0.056, n = 21, Jonckheere–Terpstra test) under noninduced as well as under rifampin-induced conditions (n = 8). Differences in intestinal PGP activity under normal steady-state conditions show a significant correlation with SNP (P = 0.006, n = 14) as well as PGP activity after induction of MDR-1 expression with rifampin (P = 0.053, n = 8).
Among the 21 samples for which PGP protein levels were determined, only one clearly deviated from the described expression vs. genotype correlation. In this homozygous T/T sample, high duodenal PGP concentrations were measured even though this genotype correlates otherwise with low MDR-1 expression and low PGP activity. The data set of this sample (113-F) is included in our statistical calculations. It is known that levels of gene expression are controlled by multiple factors, and thus the high PGP levels in this individual may be caused by other so far unidentified and possibly not genetically determined factors. An exclusion of this one sample (of 21) from our data set (with the reasonable assumption that its PGP expression is “abnormal” and influenced by other unknown factors) would increase the statistical significance of the genotype–phenotype correlation from P = 0.056 (n = 21) to a significance of P = 0.019 in the Jonckheere–Terpstra test (n = 20). Because of its location at a noncoding nonpromoter position in the MDR-1 gene, it is unlikely that this SNP directly influences MDR-1 expression. Instead, it may be linked to other so far unidentified changes in regions of the MDR-1 gene that control expression, e.g., in the promoter/enhancer region or in sequences that are important for mRNA processing. Even though this particular SNP may not be causative for expression differences, it still can differentiate alleles with distinct MDR-1 expression and activity. Further studies with different substrates are required to confirm the general correlation of the SNP with PGP levels and drug uptake.
Variations in the expression levels and in the activity of MDR-1-encoded PGP have a major impact on the therapeutic efficacy of many drugs. Therefore, the ability to detect MDR-1 alleles that are relevant for the expression or activity of PGP is of great importance for the treatment of patients with agents that are substrates of the MDR-1 gene product.
Acknowledgments
Parts of this work were supported by grants 01EC9405 (Clinical Pharmacogenetics) and 01GG984–8/14, –6/8, –5/5 (Pharmacogen Diagnostic), from the German Ministry for Education, Science, Research and Technology and by the Robert Bosch Foundation in Stuttgart (PGP expression analyses and digoxin measurements). We thank S. Escher, K. Wuppermann, A. Lihl, H. Roeck, and G. Schwake for excellent technical and scientific support, U. Meyer and E. Mutschler for useful suggestions, and the staff of the Department of Surgery of the Robert Bosch Hospital for providing surgical samples.
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050585397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050585397
References
- 1.Gottesman M M, Pastan I, Ambudkar V. Curr Opin Genet Dev. 1996;6:610–617. doi: 10.1016/s0959-437x(96)80091-8. [DOI] [PubMed] [Google Scholar]
- 2.Ambudkar S V, Dey S, Hrycyna C A, Ramachandra M, Pastan I, Gottesman M M. Annu Rev Pharmacol Toxicol. 1999;39:361–398. doi: 10.1146/annurev.pharmtox.39.1.361. [DOI] [PubMed] [Google Scholar]
- 3.Ueda K, Clark D P, Chen C J, Roninson I B, Gottesman M M, Pastan I. J Biol Chem. 1987;262:505–508. [PubMed] [Google Scholar]
- 4.Chen C J, Clark D, Ueda K, Pastan I, Gottesman M M, Roninson I B. J Biol Chem. 1990;265:506–514. [PubMed] [Google Scholar]
- 5.Schinkel A H. Int J Clin Pharmacol Ther. 1998;36:9–13. [PubMed] [Google Scholar]
- 6.Schinkel A H, Mayer U, Wagenaar E, Mol C A, van Deemter L, Smit J J, van der Valk M A, Voordouw A C, Spits H, van Tellingen O, et al. Proc Natl Acad Sci USA. 1997;94:4028–4033. doi: 10.1073/pnas.94.8.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cordon-Cardo C, O'Brien J P, Boccia J, Casals D, Bertino J R, Melamed M R. J Histochem Cytochem. 1990;38:1277–1287. doi: 10.1177/38.9.1974900. [DOI] [PubMed] [Google Scholar]
- 8.Schuetz E G, Schinkel A H, Relling M V, Schuetz J D. Proc Natl Acad Sci USA. 1996;93:4001–4005. doi: 10.1073/pnas.93.9.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greiner B, Eichelbaum M, Fritz P, Kreichgauer H P, von Richter O, Zundler J, Kroemer H K. J Clin Invest. 1999;104:147–153. doi: 10.1172/JCI6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schumacher U, Mollgard K. Histochem Cell Biol. 1997;108:179–182. doi: 10.1007/s004180050159. [DOI] [PubMed] [Google Scholar]
- 11.Rao V V, Dahlheimer J L, Bardgett M E, Snyder A Z, Finch R A, Sartorelli A C, Piwnica-Worms D. Proc Natl Acad Sci USA. 1999;96:3900–3905. doi: 10.1073/pnas.96.7.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thiebaut F, Tsuruo T, Hamada H, Gottesman M M, Pastan I, Willingham M C. J Histochem Cytochem. 1989;37:159–164. doi: 10.1177/37.2.2463300. [DOI] [PubMed] [Google Scholar]
- 13.Thiebaut F, Tsuruo T, Hamada H, Gottesman M M, Pastan I, Willingham M C. Proc Natl Acad Sci USA. 1987;84:7735–7738. doi: 10.1073/pnas.84.21.7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen C J, Chin J E, Ueda K, Clark D P, Pastan I, Gottesman M M, Roninson I B. Cell. 1986;47:381–389. doi: 10.1016/0092-8674(86)90595-7. [DOI] [PubMed] [Google Scholar]
- 15.Schoenlein P V, Shen D W, Barrett J T, Pastan I, Gottesman M M. Mol Biol Cell. 1992;3:507–520. doi: 10.1091/mbc.3.5.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knutsen T, Mickley L A, Ried T, Green E D, du Manoir S, Schrock E, Macville M, Ning Y, Robey R, Polymeropoulos M, Torres R, et al. Genes Chromosomes Cancer. 1998;23:44–54. doi: 10.1002/(sici)1098-2264(199809)23:1<44::aid-gcc7>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 17.Yoshimoto K, Iwahana H, Yokogoshi Y, Saito S, Shiraishi M, Sekiya T, Gottesman M M, Pastan I. Nucleic Acids Res. 1988;16:11850. doi: 10.1093/nar/16.24.11850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chin K V, Chauhan S S, Pastan I, Gottesman M M. Cell Growth Differ. 1990;1:361–365. [PubMed] [Google Scholar]
- 19.Chaudhary P M, Roninson I B. J Natl Cancer Inst. 1993;85:632–639. doi: 10.1093/jnci/85.8.632. [DOI] [PubMed] [Google Scholar]
- 20.Germann U A, Schoenlein P V, Zimonjic D B, Popescu N C, Pastan I, Gottesman M M. Genes Chromosomes Cancer. 1994;10:267–274. doi: 10.1002/gcc.2870100408. [DOI] [PubMed] [Google Scholar]
- 21.Long A D, Langley C H. Genome Res. 1999;9:720–731. [PMC free article] [PubMed] [Google Scholar]
- 22.Mickley L A, Lee J S, Weng Z, Zhan Z, Alvarez M, Wilson W, Bates S E, Fojo T. Blood. 1998;91:1749–1756. [PubMed] [Google Scholar]
- 23.Ramachandra M, Ambudkar S V, Gottesman M M, Pastan I, Hrycyna C A. Mol Biol Cell. 1996;7:1485–1498. doi: 10.1091/mbc.7.10.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller M, Bakos E, Welker E, Varadi A, Germann U A, Gottesman M M, Morse B S, Roninson I B, Sarkadi B. J Biol Chem. 1996;271:1877–1883. doi: 10.1074/jbc.271.4.1877. [DOI] [PubMed] [Google Scholar]
- 25.Germann U A, Chambers T C, Ambudkar S V, Licht T, Cardarelli C O, Pastan I, Gottesman M M. J Biol Chem. 1996;271:1708–1716. doi: 10.1074/jbc.271.3.1708. [DOI] [PubMed] [Google Scholar]
- 26.Loo T W, Clarke D M. J Biol Chem. 1997;272:31945–31948. doi: 10.1074/jbc.272.51.31945. [DOI] [PubMed] [Google Scholar]
- 27.Loo T W, Clarke D M. J Biol Chem. 1995;270:22957–22961. doi: 10.1074/jbc.270.39.22957. [DOI] [PubMed] [Google Scholar]
- 28.Loo T W, Clarke D M. Methods Enzymol. 1998;292:480–492. doi: 10.1016/s0076-6879(98)92037-7. [DOI] [PubMed] [Google Scholar]
- 29.Gros P, Dhir R, Croop J, Talbot F. Proc Natl Acad Sci USA. 1991;88:7289–7293. doi: 10.1073/pnas.88.16.7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dey S, Hafkemeyer P, Pastan I, Gottesman M M. Biochemistry. 1999;38:6630–6639. doi: 10.1021/bi983038l. [DOI] [PubMed] [Google Scholar]
- 31.Hafkemeyer P, Dey S, Ambudkar S V, Hrycyna C A, Pastan I, Gottesman M M. Biochemistry. 1998;37:16400–16409. doi: 10.1021/bi980871+. [DOI] [PubMed] [Google Scholar]
- 32.Hrycyna C A, Airan L E, Germann U A, Ambudkar S V, Pastan I, Gottesman M M. Biochemistry. 1998;37:13660–13673. doi: 10.1021/bi9808823. [DOI] [PubMed] [Google Scholar]
- 33.Dey S, Ramachandra M, Pastan I, Gottesman M M, Ambudkar S V. Methods Enzymol. 1998;292:318–328. doi: 10.1016/s0076-6879(98)92025-0. [DOI] [PubMed] [Google Scholar]
- 34.Dey S, Ramachandra M, Pastan I, Gottesman M M, Ambudkar S V. Proc Natl Acad Sci USA. 1997;94:10594–10599. doi: 10.1073/pnas.94.20.10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johne A, Brockmöller J, Bauer S, Maurer A, Langheinrich M, Roots I. Clin Pharmacol Ther. 1999;66:338–345. doi: 10.1053/cp.1999.v66.a101944. [DOI] [PubMed] [Google Scholar]