Abstract
Phylogenetic analyses of DNA sequences have prompted spectacular progress in assembling the Tree of Life. However, progress in constructing phylogenies among closely related species, at least for plants, has been less encouraging. We show that for plants, the rapid accumulation of DNA characters at higher taxonomic levels has not been matched by conventional sequence loci at the species level, leaving a lack of well-resolved gene trees that is hindering investigations of many fundamental questions in plant evolutionary biology. The most popular approach to address this problem has been to use low-copy nuclear genes as a source of DNA sequence data. However, this has had limited success because levels of variation among nuclear intron sequences across groups of closely related species are extremely variable and generally lower than conventionally used loci, and because no universally useful low-copy nuclear DNA sequence loci have been developed. This suggests that solutions will, for the most part, be lineage-specific, prompting a move away from ‘universal’ gene thinking for species-level phylogenetics. The benefits and limitations of alternative approaches to locate more variable nuclear loci are discussed and the potential of anonymous non-genic nuclear loci is highlighted. Given the virtually unlimited number of loci that can be generated using these new approaches, it is clear that effective screening will be critical for efficient selection of the most informative loci. Strategies for screening are outlined.
Keywords: Tree of Life, plant phylogeny, nuclear DNA sequence loci, low-copy nuclear gene, comparative anchor tagged sequence, sequence characterized amplified region
1. Introduction
Reconstructing the Tree of Life has been a central objective of evolutionary biology since phylogenetic trees were first proposed as a way of representing evolutionary relationships. Progress towards that goal has accelerated in the last two decades and assembling the Tree of Life is now the focus of more research than at any time over the past 150 years (e.g. Soltis & Soltis 2001; Donoghue & Cracraft 2004; Palmer et al. 2004). This has been prompted by theoretical and methodological advances, vastly increased computational power, and technical improvements in DNA sequencing that enable the generation of large volumes of character data relatively quickly and cheaply. It is now possible to build robust hypotheses of relationships for large numbers of taxa and to contemplate the vision of a complete Tree of Known Life (Soltis & Soltis 2001; Watanabe 2002; Donoghue & Cracraft 2004). Current interest in phylogeny reconstruction has also been driven by ever wider application of phylogenetic trees within as well as beyond the confines of systematics and evolutionary biology (e.g. Soltis et al. 1999; Savolainen & Chase 2003; Futuyma 2004; Yates et al. 2004). Phylogenetic trees provide not only the basis for classification, but also studies of character evolution (Schultheis & Baldwin 1999), hybridization (e.g. Hughes et al. 2002; Linder & Rieseberg 2004), polyploidy (e.g. Doyle et al. 2003b, 2004), biogeography (e.g. Lavin et al. 2004; Pennington et al. 2004), origins of domestication (e.g. Wang et al. 1999; Nesbitt & Tanksley 2002) and speciation and species diversification (e.g. Barraclough & Vogler 2000; Barraclough & Nee 2001). Phylogenies also provide information to develop comprehensive comparative systems in developmental biology (e.g. Doust & Kellogg 2002; Thießen et al. 2002) and comparative genomics (e.g. Bennetzen & Kellogg 1997; Eisen & Fraser 2003; Hong et al. 2003). In the last two decades, phylogenies have come of age as a universal component of comparative biology (Hillis 2004).
Questions remain about the best ways to measure progress towards estimating the Tree of Life (Donoghue 2004). Numbers of published phylogenetic trees and taxa represented therein alongside measures of levels of resolution among taxa, statistical confidence in published trees (Sanderson 1995), and discovery of paraphyly, polyphyly, and circumscription of monophyletic groups, all suggest that recent progress has indeed been impressive (reviewed in Cracraft & Donoghue 2004). However, measures of success are greatly influenced by the intended use(s) of the resultant phylogenetic trees. For the purposes of ridding classifications of non-monophyletic groups, completely resolved trees with uniformly high statistical support for all nodes are not necessarily required. Analyses of exemplar taxa that resolve a subset of well-supported nodes will often suffice for the purposes of providing robust phylogenetic classifications. There is little doubt that in this sense there has been massive progress towards assembling the Tree of Life. The angiosperms provide a good example. Despite some persistent areas of poor resolution and/or weak support (e.g. Wortley et al. 2005), the branching order of most clades of angiosperms is now relatively clear and well supported (e.g. Soltis et al. 1999; Savolainen & Chase 2003; Soltis & Soltis 2004), prompting a new classification (APGII 2003). Similar progress has been made towards delimiting genera and understanding relationships within many angiosperm families (e.g. Lavin et al. 2001). In this realm there are clear theoretical reasons to believe that further progress can be made by simply increasing the number of characters via sequencing additional readily accessible loci that can be combined in simultaneous analyses to increase resolution, accuracy (Hillis 1996, 1998) and support (Bremer et al. 1999) for critical nodes (Wortley et al. 2005). This is borne out by a number of empirical studies that have employed large scale DNA sequence datasets (Herniou et al. 2001; Bapteste et al. 2002; Matsuoka et al. 2002; Rokas et al. 2003), albeit with the provisos that increased character sampling is not compromised by sparser taxon sampling (Soltis et al. 2004) and that the methodological challenges posed by increasingly large data matrices can be adequately addressed (Sanderson & Driskell 2003).
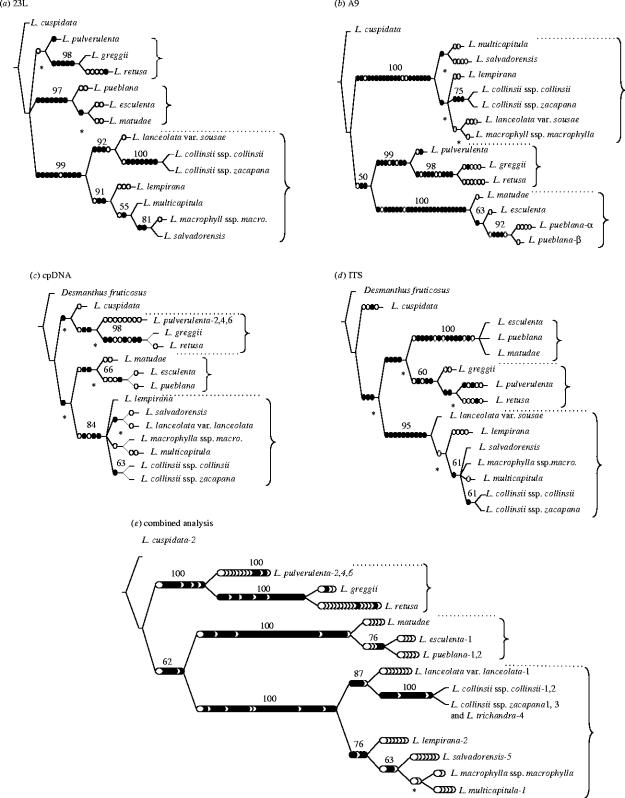
However, when we come to examine progress in constructing accurate phylogenies among closely related species the situation, at least for plants, is less encouraging. Partially resolved gene and species trees are of limited use for studies of hybridization and polyploidy, character evolution, species diversification, speciation and domestication, or as comprehensive comparative systems underpinning biology more generally. Species-level phylogenies for detailed evolutionary studies often demand not just complete or near-complete taxon sampling, but ideally multiple accessions within species, as well as complete or near-complete resolution and high statistical support. Accurate statements about hybrid and polyploid origins require that the terminal branches leading to putative hybrids and their parents on gene trees are resolved and well supported. For example, lack of resolution in gene trees derived from conventional cpDNA and nrDNA loci in the legume genus Leucaena (figure 1) meant that the parentage of hybrids and polyploid species in the genus could not be hypothesized beyond major clades using these loci (Hughes et al. 2002). The same applies to studies of domestication where gene trees are used to infer potential progenitors of crop plants (e.g. Emshwiller & Doyle 1999, 2002). Using phylogenies to understand geographical patterns of species diversification or to plot lineage diversification through time and understand speciation processes requires similarly high-quality phylogenetic trees (e.g. Barraclough & Nee 2001).
Figure 1.
Example of increased resolution and support obtained among closely related diploid species of the Mimosoid legume genus Leucaena using a SCAR-based approach (modified and extended from Bailey et al. 2004). A fully resolved and well-supported diploid species tree for Leucaena is required to investigate the origins of a set of putative hybrid and five polyploid species (Hughes et al. 2002). Solid and open circles along branches represent unique and homoplastic unambiguous character state transformations, respectively. Values above branches are strict consensus bootstrap support values; nodes that are not present in the strict consensus trees are marked with an *. (a) Anonymous SCAR-based nuclear locus 23L—one of four EMPTs; L=71; CFI=0.75; CI=0.80; RI=0.88. (b) Anonymous SCAR-based nuclear locus A9—one of six EMPTs; L=132; CFI=0.69; CI=0.75; RI=0.88. (c) Chloroplast RFLP—one of 24 EMPTs; L=63; CFI=0.46; CI=0.58; RI=0.69. (d) nrDNA ITS—one of six EMPTs; L=100; CFI=0.46; CI=0.70; RI=0.81. (e) Combined analysis of 23L, A9, cpDNA and ITS datasets—one of three EMPTs; L=339; CFI=0.92; CI=0.73; RI=0.84. EMPT=equally most parsimonious trees; L=length (number of steps); CFI=consensus fork index (calculated from strict consensus trees); CI=consistency index; RI=retention index.
Conflict attributable to lineage sorting, hybridization, polyploidy and introgression can cause gene genealogies to differ from the branching history of the organisms from which the genes were sampled (Pamilo & Nei 1988; Doyle 1992; Hillis 1995; Wendel & Doyle 1998). The impacts of these processes are thought to be more serious at species level than at higher levels, suggesting that fully resolved divergent species trees should not necessarily be expected for many plant genera (e.g. Linder & Rieseberg 2004). This means that the general desirability of employing multiple independent loci to infer accurate phylogenies can be much more critical in species-level phylogenies especially for plant groups where reticulation is common (Ferguson & Sang 2001; Raymond et al. 2002; Linder & Rieseberg 2004; Hegarty & Hiscock 2005). Employing additional DNA sequence loci to obtain greater species-tree resolution works on the premise that individual matrices will either be combined for simultaneous analysis to maximize congruence among independent sources of data (Nixon & Carpenter 1996), or, in the case of reticulation due to hybridization–introgression, that sets of congruent genes can be used to distinguish divergent from reticulate relationships. Differentiating between these alternatives requires gene trees with sufficient resolution to discriminate conflict from congruence.
Here we attempt to encapsulate how far plant species-level phylogenetics has progressed. We show that the rapid accumulation of DNA characters at higher taxonomic levels has not typically been matched in species-level analyses. It is also apparent that success in locating DNA sequence loci that can resolve relationships among species has been extremely patchy, lagging behind studies of animal taxa. Examples of densely sampled, well-resolved and supported species-level phylogenies for plants based on multiple independent loci are scarce and in many cases extremely challenging to reconstruct (see below). This means that many interesting questions in plant evolutionary biology and biogeography are currently frustrated by lack of resolution towards the tips of the tree. There has been little mention of these issues in the recent discussions about completing the Tree of Life (Cracraft & Donoghue 2004), although there have been recurrent calls for improved species-level phylogenies from those involved in evolutionary studies (e.g. Baldwin & Sanderson 1998; Barraclough & Nee 2001; Doyle et al. 2003b; Bailey et al. 2004; Futuyma 2004; Linder & Rieseberg 2004). In the last few years, new approaches to identify nuclear DNA sequence loci have emerged that have the potential to transform the current famine into a veritable feast of hypervariable species-level loci. We provide an explanatory overview that describes and compares the utility, potential benefits and pitfalls of the different approaches. Finally, we discuss the need to implement pilot studies that involve the screening of exemplar accessions to rank loci by potential utility prior to embarking on full scale sequence analysis of any one locus.
2. Survey of species-level matrices
One of the strengths of DNA sequence data for resolving relationships is the scope to select among conserved and more rapidly evolving sequence regions, the ‘tortoise and hare’ of Small et al. (1998), to address questions at different hierarchical levels (e.g. Small et al. 1998, 2004; Soltis & Soltis 1998; Yang 1998). Molecular systematists have thus been able to tailor the selection of sequence regions to the question at hand—using more slowly evolving loci to analyse higher level relationships and more rapidly evolving regions for studies of closely related species. In this way, sequences from different genomes, genes and coding/non-coding regions can potentially generate data appropriate to resolve all branches of the Tree of Life from species level (or even within species) upwards.
There are a number of different ways to measure the potential informativeness of different DNA sequence loci (Wortley & Scotland in press). Two measures, the number of parsimony informative (PI) characters and the percentage of PI characters (total number of PI characters divided by the aligned length), are widely used. Wortley & Scotland (in press) point to the minimum number of PI character state changes as a more accurate measure of potential utility. However, this measure is rarely reported and for comparisons among molecular datasets is strongly correlated with number of PI characters. Number and percentage PI characters are not independent. From a tree building perspective, the total number of PI characters is more important than the per cent variability, in that a highly variable but very short region may not provide sufficient characters. However, from a practical perspective, very long regions, while potentially contributing more characters, may be inefficient in terms of sequencing effort. In spite of falling costs of sequencing, obtaining the maximum number of characters per sequencing reaction will remain an important factor for species-level phylogeny reconstruction given the scale of taxon sampling with multiple accessions of species that is needed. Thus, both total number of PI characters and per cent PI characters are useful measures for comparing loci. These can be reported relative to the number of taxa as character–taxon ratios.
We have undertaken a two-tier survey of recent plant species-level phylogenetic analyses. First, we surveyed a sample of the most recently published studies (2003 and 2004) from American Journal of Botany, Molecular Phylogenetics and Evolution, Systematic Botany and Taxon (136 studies encompassing 345 individual data matrices). These were assessed in terms of their objectives, taxon sampling, character sampling (numbers and types of DNA sequence loci used) and the degree of resolution obtained. In order to assess resolution we used the consensus fork index (CFI; Colless 1980; the number of resolved nodes on a strict consensus tree divided by the number of possible resolved nodes, n−2, where n is the number of terminals). Second, we looked in more detail at potential informativeness of different types of sequence loci using a subset of 226 recent plant species-level data matrices from studies that include nuclear (nDNA) and non-coding chloroplast (cpDNA) and/or nuclear ribosomal (nrDNA) loci. We have restricted both surveys and discussions to sets of congeneric species, and data matrix statistics to ingroup species only. This could be viewed as somewhat arbitrary not least because genera are not uniformly applied, but also because some studies focus on subclades within genera. Numbers of PI characters can also be influenced by levels of taxon sampling within genera. Nevertheless, a broad sample of species-level studies allows for the direct comparison of variation provided by different classes of loci to evaluate the general utility of each class.
3. Chloroplast and its data
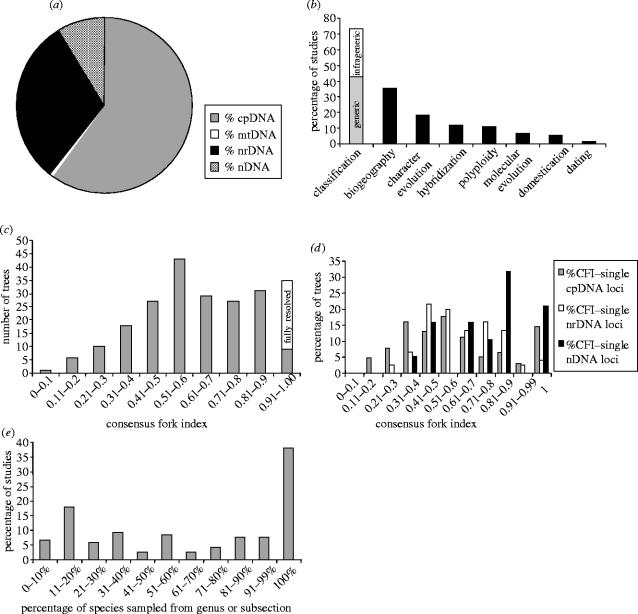
Results from the broad survey of species-level matrices are summarized in figure 2. So far, the vast majority of species-level phylogenetic analyses of plants have relied on a limited set of non-coding cpDNA loci and the nrDNA internal transcribed spacer (ITS) region (ITS 1, 5.8S and ITS 2; figure 2a; Alvarez & Wendel 2003; Shaw et al. 2005b). These loci account for 87% of 345 matrices surveyed (figure 2a) and have been favoured because they are variable at low taxonomic levels and easy to amplify using universal primers (Taberlet et al. 1991; Baldwin et al. 1995; Shaw et al. 2005b). Routine application of these loci has produced an explosion of new phylogenetic data providing insights into species relationships across a wide range of plant genera. The average resolution across studies as measured by the CFI is 0.64 (figure 2c). Even at this level of resolution, many of these analyses serve the primary purpose—i.e. form a basis for generic delimitation and infrageneric classification (figure 2b; Alvarez & Wendel 2003). However, the limitations of relying on this limited set of loci are also apparent, especially for studies with objectives beyond classification (figure 2b). Only 11% (15 trees) of our sample of recently published species-level phylogenies are fully resolved, more than half are poorly resolved (CFI<0.6), and 14% are very poorly resolved (CFI<0.4; figure 2c). Lack of resolution is, thus, a widespread problem even for morphologically diverse species representing large genera (Wojciechowski et al. 1999; Richardson et al. 2001; Malcomber 2002; Mitchell & Heenan 2002; Syring et al. in press). Usually, this lack of resolution is directly attributable to insufficient variation (Small et al. 1998; Bailey et al. 2004; Shaw et al. 2005b), rather than incongruence, although poorly resolved trees can in themselves preclude observation of incongruence. It is also worth noting that taxon sampling is extremely variable across species-level studies, with only 37% fully sampled (figure 2e).
Figure 2.
Attributes of current plant species-level phylogenetic studies based on a survey of 345 data matrices from 136 studies (see text for details). (a) Loci currently used in species-level analyses: proportions of cpDNA, nrDNA (ITS), mtDNA and nDNA loci. (b) Objectives and applications of species-level phylogenetic analyses. (c) Resolution obtained in recent species-level analyses across all loci: frequency distribution of consensus fork indices, including both combined and separate analyses. (d) Resolution obtained using different loci: frequency distribution of consensus fork indices for analyses of individual loci. (e) Variation in taxon sampling in current species-level analyses.
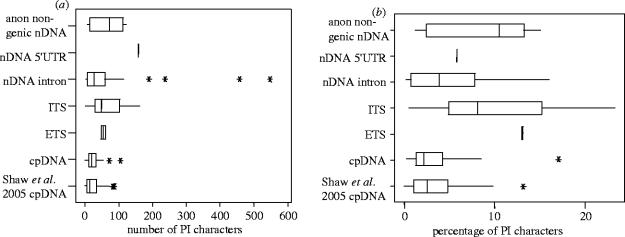
Results from the more detailed comparisons of the potential utility of different DNA sequence loci are presented in figure 3. Percentage PI character values for non-coding cpDNA range from 0.2 to 8.5 (−13.1 in Shaw et al. 2005b) and for ITS from 0.3 to 14 (−24) (figure 3). These values are an order of magnitude lower than those available in many higher level analyses (typically 20–60%). They are also lower than commonly used mitochondrial (mt) DNA loci in animals, where highly variable (typically 15–30% PI characters) DNA sequences have facilitated reconstruction of highly resolved species-level phylogenies (Avise et al. 1987; Moritz et al. 1987; Moore 1995) and speciation events (e.g. Irwin et al. 2001). The low percentages of PI characters are reflected in lower CFI values for cpDNA and ITS loci when analysed alone compared to some nuclear loci (figure 2d).
Figure 3.
Potential informativeness of cpDNA, nrDNA ITS, nrDNA ETS, nDNA intron, nDNA 5′ UTR and anonymous, SCAR-based DNA sequence loci at species level. (a) Number of PI characters per locus. (b) Percentage of PI characters for individual loci. Boxes encompass the 75% interquartiles, showing the median while outliers are indicated with an *. Data are derived from species-level phylogenetic analyses of 226 matrices from studies of plant genera. Character statistics are calculated for congeneric terminals only, excluding outgroup taxa. Tallies of characters included both PI substitutions and PI indels coded according to the simple gap coding method of Simmons & Ochoterena (2000). Plant groups included are: Sphagnum (Shaw et al. 2003), Leucaena (Bailey et al. 2004 and unpublished data), Gossypium (Cronn et al. 2002), Gaetnera (Malcomber 2002), Spaerocardamum (Bailey & Doyle 1999), Passiflora (Yockteng & Nadot 2004), Glycine—alignments assembled for nine species based on data from Doyle et al. (1996, 2003a,b) and Sakai et al. (2003), Paeonia—alignments assembled for eight species based on data from Sang et al. (1995, 1997), Ferguson & Sang (2001) and Tank & Sang (2001), Lycopersicum—alignments assembled for 13 accessions of seven species based on data from Nesbitt & Tanksley (2002), Viburnum (Winkworth & Donoghue 2004), Clarkia (Ford & Gottlieb 2003), Hibiscus (Small 2004), Neillia/Stephandra (Oh & Potter 2003) and Bursera (Weeks & Simpson 2004). The broader survey of non-coding cpDNA loci (Shaw et al. 2005b) is included as a second cpDNA bar.
In the face of insufficient variation to resolve relationships among sequences from conventional non-coding cpDNA and/or ITS loci, a number of approaches have been adopted (see below). Increasing the amount of cpDNA sequence data, potentially guided by selecting more variable non-coding cpDNA loci (Shaw et al. 2005b), has been successfully used to obtain greater resolution and support. Inevitably, this approach involves large volumes of DNA sequencing (e.g. 4–7 kb of plastid sequence used by Cronn et al. 2002; Clarkson et al. 2004; Shaw & Small 2004) and still does not guarantee well-resolved trees (e.g. Shaw & Small 2004). Other approaches have been to sequence the structurally complex external transcribed spacer (ETS; e.g. Linder et al. 2000), or resort to dominant PCR-based fragment length characters derived from amplified fragment length polymorphisms (AFLPs), inter-simple sequence repeats (ISSRs) or randomly amplified polymorphic DNAs (RAPDs) (reviewed by Wolfe & Liston 1998; Harris 1999). Contrary to the situation in the animal kingdom, mtDNA sequences have only played a minor role in phylogenetic studies of plants. This is because of structural instability, gene transfer to the nucleus and alleged sequence conservation (Wolfe et al. 1987; Palmer 1992; Soltis & Soltis 1998). However, some recent studies have found mtDNA sequence loci sufficient to build species-level phylogenies in plants (e.g. Sanjur et al. 2002).
Aside from the lack of sufficient variation in many groups, cpDNA and nrDNA loci individually are of reduced utility for reconstructing relationships among potentially reticulating species (Linder & Rieseberg 2004). Uniparental inheritance of plastids makes cpDNA essential for teasing apart maternal versus paternal contributions to putative hybrid species when compared to other data. Similarly, nrDNA multi-copy sequences that have undergone complete concerted evolution do not retain evidence of biparental inheritance and therefore reticulation (Baldwin et al. 1995; Wendel et al. 1995). The apparently unpredictable extent and direction of concerted evolution among nrDNA repeats, which can even vary among individuals of the same species (Doyle et al. 2004), can create situations where nrDNA data provide partial and potentially confusing information about reticulation. Conversely, lack of concerted evolution can create serious paralogy-related problems in some ITS datasets (Alvarez & Wendel 2003; Bailey et al. 2003; Doyle et al. 2004). These limitations mean that multiple independent bi-parentally inherited loci are essential for species-level phylogenetic studies where reticulation is a possibility (Ferguson & Sang 2001; Raymond et al. 2002; Linder & Rieseberg 2004; Hegarty & Hiscock 2005), and these can only come from the nuclear genome.
4. Strategies for selecting nuclear sequence loci
(a) Low-copy nuclear gene approaches
The most widely used approach to resolve relationships among closely related plant species where cpDNA and ITS loci fail to provide resolution has been to use sequence data from low-copy nuclear genes (LCNG), or specific members of multi-gene families and especially their introns (Doyle & Doyle 1999; Cronn et al. 2002; Sang 2002; Small et al. 2004). Initial studies using nuclear sequences were restricted by the availability of primers to a handful of LCNGs (e.g. floral development genes) under investigation in related taxa in other fields (Strand et al. 1997; Small et al. 1998; Cronn et al. 2002). This approach has succeeded in locating sporadic highly variable loci for specific plant groups (e.g. Histone H3-D in Glycine, Doyle et al. 1996; GBSSI/waxy in grasses, Mason-Gamer et al. 1998 and Rosaceae, Evans et al. 2000; GPAT in Paeonia, Tank & Sang 2001; pgiC1 and pgiC2 in Clarkia, Ford & Gottlieb 2003; cycloidea in Lupinus, Ree et al. 2004). However, LCNG loci still comprise less than 10% of published species-level matrices (figure 2a), and for the vast majority of plant genera choice of LCNG loci remains limited and access to alternatives a significant hurdle.
Also, it is increasingly clear that nuclear-encoded loci vary widely in nucleotide substitution rates among closely related species. For example, Cronn et al. (2002) found a fivefold range in substitution rates among 12 nuclear encoded loci developed for Gossypium. In combined analysis of these nuclear loci plus ITS, 53% of the PI characters were recovered from just two loci (ITS and Fad2-1). Furthermore, the ITS region provided 33% of PI sites from just 10% of the total sequence data. An even wider sevenfold difference in substitution rates was documented in a survey of 36 nuclear genes for the same species (Senchina et al. 2003). There are clear indications of similar variability in rates of evolution among intron sequences of closely related species from other genera (e.g. Doyle et al. 2003a, 2004 for Glycine; Syring et al. in press for Pinus). A wider survey of nuclear intron datasets confirms this variability which ranges from 0.21 to 16% PI characters across groups of congeneric species (figure 3). Of the 24 nuclear intron datasets surveyed here which have ITS data for the same taxa, only seven provided more PI characters than ITS (figure 3). While this variability has been recognized for some time (Small et al. 2004), what has not been widely acknowledged is that the majority of nuclear introns sequenced across groups of species, so far, are less variable (and often much less variable) and provide fewer PI characters than ITS. Only a small subset exhibits similar or higher levels of variation (figure 3). This is perhaps not surprising considering that some introns, and particularly large introns, have been shown to influence patterns of gene expression, acting as cis-regulatory elements (e.g. Sieburth & Meyerowitz 1997). A similarly wide span of variation has been found for LCNG loci used in phylogenetic analyses of animal species where most LCNG sequences are less variable than mtDNA loci (e.g. Helbig et al. 2005; Peters et al. 2005).
It is also notable that no universally useful LCNG sequence loci have been developed during the last 10 years (Sang 2002). Low-copy nuclear loci have had to be developed specifically for the taxonomic group of interest. In fact, building a phylogeny for a particular plant group has been the primary goal of many of the studies that have also discussed the potential broader utility of specific genes (e.g. Doyle et al. 1996; Galloway et al. 1998; Mason-Gamer et al. 1998; Small et al. 1998; Bailey & Doyle 1999; Emshwiller & Doyle 1999; Ree et al. 2004), a potential that has not so far been realized. Difficulties of extrapolating loci that have provided informative variation in one group can be due to failure of primers to amplify, variation in copy number, differences in intron presence or length, or differences in nucleotide substitution rates in different plant groups. Thus, up to now it seems that development of LCNG sequence loci has been somewhat of a lottery spawning the occasional lucky jackpot against a backdrop of generally disappointing results and considerable investment in developing and optimizing loci that often fail to produce much data (figure 3). This backdrop is rarely reported in the literature but is reflected in the experiences of several researchers who have acknowledged an element of good fortune in developing highly informative LCNG sequence loci, when they come to try to find similarly variable additional loci for the same group of plants (Doyle et al. 2003a; Small et al. 2004).
A further issue of concern here is the potential imbalance caused by one or a few high variability loci dominating and potentially distorting analyses. In comparisons of closely related species, the range of variation between loci is often skewed towards having few characters per locus. Having one highly variable LCNG locus alongside a set of much less variable loci may create a false sense of security when reconstructing species trees because poorly resolved trees are less likely to reveal incongruence. Furthermore, the combination of such loci in simultaneous analysis runs an increased risk of having one or a few highly variable loci that are incongruent with the underlying species tree negatively impacting the signal provided by less variable loci (Bull et al. 1993; Miyamoto & Fitch 1995; Page & Charleston 1997; Slowinski & Page 1999; Page 2000). Methods that screen multiple loci for comparable levels of informative variation prior to generating fully sampled trees (see below) will ensure that individual loci provide more equitable contributions towards combined analyses. While it is worth bearing in mind the idea that different loci may provide resolution on different parts of the tree (e.g. Pennington 1996; Ree et al. 2004; Helbig et al. 2005), this has not been convincingly demonstrated and balanced contributions of character data from a set of highly variable individual loci is likely to be more informative.
(b) Comparative anchor tagged sequence-based approaches
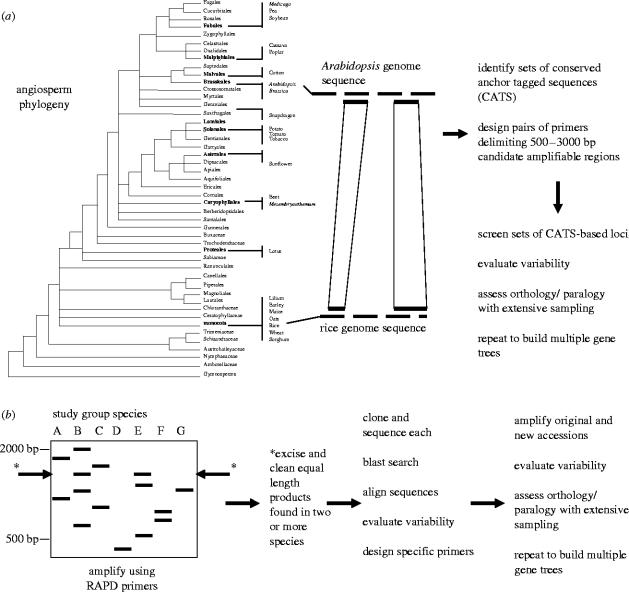
In the absence of universally accessible and informative LCNG loci, alternative approaches to locate more variable nuclear genes have been tried. The first of these involves comparisons of EST and/or complete genome sequences between model organisms to identify evolutionarily conserved regions, termed comparative anchor tagged sequences (CATS) (Lyons et al. 1997; Chandappa et al. 2005; Syring et al. in press) or conserved orthologue set (COS) markers (Fulton et al. 2002). While the primary goal of many of these studies has been comparative genome mapping, CATS markers can be used to develop sets of primers circumscribing potentially amplifiable sequence regions for phylogenetic analyses (figure 4a). For example, comparison of the tomato EST database with the Arabidopsis genome sequence identified a set of more than 1000 COS markers (Fulton et al. 2002). Similarly, screening of the Medicago trunculata EST database against the Arabidopsis genome sequence, and other available legume sequence, identified 274 loci that show strong sequence similarity. A subset of these are being screened as potential DNA sequence loci for species-level phylogenetic reconstruction of the large legume genus Astragalus (Scherson et al. in press). Similarly, Chandappa et al. (2005) used genome and EST sequence alignments to design sets of conserved markers for monocts based on rice–onion and rice–banana alignments. This approach has also been used directly with the Arabidopsis and rice genome sequences to identify conserved DNA oligomers that can be employed as primers to amplify orthologous DNA sequence loci for species-level phylogenetics (Padolina et al. 2004; Xu et al. 2004). One of the most notable features of these methods is the large number of potential primer pairs that can be generated. The few studies undertaken so far identify 100s or 1000s of possible DNA sequence loci and primer pairs. At the extreme, 13 418 candidate primer pair combinations were identified in the studies of Padolina et al. (2004) and Xu et al. (2004).
Figure 4.
Overview of CATS-based and SCAR-based approaches used to develop nuclear DNA sequence loci. (a) The CATS-based approach, where genomic or EST sequences available for (usually) divergent taxa (phylogeny showing distribution of model taxa for which genomic or EST sequence data are available, modified from Soltis & Soltis 2004), are compared to identify sets of conserved anchor tagged sequences and design primer pairs delimiting 400–3000 bp potentially amplifiable regions that are subsequently screened for variability (Syring et al. 2004; Xu et al. 2004). (b) The SCAR-based approach where screening for variability in RAPD-generated DNA fragments among closely related species precedes design of primers (modified from Bailey et al. 2004).
Development of CATS-based approaches is at an early stage and, as far as we are aware, they have yielded very few, if any hypervariable DNA sequence loci for use in species-level phylogenetics. CATS-based approaches, as currently implemented, generally compare EST/genomic sequences between widely divergent taxa (e.g. rice and Arabidopsis; figure 4a). While this may be advantageous for locating widely conserved markers (Fulton et al. 2002) thereby facilitating design of potentially ‘universal’ primers, it also means that primers typically span relatively more conserved coding regions. The simple fact that alignments can be constructed between such divergent taxa suggests that such regions are less likely to provide highly variable loci between closely related taxa. However, this is presumably a temporary limitation of our current data bases that include few taxa with many loci while many taxa have very few (or no) loci (e.g. Driskell et al. 2004). As the taxon×loci matrix becomes even a bit more densely populated, future CATS-based comparisons among more closely related taxa will become increasingly powerful for locating DNA sequence loci for specific groups of interest. For example, mapped conifer anchor loci developed from two species of pine (Brown et al. 2001) are being used to screen and select potential DNA sequence loci to resolve incongruent and incompletely resolved cpDNA and nrDNA phylogenetic trees for Pinus (Syring et al. in press). Other examples of comparisons across narrower taxonomic spans illustrate this potential (e.g. in Brassicaceae, Kuittenen et al. 2002; Lukens et al. 2003 and in monocots, Chandappa et al. 2005).
(c) Sequence characterized amplified region-based approaches
Another approach is to use RAPD or AFLP primers to generate and identify sequence characterized amplified regions (SCARs; Melotto et al. 1996) that can be screened as potentially useful sequence loci prior to development of specific primers (Shaw et al. 2003; Bailey et al. 2004). This strategy is a modified version of the AFLP-based method of McLenachan et al. (2000) for characterizing population level gel-based markers. Under this method, PCR products of equal length amplified across a subset of species using commercially available random primers are excised, cloned, sequenced and aligned to evaluate variability (figure 4b; Bailey et al. 2004). By using a range of primer combinations many candidate loci can be generated. Specific SCAR primers are then designed for potentially useful regions that show high levels of variability compared to previously used conventional loci such as ITS. As for the CATS-based approaches, the SCAR strategy can be used to screen and generate numerous candidate loci (see below). The SCAR-based approach has been used successfully to increase resolution and support for groups of closely related species of legumes (Bailey et al. 2004; see figure 1) and mosses (Shaw et al. 2003, 2005a).
There are several potential benefits of using random genomic regions rather than previously characterized genic regions. First, no prior sequence information is required to develop many loci for a group of interest. Second, random amplification of regions across the entire genome eliminates the restriction of working within known genic exon/intron regions (and potentially adjacent 3′ UTRs and 5′ promoter regions), which are likely to represent the less variable half of the nuclear genome. Given that 53–57% of the Arabidopsis genome has been classified as non-coding DNA (Initiative 2000), it presents significant untapped potential as a source of DNA characters (figure 3; Shaw et al. 2003, 2005a; Bailey et al. 2004). Third, unlike the CATS-based approaches, the initial screening process facilitates selection of loci with levels of variation needed to address species-level problems prior to development of specific primers. Fourth, by homing in on sets of loci with similar high levels of sequence divergence this reduces the chances of locus imbalance where one highly variable locus overrides signal from less variable loci. Finally, by initially selecting for regions with limited length variation this reduces the chances of encountering highly length variable regions that may be difficult to align (e.g. Syring et al. in press). This last feature may of course eliminate many potentially useful but length variable DNA sequence loci from consideration (Britten et al. 2003; Fondon & Gamer 2004).
A number of potential drawbacks associated with SCAR-based loci are immediately apparent. First, there is no indication that selected loci will be bi-parentally inherited which is critical for studies of reticulation. BLAST searches can be used to check for significant similarity to complete angiosperm cpDNA or mtDNA sequences available in GenBank, eliminating known uniparentally inherited loci from those sources. Screening via artificial or other known hybrids provides a more certain route to ascertain this. A second potential limitation is the possibility of amplifying regions of junk repeat DNA, with consequent paralogy problems. While this might suggest greater probability of encountering paralogy problems with SCAR-based loci, in practice all loci are subject to most, if not all of the same considerations. Dealing with paralogy needs to be an integral and central step in screening all DNA sequence loci for phylogenetic analysis (see below). Finally, there are clearly tradeoffs to be weighed up when looking for SCAR-based loci in terms of what span of taxa are to be included in the initial selection of amplified fragments. Inclusion of bands that amplify in just two or three closely related species runs the risk that more divergent taxa will not be amplified by primers developed from the initial sequence data, thereby potentially forfeiting inclusion of outgroup taxa (Bailey et al. 2004). Conversely, selecting fragments that amplify across a wider range of taxa than the specific clade of interest may mitigate against finding the hypervariable regions that are needed to resolve relationships within that clade. On the plus side, this is under the control of the investigator; different options can be tested.
5. Pilot studies and screening
During the short history of plant molecular systematics, the choice of biparentally inherited nuclear loci has been severely limited by the availability of useful primers. The accessibility of new approaches (discussed above) is rapidly expanding the plant systematist's ‘toolbox’ to a situation with essentially unlimited options (Shaw et al. 2003; Bailey et al. 2004; Padolina et al. 2004; Xu et al. 2004; Syring et al. in press). For those embarking on the development of new datasets or studies of new groups, these breakthroughs provide tremendous opportunities. To derive maximum benefits from the available loci, investigators need to employ pilot studies incorporating extensive screening to identify the most promising loci prior to investing heavily in any one locus (e.g. Bailey et al. 2004; Syring et al. in press). The exact order of steps in a screening process will depend on whether or not one is employing an approach that begins with the purchase of specific primers (e.g. CATS as a modified LCNG approach) or the use of random primers (e.g. SCAR-based approaches), but the critical factors that need to be considered remain essentially the same. Any screening approach should consider: (i) how many loci and what DNA samples/taxa to screen; (ii) ease and reliability of PCR amplification and sequencing; (iii) the potential orthology of sequences generated; (iv) alignment difficulties; and (v) last, but certainly not least, relative resolving power of each locus (e.g. Strand et al. 1997; Cronn et al. 2002; Bailey et al. 2004; Syring et al. in press).
The number of screened loci and DNA samples used in the selection process are largely dictated by the study group and available resources. Without doubt, selection strategies that screen larger numbers of samples and loci relative to other approaches will provide better starting points. However, these benefits need to be balanced against costs (time and money). The decreasing cost of oligonucleotide synthesis, PCR, and sequencing means that a pre-screening strategy that generates comparative data from 25 to 100 starting loci for 10 or more individuals is generally feasible and realistic. Such numbers may initially sound excessive, but considering that the goal is to discard problematic loci identified at each level of screening, the actual cost will be much lower than if all loci were carried through every step. Sampling should, whenever possible, focus on non-hybrid diploid individuals spanning the taxonomic breadth of the ultimate study group. Furthermore, the inclusion of some intraspecific samples and samples for which data already exists (e.g. ITS or cpDNA data) will sharpen subsequent screening (e.g. for orthology and variability).
Loci that are easy to amplify and sequence are far preferable to those that are not. Problems with either amplification or sequencing in many accessions suggest that significant unnecessary effort and expense may be involved in primer development, cloning and extra sequencing. Furthermore, templates that are difficult to sequence may be indicative of underlying paralogy problems (see below). Regions which are reasonably straightforward to sequence using PCR primers (plus any additional sequencing primers needed for longer fragments) will facilitate efficient data generation. Of course, there will still be some individual samples that require cloning (e.g. due to hybridization, heterozygosity, or poor primer match), but selection should aim to minimize amplification and sequencing problems.
The assessment of sequence orthology and paralogy can and should be made at several stages. First, simple observation of the number of bands amplified using the ‘locus specific’ primers can reveal obvious problems. Primer pairs that amplify multiple bands, particularly of similar sizes, should be given lower priority or discarded. Second, difficult to sequence PCR products can be indicative of problems caused by a mixture of paralogous sequencing templates (e.g. Rauscher et al. 2002). Furthermore, Scherson et al. (in press) have recently noted that repeat patterns of subset polymorphisms in otherwise clean sequencing reads are more likely to be caused by underlying paralogy than by heterozygosity. Consideration of the patterns discussed by Scherson et al. (in press) is worthy of inclusion in the screening process. With the large numbers of potential regions now available, there is little reason to intentionally embark on studies that will require the dissection of paralogous gene copies for species-tree reconstruction. Lastly, assessments of orthology/paralogy can be made using comparisons of gene trees developed from each locus in relation to previously generated data (e.g. ITS and cpDNA). In these cases, the inclusion of intraspecific samples can also help identify potential paralogy problems.
Data generated during initial screening to demonstrate reliable single band amplifications and lack of serious sequencing difficulties or obvious paralogy problems can also be used to compare sequence variability, to identify those loci that are easy to use and informative. Assessments of inter-locus variability can be made among the pool of potential new loci and between these loci and previously developed data, such as ITS and cpDNA markers. Comparisons of the former are critical for the selection of the most variable loci, while evaluation of the latter should provide some idea how much additional resolution might (or might not) be gained from a fully sampled matrix using the ‘best’ of the newly screened loci. Measures of sequence variability may be based on percentage of PI characters, pairwise divergence, or more comprehensive analysis of gene-tree comparisons. Gene-tree approaches best characterize the overall distribution of variability and therefore the relative resolving power of each locus based on the same samples (Wenzel & Siddall 1999). Additional measures of utility for each gene may also be drawn from the individual gene trees (e.g. consistency index, number of resolved nodes, average branch support, etc.).
In plant species-level phylogenetics, rarely has the issue of too much variation been a problem. In general, researchers want to select the most variable loci that fit all the other criteria discussed above. However, one additional measure of variability and potential utility that may be considered in screening is the presence of indels that can provide potentially useful characters, but also potential alignment difficulties. Clean indels contribute useful phylogenetic markers while complicated indel patterns tied to high substitution rates can limit the utility of the data generated. Ideal loci will be relatively straightforward to align while maintaining relatively high levels of information (Bailey et al. 2004).
6. Summary comments
One of the frequently cited advantages of DNA sequence data for reconstructing phylogenies is the virtually unlimited number of characters that can be generated. However, for closely related plant groups it may be extremely difficult to locate sufficiently variable DNA sequence loci to generate enough characters to provide well-resolved and robustly supported hypotheses of species relationships without generating very large volumes of sequence data. In contrast to those who study animals, for whom rapidly evolving mtDNA loci provide a rich source of species-level data, the plant molecular systematist's toolbox lacks readily accessible hypervariable sequence regions. Well-supported resolution at the gene-tree level remains elusive, suggesting limited confidence in how systematists currently infer relationships among closely related species.
Several routes to locating hypervariable nuclear DNA sequence loci for species-level phylogenetic analysis have emerged. The relative efficiency of these alternatives should increase rapidly because locating informative loci depends on how closely related the study group is to species for which significant (genome or EST) DNA sequence is available. SCAR-based approaches seem to present the most efficient strategy for many plant groups at present. However, as the gene×taxon GenBank matrix become more densely populated, CATS-type approaches based on significantly narrower taxonomic spans will become increasingly frequent and productive.
A number of recent studies have demonstrated the high utility of anonymous, presumed non-coding, non-genic nuclear regions for resolving relationships (figures 1 and 3; Shaw et al. 2003, 2005a; Bailey et al. 2004). At the same time, it is increasingly clear that although low-copy nuclear introns range greatly in variability (figure 3; Small et al. 2004), the majority are likely to be less variable (at least on a percentage basis) than ITS. Tapping into the large non-genic, or less-conserved genic fractions of plant genomes may thus be particularly productive. At present, the majority of researchers appear to be following CATS-based approaches to develop species-level nuclear DNA sequence loci (e.g. Syring et al. in press; Xu et al. 2004; Scherson et al. in press). In part this may be due to the large number of primer pairs that CATS approaches can provide, or to the attraction of locating universal or at least widely applicable markers. However, we believe that there is a need for caution to avoid the pitfalls associated with the LCNG approach that has been shown to be sub-optimal.
Whether the focus is on LCNG sequences or loci derived from SCAR-based approaches, the solutions will, for the most part, be lineage-specific. Once we stray beyond well-known nrDNA and cpDNA loci, opportunities to develop universal markers or primers appear to be extremely limited, if not non-existent (Sang 2002), suggesting a need to get away from universal gene thinking for species-level phylogenetics. A requirement for marker development, or at least substantive optimization, for specific plant lineages, and potentially for each individual study, shifts the emphasis towards approaches that involve screening prior to primer development. The need to screen candidate loci is further reinforced by the growing ease of identifying large numbers of candidate primer pairs. Thus, whatever sources of sequence data—non-coding cpDNA, LCNG, CATS-based or SCAR-based—are used, strategies for screening candidate sequence loci are key. Given the proliferation of potential sequence loci, screening can afford, and probably needs, to be more ruthless than anything envisaged before.
Finally, none of these developments denies the ongoing utility of, and need for, non-coding cpDNA and ITS data. Not only will these continue to provide useful data in their own right, but cpDNA and ITS data also form essential foundations for subsequent development of other DNA sequence loci. This will take the form of a framework to compare levels of variation, provide initial hypotheses of relationships to direct taxon sampling and assess orthology/paralogy concerns during screening and data generation for candidate nuclear DNA sequence loci.
Acknowledgments
The authors thank two anonymous reviewers for comments on an earlier version of the manuscript and John Syring, Rosita Scherson and Alex Wortley for pre-print copies of their papers. C.E.H. is supported by the Royal Society and R.J.E. by BBSRC.
GLOSSARY
- AFLP
amplified fragment length polymorphism
- CATS
comparative anchor tagged sequences
- CFI
consensus fork index
- COS
conserved orthologue set
- EMPT
equally most parsimonious tree
- EST
expressed sequence tag
- ITS
internal transcribed spacer
- LCNG
low-copy nuclear gene
- PI
parsimony informative
- RAPD
randomly amplified polymorphic DNA
- SCAR
sequence characterized amplified region
- UTR
untranscribed region
Footnotes
These authors contributed equally to this work.
References
- Alvarez I, Wendel J.F. Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylog. Evol. 2003;29:417–434. doi: 10.1016/s1055-7903(03)00208-2. doi:10.1016/S1055-7903(03)00208-2 [DOI] [PubMed] [Google Scholar]
- APGII An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APGII. Bot. J. Linn. Soc. 2003;141:399–436. doi:10.1046/j.1095-8339.2003.t01-1-00158.x [Google Scholar]
- Avise J.C, Arnold J, Bermingham E, Lamb T, Neigel J.E, Reeb C.A, Saunders N.C. Intraspecific phylogeography: mitochondrial DNA bridge between population genetics and systematics. Annu. Rev. Ecol. Syst. 1987;18:489–522. [Google Scholar]
- Bailey C.D, Doyle J.J. Potential phylogenetic utility of the low-copy nuclear gene pistillata in Dicotylednous plants: comparison to nrDNA ITS and trnL intron in Sphaerocardamum and other Brassicaceae. Mol. Phylog. Evol. 1999;13:20–30. doi: 10.1006/mpev.1999.0627. doi:10.1006/mpev.1999.0627 [DOI] [PubMed] [Google Scholar]
- Bailey C.D, Carr T.G, Harris S.A, Hughes C.E. Characterization of angiosperm nrDNA polymorphism, paralogy and pseudogenes. Mol. Phylog. Evol. 2003;29:435–455. doi: 10.1016/j.ympev.2003.08.021. doi:10.1016/j.ympev.2003.08.021 [DOI] [PubMed] [Google Scholar]
- Bailey C.D, Hughes C.E, Harris S.A. Using RAPDs to identify DNA sequence loci for species level phylogeny reconstruction: an example from Leucaena (Fabaceae) Syst. Bot. 2004;29:4–14. doi:10.1600/036364404772973483 [Google Scholar]
- Baldwin B.G, Sanderson M.J. Age and rate of diversification of the Hawaiian silversword alliance. Proc. Natl Acad. Sci. 1998;95:9402–9406. doi: 10.1073/pnas.95.16.9402. doi:10.1073/pnas.95.16.9402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin B.G, Sanderson M.J, Porter J.M, Wojciechowski M.F, Campbell C.S, Donoghue M.J. The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Ann. Missouri Bot. Gard. 1995;82:247–277. [Google Scholar]
- Bapteste E, et al. The analysis of 100 genes supports the grouping of three highly divergent amoebae: Dictyostelium, Entamoeba, and Mastigamoeba. Proc. Natl Acad. Sci. 2002;99:1414–1419. doi: 10.1073/pnas.032662799. doi:10.1073/pnas.032662799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraclough T.G, Nee S. Phylogenetics and speciation. Trends Ecol. Evol. 2001;16:391–399. doi: 10.1016/s0169-5347(01)02161-9. doi:10.1016/S0169-5347(01)02161-9 [DOI] [PubMed] [Google Scholar]
- Barraclough T.G, Vogler A.P. Detecting the geographical pattern of speciation from species-level phylogenies. Am. Nat. 2000;155:419–434. doi: 10.1086/303332. doi:10.1086/303332 [DOI] [PubMed] [Google Scholar]
- Bennetzen J.L, Kellogg E.A. Do plants have a one-way ticket to genomic obesity? Plant Cell. 1997;9:1509–1514. doi: 10.1105/tpc.9.9.1509. doi:10.1105/tpc.9.9.1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer B, Jansen R.K, Oxelman B, Backland M, Lantz H, Kim K.-J. More characters or more taxa for robust phylogeny—case study from the coffee family (Rubiaceae) Syst. Biol. 1999;48:413–435. doi: 10.1080/106351599260085. doi:10.1080/106351599260085 [DOI] [PubMed] [Google Scholar]
- Britten R.J, Rowen L, Williams J, Cameron R.A. Majority of divergence between closely related DNA samples is due to indels. Proc. Natl Acad. Sci. 2003;100:4661–4665. doi: 10.1073/pnas.0330964100. doi:10.1073/pnas.0330964100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G.R, Kadd E.E, Bassoni D.L, Kiehne K.L, Temesgen B, van Buijtenen J.P, Sewell M.H, Marshall K.A, Neale D.B. Anchored reference loci in loblolly pine (Pinus taeda L.) for integrating pine genomics. Genetics. 2001;159:799–809. doi: 10.1093/genetics/159.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull J.J, Huelsenbeck J.P, Cunningham C.W, Swofford D.L, Waddell P.J. Partitioning and combining data in phylogenetic analysis. Syst. Biol. 1993;42:384–397. [Google Scholar]
- Chandappa, L. H., Feltus, F. A., Singh, H. P. & Paterson, A. H. 2005 Conserved PCR primers from rice–banana and rice–onion alignments. XIII Plant–Animal Genome Conf. Abstract, San Diego, USA.
- Clarkson J.J, Knapp S, Garcia V.F, Olmstead R.G, Leitch A.R, Chase M.W. Phylogenetic relationships in Nicotiana (Solanaceae) inferred from multiple plastid DNA regions. Mol. Phylog. Evol. 2004;33:75–90. doi: 10.1016/j.ympev.2004.05.002. doi:10.1016/j.ympev.2004.05.002 [DOI] [PubMed] [Google Scholar]
- Colless D.H. Congruence between morphometric and allozyme data for Menidia species: a reappraisal. Syst. Zool. 1980;29:288–299. [Google Scholar]
- Cracraft J, Donoghue M.J, editors. Assembling the Tree of Life. Oxford University Press; New York: 2004. p. 576. [Google Scholar]
- Cronn R.C, Small R.L, Haselkorn T, Wendel J.F. Rapid diversification of the cotton genus (Gossypium: Malvaceae) revealed by analysis of sixteen nuclear and chloroplast genes. Am. J. Bot. 2002;89:707–725. doi: 10.3732/ajb.89.4.707. [DOI] [PubMed] [Google Scholar]
- Donoghue M.J. Immeasurable progress on the Tree of Life. In: Cracraft J, Donoghue M.J, editors. Assembling the Tree of Life. Oxford University Press; New York: 2004. pp. 548–552. [Google Scholar]
- Donoghue M.J, Cracraft J. Introduction. Charting the Tree of Life. In: Cracraft J, Donoghue M.J, editors. Assembling the Tree of Life. Oxford University Press; New York: 2004. pp. 1–4. [Google Scholar]
- Doust A.N, Kellogg E.A. Inflorescence diversification in the panicoid ‘bristle grass’ clade (Paniceae, Poaceae): evidence from molecular phylogenies and developmental morphology I. Am. J. Bot. 2002;89:1203–1222. doi: 10.3732/ajb.89.8.1203. [DOI] [PubMed] [Google Scholar]
- Doyle J.J. Gene trees and species trees: molecular systematics as one-character taxonomy. Syst. Bot. 1992;17:144–163. [Google Scholar]
- Doyle J.J, Doyle J.L. Nuclear protein-coding genes in phylogeny reconstruction and homology assessment: some examples from Leguminosae. In: Hollingsworth P.M, Bateman R.M, Gornall R.J, editors. Molecular systematics and plant evolution. Taylor & Francis; London: 1999. pp. 229–254. [Google Scholar]
- Doyle J.J, Kanazin V, Shoemaker R.C. Phylogenetic utility of histone H3 intron sequences in the perennial relatives of soybean (Glycine: Leguminosae) Mol. Phylog. Evol. 1996;6:438–447. doi: 10.1006/mpev.1996.0092. doi:10.1006/mpev.1996.0092 [DOI] [PubMed] [Google Scholar]
- Doyle J.J, Doyle J.L, Harbison C. Chloroplast expressed glutamine synthetase in Glycine and related Leguminosae: phylogeny, gene duplication and ancient polyploidy. Syst. Bot. 2003a;28:567–577. [Google Scholar]
- Doyle J.J, Doyle J.L, Rauscher J.T, Brown A.H.D. Diploid and polyploid reticulate evolution throughout the history of the perennial soybeans (Glycine subgenus Glycine) New Phytol. 2003b;161:121–132. doi:10.1046/j.1469-8137.2003.00949.x [Google Scholar]
- Doyle J.J, Doyle J.L, Rauscher J.T, Brown A.H.D. Evolution of the perennial soybean polyploidy complex (Glycine subgenus Glycine): a study of contrasts. Biol. J. Linn. Soc. 2004;82:583–597. doi:10.1111/j.1095-8312.2004.00343.x [Google Scholar]
- Driskell A.C, Ane C, Burleigh J.G, McMahon M.M, O'Meara B.C, Sanderson M.J. Prospects for building the Tree of Life from large sequence databases. Science. 2004;306:1172–1174. doi: 10.1126/science.1102036. doi:10.1126/science.1102036 [DOI] [PubMed] [Google Scholar]
- Eisen J.A, Fraser C.M. Phylogenomics: intersection of evolution and genomics. Science. 2003;300:1706–1707. doi: 10.1126/science.1086292. doi:10.1126/science.1086292 [DOI] [PubMed] [Google Scholar]
- Emshwiller E, Doyle J.J. Chloroplast-expressed glutamine synthetase (ncpGS): potential utility for phylogenetic analysis with an example from Oxalis (Oxalidaceae) Mol. Phylog. Evol. 1999;12:310–319. doi: 10.1006/mpev.1999.0613. doi:10.1006/mpev.1999.0613 [DOI] [PubMed] [Google Scholar]
- Emshwiller E, Doyle J.J. Origins of domestication and polyploidy in Oca (Oxalis tuberosa: Oxalidaceae). 2. Chloroplast-expressed glutamine synthetase data. Am. J. Bot. 2002;89:1042–1056. doi: 10.3732/ajb.89.7.1042. [DOI] [PubMed] [Google Scholar]
- Evans R.C, Alice L.A, Campbell C.S, Kellogg E.A, Dickinson T.A. The granule-bound starch synthase (GBSSI) gene in Rosaceae: multiple loci and phylogenetic utility. Mol. Phylog. Evol. 2000;17:388–400. doi: 10.1006/mpev.2000.0828. doi:10.1006/mpev.2000.0828 [DOI] [PubMed] [Google Scholar]
- Ferguson D, Sang T. Speciation through homoploid hybridization between allotetraploids in peonies (Paeonia) Proc. Natl Acad. Sci. 2001;98:3915–3919. doi: 10.1073/pnas.061288698. doi:10.1073/pnas.061288698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondon J.W, Gamer H.R. Molecular origins of rapid and continuous morphological evolution. Proc. Natl Acad. Sci. 2004;101:18 058–18 063. doi: 10.1073/pnas.0408118101. doi:10.1073/pnas.0408118101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford V.S, Gottlieb L.D. Reassessment of phylogenetic relationships in Clarkia sect. Sympherica. Am. J. Bot. 2003;90:284–292. doi: 10.3732/ajb.90.2.284. [DOI] [PubMed] [Google Scholar]
- Fulton T.M, Van der Hoeven R, Eannetta N.T, Tanksley S.D. Identification, analysis, and utilization of conserved ortholog set markers for comparative genomics in higher plants. Plant Cell. 2002;14:1457–1467. doi: 10.1105/tpc.010479. doi:10.1105/tpc.010479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futuyma D.J. The fruit of the Tree of Life. Insights into evolution and ecology. In: Cracraft J, Donoghue M.J, editors. Assembling the Tree of Life. Oxford University Press; New York: 2004. pp. 25–39. [Google Scholar]
- Galloway G.L, Malmberg R.L, Price R.A. Phylogenetic utility of the nuclear gene arginine decarboxylase: an example from Brassicaceae. Mol. Biol. Evol. 1998;15:1312–1320. doi: 10.1093/oxfordjournals.molbev.a025859. [DOI] [PubMed] [Google Scholar]
- Harris S.A. RAPDs in systematics—a useful methodology? In: Hollingsworth P.M, Bateman R.M, Gornall R.J, editors. Molecular systematics and plant evolution. Taylor & Francis; London: 1999. pp. 211–228. [Google Scholar]
- Hegarty M.J, Hiscock S.J. Hybrid speciation in plants: new insights from molecular studies. New Phytol. 2005;165:411–423. doi: 10.1111/j.1469-8137.2004.01253.x. doi:10.1111/j.1469-8137.2004.01253.x [DOI] [PubMed] [Google Scholar]
- Helbig A.J, Kocum A, Seibold I, Braun M.J. A multi-gene phylogeny of aquiline eagles (Aves: Acciptriformes) reveals extensive paraphyly at genus level. Mol. Phylog. Evol. 2005;35:147–164. doi: 10.1016/j.ympev.2004.10.003. doi:10.1016/j.ympev.2004.10.003 [DOI] [PubMed] [Google Scholar]
- Herniou E.A, Luque T, Chen X, Vlak J.M, Winstanley D, Cory J.S, O'Reilly D.R. Use of whole genome sequence data to infer baculovirus phylogeny. J. Virol. 2001;75:8117–8126. doi: 10.1128/JVI.75.17.8117-8126.2001. doi:10.1128/JVI.75.17.8117-8126.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillis D.M. Approaches for assessing phylogenetic accuracy. Syst. Biol. 1995;44:3–16. [Google Scholar]
- Hillis D.M. Inferring complex phylogenies. Nature. 1996;383:130–131. doi: 10.1038/383130a0. doi:10.1038/383130a0 [DOI] [PubMed] [Google Scholar]
- Hillis D.M. Taxonomic sampling, phylogenetic accuracy and investigator bias. Syst. Biol. 1998;47:3–8. doi: 10.1080/106351598260987. doi:10.1080/106351598260987 [DOI] [PubMed] [Google Scholar]
- Hillis D.M. The Tree of Life and the grand synthesis in biology. In: Cracraft J, Donoghue M.J, editors. Assembling the Tree of Life. Oxford University Press; New York: 2004. pp. 545–547. [Google Scholar]
- Hong R.L, Hamaguchia M, Maximilian A.B, Weigel D. Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing. Plant Cell. 2003;15:1296–1309. doi: 10.1105/tpc.009548. doi:10.1105/tpc.009548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes C.E, Bailey C.D, Harris S.A. Divergent and reticulate species relationships in Leucaena (Fabaceae) inferred from multiple data sources: insights into polyploid origins and nrDNA polymorphism. Am. J. Bot. 2002;89:1057–1073. doi: 10.3732/ajb.89.7.1057. [DOI] [PubMed] [Google Scholar]
- Initiative T.A.G. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. doi:10.1038/35048692 [DOI] [PubMed] [Google Scholar]
- Irwin D.E, Bensch S, Price T.D. Speciation in a ring. Nature. 2001;409:333–337. doi: 10.1038/35053059. doi:10.1038/35053059 [DOI] [PubMed] [Google Scholar]
- Kuittenen H, et al. Primers for 22 candidate genes for ecological adaptations in Brassicaceae. Mol. Ecol. Notes. 2002;2:258–262. doi:10.1046/j.1471-8286.2002.00210.x [Google Scholar]
- Lavin M, Pennington R.T, Klitgaard B.B, Sprent J.I, Cavalcante de Lima H, Gasson P.E. The dalbergioid legumes (Fabaceae): delimitation of a pantropical monophyletic clade. Am. J. Bot. 2001;88:503–533. [PubMed] [Google Scholar]
- Lavin M, Schrire B.P, Lewis G, Pennington R.T, Delgado-Salinas A, Thulin M, Hughes C.E, Matos A.B, Wojciechowski M.F. Metacommunity process rather than continental tectonic history better explains geographically structured phylogenies in legumes. Phil. Trans. R. Soc. B. 2004;359:1509–1522. doi: 10.1098/rstb.2004.1536. doi:10.1098/rstb.2004.1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder C.R, Rieseberg L.H. Reconstructing patterns of reticulate evolution in plants. Am. J. Bot. 2004;91:1700–1708. [PMC free article] [PubMed] [Google Scholar]
- Linder C.R, Goertzen L.R, Heuval B.V, Francisco-Ortega J, Jansen R.K. The complete external transcribed spacer of 18S-26S rDNA: amplification and phylogenetic utility at low taxonomic levels in Asteraceae and closely allied families. Mol. Phylog. Evol. 2000;14:285–303. doi: 10.1006/mpev.1999.0706. doi:10.1006/mpev.1999.0706 [DOI] [PubMed] [Google Scholar]
- Lukens L, Zou F, Lydiate D, Parkin I, Osborn T. Comparison of a Brassica oleracea genetic map with the genome of Arabidopsis thaliana. Genetics. 2003;146:359–372. doi: 10.1093/genetics/164.1.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons L.A, Laughlin T.F, Copeland N.G, Jenkins N.A, Womack J.E, O'Brien S.J. Comparative anchor tagged sequences (CATS) for integrative mapping of mammalian genomes. Nat. Genet. 1997;15:47–56. doi: 10.1038/ng0197-47. doi:10.1038/ng0197-47 [DOI] [PubMed] [Google Scholar]
- Malcomber S.T. Phylogeny of Gaetnera Lam. (Rubiaceae) based on multiple DNA markers: evidence of a rapid radiation in a widespread, morphologically diverse genus. Evolution. 2002;56:42–57. doi: 10.1111/j.0014-3820.2002.tb00848.x. [DOI] [PubMed] [Google Scholar]
- Mason-Gamer R.J, Weil C.F, Kellogg E.A. Granule-bound starch synthase: structure, function and phylogenetic utility. Mol. Biol. Evol. 1998;15:1658–1673. doi: 10.1093/oxfordjournals.molbev.a025893. [DOI] [PubMed] [Google Scholar]
- Matsuoka Y, Yamazaki Y, Ogihara Y, Tsunewaki K. Whole chloroplast genome comparison of rice, maize, and wheat: implications for chloroplast gene diversification and phylogeny of cereals. Mol. Biol. Evol. 2002;19:2084–2091. doi: 10.1093/oxfordjournals.molbev.a004033. [DOI] [PubMed] [Google Scholar]
- McLenachan P.A, Stockler K, Winkworth R.C, McBreen K, Zauner S. Markers derived from amplified fragment length polymorphism gels for plant ecology and evolution studies. Mol. Ecol. 2000;9:1899–1903. doi: 10.1046/j.1365-294x.2000.01075.x. doi:10.1046/j.1365-294x.2000.01075.x [DOI] [PubMed] [Google Scholar]
- Melotto M, Afanador L, Kelly J.D. Development of a SCAR marker linked to the I gene in common bean. Genome. 1996;39:1216–1219. doi: 10.1139/g96-155. [DOI] [PubMed] [Google Scholar]
- Mitchell A.D, Heenan P.B. Sophora sect Edwardsia (Fabaceae): further evidence from nrDNA sequence data of a recent and rapid radiation around the Southern Oceans. Bot. J. Linn. Soc. 2002;140:435–441. doi:10.1046/j.1095-8339.2002.00101.x [Google Scholar]
- Miyamoto M.M, Fitch W.M. Testing species phylogenies and phylogenetic methods with congruence. Syst. Biol. 1995;44:64–76. [Google Scholar]
- Moore W.S. Inferring phylogenies from mtDNA variation: mitochondrial-gene versus nuclear-gene trees. Evolution. 1995;49:718–726. doi: 10.1111/j.1558-5646.1995.tb02308.x. [DOI] [PubMed] [Google Scholar]
- Moritz C, Dowling T.E, Brown W.M. Evolution of animal mitochondrial DNA: relevance for population biology and systematics. Annu. Rev. Ecol. Syst. 1987;18:269–292. doi:10.1146/annurev.es.18.110187.001413 [Google Scholar]
- Nesbitt T.C, Tanksley S.D. Comparative sequencing in the genus Lycopersicon: implications for the evolution of fruit size in the domestication of cultivated tomatoes. Genetics. 2002;162:365–379. doi: 10.1093/genetics/162.1.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon K.C, Carpenter J.M. On simultaneous analysis. Cladistics. 1996;12:221–241. doi: 10.1111/j.1096-0031.1996.tb00010.x. doi:10.1111/j.1096-0031.1996.tb00010.x [DOI] [PubMed] [Google Scholar]
- Oh S.-H, Potter D. Phylogenetic utility of the second intron of LEAFY in Neillia and Stephanandra (Rosaceae) and implications for the origin of Stephanandra. Mol. Phylog. Evol. 2003;29:203–215. doi: 10.1016/s1055-7903(03)00093-9. doi:10.1016/S1055-7903(03)00093-9 [DOI] [PubMed] [Google Scholar]
- Padolina J, Timme R, Linder R, Briggs W, Xu W, Liu W, Miranker D. Identification of broadly applicable nuclear DNA regions for phylogenetic reconstruction in angiosperms. Bot. Soc. Am. 2004;2004:528. (Botany 2004 Meeting Abstract 528.) [Google Scholar]
- Page R.D.M. Extracting species trees from complex gene trees: reconciled trees and vertebrate phylogeny. Mol. Phylog. Evol. 2000;14:89–106. doi: 10.1006/mpev.1999.0676. doi:10.1006/mpev.1999.0676 [DOI] [PubMed] [Google Scholar]
- Page R.D.M, Charleston M.A. From gene to organismal phylogeny: reconciled trees and the gene-tree species-tree problem. Mol. Phylog. Evol. 1997;7:231–240. doi: 10.1006/mpev.1996.0390. doi:10.1006/mpev.1996.0390 [DOI] [PubMed] [Google Scholar]
- Palmer J.D. Mitochondrial DNA in plant systematics: applications and limitations. In: Soltis P.S, Soltis D.E, Doyle J.J, editors. Molecular systematics of plants. Chapman & Hall; New York: 1992. pp. 36–49. [Google Scholar]
- Palmer J.D, Soltis D.E, Chase M.W. The plant Tree of Life: an overview and some points of view. Am. J. Bot. 2004;91:1437–1445. doi: 10.3732/ajb.91.10.1437. [DOI] [PubMed] [Google Scholar]
- Pamilo P, Nei M. Relationships between gene trees and species trees. Mol. Biol. Evol. 1988;5:568–583. doi: 10.1093/oxfordjournals.molbev.a040517. [DOI] [PubMed] [Google Scholar]
- Pennington R.T. Molecular and morphological data provide resolution at different levels in Andira. Syst. Biol. 1996;45:496–515. [Google Scholar]
- Pennington R.T, Cronk Q.C.B, Richardson J.A. Introduction and synthesis: plant phylogeny and the origin of major biomes. Phil. Trans. R. Soc. B. 2004;359:1455–1464. doi: 10.1098/rstb.2004.1539. doi:10.1098/rstb.2004.1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.L, McCraken K.G, Zhuravlev Y.N, Wilson R.E, Johnson K.P, Omland K.E. Phylogenetics of wigeons and allies (Anatidae: Aves): the importance of sampling multiple loci and multiple individuals. Mol. Phylog. Evol. 2005;35:209–224. doi: 10.1016/j.ympev.2004.12.017. doi:10.1016/j.ympev.2004.12.017 [DOI] [PubMed] [Google Scholar]
- Rauscher J.T, Doyle J.J, Brown A.D.H. Internal transcribed spacer repeat-specific primers and the analysis of hybridization in the Glycine tomentella (Leguminosae) polyploidy complex. Mol. Ecol. 2002;11:2691–2702. doi: 10.1046/j.1365-294x.2002.01640.x. doi:10.1046/j.1365-294X.2002.01640.x [DOI] [PubMed] [Google Scholar]
- Raymond O, Piola F, Sanlaville-Boisson C. Inferences of reticulation in outcrossing allopolyploid taxa: caveats, likelihood and perspectives. Trends Ecol. Evol. 2002;17:3–6. doi:10.1016/S0169-5347(01)02378-3 [Google Scholar]
- Ree R.H, Citerne H.L, Lavin M, Cronk Q.C.B. Heterogeneous selection on LEGCYC paralogs in relation to flower morphology and the phylogeny of Lupinus (Leguminosae) Mol. Biol. Evol. 2004;21:321–331. doi: 10.1093/molbev/msh022. doi:10.1093/molbev/msh022 [DOI] [PubMed] [Google Scholar]
- Richardson J.E, Pennington R.T, Pennington T.D, Hollingsworth P.M. Rapid diversification of a species-rich genus of Neotropical rainforest trees. Science. 2001;293:2242–2245. doi: 10.1126/science.1061421. doi:10.1126/science.1061421 [DOI] [PubMed] [Google Scholar]
- Rokas A, Williams B.L, King N, Carroll S.B. Genome scale approaches to resolving incongruence in molecular phylogenies. Nature. 2003;425:798–804. doi: 10.1038/nature02053. doi:10.1038/nature02053 [DOI] [PubMed] [Google Scholar]
- Sakai M, Kanazawa A, Fujii A, Thseng F.S, Abe J, Shimamoto Y. Phylogenetic relationships of the chloroplast genomes in the genus Glycine inferred from four intergenic spacer regions. Plant Syst. Evol. 2003;239:29–54. doi:10.1007/s00606-002-0226-9 [Google Scholar]
- Sanderson M.J. Objections to bootstrapping phylogenies: a critique. Syst. Biol. 1995;44:299–320. [Google Scholar]
- Sanderson M.J, Driskell A.C. The challenge of constructing large phylogenetic trees. Trends Plant Sci. 2003;8:374–379. doi: 10.1016/S1360-1385(03)00165-1. doi:10.1016/S1360-1385(03)00165-1 [DOI] [PubMed] [Google Scholar]
- Sang T. Utility of low-copy nuclear gene sequences in plant phylogenetics. Crit. Rev. Biochem. Mol. Biol. 2002;37:121–147. doi: 10.1080/10409230290771474. doi:10.1080/10409230290771474 [DOI] [PubMed] [Google Scholar]
- Sang T, Crawford D.J, Stuessy T.F. Documentation of reticulate evolution in peonies (Paeonia) using internal transcribed spacer sequences of nuclear ribosomal DNA: implications for biogeography and concerted evolution. Proc. Natl Acad. Sci. 1995;92:6813–6817. doi: 10.1073/pnas.92.15.6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang T, Donoghue M.J, Zhang D. Evolution of alcohol dehydrogenase genes in peonies (Paeonia): phylogenetic relationships of putative nonhybrid species. Mol. Biol. Evol. 1997;14:994–1007. doi: 10.1093/oxfordjournals.molbev.a025716. [DOI] [PubMed] [Google Scholar]
- Sanjur O.T, Piperno D.R, Andres T.C, Wessel-Beaver L. Phylogenetic relationships among domesticated and wild species of Cucurbita (Cucurbitaceae) inferred from a mitochondrial gene: implications for crop evolution and areas of origin. Proc. Natl Acad. Sci. 2002;99:535–540. doi: 10.1073/pnas.012577299. doi:10.1073/pnas.012577299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen V, Chase M.W. A decade of progress in plant molecular phylogenies. Trends Genet. 2003;19:717–724. doi: 10.1016/j.tig.2003.10.003. doi:10.1016/j.tig.2003.10.003 [DOI] [PubMed] [Google Scholar]
- Scherson, R., Choi, H.-K., Cook, D. & Sanderson, M. J. In press. Phylogenetics of New World Astragalus: the utility of genomics technology in reconstructing phylogenies at low taxonomic levels. Brittonia
- Schultheis L.M, Baldwin B.G. Molecular phylogenetics of Fouquieriaceae: evidence from nuclear rDNA ITS studies. Am. J. Bot. 1999;86:578–589. [PubMed] [Google Scholar]
- Senchina D.S, et al. Rate variation among nuclear genes and the age of polyploidy in Gossypium. Mol. Biol. Evol. 2003;20:633–643. doi: 10.1093/molbev/msg065. doi:10.1093/molbev/msg065 [DOI] [PubMed] [Google Scholar]
- Shaw A.J, Cox C.J, Boles S.B. Polarity of peatmoss (Sphagnum) evolution: who says bryophytes have no roots. Am. J. Bot. 2003;90:1777–1787. doi: 10.3732/ajb.90.12.1777. [DOI] [PubMed] [Google Scholar]
- Shaw A.J, Cox C.J, Boles S.B. Phylogeny, species delimitation and recombination in Spahgnum Section Acutifolia. Syst. Bot. 2005a;30:16–33. doi:10.1600/0363644053661823 [Google Scholar]
- Shaw J, Small R.L. Addressing the ‘hardest puzzle in American pomology’: phylogeny of Prunus Sect. Prunocerasus (Rosaceae) based on seven non-coding chloroplast DNA regions. Am. J. Bot. 2004;91:985–996. doi: 10.3732/ajb.91.6.985. [DOI] [PubMed] [Google Scholar]
- Shaw J, et al. The tortoise and the hare II: relative utility of 21 non-coding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 2005b;92:142–166. doi: 10.3732/ajb.92.1.142. [DOI] [PubMed] [Google Scholar]
- Sieburth L.E, Meyerowitz E.M. Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell. 1997;9:355–365. doi: 10.1105/tpc.9.3.355. doi:10.1105/tpc.9.3.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons M.P, Ochoterena H. Gaps as characters in sequence-based phylogenetic analyses. Syst. Biol. 2000;49:369–381. doi:10.1080/10635159950173889 [PubMed] [Google Scholar]
- Slowinski J.B, Page R.D.M. How should species phylogenies be inferred from sequence data. Syst. Biol. 1999;48:814–825. doi: 10.1080/106351599260030. doi:10.1080/106351599260030 [DOI] [PubMed] [Google Scholar]
- Small R.L. Phylogeny of Hibiscus sect. Muenchhusia (Malvaceae) based on chloroplast rpl16 and ndhF and nuclear ITS and GBSSI sequences. Syst. Bot. 2004;29:385–392. doi:10.1600/036364404774195575 [Google Scholar]
- Small R.L, Ryburn J.A, Cronn R.C, Seelanan T, Wendel J.F. The tortoise and the hare: choosing between non-coding plastome and nuclear Adh sequences for phylogeny reconstruction in a recently diverged plant group. Am. J. Bot. 1998;85:1301–1315. [PubMed] [Google Scholar]
- Small R.L, Cronn R.C, Wendel J.F. Use of nuclear genes for phylogeny reconstruction in plants. Aust. Syst. Bot. 2004;17:145–170. doi:10.1071/SB03015 [Google Scholar]
- Soltis D.E, Soltis P.S. Choosing an approach and an appropriate gene for phylogenetic analysis. In: Soltis D.E, Soltis P.S, Doyle J.J, editors. Molecular systematics of plants II. DNA sequencing. Kluwer Academic; Boston, MA: 1998. pp. 1–42. [Google Scholar]
- Soltis P.S, Soltis D.E. Molecular systematics: assembling and using the Tree of Life. Taxon. 2001;50:663–677. [Google Scholar]
- Soltis P.S, Soltis D.E. The origin and diversification of angiosperms. Am. J. Bot. 2004;91:1614–1626. doi: 10.3732/ajb.91.10.1614. [DOI] [PubMed] [Google Scholar]
- Soltis P.S, Soltis D.E, Chase M.W. Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology. Nature. 1999;402:402–404. doi: 10.1038/46528. doi:10.1038/46528 [DOI] [PubMed] [Google Scholar]
- Soltis D.E, et al. Genome-scale data, angiosperm relationships, and ‘ending incongruence’: a cautionary tale in phylogenetics. Trends Plant Sci. 2004;9:477–483. doi: 10.1016/j.tplants.2004.08.008. doi:10.1016/j.tplants.2004.08.008 [DOI] [PubMed] [Google Scholar]
- Strand A.E, Leebens-Mack J, Milligan B.G. Nuclear DNA-based markers for plant evolutionary biology. Mol. Ecol. 1997;6:113–118. doi: 10.1046/j.1365-294x.1997.00153.x. doi:10.1046/j.1365-294X.1997.00153.x [DOI] [PubMed] [Google Scholar]
- Syring, J. V., Willyard, A., Cronn, R. C. & Liston, A. In press. Evolutionary relationships among Pinus (Pinaceae) subsections inferred from multiple low-copy nuclear loci. Am. J. Bot [DOI] [PubMed]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 1991;17:1105–1110. doi: 10.1007/BF00037152. doi:10.1007/BF00037152 [DOI] [PubMed] [Google Scholar]
- Tank D.C, Sang T. Phylogenetic utility of the glycerol-3-phosphate acyltransferase gene: evolution and implications in Paeonia (Paeoniaceae) Mol. Phylog. Evol. 2001;19:421–429. doi: 10.1006/mpev.2001.0931. doi:10.1006/mpev.2001.0931 [DOI] [PubMed] [Google Scholar]
- Thießen G, Becker A, Winter K.-U, Münster T, Kirchner C, Saedler H. How the land plants learned their floral ABCs: the role of MADS-box genes in the evolutionary origin of flowers. In: Cronk Q.C.B, Bateman R.M, Hawkins J.A, editors. Developmental genetics and plant evolution. Taylor & Francis; London: 2002. pp. 173–205. [Google Scholar]
- Wang R.L, Stec A, Hey J, Lukens L, Doebley J. The limits of selection during maize domestication. Nature. 1999;398:236–239. doi: 10.1038/18435. doi:10.1038/18435 [DOI] [PubMed] [Google Scholar]
- Watanabe M. Describing the ‘Tree of Life’: attainable goal or stuff of dreams? BioScience. 2002;52:875–880. [Google Scholar]
- Weeks A, Simpson B.B. Molecular genetic evidence for interspecific hybridization among endemic Hispaniolan Bursera (Burseraceae) Am. J. Bot. 2004;91:976–984. doi: 10.3732/ajb.91.6.976. [DOI] [PubMed] [Google Scholar]
- Wendel J.F, Doyle J.J. Phylogenetic incongruence: window into genome history and molecular evolution. In: Soltis D.E, Soltis P.S, Doyle J.J, editors. Molecular systematics of plants II. DNA sequencing. Kluwer Academic; Boston, MA: 1998. pp. 265–296. [Google Scholar]
- Wendel J.F, Schnabel A, Seelanan T. Bi-directional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium) Proc. Natl Acad. Sci. 1995;92:280–284. doi: 10.1073/pnas.92.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel J.W, Siddall M.E. Noise. Cladistics. 1999;15:51–64. doi:10.1111/j.1096-0031.1999.tb00394.x [Google Scholar]
- Winkworth R.C, Donoghue M.J. Viburnum phylogeny: evidence from the duplicated nuclear gene GBSSI. Mol. Phylog. Evol. 2004;33:109–126. doi: 10.1016/j.ympev.2004.05.006. doi:10.1016/j.ympev.2004.05.006 [DOI] [PubMed] [Google Scholar]
- Wojciechowski M.F, Sanderson M.J, Hu J.M. Evidence on the monophyly of Astragalus and its major subgroups based on nrDNA ITS and cpDNA trnL intron data. Syst. Bot. 1999;24:408–437. [Google Scholar]
- Wolfe A.D, Liston A. Contributions of PCR-based methods to plant systematics and evolutionary biology. In: Soltis D.E, Soltis P.S, Doyle J.J, editors. Molecular systematics of plants II. DNA sequencing. Kluwer Academic; Boston, MA: 1998. pp. 43–86. [Google Scholar]
- Wolfe K.H, Li W.-H, Sharp P.M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast and nuclear DNAs. Proc. Natl Acad. Sci. 1987;84:9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortley, A. H. & Scotland, R.W. 2005 Determining the potential utility of datasets for phylogeny reconstruction. Taxon In press.
- Wortley A.H, Rudall P.J, Harris D.J, Scotland R.W. How much data is needed to resolve a difficult phylogeny? Case study in Lamiales. Syst. Biol. 2005;54:697–709. doi: 10.1080/10635150500221028. [DOI] [PubMed] [Google Scholar]
- Xu W, Briggs W.J, Padolina J, Timme R.E, Liu W, Linder C.R, Miranker D.P. Using MoBIoS scalable genome join to find conserved primer pair candidates between two genomes. Bioinformatics. 2004;20(Suppl. 1):i355–i362. doi: 10.1093/bioinformatics/bth929. doi:10.1093/bioinformatics/bth929 [DOI] [PubMed] [Google Scholar]
- Yang Z. On the best evolutionary rate for phylogenetic analysis. Syst. Biol. 1998;47:125–133. doi: 10.1080/106351598261067. doi:10.1080/106351598261067 [DOI] [PubMed] [Google Scholar]
- Yates T.L, Salazar-Bravo J, Dragoo J.W. The importance of the Tree of Life to society. In: Cracraft J, Donoghue M.J, editors. Assembling the Tree of Life. Oxford University Press; New York: 2004. pp. 7–17. [Google Scholar]
- Yockteng R, Nadot S. Infrageneric phylogenies: a comparison of chloroplast-expressed glutamine synthetase, cytosol-expressed glutamine synthetase and cpDNA maturase K in Passiflora. Mol. Phylog. Evol. 2004;31:397–402. doi: 10.1016/S1055-7903(03)00276-8. doi:10.1016/S1055-7903(03)00276-8 [DOI] [PubMed] [Google Scholar]