Abstract
Recognition of pathogens by plants involves the coordinated efforts of molecular chaperones, disease resistance (R) proteins, and components of disease resistance signaling pathways. Characterization of events associated with pathogen perception in Arabidopsis thaliana has advanced understanding of molecular genetic mechanisms associated with disease resistance and protein interactions critical for the activation of resistance signaling. Regulation of R protein–mediated signaling in response to the bacterial pathogen Pseudomonas syringae in Arabidopsis involves the physical association of at least two R proteins with the negative regulator RPM1 INTERACTING PROTEIN4 (RIN4). While the RIN4-RPS2 (for RESISTANCE TO P. SYRINGAE2) and RIN4-RPM1 (for RESISTANCE TO P. SYRINGAE PV MACULICOLA1) signaling pathways exhibit differential mechanisms of activation in terms of effector action, the requirement for NON-RACE-SPECIFIC DISEASE RESISTANCE1 (NDR1) is shared. Using a yeast two-hybrid screen, followed by a series of coimmunoprecipitation experiments, we demonstrate that the RIN4–NDR1 interaction occurs on the cytoplasmically localized N-terminal portion of NDR1 and that this interaction is required for the activation of resistance signaling following infection by P. syringae expressing the Cys protease Type III effector protein AvrRpt2. We demonstrate that like RPS2 and RPM1, NDR1 also associates with RIN4 in planta. We suggest that this interaction serves to further regulate activation of disease resistance signaling following recognition of P. syringae DC3000-AvrRpt2 by Arabidopsis.
INTRODUCTION
The activation of disease resistance signaling in plants is regulated by multiple disease resistance components, including the association of disease resistance (R) proteins with negative regulators (Mackey et al., 2002, 2003; Axtell and Staskawicz, 2003; Day et al., 2005), molecular chaperones (reviewed in Hubert et al., 2003; Shirasu and Schulze-Lefert, 2003; Takahashi et al., 2003; Coaker et al., 2005), and proteins that contribute to the subsequent activation of signaling cascades required for the initiation of defense responses (reviewed in Dangl and Jones, 2001; Belkhadir et al., 2004a).
The identification of RPM1 INTERACTING PROTEIN4 (RIN4) as a molecular switch controlling R protein activation greatly enhanced our understanding of the genetic and cellular events associated with pathogen perception and disease resistance activation as well as our understanding of the protein dynamics required for disease signaling. While the exact mechanisms associated with the perception of plant pathogens are poorly understood, our understanding of the downstream events required for resistance signaling is mounting. It is widely accepted that the molecular-genetic basis for R protein–mediated bacterial disease resistance in plants involves the direct or indirect recognition of pathogen-derived virulence effectors, resulting in the induction of plant disease resistance (reviewed in Van der Biezen and Jones, 1998; Chisholm et al., 2006). Following the delivery of bacterial Type III effector proteins into the plant cytosol via the Type III secretion system, recognition by the host plant results in the activation of defense signaling leading to resistance. Gene-for-gene resistance occurs when effector proteins are recognized by the host plant, thus initiating disease resistance responses. Subsequent signaling culminates in the abrogation of bacterial growth mediated by R protein signaling pathways. In the absence of R protein–mediated recognition of a Type III secretion system–delivered effector protein(s), host susceptibility prevails and pathogen growth increases, resulting in disease, and, ultimately, cell death (reviewed in Dangl and Jones, 2001).
The question remains how various protein components of these signaling pathways are assembled, activated, and subsequently regulated in response to the recognition of invading pathogens (reviewed in Dangl and Jones, 2001; Van der Hoorn et al., 2002; Chisholm et al., 2006). R protein complex assembly and activation is emerging as a model for defining the underlying mechanisms for the molecular basis of plant disease resistance (reviewed in Shirasu and Schulze-Lefert, 2003).
Experimental evidence in support of the indirect recognition model for effector–R protein association first came from the work of Mackey et al. (2002), which identified a component of the RESISTANCE TO P. SYRINGAE PV MACULICOLA1 (RPM1)-mediated disease resistance pathway. This protein, RIN4, was shown to be phosphorylated in the presence of AvrB or AvrRpm1, which in turn leads to the activation of the RPM1-mediated resistance. In the absence of Pseudomonas syringae expressing either AvrB or AvrRpm1, RIN4 functions as a negative regulator of RPM1 function, keeping it in an inactive state likely via its association with the resistance protein. Two independent studies further demonstrated that RIN4 is required for regulation and activation of a second nucleotide binding leucine-rich repeat (NB-LRR) protein, RESISTANCE TO P. SYRINGAE2 (RPS2), which confers resistance to P. syringae expressing AvrRpt2 (Axtell and Staskawicz, 2003; Mackey et al., 2003). As in the case of RPM1, RIN4 also functions as a negative regulator of RPS2 activation. The RIN4-RPS2 association appears to function differently from the RIN4–RPM1 interaction. Rather than phosphorylation of RIN4 leading to activation, as is the case with RPM1, RPS2 activity requires the AvrRpt2-mediated proteolysis of RIN4 (Coaker et al., 2005). This suggests that a physical association between RPS2 and RIN4, whether direct or indirect, serves to hold RPS2 in an inactive state. Indeed, evidence in support of this hypothesis was obtained by demonstrating that the physical association of RIN4 with RPS2 is required for the negative regulation of RPS2-mediated signaling and that this association requires the C-terminal, plasma membrane–associated domain of RIN4 (Day et al., 2005; Kim et al., 2005a). Additional studies characterizing the mechanisms associated with the elimination of RIN4 further defined not only the physical and structural requirements for RIN4 elimination but also the mechanisms required for effector activation and function (Chisholm et al., 2005; Coaker et al., 2005; Kim et al., 2005a). Taken together with the results of Mackey et al. (2002, 2003), RIN4 appears to play the role of a broad spectrum molecular switch regulating at least two independent R protein–mediated defense pathways in Arabidopsis thaliana. Interestingly, Belkhadir et al. (2004b) suggested that the activation of RPS2 is NON-RACE-SPECIFIC DISEASE RESISTANCE1 (NDR1) independent, in contrast with the established requirement for NDR1 during AvrRpt2-dependent RPS2 activation. In this study, the authors hypothesized that RIN4 may function cooperatively with NDR1 to negatively regulate RPS2 in the absence of pathogen.
In this study, we report the identification of another protein association required for RIN4-mediated disease resistance signaling in Arabidopsis: the RIN4–NDR1 interaction. NDR1 was first identified in a genetic screen aimed at identifying genetic loci required for disease resistance signaling in Arabidopsis in response to infection by P. syringae (Century et al., 1995, 1997). NDR1 is a plasma membrane, glycophosphatidyl-inositol (GPI)-anchored protein required for the activation of disease resistance signaling mediated by members of the largest class of disease resistance proteins in Arabidopsis (Coppinger et al., 2004). Previous work addressed the genetic requirement for NDR1 in the activation of resistance signaling mediated by the coiled-coil (CC) NB-LRR class of resistance proteins; yet to date, the mechanism of NDR1 function in disease resistance signaling remains elusive (Century et al., 1995, 1997).
The proposed topology of NDR1 within the plasma membrane suggests that an approximate 18–amino acid portion lies within the cytoplasm, while the remainder of the NDR1 protein resides on the outside surface (i.e., apoplast) of the plasma membrane (Coppinger et al., 2004). We set out to determine the domain architecture required for NDR1–RIN4 interaction and, moreover, to determine how this protein–protein interaction contributes to disease resistance signaling following P. syringae perception. We suggest that the NDR1–RIN4 interaction may function as an additional layer of regulation, modulating the activation of RPS2.
RESULTS
NDR1 and RIN4 Interact in a Yeast Two-Hybrid Screen
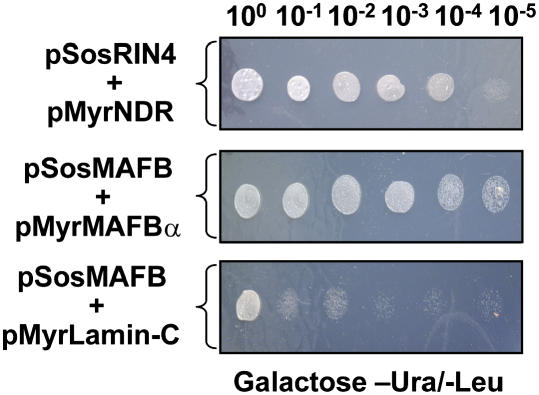
RIN4 was identified in a yeast two-hybrid screen using the bacterial effector protein AvrB as bait and subsequently shown to interact with RPM1 (Mackey et al., 2002). Given the involvement of RIN4 in disease resistance signaling in Arabidopsis, we sought to identify RIN4-interacting proteins through screening a CytoTrap Arabidopsis cDNA library, with the aim of uncovering additional proteins required for disease resistance. Using RIN4 as a C-terminal bait fusion protein (i.e., pSos-RIN4), we screened 106 yeast colonies comprising an Arabidopsis cDNA library generated from various combinations of pathogen and mock-inoculated Arabidopsis genotypes. As shown in Figure 1, we identified a specific interaction between RIN4 and NDR1. Isolation of rescued cDNAs and recapitulation of the interaction was confirmed by reconstructing the NDR1 (i.e., prey) target plasmid (top panel). Serial dilutions of yeast clones expressing the pSos-RIN4 bait construct and pMyrNDR1 prey construct revealed a specific interaction when compared with MAFB/α positive and MAFB/Lamin-C negative controls (Figure 1, middle and bottom panels, respectively).
Figure 1.
Identification of the NDR1 and RIN4 Interaction by a Yeast Two-Hybrid Screen.
A specific interaction between NDR1 and RIN4 was identified using the CytoTrap two-hybrid system. Approximately 106 S. cerevisiae cdc25H cells comprising an Arabidopsis cDNA library made from pathogen and non-pathogen-treated tissues were screened using RIN4 (pSosRIN4) as a bait. Recapitulation of the interaction was performed on galactose-containing minimal media in the absence of uracil and leucine at 37°C (top panel). Control interactions, consisting of pSosMAFB+pMyrMAFBα (center panel, positive control) and pSOSMAFB+pMyrLamin-C (bottom panel, negative control) confirmed the NDR1–RIN4 interaction as specific.
NDR1 and RIN4 Interact in Planta
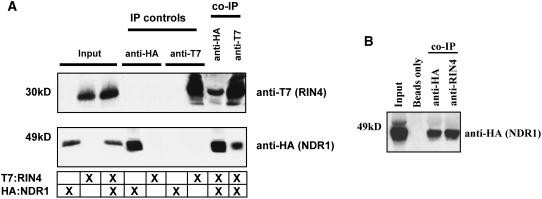
While successful in identifying a specific interaction between two well-characterized components of resistance signaling in Arabidopsis by a yeast two-hybrid screen, the significance of this association requires in planta confirmation. To do this, we chose to further investigate the NDR1–RIN4 interaction by the expression of epitope-tagged NDR1 and RIN4 constructs using Agrobacterium tumefaciens–mediated transient gene expression in Nicotiana benthamiana. Previous studies have demonstrated the use of this heterologous expression system in its applicability toward characterizing a number of disease-associated proteins in a variety of plant-pathogen systems (Jin et al., 2002; Moffett et al., 2002; Escobar et al., 2003; He et al., 2004; Zhang et al., 2004; Day et al., 2005). As shown in Figure 2A, following transient coexpression of both genes for 40 h in N. benthamiana leaves by Agrobacterium infection, we observed that both HA:NDR1 and T7:RIN4 can be shown to associate in a series of coimmunoprecipitation experiments. Moreover, using HA:NDR1 epitope–tagged complemented ndr1-1 mutant Arabidopsis plants (Coppinger et al., 2004), we were successful in demonstrating a specific interaction between RIN4 and NDR1 (Figure 2B). The in planta coimmunoprecipitation experiment validates our initial yeast two-hybrid results demonstrating that NDR1 and RIN4 physically associate in planta.
Figure 2.
RIN4 and NDR1 Interact in Planta.
Coimmunoprecipitation of RIN4 and NDR1 in N. benthamiana and Arabidopsis.
(A) Transient coexpression in wild-type N. benthamiana plants. Immunoblot of anti-HA and anti-T7 immunoprecipitated proteins isolated 40 h after inoculation from wild-type N. benthamiana leaves hand-infiltrated with Agrobacterium strains expressing HA:NDR1 and T7:RIN4. Total protein extracts were immunoprecipitated with anti-HA (NDR1) and anti-T7 (RIN4) antibodies. Immunoprecipitated proteins were detected by immunoblotting anti-T7 (top panel) and anti-HA (bottom panel) immunoblots. Protein sizes are indicated at the left side of the immunoblots.
(B) Expression and coimmunoprecipitation of HA:NDR1 and native RIN4 in Arabidopsis. Total protein extracts were immunoprecipitated with either anti-HA (positive control) or anti-RIN4 antibodies, followed by protein gel blot analysis with anti-HA. “Beads only” indicates a negative control whereby total extracts were incubated at 4°C in the absence of antibody.
The NDR1–RIN4 Interaction Occurs within the C-Terminal Half of RIN4
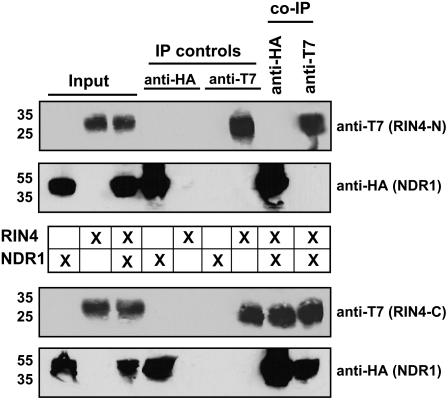
Previous work identified the C terminus of RIN4 as being required for both regulation of and association with RPS2 (Day et al., 2005). To further characterize the NDR1–RIN4 interaction, we used the RIN4 deletion constructs described in our previous study to identify regions of RIN4 that are required for its interaction with NDR1. Using in planta coimmunoprecipitation experiments as an assay to monitor the NDR1–RIN4 interaction, we determined that only the C terminus of RIN4 was required for its association with NDR1, as was observed in the case of the RIN4–RPS2 interaction (Day et al., 2005). As shown in Figure 3, transient expression and coimmunoprecipitation of epitope-tagged NDR1 and RIN4 deletion constructs (i.e., RIN4-N and RIN4-C) in N. benthamiana by transient Agrobacterium infection revealed a reciprocal coimmunoprecipitation with HA and T7 antibodies specific to NDR1 and RIN4-C, respectively. Coinfiltration and expression of HA:NDR1 and T7:RIN4-N by Agrobacterium in N. benthamiana did not result in the reciprocal coimmunoprecipitation of either protein (Figure 3, top two panels).
Figure 3.
The C Terminus of RIN4 Is Required for NDR1 Interaction.
Coimmunoprecipitation of HA:NDR1 and the N-terminal half (i.e., T7:RIN4-N) and C-terminal half (i.e., T7:RIN4-C) of RIN4. Immunoblot of anti-HA and anti-T7 immunoprecipitated proteins isolated 40 h after inoculation from wild-type N. benthamiana leaves hand-infiltrated with Agrobacterium strains expressing HA:NDR1 and the N- and C-terminal halves (separately) of RIN4. Total protein extracts were immunoprecipitated with anti-HA (NDR1) and anti-T7 (RIN4) antibodies. Immunoprecipitated proteins were detected by immunoblotting with anti-HA (NDR1) and anti-T7 (RIN4) antibodies. The top two panels represent the absence of HA:NDR1-T7:RIN-N coimmunoprecipitation. The bottom two panels represent the HA:NDR1-T7:RIN4-C coimmunoprecipitation.
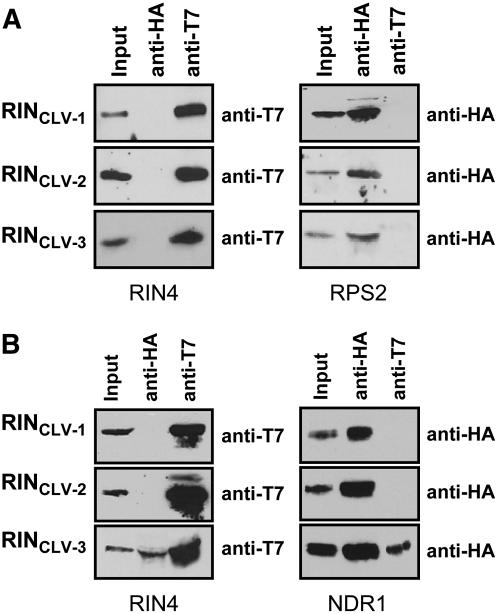
NDR1 Associates with the C-Terminal AvrRpt2-Generated RIN4 Cleavage Product
Previous work in our laboratory showed that the terminal nine amino acid residues in RIN4 are required for its association with RPS2 (Day et al., 2005). Additional work in other laboratories has since revealed that the requirement for these residues extends beyond the physical coordination of the protein–protein interaction and likely involves posttranslational modifications that function to target RIN4 to the plasma membrane (Jones and Takemoto, 2004; Kim et al., 2005a). As an extension of these experiments, we have also demonstrated that epitope-tagged peptides resembling the RIN4 cleavage products generated by exposure to AvrRpt2 did not inhibit the RPS2 hypersensitive response (Day et al., 2005). To determine if the mechanism of RIN4–NDR1 association/disassociation is similar to that of RPS2, we investigated whether any of the RIN4 cleavage products were capable of associating with NDR1 in planta. To test this model, we transiently coexpressed, individually, the three T7-epitope tagged proteins resembling the RIN4 cleavage products (i.e., CLV-1, CLV-2, and CLV-3; Figure 4A) together with NDR1 in N. benthamiana by Agrobacterium transient expression and performed coimmunoprecipitation experiments to determine the extent of protein–protein associations. As shown in Figure 4B, none of the RIN4 cleavage products immunoprecipitated RPS2 in planta, consistent with our results in the RIN4-RPS2 abrogation experiments (Day et al., 2005). However, as shown in Figure 4B, coexpression of the third RIN4 cleavage product together with NDR1 resulted in a reciprocal pull-down by coimmunoprecipitation. Neither CLV-1 nor CLV-2 was able to immunoprecipitate NDR1 in planta (Figure 4B).
Figure 4.
The C-Terminal RIN4 Cleavage Products Differentially Associate with RPS2 and NDR1.
Coimmunoprecipitation of RPS2:HA and HA:NDR1 with T7:RIN4CLV-1, T7:RIN4CLV-2, and T7:RIN4CLV-3. Immunoblot of anti-HA and anti-T7 immunoprecipitated proteins isolated 24 h and 40 h after inoculation from wild-type N. benthamiana leaves hand-infiltrated with Agrobacterium strains expressing RPS2:HA, HA:NDR1, and RIN4-like cleavage products. Total protein extracts were immunoprecipitated with anti-HA (RPS2 and NDR1) and anti-T7 (RIN4) antibodies. Immunoprecipitated proteins were detected by immunoblotting with anti-T7 (left panels) and anti-HA (right panels) antibodies.
(A) RPS2:HA + T7:RIN4 cleavage products.
(B) HA:NDR1 + T7:RIN4 cleavage products.
The N Terminus of NDR1 Is Required for RIN4 Association
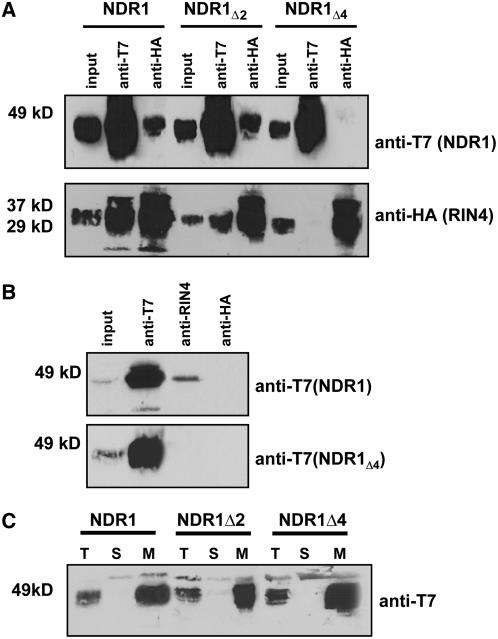
Our working hypothesis is that NDR1 assumes a double anchor membrane conformation: anchored at the N terminus by a transmembrane domain (residues 19 to 34) and at the C terminus by the addition of the GPI anchor (Coppinger et al., 2004). If our working model for NDR1's topology is correct, the 18 N-terminal amino acids of NDR1 are the only residues that lie within the cytoplasm and thus are positioned to interact with the cytoplasmically facing plasma membrane–localized RIN4. To test this hypothesis, we performed a stepwise deletion analysis of NDR1 to monitor for a loss in RIN4 interaction. As shown in Figure 5A, deletion of the first two residues (i.e., Met-Asn; NDR1Δ2) did not affect the ability of NDR1 to interact with RIN4. However, when the first four residues were deleted (i.e., Met-Asn-Asn-Gln; NDR1Δ4) from NDR1, the ability to interact with RIN4 was lost, as monitored by coimmunoprecipitation experiments in N. benthamiana transient expression assays. Additional deletions of Δ8, Δ10, Δ16, and Δ20 amino acids from the N terminus of NDR1 revealed an absence of coimmunoprecipitation with RIN4 (data not shown). Using homologous expression of T7-tagged NDR1 and NDR1Δ4 in Arabidopsis showed a loss of coimmunoprecipitation with RIN4 when the first four amino acids were deleted from the N terminus, again consistent with the data obtained in the heterologous N. benthamiana expression system (Figure 5B). Furthermore, localization of NDR1Δ2 and NDR1Δ4 mirrored wild-type membrane localization, thereby removing the possibility that errant localization was responsible for a loss in protein interaction (Figure 5C).
Figure 5.
The NDR1–RIN4 Interaction Occurs within the First Four Amino Acids of NDR1.
Stepwise deletions of two and four amino acids from the N terminus of NDR1 results in a differential pattern of association with RIN4.
(A) Coimmunoprecipitation and protein gel blot analysis of T7:NDR1 and HA:RIN4 from total protein extracts isolated from transiently expressed N. benthamiana. Loss of the NDR1–RIN4 interaction occurs following deletion of four amino acids from the N terminus of NDR1.
(B) Homologous expression of the NDR1Δ4 deletion mutant in Arabidopsis. Native promoter expression of T7:NDR1 and T7:NDR1Δ4 cDNAs in the Arabidopsis ndr1-1 mutant background confirms results of transient heterologous expression. Native RIN4 antibodies coimmunoprecipitate T7:NDR1 yet fail to coimmunoprecipitate T7:NDR1Δ4.
(C) Ultracentrifugation and localization of NDR1 and NDR1 deletion constructs confirm membrane localization. Total protein extracts from Arabidopsis plants (ndr1-1/HA:NDR1) expressing wild-type and deletion constructs were subjected to ultracentrifugation and separation by SDS-PAGE. Protein gel blot analysis (anti-T7) confirms plasma membrane localization for wild-type, Δ2, and Δ4 T7:NDR1 constructs.
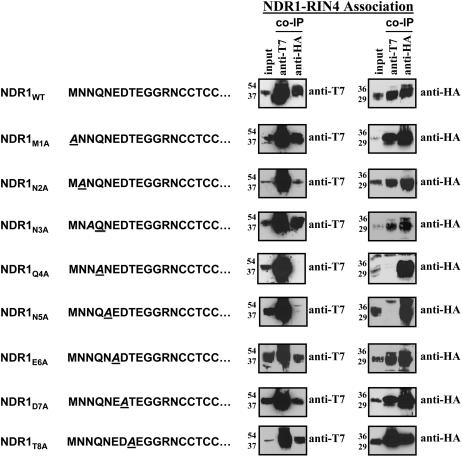
Gln and Asn at Positions 3 and 4 in NDR1 Are Required for Interaction with RIN4
Once we had determined that the first four residues within the N terminus of NDR1 were required for RIN4 association, we undertook a PCR-based Ala-scanning approach to define the precise location of the required residues. As shown in Figure 6, individual changes within the first eight amino acids of NDR1 to Ala revealed a pattern of RIN4 interaction consistent with the results of our deletion analyses described above. Infiltration of N. benthamiana with Agrobacterium expressing T7:NDR1 and HA:RIN4 constructs with the indicated amino acid changes in NDR1 suggests a requirement for the Gln residue at position four (i.e., Q4) and Asn at position five (i.e., N5) for association with RIN4. Individual changes to Ala in residues 1 to 3 and 6 to 8 did not affect coimmunoprecipitation of NDR1 with RIN4. As expected, changes of both Gln and Asn to Ala residues resulted in a loss of RIN4 interactions (data not shown).
Figure 6.
Gln and Asn at Positions 4 and 5 in NDR1 Are Required for RIN4 Association.
Ala scanning of the first eight amino acids of NDR1 by QuickChange PCR mutagenesis. Sequential, independent changes in the native amino acid residues within the N-terminal tail of NDR1 demonstrate that the Gln and Asn residues at positions 4 and 5, respectively, are required for association with RIN4, as determined by coimmunoprecipitation experiments. NDR1 mutant constructs were transiently coexpressed with wild-type RIN4 in N. benthamiana for 40 h and processed as described previously. The sequence changes, where applicable, are indicated to the left of the protein gel blots as italicized and underlined letters. Anti-T7 blots (NDR1) are shown in the left column. Reciprocal, anti-HA blots (RIN4) are shown at the right.
The NDR1–RIN4 Interaction Is Required for Successful Activation of RPS2-Mediated Resistance following Delivery of AvrRpt2
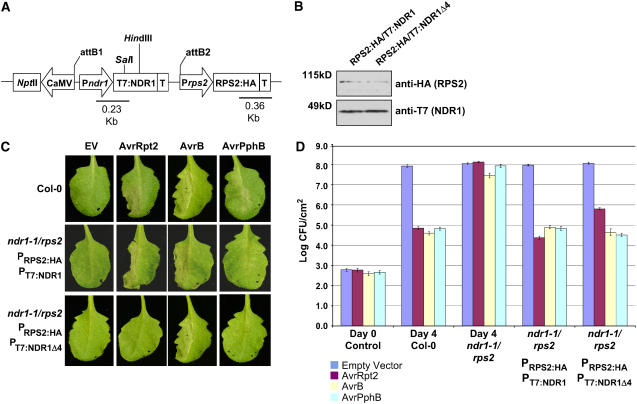
NDR1 has been shown to be required for the activation of resistance mediated by at least three members of the CC-NB-LRR class of resistance proteins in Arabidopsis: RPS2, RPM1, and RPS5. RIN4, however, is required for the activity of only two: RPS2 and RPM1. To investigate the requirement of the NDR1–RIN4 association and to determine if this interaction influences disease resistance meditated by RPS2, we monitored the activation of resistance signaling following P. syringae infection using the complementation strategy outlined in Figure 7. A dual-promoter binary vector system (Figure 7A) was constructed whereby native promoter-driven RPS2:HA and various T7:NDR1 derivatives described above could be transformed into rps2/ndr1-1 plants and tested for their ability to activate resistance to P. syringae expressing AvrRpt2 as well as the cognate bacterial effector proteins of the resistance proteins RPS5 and RPM1. Confirmation of T-DNA integration (Figure 7B), induction of the hypersensitive response following pathogen infection (Figure 7C), and induction of R protein–mediated disease resistance (Figure 7D) collectively validates use of the dual-promoter epitope-tag Arabidopsis system for further studying the RIN4-RPS2-NDR1 protein associations. As shown in Figure 7C, the complemented (i.e., RPS2:HA/T7:NDR1) ndr1-1/rps2 mutant was capable of initiating a hypersensitive response associated with a high-density P. syringae inoculation of and the corresponding recognition of AvrRpt2 (RPS2), AvrB (RPM1), and AvrPphB (RPS5), all three of which require the function of NDR1. Interestingly, in agreement with our coimmunoprecipitation data (Figure 5), the RPS2:HA/T7:NDR1Δ4 construct did not complement the ndr1-1/rps2 mutation, as indicated by the absence of an AvrRpt2-RPS2–induced hypersensitive response.
Figure 7.
Construction of a Homologous Expression System for Characterizing the RPS2–RIN4–NDR1 Interaction.
(A) rps2/ndr1-1 mutant Arabidopsis plants were transformed with a dual promoter construct expressing RPS2:HA and T7:NDR1/T7:NDR1Δ4 expressed under the control of their respective native promoters.
(B) Protein gel blot analysis of RPS2:HA/T7:NDR1 and RPS2:HA/T7:NDR1Δ4 transgenic plants. Total protein extracts were separated by SDS-PAGE and probed with anti-HA antibodies (RPS2) and anti-T7 (NDR1) to determine protein expression levels in complemented Arabidopsis lines.
(C) High-density (105 colony-forming units [cfu]/mL) inoculation of wild-type Col-0 (top panels), RPS2:HA/T7:NDR1 (middle panels), and RPS2:HA/T7:NDR1 (bottom panels) with P. syringae DC3000 expressing empty vector (EV; control), AvrRpt2, AvrB, and AvrPphB effector proteins.
(D) Bacterial growth curve analysis of native promoter expression lines of Arabidopsis RPS2:HA/T7:NDR1- and RPS2:HA/T7:NDR1Δ4-complemented plants. Bacterial counts were determined at days 0 and 4 following inoculation with P. syringae strains (104 cfu/mL) expressing the indicated effector protein. Day 0 controls were plated 1 h after inoculation and represent the average bacterial growth count of all plant genotypes tested. The Arabidopsis ndr1-1/rps2 genotype corresponds to the parent genotype of the complemented expression system shown in (A). Data shown are the mean of four independent lines for both RPS2:HA/T7:NDR1 and RPS2:HA/T7:NDR1Δ4 constructs. Error bars indicate standard deviation calculated from the mean of replicate samples.
To quantify bacterial growth, and thereby assess the activation of resistance following pathogen perception in the wild type and NDR1Δ4 complemented lines described above, wild-type Columbia (Col-0), RPS2:HA/T7:NDR1, and RPS2:HA/T7:NDR1Δ4 transgenic plants were infected with P. syringae expressing the cognate effector proteins of the R proteins requiring NDR1 function (i.e., RPS2, RPM1, and RPS5). As shown in Figure 7D, wild-type Col-0 plants showed a typical pattern of resistance 4 d after inoculation, whereas ndr1-1/rps2 plants were fully susceptible to P. syringae expressing AvrRpt2, AvrB, and AvrPphB. Full complementation of wild-type levels of resistance were observed in plants transformed with the RPS2:HA/T7:NDR1 construct. Interestingly, Arabidopsis plants expressing RPS2:HA/T7:NDR1Δ4 showed a degree of susceptibility corresponding to 1 log higher growth when inoculated with P. syringae expressing AvrRpt2. This pattern of susceptibility is consistent with the RIN4 overexpression phenotype observed by Mackey et al. (2003), which has been suggested to correspond to a saturation in the negative regulation of the corresponding R protein, resulting in a lag in the activation of resistance (Axtell and Staskawicz, 2003; Mackey et al., 2003; Day et al., 2005). As shown in Figure 7D, resistance to P. syringae expressing AvrB (RPM1) and AvrPphB (RPS5) was unaffected, irrespective of the mutation status (i.e., Δ4) of NDR1.
DISCUSSION
In this study, we demonstrate that NDR1 interacts with RIN4 in a yeast two-hybrid assay; furthermore, this association is also observed in planta. These data provide direct molecular evidence for the physical involvement of NDR1 in disease resistance signaling in Arabidopsis. To date, the requirement for NDR1 in the activation of disease resistance signaling mediated by members of the CC-NB-LRR class of resistance proteins has been well documented (Century et al., 1995, 1997; Belkhadir et al., 2004b; Coppinger et al., 2004). However, its biochemical function within the cell has remained elusive.
The impetus for this study was to identify proteins that interact with RIN4 and that thus may play a role in disease resistance signaling in Arabidopsis following P. syringae perception. The identification of a specific interaction between RIN4 and NDR1 by yeast two-hybrid assay established a testable model for which we could pursue our previous work in characterizing the protein–protein interactions that specify disease resistance to P. syringae expressing the bacterial effector protein AvrRpt2. Based on previous results in our laboratory (Coppinger et al., 2004), we hypothesized that NDR1's role in disease resistance may largely be that of one of surveillance, particularly given its position on the outside of the plasma membrane. Interestingly, however, genetic evidence suggests that NDR1 is likely downstream of many of the early signaling events associated with P. syringae perception and complex assembly (Belkhadir et al., 2004b).
The association of RIN4 with both RPS2 and NDR1 presents several interesting possibilities in terms of the stoichiometry associated with both the regulation and activation of resistance in response to pathogen perception. At the onset of this study, our model was that cleavage of RIN4 by the P. syringae Type III effector protein AvrRpt2 (and ultimate elimination of these cleavage products) releases the negative regulation placed on RPS2 by RIN4. Based on our results in this study, we now propose that NDR1 may function to positively regulate the resistance response, at least in part, by its association with RIN4 and subsequent titration of the negative regulator away from its associated R protein. Several lines of evidence support this hypothesis. First, overexpression of RIN4 abrogates the activation of resistance mediated by RPS2 (Axtell and Staskawicz, 2003; Mackey et al., 2003), presumably due to a saturation in the negative regulation circuitry. Secondly, overexpression of NDR1 hyperactivates RPS2-mediated resistance, enhancing the level of resistance following infection with P. syringae expressing the Cys protease AvrRpt2 (Coppinger et al., 2004). In this study, we demonstrate that by interfering with the RIN4–NDR1 interaction (i.e., NDR1Δ4), we observe an enhanced susceptibility in Arabidopsis plants (RPS2:HA/T7:NDR1Δ4) inoculated with P. syringae expressing AvrRpt2. This is likely due to a disruption in the RIN4–NDR1 interaction, which results in an increase in the amount of free RIN4 that is now available to associate with RPS2. Taken together, we propose that by changing the relative concentration of the RIN4–NDR1 interaction, modulation of the activation status of RPS2 in the absence of bacterial pathogen occurs as a result of shifting the balance of RIN4 from RPS2 to NDR1 and vice versa. While much work remains to be done to fully understand the stoichiometry of the protein–protein interactions associated with resistance signaling in plant defense, the association of RIN4 with multiple protein components required for resistance signaling lends itself to the possibility that RIN4 may exert its activity as a molecular switch by regulating various proteins involved in a number of effector-mediated and basal defense responses (Kim et al., 2005b).
As described above, our data demonstrate a difference in the activation of RPS2, RPM1, and RPS5 in Arabidopsis plants (RPS2:HA/T7:NDR1Δ4) inoculated with P. syringae expressing AvrRpt2, AvrB, and AvrPphB. The simplest explanation for these results may in fact lie in the previously characterized differential activation of the resistance proteins themselves. Mackey et al. (2002, 2003) showed that RPM1 activity required the phosphorylation of RIN4 following delivery of the effector proteins AvrB or AvrRpm1. This phosphorylation event served to remove the negative regulation imposed on RPM1 by RIN4. Conversely, research has shown that cleavage of RIN4 by the Cys protease AvrRpt2 results in the activation of RPS2 (Axtell and Staskawicz, 2003; Day et al., 2005). Given the differential mechanisms by which each of these R proteins is activated, we propose that NDR1 may also respond and function differentially depending on which effector protein is delivered into the host cell. Support for this hypothesis lies in the work of Axtell and Staskawicz (2003) and Belkhadir et al. (2004a). Axtell and Staskawicz demonstrated that the avirulence function of the effector protein AvrRpt2 was NDR1 dependent. That is, activation of disease resistance signaling mediated by RPS2 requires NDR1. Conversely, the avirulence activity (i.e., the HR-inducing activity) of AvrRpt2 is NDR1 independent. These data suggest that at least two independent signaling events involving AvrRpt2 perception by Arabidopsis exist. This is likely a mechanism to regulate the activity of host resistance responses upstream of NB-LRR activation. Given that NDR1 appears to function, genetically, upstream of R protein activation (Belkhadir et al., 2004a), we hypothesize that additional layers of regulation may exist to prime the defense signaling machinery, depending upon the specific effector proteins delivered. As such, NDR1 may fulfill this regulatory role, in part, through its association with RIN4 as well as with yet identified proteins also required for the activation of resistance responses in Arabidopsis.
Using a twofold approach of mutational analysis coupled with a genetic analysis of disease resistance, we were able to define the structural components of both NDR1 and RIN4 required for their association and for the activation of resistance signaling following P. syringae infection. We previously established that NDR1 is a GPI-anchored protein with a noncleaved N-terminal signal peptide (Coppinger et al., 2004). As such, the most likely conformation that NDR1 assumes within the plasma membrane is that of a double anchor; the N terminus is anchored by a transmembrane domain, while the GPI moiety tethers the C terminus of the protein to the plasma membrane. Given this confirmation, the candidate residues for RIN4 interaction are few: the 18 amino acids at the extreme N terminus that protrude into the cytoplasm. Indeed, as shown in Figures 5 and 6, our results are consistent with this hypothesis and demonstrate that NDR1 association with RIN4 requires the Gln and Asn residues at positions four and five, respectively.
Here, we provide a detailed analysis of the RIN4–NDR1 interaction, defining the residues in NDR1 required for this association and, moreover, providing data that are in agreement with the likely double anchor model for NDR1's topology within the plasma membrane. Our data clearly demonstrate that the RIN4–NDR1 interaction is specific and likely includes an association following RIN4 cleavage by AvrRpt2, as monitored by coimmunoprecipitation experiments with NDR1 and the RIN4 cleavage products (Figure 4). However, the function of NDR1 remains elusive. It is interesting to speculate that one of the functions of NDR1 may in fact be as a mechanism to control the amount of available free RIN4 within the plant cell, thereby regulating the amount of unbound, negative regulator within the cell. Ultimately, the future goals of this research are to understand the function not only of NDR1, but also the proteins required for NDR1-dependent resistance signaling in the RPS2-specified disease resistance pathway.
METHODS
Strains and Growth
Escherichia coli DH5α strains were grown on Luria-Bertani agar medium, as were binary constructs mobilized in Agrobacterium tumefaciens strain C58-C1, at 37°C and 28°C, respectively. Binary vector constructs were mobilized into A. tumefaciens by triparental matings according to standard protocols.
Saccharomyces cerevisiae strain cdc25H was maintained on YPAD medium, except when harboring two-hybrid plasmids, in which case strains were handled according to the manufacturer's specifications (Stratagene).
Nicotiana benthamiana plants were grown at 24°C in a growth cabinet under a 16-h-light/8-h-dark cycle.
Yeast Two-Hybrid Assays
Library Construction
Total RNA used in the construction of the Arabidopsis thaliana cDNA library was isolated from a variety of source tissue, including both pathogen-inoculated and water (mock)-inoculated plant tissues. Arabidopsis genotypes used as source material for total RNA included wild-type Col, ndr1-1, rps2/101c, rpm1-1, rps5-1, and rps4. Pathogen treatments included 0-, 2-, 4-, 6-, 8-, 24-, and 48-h low-density (i.e., 104 cfu/mL) inoculations, individually, with Pseudomonas syringae expressing empty vector (pVSP61), AvrRpt2, AvrRpm1, AvrB, AvrPphB, and AvrRpm1. Total RNA isolated from each genotype (either treated or mock inoculated) was pooled and used as a source for mRNA.
An Arabidopsis CytoTrap yeast two-hybrid library was constructed according to the manufacturer's specifications (Stratagene). Total RNA was isolated using Trizol reagent (Invitrogen). Total RNA was further processed, and template mRNA was isolated using an mRNA isolation kit (Stratagene).
Bait Construction
A full-length cDNA clone corresponding to the open reading frame of RIN4 was amplified by PCR with a 5′ BamHI site and a 3′ NotI site to facilitate cloning into the bait vector pSos. The resultant plasmid was a 5′, in-frame fusion with hSos. Expression of the hSos-RIN4 fusion was confirmed by protein gel blot analysis using a monoclonal antibody raised against hSos (BD Transduction Laboratories; data not shown).
Two-Hybrid Screen
Library transformation, replication, and yeast two-hybrid screening was performed according to the manufacturer's specifications. The 1 × 106 yeast colonies were screened. Confirmation of positive interactions was verified by plasmid isolation, sequencing, reconstruction of the rescued prey construct, and recapitulation of the yeast transformation and screening by directed (bait + rescued prey) analysis. Positive and negative control plasmids were used according to the manufacturer's specifications.
Generation of NDR1 Ala Scanning Mutants
QuickChange PCR was performed as previously described (Day et al., 2005). DNA primers used for NDR1 and RIN4 Ala scanning PCR were as follows: NDR1M1A forward 5′-GCTAATAATCAAAATGAAGACACAGAAGGT-3′ and reverse 5′-ACCTTCTGTGTCTTCATTTTGATTATTAGC-3′; NDR1N2A forward 5′-ATGGCTAATCAAAATGAAGACACAGAAGGTGGT-3′ and reverse 5′-ACCACCTTCTGTGTCTTCATTTTGATTAGCCAT-3′; NDR1N3A forward 5′-ATGAATGCTCAAAATGAAGACACAGAAGGTGGTCGA-3′ and reverse 5′-TCGACCACCTTCTGTGTCTTCATTTTGAGCATTCAT-3′; NDR1Q4A forward 5′-ATGAATAATACTAATGAAGACACAGAAGGTGGTCGAAACTGT-3′ and reverse 5′-ACAGTTTCGACCACCTTCTGTGTCTTCATTAGTATTATTCAT-3′; NDR1N5A forward 5′-ATGAATAATCAAGCTGAAGACACAGAAGGTGGTCGAAACTGTTGT-3′ and reverse 5′-ACAACAGTTTCGACCACCTTCTGTGTCTTCAGCTTGATTATTCAT-3′; NDR1E6A forward 5′-ATGAATAATCAAAATGCTGACACAGAAGGTGGTCGAAACTGTTGTACTTG-3′ and reverse 5′-CAAGTACAACAGTTTCGACCACCTTCTGTGTCAGCATTTTGATTATTCAT-3′; NDR1D7A forward 5′-AATCAAAATGAAGCTACAGAAGGTGGTCGAAACTGTTGTACTTGC-3′ and reverse 5′-GCAAGTACAACAGTTTCGACCACCTTCTCTAGCTTCATTTTGATT-3′; and NDR1T8A forward 5′-CAAAATGAAGACGCTGAAGGTGGTCGAAACTGTTGTACTTGC-3′ and reverse 5′-GCAAGTACAACAGTTTCGACCACCTTCAGCGTCTTCATTTTG-3′.
Sequence of all DNA constructs was confirmed by automated DNA sequencing using an ABI-3100 capillary DNA sequencer (Applied Biosystems).
Agrobacterium-Mediated Transient Expression
Agrobacterium strain C58-C1 (pCH32) carrying the gene of interest was infiltrated into leaves of N. benthamiana essentially as described by Tai et al. (1999). Agrobacterium was grown overnight at 28°C on Luria-Bertani agar containing 100 μg/mL rifampicin, 25 μg/mL kanamycin, and 5 μg/mL tetracycline. Cells were resuspended in induction medium (10 mM MES, pH 5.6, 10 mM MgCl2, and 150 μM acetosyringone) and incubated at room temperature for 1.5 h before inoculation. For HA- and T7-tagged NDR1 and RIN4 constructs, Agrobacterium was infiltrated at a final OD600 of 0.4. Agrobacterium RPS2:HA constructs were infiltrated at a final OD600 of 0.075.
For transient expression, RPS2, NDR1, RIN4, and the NDR1/RIN4 deletion constructs were expressed from a modified pE1776 with a chimeric octopine and manopine synthase promoter engineered into a pBIN derivative (Ni et al., 1995).
Generation of Native Promoter, Epitope-Tagged, Complementation Lines of Arabidopsis
The RPS2:HA/T7:NDR1 T-DNA construct pRPS2:HA/T7:NDR1 was constructed by first cloning the Gateway (Invitrogen) ccdB cassette into the T4 DNA polymerase–filled single SacI site flanking the promoter end of the RPS2:HA complementing construct p4104-HA (Axtell and Staskawicz, 2003) to create the destination vector pRPS2:HA-attR1-attR2. For the construction of the NDR1 portion of the construct, a native promoter-driven, epitope-tagged (i.e., T7) NDR1 construct was cloned into the Gateway entry vector pENTR/D TOPO. To generate this construct, a C-terminal region including the native terminator located on a 1401-bp HindIII and DraI fragment from an NDR1-containing cosmid (Century et al., 1997) was cloned into pBlueScript KS+ HindIII and T4 filled-in SalI sites. This allowed the fragment to be recloned as a NotI-XhoI fragment into a modified pENTR/D TOPO containing an internal XhoI site. The native NDR1 promoter clone with the NdeI site introduced at the start codon (Coppinger et al., 2004) was cloned from pCR2.1 as a NotI-EcoRI fragment and moved into the NotI and EcoRI sites in pENTR/D TOPO. The T7 epitope tag (MASMTGGQQMG) was constructed as an NdeI and SalI fragment and cloned into the NdeI and SalI sites in pENTR/D TOPO, with the two NDR1 fragments, to create pENTR/D T7-SalI-HindIII-NDR1. The NDR1 N-terminal SalI and HindIII fragments from both pMD1-T7:NDR1 and pMD1-T7:NDR1Δ4 were recloned in pENTR/D T7-SalI-HindIII-NDR1 to create pENTR/D T7:NDR1 and pENTR/D T7:NDR1Δ4. These two NDR1 native constructs were then moved into the native RPS2:HA destination construct with LR clonase (Invitrogen) to create the two T-DNA constructs pRPS2:HA/T7:NDR1 and pRPS2:HA/T7:NDR1Δ4. Both constructs were introduced to Agrobacterium C58-C1 using triparental mating (Figurski and Helinski, 1979) and transformed into rps2 ndr1-1 double mutant Arabidopsis plants (Belkhadir et al., 2004b) by the floral dip method (Clough and Bent, 1998). Kanamycin-resistant T1 plants were selected on Murashige and Skoog agar containing 75 μg/mL kanamycin. Four independent lines for each construct were identified as homozygous for a single transgene locus by segregation of kanamycin resistance. Subsequent lines were screened for complementation by pathogen inoculation and protein expression by anti-HA and anti-T7 immunoblots. PCR was used to confirm that these lines were homozygous for the ndr1-1 mutation using NDR1-specific primers as follows: NDR1 forward minus nucleotide 145 (5′-GTGTGTCCTACTGAGTC-3′) and NDR1 reverse plus nucleotide 103 (5′-AGGTGAGACCAGCTGTGA-3′). Integration of the RPS2:HA transgene was monitored by PCR using the following primers: RPS2fw2550 (5′-CTAGGGATCTGCCAGAACT-3′) and RPS2term-137 (5′-TCCTGCTACTTATGAATGGACA-3′).
Coimmunoprecipitation
Following Agrobacterium-mediated transient expression for 22 h, N. benthamiana leaves (∼0.3 g) were harvested and ground to a powder in liquid nitrogen. Ground tissues were resuspended in 3.0 mL of IP buffer (50 mM HEPES, pH 7.5, 50 mM NaCl, 10 mM EDTA, 5 mM DTT, 0.1% Triton X-100, and 1× Complete Protease Inhibitor [Roche]). The crude lysates were then spun at 20,000g for 15 min at 4°C. Following centrifugation, 1 mL of supernatant was used for each immunoprecipitation. Five microliters of either anti-HA (Covance) or anti-T7 (Novagen) antibody was used to capture the epitope-tagged proteins. Following 1 h of incubation at 4°C, immunocomplexes were collected by the addition of 50 mL of protein G Sepharose-4 fast flow beads (Amersham) and incubation end-over-end for 4 h at 4°C. Immunocomplexes were then washed four times with 1 mL wash buffer (IP buffer + 0.2% Triton X-100). After washing, the beads were resuspended in 50 mL of 3× SDS-PAGE loading buffer, boiled for 5 min, and briefly centrifuged, and the supernatant was removed for SDS-PAGE and protein gel blot analysis.
SDS-PAGE and Immunoblotting
Protein samples analyzed for experiments other than coimmunoprecipitations were isolated by homogenizing corresponding leaf disks in 3× Laemmli buffer (Laemmli, 1970) in a microfuge tube using a Kontes pestle (Fisher Scientific). Protein samples were separated by SDS-PAGE on 12% polyacrylamide gels and transferred for immunoblot analysis by electroblotting to nitrocellulose membranes according to standard protocols. Membranes were probed with anti-HA-HRP (Roche) or anti-T7-peroxidase (Novagen) to detect HA- and T7-epitope tagged proteins, respectively. All antibodies were used as recommended by their respective manufacturers. RIN4 polyclonal antiserum, where noted, was used at a 1:5000 dilution, as previously described (Axtell and Staskawicz, 2003; Mackey et al., 2003; Day et al., 2005).
Accession Numbers
Sequence data for all cDNAs from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: RIN4, NM_113411; RPS2, NM_118742; NDR1, AF021346; AvrRpt2, Z21715; AvrPphB; Q4LBP1; AvrRpm1, YP_233840.1; and AvrRPS4, AAB51082.
Acknowledgments
B.J.S. is funded by the U.S. Department of Energy (DE-FG03-88ER13917) and the National Institutes of Health (R01-GM069680). B.D. was supported by a National Institutes of Health–National Research Service Award postdoctoral fellowship (F32GM067520-03). We thank Gitta Coaker, Maike Rentel, and Jeff Dangl for critical reading of the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Brian J. Staskawicz (stask@nature.berkeley.edu).
References
- Axtell, M.J., and Staskawicz, B.J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112 369–377. [DOI] [PubMed] [Google Scholar]
- Belkhadir, Y., Nimchuk, Z., Hubert, D.A., Mackey, D., and Dangl, J.L. (2004. a). Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell 16 2822–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir, Y., Subramaniam, R., and Dangl, J.L. (2004. b). Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr. Opin. Plant Biol. 7 391–399. [DOI] [PubMed] [Google Scholar]
- Century, K.S., Holub, E.B., and Staskawicz, B.J. (1995). NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc. Natl. Acad. Sci. USA 92 6597–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century, K.S., Shapiro, A.D., Repetti, P.P., Dahlbeck, D., Holub, E., and Staskawicz, B.J. (1997). NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278 1963–1965. [DOI] [PubMed] [Google Scholar]
- Chisholm, S.T., Coaker, G., Day, B., and Staskawicz, B.J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124 803–814. [DOI] [PubMed] [Google Scholar]
- Chisholm, S.T., Dahlbeck, D., Krishnamurthy, N., Day, B., Sjolander, K., and Staskawicz, B.J. (2005). Molecular characterization of proteolytic cleavage sites of the Pseudomonas syringae effector AvrRpt2. Proc. Natl. Acad. Sci. USA 102 2087–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Coaker, G., Falik, A., and Staskawicz, B.J. (2005). Activation of a phytopathogenic bacterial effector protein by a eukaryotic cyclophilin. Science 308 548–550. [DOI] [PubMed] [Google Scholar]
- Coppinger, J.P., Repetti, P., Day, B., Dahlbeck, D., Mehlert, A., and Staskawicz, B. (2004). Overexpression of the plasma membrane-localized NDR1 protein results in enhanced bacterial disease resistance in Arabidopsis thaliana. Plant J. 40 225–237. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411 826–833. [DOI] [PubMed] [Google Scholar]
- Day, B., Dahlbeck, D., Huang, J., Chisholm, S.T., Li, D., and Staskawicz, B.J. (2005). Molecular basis for the RIN4 negative regulation of RPS2 disease resistance. Plant Cell 17 1292–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar, C., Hernandez, L.E., Jimenez, A., Creissen, G., Ruiz, M.T., and Mullineaux, P.M. (2003). Transient expression of Arabidopsis thaliana ascorbate peroxidase 3 in Nicotiana benthamiana plants infected with recombinant potato virus X. Plant Cell Rep. 21 699–704. [DOI] [PubMed] [Google Scholar]
- Figurski, D.H., and Helinski, D.R. (1979). Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76 1648–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X., Anderson, J.C., del Pozo, O., Gu, Y.Q., Tang, X., and Martin, G.B. (2004). Silencing of subfamily I of protein phosphatase 2A catalytic subunits results in activation of plant defense responses and localized cell death. Plant J. 38 563–577. [DOI] [PubMed] [Google Scholar]
- Hubert, D.A., Tornero, P., Belkhadir, Y., Krishna, P., Takahashi, A., Shirasu, K., and Dangl, J.L. (2003). Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J. 22 5679–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H., Axtell, M.J., Dahlbeck, D., Ekwenna, O., Zhang, S., Staskawicz, B., and Baker, B. (2002). NPK1, an MEKK1-like mitogen-activated protein kinase kinase kinase, regulates innate immunity and development in plants. Dev. Cell 3 291–297. [DOI] [PubMed] [Google Scholar]
- Jones, D.A., and Takemoto, D. (2004). Plant innate immunity - Direct and indirect recognition of general and specific pathogen-associated molecules. Curr. Opin. Immunol. 16 48–62. [DOI] [PubMed] [Google Scholar]
- Kim, H.S., Desveaux, D., Singer, A.U., Patel, P., Sondek, J., and Dangl, J.L. (2005. a). The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc. Natl. Acad. Sci. USA 102 6496–6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M.G., da Cunha, L., McFall, A.J., Belkhadir, Y., DebRoy, S., Dangl, J.L., and Mackey, D. (2005. b). Two Pseudomonas type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell 121 749–759. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Belkhadir, Y., Alonso, J.M., Ecker, J.R., and Dangl, J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112 379–389. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Holt, B.F., Wiig, A., and Dangl, J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108 743–754. [DOI] [PubMed] [Google Scholar]
- Moffett, P., Farnham, G., Peart, J., and Baulcombe, D.C. (2002). Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 21 4511–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, M., Cui, D., Einstein, J., Narasimhulu, S., Vergara, C.E., and Gelvin, S.B. (1995). Strength and tissue specificity of chimeric promoters derived from the octopine and mannopine synthase genes. Plant J. 7 661–676. [Google Scholar]
- Shirasu, K., and Schulze-Lefert, P. (2003). Complex formation, promiscuity and multi-functionality: Protein interactions in disease-resistance pathways. Trends Plant Sci. 8 252–258. [DOI] [PubMed] [Google Scholar]
- Tai, T.H., Dahlbeck, D., Clark, E.T., Gajiwala, P., Pasion, R., Whalen, M.C., Stall, R.E., and Staskawicz, B.J. (1999). Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc. Natl. Acad. Sci. USA 96 14153–14158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, A., Casais, C., Ichimura, K., and Shirasu, K. (2003). HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. USA 100 11777–11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Biezen, E.A., and Jones, J.D.G. (1998). Plant disease resistance proteins and the “gene-for-gene” concept. Trends Biochem. Sci. 23 454–456. [DOI] [PubMed] [Google Scholar]
- Van der Hoorn, R.A., De Wit, P.J., and Joosten, M.H. (2002). Balancing selection favors guarding resistance proteins. Trends Plant Sci. 7 67–71. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Dorey, S., Swiderski, M., and Jones, J.D.G. (2004). Expression of RPS4 in tobacco induces an AvrRps4-independent HR that requires EDS1, SGT1 and HSP90. Plant J. 40 213–224. [DOI] [PubMed] [Google Scholar]