Abstract
Coordination between cell proliferation and differentiation is essential to create organized and functional tissues. Arabidopsis thaliana stomata are created through a stereotyped series of symmetric and asymmetric cell divisions whose frequency and orientation are informed by cell–cell interactions. Receptor-like proteins and a mitogen-activated protein kinase kinase kinase were previously identified as negative regulators of stomatal development; here, we present the characterization of a bona fide positive regulator. FAMA is a putative basic helix-loop-helix transcription factor whose activity is required to promote differentiation of stomatal guard cells and to halt proliferative divisions in their immediate precursors. Ectopic FAMA expression is also sufficient to confer stomatal character. Physical and genetic interaction studies combined with functional characterization of FAMA domains suggest that stomatal development relies on regulatory complexes distinct from those used to specify other plant epidermal cells. FAMA behavior provides insights into the control of differentiation in cells produced through the activity of self-renewing populations.
INTRODUCTION
Stomata are structures in the epidermis of plant leaves that serve as the major conduit for the exchange of water vapor and carbon dioxide between the plant and the environment. Minimally, stomata consist of a pair of sister epidermal cells (guard cells) that flank a pore overlying an airspace in the photosynthetic tissue below. Several aspects of stomatal development make this an attractive model for understanding patterning and cell fate. Stomatal guard cells are the terminal product of a unique plant cell specification program that features a stereotyped series of asymmetric and symmetric cell divisions (Figure 1A). A fraction of the protodermal cells initiate the stomatal lineage by undergoing asymmetric entry divisions. The daughters of this type of division are the meristemoid and a larger sister cell. It is not known what provides a cell the ability to undergo an entry division. The meristemoid then undergoes a series of asymmetric amplifying divisions that recreate the meristemoid and produce additional sister cells. The sisters may become epidermal pavement cells or may undergo entry divisions later. The meristemoid is the precursor of the stomatal guard cells; however, it does not differentiate directly into this cell type but first becomes another specialized precursor type, the guard mother cell (GMC). The GMC divides only a single time, symmetrically, to form two guard cells. The cell wall laid down during GMC cytokinesis is later thickened and partially degraded to form the stomatal pore between the two guard cells. The overall pattern of Arabidopsis thaliana stomata is established by cell communication between early developing stomata and their neighbor cells, and oriented spacing divisions ensure that the meristemoids produced from later divisions of sister cells are not created in contact with existing stomata (reviewed in Nadeau and Sack, 2002a).
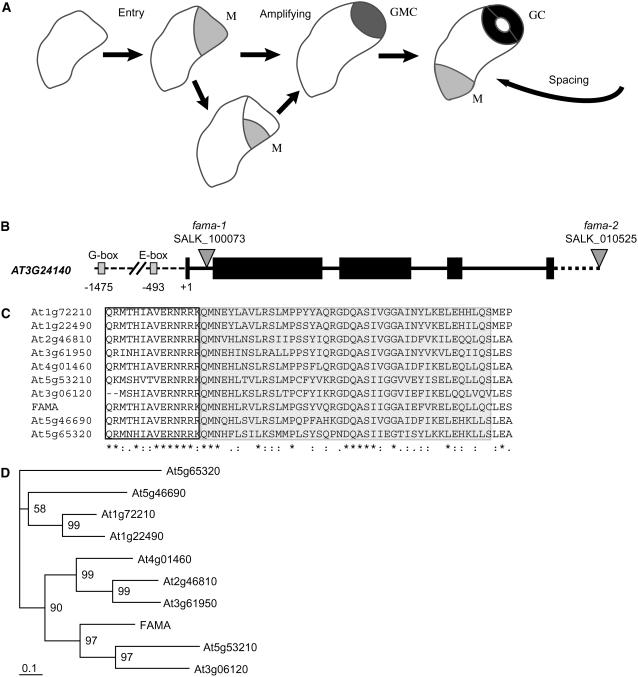
Figure 1.
Stomatal Development Pathway and FAMA Gene Structure.
(A) Diagram of events in stomatal differentiation. A protodermal cell makes an asymmetric entry division to create a meristemoid (M, light gray) and larger sister cell. Meristemoids undergo up to three rounds of proliferative asymmetric division before differentiating into GMCs (medium gray). GMCs divide once to become the two guard cells (GC, dark gray) of the mature stoma.
(B) Locus structure of FAMA. Exons are indicated by black boxes and location of T-DNA insertions by gray triangles.
(C) Alignment of bHLH domain among FAMA clade sequences. Boxed region is basic domain, and shaded region is the HLH domain.
(D) Neighbor-joining tree using full-length proteins in FAMA clade. Branch lengths are proportional to distances between sequences. Numbers between branches indicate percentage of support in bootstrap analyses (1000 replicates).
Several genes encoding components of cell signaling systems regulate the divisions in the stomatal lineage. Mutations in putative receptors TOO MANY MOUTHS (TMM) (Nadeau and Sack, 2002b) and the ERECTA (ER) family kinases (Shpak et al., 2005) affect all types of asymmetric cell divisions in the stomatal lineage, as does elimination of the mitogen-activated protein kinase kinase kinase YODA (Bergmann et al., 2004). In leaves, all of these proteins act as negative regulators of stomatal formation. The signaling pathway genes are important for the initial choice of a cell to become a stomatal precursor, but they do not appear to be required for differentiation once a cell has committed to stomatal fate (Berger and Altmann, 2000; Nadeau and Sack, 2002b). Two partially redundant R2R3 MYB-type transcription factors, FOUR LIPS (FLP) and MYB88, were recently shown to be required for progression from meristemoid to guard cell. In flp-1 mutants, stomata often consist of three or four guard cells, as if the GMCs continued to divide (Yang and Sack, 1995; Lai et al., 2005). MYB88 has no mutant phenotype alone but enhances the proliferation defect in flp-1 GMCs (Lai et al., 2005).
In this article, we describe FAMA (FMA), named after the goddess of rumor. FAMA encodes a putative basic helix-loop-helix (bHLH) transcription factor (bHLH097), whose transcript and protein are specifically expressed in the stomatal lineage. FAMA regulates a critical switch between division and differentiation. Like FLP/MYB88, FAMA is required for halting divisions at the end of the stomatal lineage, but, in addition, FAMA has an instructive role in promoting guard cell fate. Tests of genetic and physical interactions between FAMA and FLP/MYB88 indicate that these proteins are likely to influence the GMC–guard cell transition independently. Functional characterization of FAMA protein domains, however, suggests that FAMA interacts with other partners during stomatal development.
RESULTS
Identification of FAMA and Phenotype of fama Loss-of-Function Mutants
FAMA was identified through a genome-wide survey of genes that were more highly expressed in plants with an enriched stomatal population than in plants lacking stomata (Bergmann et al., 2004). The FAMA gene encodes a 414–amino acid protein (Figure 1B) similar to bHLH transcription factors. These transcriptional regulators have a conserved basic domain that may contact DNA, an HLH domain for homo- or heterodimerization, and additional motifs that mediate interactions with other proteins (Grandori et al., 2000; Massari and Murre, 2000). At least 160 bHLHs are predicted in the Arabidopsis genome. Other members of the bHLH family have been implicated in responses to light and hormones and in other epidermal cell fate decisions (Heim et al., 2003; Toledo-Ortiz et al., 2003). FAMA is a member of a small subclass of bHLH proteins that contains a short conserved region preceding the bHLH domain and a longer, highly conserved C-terminal domain of unknown function (Figures 1C and 1D; see Supplemental Figure 4 online).
Negligible FAMA transcript is produced in lines homozygous for the SALK_100073 T-DNA insertion (fama-1) (Figure 2A). fama-1 plants completely lack recognizable stomata but instead produce clusters of small, narrow epidermal cells in the normal location of stomata (Figure 2C). These collections of cells will be referred to as fama tumors. The overall morphology of fama-1 mutant embryos and seedlings is not dramatically different from the wild type (data not shown); however, later development is compromised, and fama-1 mutants arrest as pale plants with small rosette leaves and bushy inflorescences topped by one to three sterile flowers (Figure 2D). In fama-1, epidermal tumors are found on all organs that normally make stomata, including cotyledons, rosette leaves, cauline leaves, stems, pedicles, sepals, and siliques (see Supplemental Figure 1 online). On cotyledons, fama-1 tumors are larger on the adaxial surface (6.3 ± 0.15 se cells/tumor) than on the abaxial surface (3.4 ± 0.067 se cells/tumor). A second T-DNA insertion allele (SALK_010525) near the 3′ end of the FAMA gene (fama-2) results in healthy, fertile plants that contain occasional three- and four-celled stomata, similar to those in a flp-1 mutant (see Supplemental Figures 1F and 1G online).
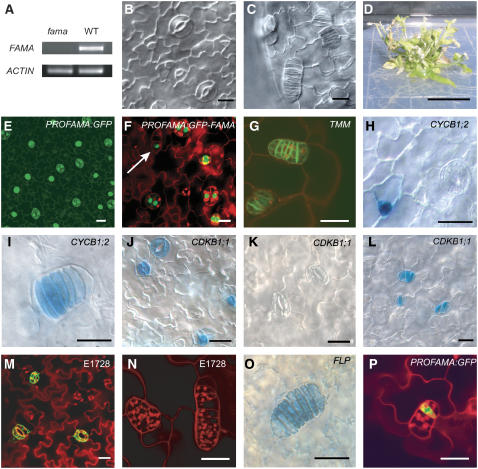
Figure 2.
FAMA Expression Pattern and Loss-of-Function Phenotypes.
(A) RT-PCR of FAMA from fama-1 allele and Col control.
(B) and (C) Differential interference contrast (DIC) images of wild-type and fama-1 cotyledon epidermis 7 d after germination (DAG).
(D) Two-month-old fama-1 plant. Bar = 1 cm.
(E) and (F) Expression of PROFAMA:GFP and PROFAMA:GFP-FAMA, respectively, in Col cotyledon epidermis. Arrow in (F) points to earliest expression in GMCs.
(G) to (P) Expression of cell fate and cell division markers in fama-1. Bar = 10 μm.
(G) PROTMM:TMM-GFP.
(H) and (I) CYCB1;2:GUS in Col and fama-1, respectively.
(J) to (L) CDKB1;1:GUS in Col at 5 DAG (J) and 10-DAG (K) and fama-1 at 10 DAG (L). Note the prolonged expression of this marker in fama-1.
(M) and (N) Mature stomata marker E1728:GFP in Col and fama-1, respectively.
(O) PROFLP:GUS/GFP in fama-1.
(P) PROFAMA:GFP in fama-1.
Expression of the FAMA cDNA preceded by 2.5 kb of genomic sequence 5′ of the predicted translational start rescues the fama-1 mutant phenotype (see Methods). To examine the specific spatial and temporal patterns of gene expression at cellular resolution, a reporter construct consisting of this 2.5-kb FAMA upstream region was fused to green fluorescent protein (GFP) or β-glucuronidase (GUS). PROFAMA:GFP expression is restricted to the stomatal lineages. It is not expressed in meristemoids but is strongly expressed in GMCs and in young guard cells (Figure 2E). Expression of PROFAMA:GUS was observed in similar patterns. No GUS expression was seen in internal tissue layers or in primary or lateral roots (see Supplemental Figures 2A to 2E online). FAMA protein localization was assayed with GFP-tagged FAMA constructs driven by the 2.5-kb FAMA upstream region. These transgenes are capable of rescuing fama-1 mutants (see Methods). Both N- and C-terminal fusions of GFP to FAMA were predominantly nuclear localized and were expressed in the same spatial and temporal pattern. PROFAMA:GFP-FAMA was more easily visualized and thus was used for subsequent analysis. The PROFAMA:GFP-FAMA reporter was strongly expressed in GMCs and young guard cells of aerial organs (Figure 2F), similar to the transcriptional fusion, suggesting that FAMA protein does not traffic from its site of synthesis.
The identity of cells in the fama tumors was determined by the analysis of several cell fate markers. As cells enter the stomatal development pathway, they express the receptor-like protein TMM (PROTMM:TMM-GFP) (Nadeau and Sack, 2002b). Reporters for the two cell cycle regulators PROcyclinB1;2:GUS (Donnelly et al., 1999) and PROCDKB1;1:GUS (Boudolf et al., 2004) are expressed in dividing cells. In the leaf epidermis, these markers are expressed strongly in meristemoids and GMCs, moderately in young guard cells, and weakly in mature guard cells (Figures 2H, 2J, and 2K). These three markers are normally downregulated upon a cell reaching its terminal differentiated state. Cells in fama tumors express all of these markers and continue to express them into the stages of leaf development when they are normally turned off (Figures 2G to 2L). Conversely, cells in fama tumors fail to express the PROKAT1:GUS (Nakamura et al., 1995) and E1728:GFP markers that are normally expressed in mature guard cells (Figures 2M and 2N; see Supplemental Figures 2F and 2G online; data not shown). The expression of PROFLP:GUS, a marker of GMC cells (Lai et al., 2005), in tumors suggests that fama tumor cells progress to at least the GMC stage (Figure 2O).
FAMA Is Not Required for Its Own Expression
Many transcription factors are regulated by feedback loops in which they regulate their own expression. Upstream of FAMA are several consensus bHLH binding sites (Figure 1B), suggesting it was possible that FAMA also regulated its own transcription. Many cells in the fama-1 tumors express the PROFAMA:GFP reporter (Figure 2P). This indicates that FAMA is not absolutely required to promote its own expression. Because not all tumor cells express PROFAMA:GFP, however, it is possible that FAMA has an autoregulatory role in promoting or maintaining its expression.
FAMA Overexpression Phenotypes Are Opposite of Loss of Function
Lack of FAMA prevents cells from progressing to the guard cell stage, suggesting that FAMA is necessary both to prevent cell division and to promote guard cell fate. To determine whether FAMA was sufficient to induce guard cell fate, we ectopically expressed FAMA using an estrogen inducible system (Zuo et al., 2000). Induced overexpression of FAMA generated an epidermal phenotype that could easily be interpreted as opposite to the loss-of-function phenotype. In these plants, the cotyledon epidermis consisted of cells that have the kidney-shaped morphology of guard cells (Figures 3A to 3D). These cells produce a cell wall thickening containing biorefringent material reminiscent of stomatal pore sites, and some cells produce structures that morphologically resemble pores (Figures 3A and 3B, arrows). The cells also expressed two molecular markers, PROKAT1:GUS and E1728:GFP (Figures 3C and 3D), providing additional evidence that these cells posses guard cell identity. In contrast with guard cells of normal stomata, however, the cells resulting from FAMA overexpression are unpaired.
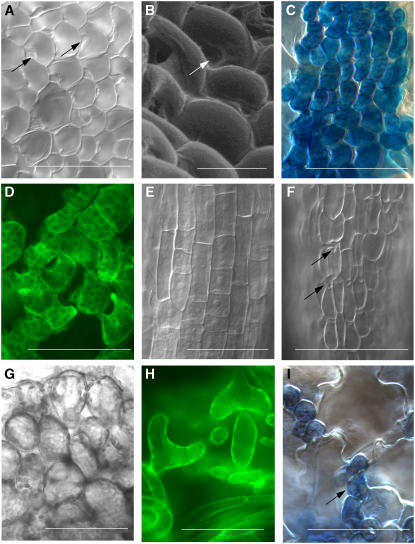
Figure 3.
FAMA Gain-of-Function Phenotypes.
(A) to (F) The 7-DAG PROEST:FAMA seedling germinated on estrogen.
(A) and (B) DIC and scanning electron microscopy images of cotyledon epidermis; arrows indicate stomatal pores. Scanning electron micrographs taken at ×480 magnification.
(C) PROKAT1:GUS in cotyledon epidermis.
(D) E1728:GFP in cotyledon epidermis.
(E) and (F) DIC image induced of Col and PROEST:FAMA root epidermis, respectively.
(G) and (H) Col and PROEST:FAMA leaf mesophyll, respectively. In PROEST:FAMA lines, mesophyll morphology is crescent-shaped instead of spherical and the cells express E1728:GFP.
(I) Ectopic PROFAMA:FAMA-induced guard cells expressing KAT1:GUS. Unaffected pavement cells are visible as large crenulated cells without GUS expression.
Bars = 100 μm except in (B) (20 μm) and (E) (50 μm).
FAMA overexpression is also able to convert nonstomatal lineage cells to guard cells. In strong PROEST:FAMA lines germinated on estrogen, induction caused guard cell transdifferentiation in almost all epidermal cells, including cells in nonstomatal files of the hypocotyls (see Supplemental Figures 1E and 1F online) and in roots (Figures 3E and 3F). FAMA overexpression may also be able to induce partial transdifferentiation of nonepidermal tissues. Cells presumed to be mesophyll by their location have altered cell shape and express guard cell markers E1728:GFP (Figure 3H) and PROKAT1:GUS (data not shown). The change in cell morphology is visible within 48 h after germination (on 5 μM estrogen), coincident with the time the first guard cells normally appear. Seedlings that produce excess, presumably nonfunctional, unpaired guard cells in multiple cell layers were not viable.
Expression of FAMA in wild-type plants under the control of tissue-specific promoters also promoted ectopic or premature guard cell formation. Expression of FAMA with the stomatal lineage-specific promoter TMM (PROTMM:FAMA) led to the formation of unpaired guard cells in places where TMM is normally expressed (see Supplemental Figure 5C online). Likewise, some lines expressing FAMA in GMCs (PROFAMA:FAMA) could also induce the production of some single guard cells in the epidermis (Figure 3I). In these lines, FAMA-GFP was seen in the nucleus of morphologically normal GMCs, and weak expression was visible in nuclei of both normal and unpaired (ectopic) guard cells (n > 200 cells in six leaves).
Despite possessing many characteristics of guard cells, the cells created by FAMA overexpression are not correctly organized into pairs flanking a pore. One interpretation of this phenotype is that FAMA overexpression not only drives differentiation but inhibits cell division so that the GMC converts directly into a guard cell without undergoing cytokinesis. This possibility was tested in the PROFAMA:GFP-FAMA lines where the fate of individual GFP positive cells was followed over 48 h (see Methods). Twelve GFP positive cells with the morphology of GMCs were identified in leaf 2 of 9-d-old plants. Two days later, single guard cells were identified in the same locations as these GMCs (8/12). Multiple guard cells (2/12) or ambiguous cells (2/12) were also occasionally observed. These data suggest that overexpression of FAMA can force GMCs to differentiate into guard cells without undergoing the normal symmetrical cell division.
FAMA Acts as a Master Regulator of Stomatal Differentiation
The expression pattern of FAMA and its loss- and gain-of-function phenotypes are all consistent with FAMA regulating the final stage in stomatal development. FAMA appears to be absolutely required for stomatal development in wild-type plants. To determine whether this requirement could be overcome by generating a larger number of stomatal lineage cells, loss-of-function mutations in the negative regulators TMM1, SDD1, YODA, and the ER family (ER;ERL1;ERL2) were combined with a fama-1 mutation (see Supplemental Figure 3 online). In no case were guard cells formed, indicating that there is an absolute requirement for FAMA in guard cell differentiation. The number of tumors and their orientation and spacing relative to each other reflect the phenotype of the second mutation (for example, with yoda; Figures 4A to 4C).
Figure 4.
Genetic and Physical Interactions between FAMA and Other Stomatal Regulators.
(A) to (F) DIC images of cotyledon epidermis 10 DAG, all at the same magnification. (A) fama-1; (B) yda; (C) fama-1 yda; (D) flp-1; (E) flp-1 fama-1; (F) fama-1 myb88; and (G) expression of PROFAMA:GFP in flp-1 mutant clusters. Bars = 100 μm.
(H) and (I) Fluorescence images of tobacco epidermal cells transiently transformed with BiFC constructs. Green indicates protein interaction in nucleus, and red is false color of cell outlines. Positive BiFC interaction between FAMA and bHLH093 provided (H) compared to negative BiFC interaction between FLP and FAMA (I).
FAMA and FLP Appear to Act in Independent Pathways
In other epidermal cell fate decisions, a complex of an R2R3-type MYB, a bHLH, and a WD-repeat protein is at the top of a regulatory hierarchy that leads to cell fate specification (Lee and Schiefelbein, 1999; Zimmermann et al., 2004; Bernhardt et al., 2005). In stomatal development, two R2R3-type MYBs, FLP and MYB88, are expressed in GMCs and are required to regulate the transition from GMC to guard cell (Lai et al., 2005). Because FAMA and FLP/MYB88 have overlapping expression patterns and similar mutant phenotypes, genetic, physical, and regulatory relationships between them were tested. fama and flp-1 form clusters of GMCs, although GMCs in flp-1 eventually become guard cells (Figure 4D). The fama-1 differentiation phenotype is epistatic in double mutants of flp fama, but the proliferation phenotype is additive (number of cells/stomatal unit on abaxial side of cotyledon: flp-1 = 2.6 ± 0.08 se; fama-1 = 3.4 ± 0.067 se; flp fama = 6.1 ± 0.38 se) (Figure 4E). myb88 (SALK_068691) single mutants are indistinguishable from the wild type but enhance the flp mutant phenotype. Double mutant combinations of myb88 fama-1 resemble fama-1 (Figure 4F). These genetic interactions argue against a model that FAMA and FLP or MYB88 form obligate heterodimers. The fama-1 phenotype is more severe than flp or flp myb88, suggesting that FAMA acts prior to FLP or that it is required for a broader range of activities. To determine whether FAMA is required for FLP expression, a PROFLP:GUS-GFP reporter was examined in fama tumors. PROFLP:GUS is expressed in fama-1 (Figure 2O). The PROFAMA:GFP reporter is expressed in flp-1 (Figure 4G), suggesting that neither protein is required for the transcriptional activation of the other. Neither FLP or MYB88 nor FAMA possess the domains known to promote MYB/bHLH dimerization (Zimmermann et al., 2004). Nevertheless, FAMA was tested for its ability to interact with FLP in yeast two-hybrid assays and with an in planta split GFP assay. FAMA failed to interact with FLP in these assays (Figures 4H and 4I; data not shown). Taken together, the evidence does not support a model that FAMA and FLP/MYB88 form a complex equivalent to that used in other epidermal cell fate decisions.
FAMA Is Likely to Act as a Transcription Factor
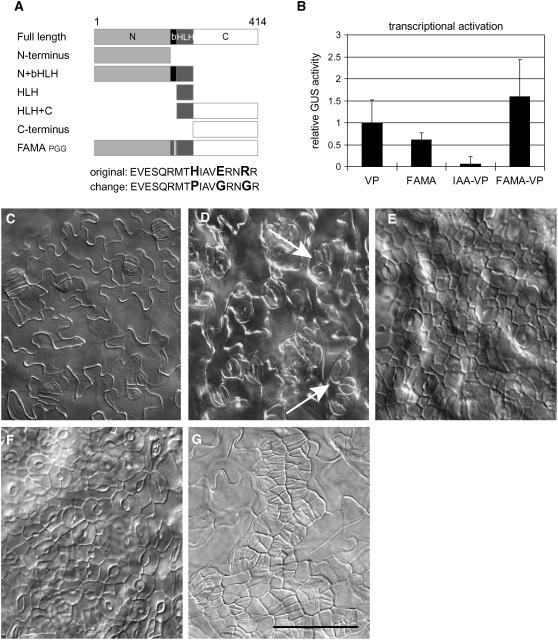
FAMA acts as a major regulator of the decision to proliferate or differentiate. The nuclear localization and presence of conserved domains belonging to bHLH transcription factors suggest that it is likely that FAMA plays this role by regulating gene expression. FAMA contains the conserved residues H-E-R in the putative DNA binding domain. This triad is characteristic of proteins that bind DNA at the G-box (CACGTG) motif (Toledo-Ortiz et al., 2003). Substituting noncharged residues at the H-E-R sites (PROEST:FAMAPGG) (Figure 5A) resulted in a protein that failed to rescue fama-1. Attaching a repression domain (EAR domain) to the C terminus of FAMA created a dominant negative phenotype (Figure 5C), also consistent with FAMA acting as a transcriptional regulator. Plant bHLH transcription factors may act as transcriptional activators or transcriptional repressors (Grandori et al., 2000; Massari and Murre, 2000). Because the conversion from GMC to guard cells could conceivably involve both the repression and activation of targets, FAMA was tested in a protoplast-based transcriptional activation/repression assay (Tiwari et al., 2004; He et al., 2005). To test for transcriptional activation, full-length FAMA was fused to the yeast GAL4 DNA binding domain and transiently coexpressed with a reporter containing GAL4 and LexA binding sites in front of GUS. FAMA was a robust activator of GUS activity, indicating that it can act as a transcriptional activator (Figure 5B). To test if FAMA has transcriptional repression activity, FAMA-GAL4-DB was cotransformed with a constitutive transcriptional activator, VP16-LexA-DB, and the GAL4-LexA-GUS reporter. Coexpression of VP16 with a known repressor (IAA-GAL4) can eliminate the VP16 activation; however, coexpression of FAMA and VP16 increased GUS activity above the level achieved when either protein was expressed alone (Figure 5B), again suggesting that FAMA behaves primarily as a transcriptional activator.
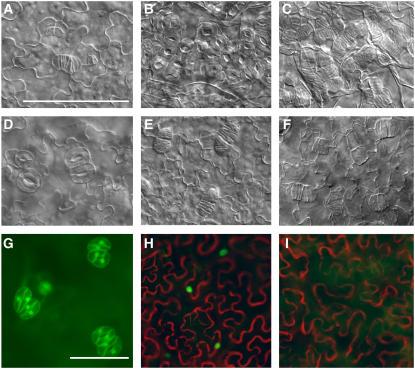
Figure 5.
Behavior of FAMA Domains in Transcriptional Activity Assay and in Planta.
(A) Diagram of deletion and DNA binding site mutation constructs tested.
(B) Protoplast-based transcriptional activation assay. Effector constructs indicated on x axis. All values were normalized to VP16-driven reporter activation values. Error bars indicate standard deviations. n = 6 independent transformations.
(C) to (G) DIC images of cotyledons of plants transformed with PROEST:FAMA protein variants. All images are at the same magnification.
(C) Col plants expressing PROEST:FAMA-EAR.
(D) Col plants expressing PROEST:FAMA C terminus alone. The arrow indicates paired stomata.
(E) and (F) Col plants expressing PROEST:FAMAPGG at 4 and 10 d, respectively.
(G) PROEST:FAMAPGG in fama-1. Bar = 50 μm.
Expression of ΔN-FAMA Suggests the Existence of Other Stomatagenic Factors
In addition to the bHLH domain, the FAMA clade of bHLHs share previously uncharacterized C- and N-terminal domains (see Supplemental Figure 4 online). Functional characterization of FAMA domains was assayed by expression of deletion constructs under the control of an estrogen-inducible promoter in Columbia (Col) and in fama-1 (Figure 5A). Expression of the N terminus alone (1 to 208 amino acids) or the HLH domain alone (208 to 250 amino acids) neither rescued fama-1 nor recapitulated the gain-of-function phenotype (data not shown). Expression of the C-terminal domain (250 to 414 amino acids) also failed to rescue fama-1 but did lead to a slight increase in stomatal density and stomatal cluster formation in a wild-type background (Figure 5C).
Expression of another variant of FAMA containing the HLH domain and C terminus (208 to 414 amino acids) produced an unexpected phenotype. In 4-DAG Col plants, PROEST:ΔNFAMA induction caused the overproliferation of small cells in the epidermis (similar to Figure 5E). This phenotype is similar to the effect of overexpressing CYCD3 or other positive regulators of cell division (De Veylder et al., 2002; Dewitte et al., 2003). In contrast with the cells produced by overexpressing cell cycle genes, however, the cells produced by PROEST:ΔNFAMA all differentiated into stomata by 10 DAG. Reexamination of the epidermis of PROEST:FAMAPGG Col plants at 10 DAG revealed that induction of FAMAPGG could also cause the entire leaf epidermis to form morphologically normal (paired) guard cells (Figures 5E and 5F). This phenotype was dependent on a functional genomic copy of FAMA. In fama-1 mutants, PROEST:ΔNFAMA and PROEST:FAMAPGG produced excess GMCs but no stomata (Figure 5G; data not shown).
Expression of ΔN-FAMA or FAMAPGG with the estrogen or 35S promoter converts all cotyledon epidermal cells into stomata. Expression of PROTMM:FAMAPGG also induces ectopic stomata but in smaller domains of the epidermis, consistent with the TMM expression pattern (see Supplemental Figures 5A to 5C online). When expressed in GMCs under the FAMA promoter, however, FAMAPGG had no effect (see Supplemental Figures 5D and 5E online), suggesting that the altered FAMA protein exerts its stomata-promoting effects by either interfering with proteins other than FAMA and/or acting before GMC formation.
Other bHLHs May Be Partners of FAMA
The behavior of ΔN-FAMA suggested that FAMA might interfere with or mimic the activity of other stomatal regulators. To identify candidate partners of FAMA, a yeast two-hybrid screen was performed using an N-terminal deleted version of FAMA that contained the full bHLH domain and C terminus (190 to 414 amino acids). Two bHLH proteins, bHLH071 (At5g46690) and bHLH093 (At5g65640) (Heim et al., 2003; Toledo-Ortiz et al., 2003), were identified in multiple interacting clones. Both proteins share significant sequence similarity with FAMA in the bHLH domain, and bHLH071 is in the same subgroup as FAMA (Figures 1C and 1D; see Supplemental Figure 4 online). Bimolecular fluorescence complementation (BiFC) was used to test in planta protein–protein interactions between these bHLHs and FAMA. FAMA, bHLH071, bHLH093, and a distantly related bHLH were tested for pairwise interactions (Figures 6A to 6E): FAMA interacted strongly with bHLH093 (Figure 6A) and bHLH071 (Figure 6B) and moderately with itself (Figure 6C) but not with the distantly related bHLH (Figure 6D). To determine whether the neomorphic FAMAPGG protein variant had altered dimerization qualities, FAMAPGG was also tested in this BiFC assay. Interactions of FLP, FAMA, bHLH071, and bHLH093 with FAMAPGG were comparable to their interactions with wild-type FAMA (Figure 6E; data not shown), suggesting that the altered version can dimerize with other bHLHs.
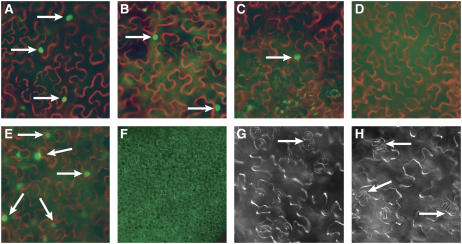
Figure 6.
Newly Identified FAMA Interaction Partners.
(A) to (E) Micrographs of BiFC interactions between FAMA and other bHLHs. Positive interactions produce GFP fluorescence (green) in nuclei. Cell outlines were visualized in DIC mode and false-colored red. Representative images of FAMA-bHLH093 (A), FAMA-bHLH071 (B), FAMA-FAMA (C), FAMA-distant bHLH (D), and FAMAPGG-bHLH093 (E) are shown.
(F) Fluorescent image of ubiquitous PRObHLH093:GFP in cotyledon epidermis.
(G) and (H) DIC images of 35S-driven overexpression of bHLH071 and bHLH093, respectively, in Col cotyledon epidermis. Arrows point to fama-2–like clustered stomata.
Both bHLH071 and bHLH093 appear to be broadly expressed (Figure 6F; data not shown). Their functional involvement in stomatal development was tested in both loss-of-function and gain-of-function assays. Loss of function was assayed by the addition of a dominant repression domain (bHLH071-EAR and bHLH093-EAR) (see Methods) and by characterizing the epidermis in plants homozygous for T-DNA insertions in bHLH071 (SALK_130027 and SALK_074601) and bHLH093 (SAIL_747_A08). In none of these cases was an obvious defect in stomatal formation observed (data not shown). Overexpression of bHLH093 or bHLH071 with the 35S promoter resulted in a weak, fama-2 (or flp-1) phenotype (Figures 6G and 6H) in a small fraction (>10%) of the lines. The lack of strong phenotype of loss or gain of function of bHLH071 and bHLH093 may indicate redundancy or may indicate that these proteins, although capable of interacting with FAMA, are not its normal partners. The strong physical interaction between FAMA and these bHLHs contrasts with the undetectable interaction between FAMA and the MYB-type transcription factor FLP, strengthening the argument against a FAMA/FLP complex and suggesting that if FAMA forms heteromeric complexes, it may do so with other bHLH proteins.
DISCUSSION
FAMA controls a critical decision in the life of the plant: whether to continue cell division or to exit mitosis and begin a terminal differentiation process. To create the two-celled stomata characteristic of Arabidopsis (and probably of all plants), the single symmetric division of the GMC must be coordinated with the differentiation of its daughters. Various genetic and pharmacological manipulations, however, have revealed that the two processes are surprisingly independent. For example, GMCs in flp mutants undergo additional cell divisions, and manipulating cell cycle regulators can prevent GMC cytokinesis, but the daughters of these aberrant divisions exhibit guard cell traits (Boudolf et al., 2004; Lai et al., 2005).
Because division and differentiation are contemporaneous, but not necessarily dependent on each other, the fact that both are affected by loss or gain of FAMA function suggests that FAMA acts early in the GMC to guard cell transition and that FAMA regulates many targets.
Because FAMA can induce guard cell traits in nonstomatal lineages, FAMA must be able to actively turn on differentiation genes. Whether these components of functional guard cells (such as KAT1) are direct targets of transcriptional regulation by FAMA or whether FAMA acts through intermediate factors is currently being investigated.
FAMA-GFP expression is first seen in the nuclei of premitotic GMCs and is downregulated as guard cells mature (Figures 2E and 2F). FAMA is present at an appropriate time to limit cell division by directly regulating cell cycle genes. The one-celled stomata produced by ectopic expression of FAMA are similar to the stomata produced by expression of a dominant negative form of the cell cycle regulator CDKB1;1 (Boudolf et al., 2004). CDKB1;1 is normally expressed exclusively within the stomatal lineage of the leaf epidermis and is downregulated when epidermal cell divisions cease (Boudolf et al., 2004). CDKB1;1:GUS expression is maintained in the tumors of fama mutants, suggesting that FAMA may negatively regulate CDKB1;1 to halt cell division (Figure 7). Because FAMA behaves as an activator in protoplast transcriptional assays (Figure 5B), its negative effect on CDKB1;1 expression could be a downstream consequence of FAMA's role in guard cell fate specification. Direct control of CDKB1;1 expression could also be mediated by a heteromeric complex between FAMA and a transcriptional repressor partner.
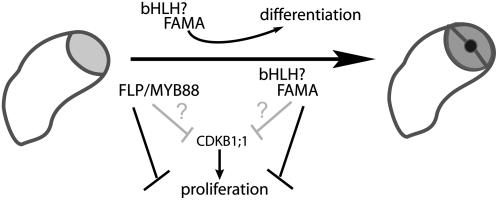
Figure 7.
Proposed Model for Transcription Factor Control of Stomatal Guard Cell Differentiation.
Genes that promote guard cell identify and structure must be upregulated while cell division promoting processes are coordinately downregulated. The transcription factors FAMA, FLP, and MYB88 are all required for the production of the normal two-celled stoma, but the processes they regulate may be different. FLP/MYB88 may work primarily to stop excessive cell divisions. FAMA may activate differentiation genes and either secondarily feedback on the cell cycle or require a partner to directly influence cell cycle gene transcription.
Mechanism of FAMA Action
FAMA encodes a bHLH-domain protein that resides in the nucleus and has transcriptional activation activity in a heterologous system. More than 160 different bHLH-containing proteins are predicted in the Arabidopsis genome, but only a small minority of Arabidopsis bHLH proteins have been characterized in terms of DNA binding, protein complex formation, or biological function (Heim et al., 2003; Toledo-Ortiz et al., 2003). No functional characterization has been reported for any other member of FAMA's subclass. The FAMA clade proteins do not contain the canonical R2R3-MYB interacting domains. We found that FAMA is capable of binding related bHLH proteins but not an R2R3-type MYB (Figures 4H and 4I). The DNA binding capabilities of plant bHLHs vary. Some bHLHs appear to require interaction with other proteins for interaction with DNA (Zimmermann et al., 2004), whereas others, notably the phytochrome interacting factor proteins involved in phytochrome-mediated light responses, have been shown to bind DNA directly (Toledo-Ortiz et al., 2003).
Deletion or alteration of DNA binding sites in animal bHLHs can produce a dominant negative phenotype (Davis et al., 1990). However, these alterations to FAMA produce a neomorphic excess complete stomata phenotype (Figures 5E and 5F), suggesting that the protein either (1) retains partial FAMA function that promotes differentiation without repressing division, (2) acts as a dominant negative with a negative regulator of stomatal differentiation, or (3) mimics the activity of a positive regulator of stomatal development.
It is unlikely that the ΔN-FAMA and FAMAPGG variants possess partial activity because they cannot rescue the differentiation (or the cell division) defects of the fama-1 mutants (Figure 5G). Expression of FAMAPGG or ΔN-FAMA with the FAMA promoter in GMCs does not produce the neomorphic phenotype (see Supplemental Figure 5 online), suggesting that these FAMA variants do not interfere with FAMA itself and probably act at earlier stages in stomatal development.
Because the neomorphic FAMA variants retain the same heterodimerization properties as wild-type FAMA protein in BiFC assays (Figure 6C), it is conceivable that they behave like the Drosophila HLH (non-DNA binding) extramacrochaete protein (Younger-Shepherd et al., 1992) or mammalian Im-f (Chen et al., 1996). These proteins negatively regulate bHLH function by forming nonfunctional heterodimers (Younger-Shepherd et al., 1992). Possible targets of a trans-acting FAMA dominant negative are the broadly expressed bHLH071 and bHLH093 proteins that were identified as proteins that could interact with FAMA in a yeast two-hybrid screen and in planta with BiFC. Overexpression of either gene produces a mild fama-2–like phenotype (Figures 6G and 6H), suggesting that these genes normally promote cell divisions of the GMCs, a phenotype opposite of FAMA overexpression. FAMA may act antagonistically toward these proteins in normal development. However, 35S- or estrogen-driven expression of bHLH071 or bHLH093 does not produce the same phenotype as PROEST:FAMAPGG plants, indicating that interference with bHLH071 and bHLH093 is not sufficient to explain the overproliferation of complete stomata.
The third possibility is that overexpression of FAMAPGG produces a gain-of-function phenotype that resembles the gain of function of another transcriptional regulator. A variation on this theme is that FAMAPGG could serve as a neutral partner of a poorly expressed positive regulator of stomatal development. If the other regulator provides a DNA binding domain, the FAMAPGG protein could simply provide stability or an activation domain and therefore increase the effective dosage of the second protein. This last idea is supported by the dose sensitivity of FAMA itself. The same PROFAMA:GFP-FAMA line that rescues fama-1 can produce a mild gain-of-function phenotype in plants homozygous for the wild-type allele of FAMA.
Comparisons between Stomata and Animal Cell Fate Regulation
The final stages in stomata formation resemble animal neuronal and muscle development both in the requirement for a switch from mitotic competence to terminal differentiation and in the molecules employed to control that switch.
Both myogenesis and neurogenesis feature families of tissue-specific bHLH transcription factors that have been called master regulators of cell fates by virtue of their gain-of-function phenotypes (Weintraub et al., 1991; Jan and Jan, 1993; Weintraub, 1993). During normal development, however, these bHLHs have very restricted expression domains and share the role of regulating cell fates among themselves and in combination with complex cell–cell signaling networks. The regulation of downstream targets requires heterodimerization of these bHLHs with a group of ubiquitously expressed bHLHs (Jan and Jan, 1993). The loss- and gain-of-function FAMA phenotypes and tissue-restricted expression pattern firmly place FAMA among the master regulator class. We have also demonstrated that FAMA has the capacity to form heterodimers with broadly expressed plant bHLHs.
It is a general feature of multicellular development that proliferation and differentiation are mutually exclusive. Stomatal development is a simple system in which to study the transition between these phases. The expansion of the bHLH family of transcription factors was independent in the plant and animal lineages. In addition to its importance in creating an essential plant cell type, FAMA is useful as a tool to identify conserved mechanisms by which bHLH proteins coordinate major developmental events.
METHODS
Plant Materials and Growth Conditions
Markers, mutants, and previously published transgenic lines are as follows. Markers of the stomatal lineage were FLP:GUS/GFP (Lai et al., 2005), TMM:GUS/GFP (Nadeau and Sack, 2002b), CYCB1;2:GUS (Donnelly et al., 1999), CDKB1;1:GUS (Boudolf et al., 2004), and KAT1:GUS (Nakamura et al., 1995). Enhancer trap line E1728 Poethig lab (http://enhancertraps.bio.upenn.edu) was used as a stomatal marker by Bergmann et al. (2004). GUS staining and GFP observations were performed using standard protocols and visualized on a Leica DM5000 microscope or a Bio-Rad 1024 confocal microscope. Mutants were as follows: flp-1 and tmm-1 (Yang and Sack, 1995), sdd-1 (Berger and Altmann, 2000), yodaY295 and yda1 (Bergmann et al., 2004), and er;erl1/+;erl2 (Shpak et al., 2005). T-DNA insertion alleles for FAMA, MYB88, bHLH093, and bHLH071 were obtained from the ABRC stock center. Col-0 was used as the wild type in all studies unless otherwise noted. T-DNA inserts were confirmed using primers designed by iSECT tools (http://salk.signal.edu). Detection of FAMA transcripts was done by RT-PCR (Invitrogen) with FAMA-specific primers F (5′-GCTCGAGCAACTCCTACAATG-3′) and R (5′-GGAACTTGCTATGTCTTCTGC-3′). Amplification of actin cDNA was done using primers F (5′-GGCGATGAAGCTCAATCCAAACG-3′) and R (5′-GGTCACGACCAGCAAGATCAAGACG-3′). Plants were grown initially on half-strength Murashige and Skoog (MS) agar plates in a Percival incubator with 24 h light for 7 d and then transferred to soil in a 22°C growth chamber with 16-h-light/8-h-dark cycles.
Double mutants between viable mutants (flp-1, tmm-1, and sdd-1) and fama-1 were constructed by crossing and planting the F1 and then identifying F2s by viable mutant phenotype. Individual F2 lines were then tested for segregation of fama-1. Double mutants between fama and inviable mutants (yoda and er;erl1;erl2) or T-DNA insert lines (myb88) were obtained in the F2 progeny and confirmed by PCR-based genotyping using previously described primers (Lukowitz et al., 2004; Shpak et al., 2005).
Observations of guard cell development in FAMA overexpression lines were as follows. Overlapping bright-field and fluorescent (GFP) photographs were made of the entire adaxial surface of second leaves of 9-d-old PROFAMA:GFP-FAMA plants (four plants, ∼20 to 30 GFP positive cells/plant) using a ×40 objective on a Leica DM5000 microscope. For imaging, whole MS agar–grown PROFAMA:GFP-FAMA seedlings were mounted in water and returned to MS agar immediately after imaging to recover. Similar photographs were taken at ∼12-h intervals over the next 2 d, at which stage many of the cells had guard cell morphology. Cells were traced back over time through the images by matching patterns of neighbor cells. DNA content of terminal stage cells was determined by 4′,6-diamidino-2-phenylindole intensity using methods described by Boudolf et al. (2004). Scanning electron microscopy images were taken of fresh (unfixed and unstained) 5- to 7-DAG seedlings mounted in cryo-gel (Ted Pella catalog No. 27221) in environmental scanning mode (60 Pa) on a FEI Quanta 200 machine.
Plant Constructs
Plant binary vectors based on Gateway cloning technology (Invitrogen) were used for most manipulations. These include pMDC7, pMDC32, pMDC99, pMDC107 (Curtis and Grossniklaus, 2003), pBGYN, and pGEAR (Kubo et al., 2005).
Rescuing and Reporter Constructs
FAMA cDNAs containing the entire coding region (± the stop codon) were generated by RT-PCR from Col RNA and cloned into pENTR/D-TOPO according to the manufacturer's protocols. A NotI site within the attB sites was used to insert 5′ regulatory sequences when appropriate. A FAMA rescuing construct was created by PCR amplifying 2.5-kb of genomic sequence upstream of FAMA (Col-0 template DNA). This FAMAproFAMA construct was recombined into pMDC99 for rescue and pMDC107 to create a translational fusion reporter line. A transcriptional reporter line (PROFAMA:FAMA-GFP) was created by recombining the 2.5-kb FAMA promoter into pMDC107. A second translational fusion (PROFAMA:GFP-FAMA) was created by digesting pMDC43-FAMA with PstI and KpnI to remove the 35S and replacing with it with a PCR-amplified FAMA 2.5-kb promoter with 5′ PstI and 3′ KpnI ends. PROFAMA:GFP-FAMA signal was brighter than PROFAMA:FAMA-GFP, but both rescued fama-1 and showed equivalent cell type specificity in expression. Rescue of fama-1 was confirmed by PCR genotyping using primers FMARESF (5′-TTTGAACGTAGGAGCCAGGCA-3′) and FMARESR (5′-GCAAATCATACAAGGTCAGTCCC-3′) that distinguish the genomic copy of FAMA (1136 bp) from the transgene (1020 bp).
Overexpression Constructs
Estrogen-inducible and cauliflower mosaic virus 35S transgenes were made by PCR amplifying cDNAs of FAMA, bHLH093, bHLH071, At2g27320 (distant bHLH used as specificity control), FLP, and domains of FAMA (N, 1 to 190 amino acids; N+bHLH, 1 to 250 amino acids; HLH, 208 to 250 amino acids; HLH+C, 208 to 414 amino acids; and C, 250 to 414 amino acids), cloning into pENTR-D-TOPO, and recombining into pMDC7 and pMDC32. Transgene silencing was prevalent in the 35S lines. Transformation of 35S:FAMA into a silencing defective rdr6 mutant background (Peragine et al., 2004) alleviates the silencing and creates an epidermal phenotype similar to the induced PROEST:FAMA lines. Because of the additional complicating phenotypes of the rdr6 mutants, estrogen-inducible lines were used for all gain-of-function analysis.
Dominant Negative Constructs
Estrogen-inducible dominant repression FAMA was created by recombining FAMA (no stop) into pEAR, PCR amplifying the resultant fusion protein, recloning into pENTR/D/TOPO, and recombining into pMDC7. The EAR domain is a transacting repression domain first identified in ERF and SUPERMAN transcription factors of Arabidopsis thaliana (Hiratsu et al., 2003). The original DNA binding site sequence HIAVERNR (5′-CAT-ATCGCGGTCGAAAGAAACCGT-3′) was converted to PIAVGRNG (5′-CCTATCGCGGTCGGAAGAAACGGT-3′) using the QuikChange II-E site-directed mutagenesis kit (Stratagene). Plants were stably transformed using Agrobacterium tumefaciens–mediated transformation (strain GV3101) with standard protocols. Transgenic lines were selected on half-strength MS medium containing 25 to 50 mg/L hygromycin. At least 10 T1 transgenic lines were analyzed for each construct.
Two-Hybrid Screen with FAMA bHLH Domain
An N-terminally deleted FAMA clone (190 to 414 amino acids) was PCR amplified and cloned into bait vector pGBK (Clontech). Yeast strain AH109 transformed with pGBK-FAMA was retransformed with a prey library made from 3-d-old seedlings in pACT (ABRC stock CD4-22). bHLH071-ACT and bHLH093-ACT complemented the his and ade auxotrophies and expressed of β-galactosidase when isolated and retransformed into yeast containing pGBK-FAMA.
BiFC
For BiFC studies, full-length FAMA, bHLH071, bHLH093, and FLP pENTR clones were recombined into four vectors that fused each half of GFP to either the N or C terminus of the test protein (Walter et al., 2004). Nicotiana benthamiana leaves were transformed by injection of Agrobacterium GV3101/pMP90 cells harboring the appropriate plasmids as previously described (Lavy et al., 2002). GFP expression was examined 24 h after Agrobacterium injection on a Leica DR5000 microscope, and digital images were captured on a Retiga Exi CCD camera. Two 14-3-3 proteins were used as a positive control. Each interaction was tested at least three times, and images of each pairwise interaction (FAMA, bHLH071, bHLH093, FLP, and controls) from a single round of experiments were taken with a common exposure time. Images were false-colored in Adobe Photoshop.
Transcriptional Activity in Protoplasts
Tobacco BY-2 protoplasts were prepared from suspension culture 4 d after passaging. Cell walls were digested in a solution containing 1% cellulase Onozuka R-10, 0.1% pectinase (Sigma-Aldrich), 0.5% Macerozymes RS, and 0.25 M mannnitol at room temperature for 3 h. The isolated protoplasts were transformed with 7.5 μg each of reporter and effector construct using a standard polyethylene glycol method. The reporter constructs (Gal-GUS and Lex-Gal-GUS) and effector constructs (VP16 and IAA) were described previously by Tiwari et al. (2004). The FAMA effector plasmid was made from a plant expression vector containing full-length FAMA fused to the yeast GAL4 DNA binding domain. Cotransfection of the reporter, the test protein GAL4BD, and a constitutive transcriptional activator, VP16 fused to a LexA-DNA binding domain, allows the test protein to be assayed for transcriptional repression (Tiwari et al., 2004; He et al., 2005). 35SLUC was cotransformed as an internal control to normalize the GUS reporter expression. GUS expression was measured at t = 0 h and t = 24 h. Relative transcriptional activation activity was calculated as [(t24 − t 0) − (t24back − t0back)]/LUC. Data from four to five independent samples for each construct were pooled.
Accession Numbers
The Arabidopsis Genome Initiative locus identifiers for genes mentioned in this article are as follows: FAMA (At3g24140), FLP (At1g14350), MYB88 (At2g02820), CDBK1;1 (At3g54180), bHLH071 (At5g46690), and bHLH093 (At5g65640). The GenBank accession number for FAMA cDNA is NM_113319 and that for the fama-1 insertion allele site is BH865915.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. DIC Images of fama Loss- and Gain-of-Function Phenotypes in Aerial Organs.
Supplemental Figure 2. Full Expression Pattern of FAMA and E1728.
Supplemental Figure 3. Genetic Interactions between fama and Mutations in Other Stomatal Regulators.
Supplemental Figure 4. Alignment of bHLHs in FAMA Clade.
Supplemental Figure 5. Phenotypes of Tissue-Specific Expression of FAMA Protein Variants.
Supplementary Material
Acknowledgments
We thank members of our laboratory for discussions and comments on the manuscript, Fred Sack and Keiko Torii for our ongoing conversations about stomatal development, and Z. Wang, J. Gendron, and S. Gampala (Carnegie Department of Plant Biology) for help with transcriptional activation assays. Research in the lab is supported by the U.S. National Science Foundation (NSF-IOB-0544895). The authors declare that they have no competing financial interests.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Dominique C. Bergmann (dbergmann@stanford.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Berger, D., and Altmann, T. (2000). A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 14 1119–1131. [PMC free article] [PubMed] [Google Scholar]
- Bergmann, D.C., Lukowitz, W., and Somerville, C.R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304 1494–1497. [DOI] [PubMed] [Google Scholar]
- Bernhardt, C., Zhao, M., Gonzalez, A., Lloyd, A., and Schiefelbein, J. (2005). The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132 291–298. [DOI] [PubMed] [Google Scholar]
- Boudolf, V., Barroco, R., de Almeida Engler, J., A., Verkest, A., Beeckman, T., Naudts, M., Inze, D., and De Veylder, L. (2004). B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana. Plant Cell 16 945–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.M., Kraut, N., Groudine, M., and Weintraub, H. (1996). I-mf, a novel myogenic repressor, interacts with members of the MyoD family. Cell 86 731–741. [DOI] [PubMed] [Google Scholar]
- Curtis, M.D., and Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, R.L., Cheng, P.F., Lassar, A.B., and Weintraub, H. (1990). The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell 60 733–746. [DOI] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T., de Almeida Engler, J., Ormenese, S., Maes, S., Naudts, M., Van Der Schueren, E., Jacqmard, A., Engler, G., and Inze, D. (2002). Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J. 21 1360–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte, W., Riou-Khamlichi, C., Scofield, S., Healy, J.M., Jacqmard, A., Kilby, N.J., and Murray, J.A. (2003). Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly, P.M., Bonetta, D., Tsukaya, H., Dengler, R.E., and Dengler, N.G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215 407–419. [DOI] [PubMed] [Google Scholar]
- Grandori, C., Cowley, S.M., James, L.P., and Eisenman, R.N. (2000). The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16 653–699. [DOI] [PubMed] [Google Scholar]
- He, J.X., Gendron, J.M., Sun, Y., Gampala, S.S., Gendron, N., Sun, C.Q., and Wang, Z.Y. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307 1634–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim, M.A., Jakoby, M., Werber, M., Martin, C., Weisshaar, B., and Bailey, P.C. (2003). The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20 735–747. [DOI] [PubMed] [Google Scholar]
- Hiratsu, K., Matsui, K., Koyama, T., and Ohme-Takagi, M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34 733–739. [DOI] [PubMed] [Google Scholar]
- Jan, Y.N., and Jan, L.Y. (1993). HLH proteins, fly neurogenesis, and vertebrate myogenesis. Cell 75 827–830. [DOI] [PubMed] [Google Scholar]
- Kubo, M., Udagawa, M., Nishikubo, N., Horiguchi, G., Yamaguchi, M., Ito, J., Mimura, T., Fukuda, H., and Demura, T. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, L.B., Nadeau, J.A., Lucas, J., Lee, E.K., Nakagawa, T., Zhao, L., Geisler, M., and Sack, F.D. (2005). The Arabidopsis R2R3 MYB proteins FOUR LIPS and MYB88 restrict divisions late in the stomatal cell lineage. Plant Cell 17 2754–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy, M., Bracha-Drori, K., Sternberg, H., and Yalovsky, S. (2002). A cell-specific, prenylation-independent mechanism regulates targeting of type II RACs. Plant Cell 14 2431–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M.M., and Schiefelbein, J. (1999). WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99 473–483. [DOI] [PubMed] [Google Scholar]
- Lukowitz, W., Roeder, A., Parmenter, D., and Somerville, C. (2004). A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116 109–119. [DOI] [PubMed] [Google Scholar]
- Massari, M.E., and Murre, C. (2000). Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau, J. A. and Sack, F. D. (2002. a). Stomatal development in Arabidopsis. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.0066, http://www.aspb.org/publications/arabidopsis/. [DOI] [PMC free article] [PubMed]
- Nadeau, J.A., and Sack, F.D. (2002. b). Control of stomatal distribution on the Arabidopsis leaf surface. Science 296 1697–1700. [DOI] [PubMed] [Google Scholar]
- Nakamura, R.L., McKendree, W.L., Jr., Hirsch, R.E., Sedbrook, J.C., Gaber, R.F., and Sussman, M.R. (1995). Expression of an Arabidopsis potassium channel gene in guard cells. Plant Physiol. 109 371–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H.L., and Poethig, R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak, E.D., McAbee, J.M., Pillitteri, L.J., and Torii, K.U. (2005). Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309 290–293. [DOI] [PubMed] [Google Scholar]
- Tiwari, S.B., Hagen, G., and Guilfoyle, T.J. (2004). Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz, G., Huq, E., and Quail, P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15 1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, M., Chaban, C., Schutze, K., Batistic, O., Weckermann, K., Nake, C., Blazevic, D., Grefen, C., Schumacher, K., Oecking, C., Harter, K., and Kudla, J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40 428–438. [DOI] [PubMed] [Google Scholar]
- Weintraub, H. (1993). The MyoD family and myogenesis: Redundancy, networks, and thresholds. Cell 75 1241–1244. [DOI] [PubMed] [Google Scholar]
- Weintraub, H., Davis, R., Tapscott, S., Thayer, M., Krause, M., Benezra, R., Blackwell, T.K., Turner, D., Rupp, R., and Hollenberg, S. (1991). The myoD gene family: Nodal point during specification of the muscle cell lineage. Science 251 761–766. [DOI] [PubMed] [Google Scholar]
- Yang, M., and Sack, F.D. (1995). The too many mouths and four lips mutations affect stomatal production in Arabidopsis. Plant Cell 7 2227–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younger-Shepherd, S., Vaessin, H., Bier, E., Jan, L.Y., and Jan, Y.N. (1992). Deadpan, an essential pan-neural gene encoding an HLH protein, acts as a denominator in Drosophila sex determination. Cell 70 911–922. [DOI] [PubMed] [Google Scholar]
- Zimmermann, I.M., Heim, M.A., Weisshaar, B., and Uhrig, J.F. (2004). Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40 22–34. [DOI] [PubMed] [Google Scholar]
- Zuo, J., Niu, Q.W., and Chua, N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.