Abstract
During plastid division, the dynamin-related protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS5 (ARC5) is recruited from the cytosol to the surface of the outer chloroplast envelope membrane. In Arabidopsis thaliana arc5 mutants, chloroplasts arrest during division site constriction. Analysis of mutants similar to arc5 along with map-based cloning identified PLASTID DIVISION1 (PDV1), an integral outer envelope membrane protein, and its homolog PDV2 as components of the plastid division machinery. Similar to ARC5, PDV1 localized to a discontinuous ring at the division site in wild-type plants. The midplastid PDV1 ring formed in arc5 mutants and the ARC5 ring formed in pdv1 and pdv2 mutants, but not in pdv1 pdv2. Stromal FtsZ ring assembly occurred in pdv1, pdv2, and pdv1 pdv2, as it does in arc5. Topological analysis showed that the large N-terminal region of PDV1 upstream of the transmembrane helix bearing a putative coiled-coil domain is exposed to the cytosol. Mutation of the conserved PDV1 C-terminal Gly residue did not block PDV1 insertion into the outer envelope membrane but did abolish its localization to the division site. Our results indicate that plastid division involves the stepwise localization of FtsZ, PDV1, and ARC5 at the division site and that PDV1 and PDV2 together mediate the recruitment of ARC5 to the midplastid constriction at a late stage of division.
INTRODUCTION
All plastids trace their origins to a primary endosymbiotic event in which a previously nonphotosynthetic protist engulfed and enslaved a cyanobacterium. Over time, most of the genes once present in the endosymbiont have been lost or transferred to the host nuclear genome; those still used by the plastid are translated by the host and targeted back into the organelle to express their functions (Bhattacharya et al., 2004; reviewed in Cavalier-Smith, 2004). Consistent with this scenario, plastids are never synthesized de novo and they cannot multiply independently. Their continuity is maintained by the division of preexisting plastids, which is performed and controlled by proteins encoded in the nuclear genome (reviewed in Boffey and Lloyd, 1988; Kuroiwa et al., 1998; Miyagishima et al., 2003a; Osteryoung and Nunnari, 2003; Aldridge et al., 2005).
Consistent with the endosymbiotic origin of chloroplasts, molecular genetic studies in Arabidopsis thaliana have defined several nucleus-encoded homologs of cyanobacterial cell division proteins that function in plastid division in photosynthetic eukaryotes. These include the tubulin-like FtsZ proteins, which assemble into a ring structure on the stromal side of the division site similar to their bacterial counterparts (Osteryoung and Vierling, 1995; Osteryoung et al., 1998; Strepp et al., 1998; McAndrew et al., 2001; Mori et al., 2001; Vitha et al., 2001; Kuroiwa et al., 2002; reviewed in Margolin, 2005), MinD (Colletti et al., 2000) and MinE (Itoh et al., 2001; Maple et al., 2002), which regulate the positioning of the FtsZ ring, and ACCUMULATION AND REPLICATION OF CHLOROPLASTS6 (ARC6), which stabilizes the plastid FtsZ ring (Vitha et al., 2003). Mutations in several other cyanobacteria-derived genes, such as Giant Chloroplast1 (Maple et al., 2004; Raynaud et al., 2004) and Crumpled Leaf (Asano et al., 2004), also cause defects in plastid division, although their effects on the division process may be indirect. However, recent work suggests that the majority of genes regulating cyanobacterial cell division were lost after endosymbiosis (Miyagishima et al., 2005) but that other genes of eukaryotic origin (Shimada et al., 2004; Raynaud et al., 2005; Haswell and Meyerowitz, 2006) have been recruited to function in plastid division. Most notable among these is ARC5, a member of the dynamin superfamily of eukaryotic membrane–remodeling GTPases (Gao et al., 2003; Miyagishima et al., 2003b). ARC5 and its orthologs are recruited during plastid division from patches in the cytosol to the outer envelope surface at the division site. Unlike FtsZ and related factors, ARC5 is required for division only after most of the division site constriction has been accomplished (Gao et al., 2003; Miyagishima et al., 2003b). Together, these findings suggest that plastid division is performed by distinct but coordinated activities that derive partly from the endosymbiont and partly from the eukaryotic host. Localization studies showing FtsZ in the stroma and ARC5 in the cytosol, as well as cytological studies showing the presence of stroma-localized inner plastid–dividing and cytosolic outer plastid–dividing rings (Hashimoto, 1986; Mita et al., 1986; Miyagishima et al., 1998; summarized in Kuroiwa et al., 1998), both of unknown composition, have also shown that plastid division involves the coordinated biochemical activities of components localized both outside and inside the plastids (reviewed in Miyagishima et al., 2003a; Osteryoung and Nunnari, 2003; Aldridge et al., 2005).
Here, we report the identification and characterization of homologous nucleus-encoded proteins required for plastid division, PLASTID DIVISION1 (PDV1) and PDV2. We show that PDV1 is an integral outer envelope protein and that PDV1 and PDV2 are required for ARC5 localization at the division site. We also show that FtsZ, PDV1/PDV2, and ARC5 function in this order, suggesting the possibility that PDV1 and PDV2 mediate the transmission of topological information from the inside to the outside of the organelle during plastid division.
RESULTS
Identification of Mutations That Cause Late-Stage Plastid Division Defects, as Do Mutations in the Dynamin-Related ARC5 Gene
Among the previously identified Arabidopsis mutants with defects in chloroplast division (summarized in Marrison et al., 1999; Pyke, 1999), mutations in the dynamin-related gene ARC5 confer a unique phenotype. In arc5-1, previously described in the Landsberg erecta background, chloroplasts constrict but do not separate, giving them a dumbbell-shaped appearance (Pyke and Leech, 1994; Robertson et al., 1996; Gao et al., 2003) (Figure 1). A newly identified allele of arc5 in ecotype Columbia (Col-0), arc5-2, which bears a T-DNA insertion in the eighth intron (SAIL 71D_11), has a similar phenotype with large, centrally constricted chloroplasts (Figure 1B). This phenotype, together with the localization of ARC5 at the midplastid division site (Gao et al., 2003; Miyagishima et al., 2003b), suggests that ARC5 is required to complete the separation process during plastid division.
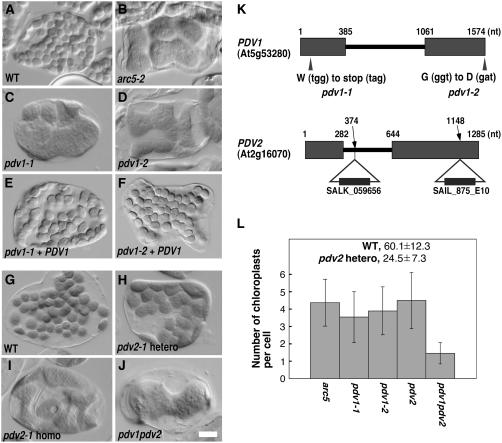
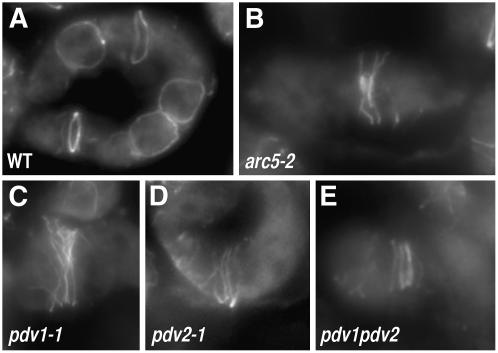
Figure 1.
Phenotypes of arc5, pdv1, pdv2, and pdv1 pdv2 Mutants, and Complementation of the Phenotypes Conferred by pdv1.
Chloroplasts in leaf mesophyll cells were observed by Nomarski optics, and a single cell is shown for each line. All mutants are Col-0 ecotype. Bar in (J) = 10 μm.
(A) and (B) Wild-type Col-0 (A) and arc5-2 (SAIL_71D11) (B) for comparison.
(C) to (F) Mutant phenotypes of pdv1-1 (C) and pdv1-2 (D) were complemented by a wild-type PDV1 transgene (pdv1-1 [E] and pdv1-2 [F]).
(G) to (I) The wild type (G) and heterozygous (H) and homozygous (I) segregants of pdv2-1.
(J) pdv1-1 pdv2-1.
(K) Mutation sites of pdv1-1 and pdv1-2, and positions of T-DNA insertion of pdv2-1 and pdv2-2. Exons are depicted as rectangles, and nucleotide numbers from the annotated start codon are shown above the genes. nt, nucleotides.
(L) Statistical comparison of the number of chloroplasts per mesophyll cell. Gray bars show mean numbers of chloroplasts, and error bars represent sd. n = 40 for the wild type and pdv2-1 heterozygous segregants; n = 80 for all others.
We predicted that identification of additional mutants with phenotypes similar to that of arc5 would lead to the identification of plastid division proteins having functional relationships with ARC5. To this end, we screened 10,000 ethyl methanesulfonate–mutagenized M2 plants (Col-0) by microscopic observation of mesophyll cell chloroplasts. Eighteen mutant lines had chloroplasts that were fewer in number and larger or more variable in size within single cells than in the wild type. Among these mutants, chloroplasts in two mutant lines were frequently constricted and larger than those in wild-type plants, similar to arc5 chloroplasts (Figures 1C and 1D). We named these two mutants pdv1-1 and pdv1-2 based on the results described below.
We examined the genetic properties of pdv1-1 and pdv1-2 after crossing the mutant lines with wild-type plants and determining the segregation of the chloroplast-division phenotype in F1 and F2 progeny. All of the F1 progeny showed wild-type chloroplast morphology, indicating that the chloroplast-division phenotypes in the two mutant lines were recessive. In F2 progeny, the chloroplast-division phenotypes segregated ∼3:1 (for pdv1-1, wild type = 106, mutant = 38 [χ2 = 0.004, P > 0.05]; for pdv1-2, wild type = 113, mutant = 31 [χ2 = 0.028, P > 0.05]), again confirming the recessive character of the chloroplast-division phenotypes and demonstrating their segregation as single Mendelian loci. When the two mutant lines were crossed, all progeny showed the chloroplast-division phenotype, indicating that the two mutations are allelic.
By map-based cloning, we determined that PDV1 corresponds to At5g53280, which was annotated as an expressed gene of unknown function. The predicted gene product (described further below) is 272 amino acids in length and 30.7 kD in mass. Sequence analysis showed that single nucleotide substitutions in pdv1-1 and pdv1-2 convert Trp-17 and Gly-272 to stop and aspartate codons, respectively (Figure 1K). A genomic copy of At5g53280 from wild-type Col-0, flanked by 0.8 kb at the 5′ end and 0.4 kb at the 3′ end, complemented the chloroplast-division defect in both pdv1-1 and pdv1-2 (Figures 1E and 1F). These results show that At5g53280 is identical to PDV1.
PDV1 and Its Homolog PDV2 Have Partially Overlapping Functions in the Late Stage of Plastid Division
A BLAST search (Altschul et al., 1997) identified a homolog of PDV1 in the Arabidopsis genome, At2g16070.2, also annotated as a gene of unknown function. We named the gene PDV2 (expected value of similarity between PDV1 and PDV2 was 4e-06). To test whether PDV2 is also involved in plastid division, we observed chloroplasts in two pdv2 mutants, pdv2-1 (SALK_059656) and pdv2-2 (SAIL_875E10), which have T-DNA insertions in the first intron and second exon, respectively (Figure 1K). In both pdv2-1 (Figure 1I) and pdv2-2 (data not shown), chloroplasts in mesophyll cells were frequently constricted and larger than those in the wild type, suggesting that PDV2 has a function that is similar to but not completely redundant with that of PDV1. Both pdv2 alleles exhibited semidominance; the phenotypes of pdv2/PDV2 plants were intermediate between those of wild-type and pdv2/pdv2 plants (Figures 1H to 1L), although the reason is unclear.
To further examine the genetic relationship between PDV1 and PDV2, we generated a pdv1-1 pdv2-1 homozygous double mutant and compared the phenotypes with those in the single mutants. The pdv1 pdv2 mutant showed a more severe phenotype than either pdv1 or pdv2, and each leaf mesophyll cell in the double mutant contained only one or two chloroplasts (Figures 1J and 1L). As in each single mutant, chloroplasts in the double mutant usually were constricted in the middle. These results suggest that PDV1 and PDV2 have partially overlapping functions that are required at a late stage of plastid division.
PDV1 and PDV2 Are Membrane-Spanning Proteins Bearing Putative Coiled-Coil Domains
To gain insight into the functions of the PDV1 and PDV2 proteins, we subjected the primary structures to analysis by several sequence-alignment and protein-prediction programs. Using BLAST, we identified probable orthologs of PDV1 and/or PDV2 in rice (Oryza sativa) and in the moss Physcomitrella patens (Figure 2), but we could not find obvious homologs in algae or other organisms, even with PSI-BLAST (Altschul et al., 1997). However, similarities among PDV1, PDV2, and their homologs were low (Figure 2). For example, the identity between Arabidopsis PDV1 and rice PDV1 was 39%, suggesting that the primary structures of PDV1 and PDV2 have been diverging relatively rapidly. Therefore, for these particular proteins, comparison based on the primary structures may not be sufficient to identify orthologs or homologs in distantly related organisms.
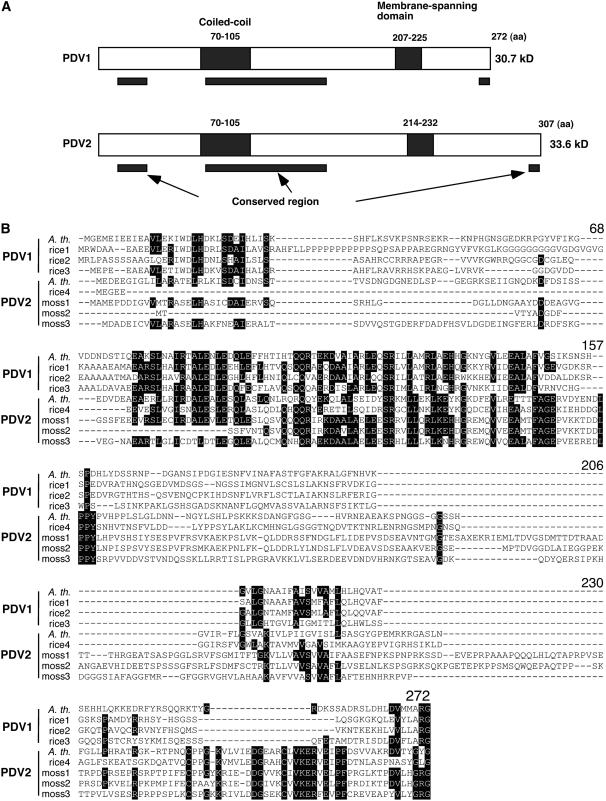
Figure 2.
Predicted Structures of PDV1 and PDV2 Proteins, and Sequence Alignment of PDV1, PDV2, and Homologous Proteins.
(A) Predicted structures of PDV1 and PDV2 proteins. Putative coiled-coil domains and transmembrane domains are depicted as closed rectangles, and their positions within the amino acid sequences are indicated. Black bars below the diagrams delineate the regions that are relatively well conserved among PDV1, PDV2, and related proteins based on the sequence alignment. aa, amino acids.
(B) Sequence alignment of PDV1, PDV2, and related proteins. Black shading indicates residues identical in five or more sequences. The numbers of amino acids from the N terminus of Arabidopsis PDV1 are indicated above the alignment. A. th., Arabidopsis thaliana; rice, Oryza sativa; moss, Physcomitrella patens. Accession numbers for each sequence are listed in Methods. The sequences were aligned with ClustalW (Thompson et al., 1994).
Alignment of PDV1 and PDV2 proteins revealed three regions that were relatively well conserved (Figure 2). Two short conserved regions exist near the N and C termini of PVD1, PVD2, and their homologs, and a Gly residue is conserved at their C termini, where the pdv1-2 mutation was located (Figures 1K and 2). A longer conserved region in the middle of the proteins was predicted by MultiCoil (Wolf et al., 1997) and Coiled-Coil Prediction (Lupas et al., 1991) to contain a coiled-coil motif, suggesting that they might oligomerize (Figure 2). In the database ARAMEMNON (Schwacke et al., 2003), one membrane-spanning region was predicted in both PDV1 and PDV2 (Figure 2). No transit peptides or other motifs were indicated in either protein by any of the prediction programs we used.
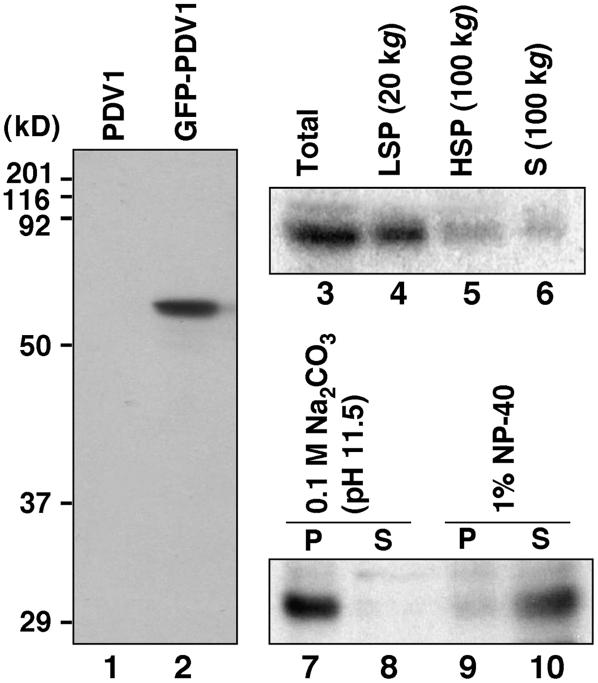
To test whether PDV1 is an integral membrane protein as predicted, we expressed it as a green fluorescent protein (GFP) fusion protein under the control of its own promoter in the pdv1-1 mutant. Because the phenotype conferred by pdv1-2 indicated that the C-terminal Gly residue is important for PDV1 function, GFP was fused to the N terminus of PDV1. The GFP-PDV1 transgene complemented the mutant phenotype in pdv1-1 (described below), indicating that GFP-PDV1 is functional. Immunoblotting of flower bud extracts (see below) with anti-GFP antibody detected protein of the estimated size of GFP-PDV1 in complemented lines (Figure 3, lane 2) but showed no reaction in pdv1-1 plants expressing the untagged PDV1 transgene (Figures 1E and 3, lane 1), indicating that the antibody specifically recognized GFP-PDV1. The lack of fusion protein cleavage downstream of the GFP is consistent with the finding that PDV1 is not predicted to have a cleavable N-terminal transit peptide. After fractionation of the extracts by centrifugation, GFP-PDV1 was detected primarily in the low-speed pellet (Figure 3, lanes 3 to 6). The protein in the pellet was not solubilized by alkaline treatment with sodium carbonate but was solubilized by the detergent Nonidet P-40 (Figure 3, lanes 7 to 10). These results indicated that PDV1 is integrated into the membrane of relatively large organelles.
Figure 3.
Expression and Rough Fractionation of GFP-PDV1.
Immunoblot analyses using anti-GFP antibody. Homogenates of flower buds from pdv1-1 plants expressing a PDV1 transgene (lane 1) and a GFP-PDV1 transgene (lane 2) were blotted. The homogenate containing GFP-PDV1 (Total; lane 3) was centrifuged at 20,000g to sediment the low-speed pellet fraction (LSP; lane 4). The supernatant fraction was fractionated further into the high-speed pellet (HSP; lane 5) and supernatant (S; lane 6) fractions at 100,000g. The low-speed pellet fraction was treated with 0.1 M sodium carbonate (lanes 7 and 8) or 1% Nonidet P-40 (NP-40; lanes 9 and 10), and then soluble (S; lanes 8 and 10) and insoluble (P; lanes 7 and 9) fractions were separated at 100,000g.
PDV1 Forms a Discontinuous Ring Structure at the Plastid Division Site, as Does ARC5
Before performing further fractionation studies, we investigated the localization of GFP-PDV1 in pdv1-1 plants expressing GFP-PDV1. Optical microscopy (data not shown) and fluorescence microscopy (Figure 4A) showed that, as with the wild-type PDV1 gene (Figure 1E), expression of GFP-PDV1 complemented the chloroplast division defect in pdv1-1. GFP fluorescence was detected at the chloroplast division sites in cells of young emerging leaves (∼5 mm long), and the fusion protein formed a discontinuous ring structure encircling the division sites (Figure 4A). The signals were stronger in fully constricted division sites, in which only one or two bright speckles were detected (Figures 4A and 4C to 4F), suggesting that PDV1 becomes locally concentrated as the division site constricts. The fluorescence signals were also observed in root tips, shoot apices, and flower buds (sepals, petals, stamens, and pistils; an image from a petal is shown in Figure 4B), whereas we rarely detected the signals in fully developed tissues, including expanded leaves. The density of signals on the field was highest in flower buds (Figure 4B).
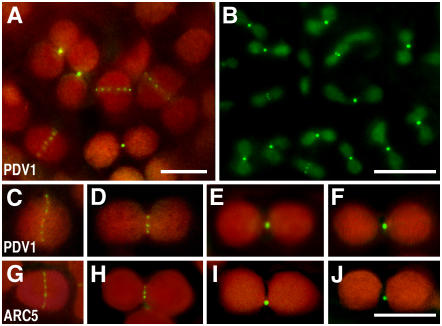
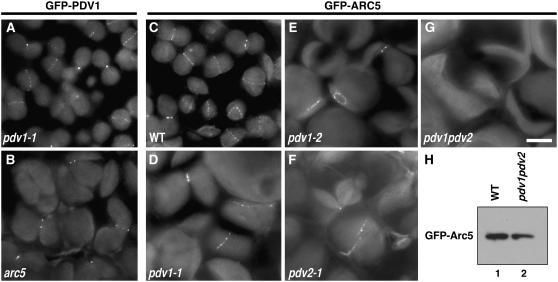
Figure 4.
Localization of GFP-PDV1 and GFP-ARC5.
A GFP-PDV1 transgene was expressed in pdv1-1 plants, and GFP fluorescence was observed by fluorescence microscopy. The fluorescence signal of GFP is green, and the autofluorescence of chlorophyll is red. The dim green fluorescence in (B) is background, because similar fluorescence was observed without the GFP-PDV1 transgene. Expression of the GFP-PDV1 transgene complemented the mutant phenotype of pdv1-1. Bars in (A), (B), and (J) = 5 μm. (C) to (J) are shown at the same magnification.
(A) Chloroplasts in mesophyll cells of a young leaf expressing GFP-PDV1.
(B) Plastids in fringes of petals from flower buds expressing GFP-PDV1.
(C) to (F) Comparison of GFP-PDV1 localization among chloroplasts at various stages of division.
(G) to (J) Comparison of GFP-ARC5 localization among chloroplasts at various stages of division.
The discontinuous midplastid localization of PDV1 differed from the patterns observed for FtsZ1, FtsZ2 (McAndrew et al., 2001; Mori et al., 2001; Vitha et al., 2001; Kuroiwa et al., 2002), ARC6 (Vitha et al., 2003), and ARC3 (Shimada et al., 2004), which have been localized at the division site as continuous rings. However, the localization pattern of PDV1 was very similar to that of ARC5 and its ortholog Cm Dnm2 in the red alga Cyanidioschyzon merolae (Gao et al., 2003; Miyagishima et al., 2003b), although these dynamin-related proteins are cytosolic proteins and PDV1 is an integral membrane protein. These observations, together with the similarity between the phenotypes conferred by pdv1 and arc5 (Figures 1B to 1D), suggest that PDV1 has a close functional relationship with ARC5 in plastid division.
PDV1 and PDV2 Are Required for the Localization of ARC5 at the Division Site after FtsZ Ring Formation
To investigate the functional relationship between PDV1 and ARC5, we first compared the effects of pdv1, pdv2, and arc5 mutations on the localization of FtsZ by immunofluorescence microscopy using an antibody against FtsZ2. In the wild type, FtsZ localizes in a single ring at the chloroplast division site (Vitha et al., 2001) (Figure 5A). As observed previously in arc5-1 (Vitha et al., 2001), FtsZ also localized at the constriction of the enlarged chloroplast in arc5-2 but appeared as multiple rings or spirals (Figure 5B). A similar localization pattern was also observed in pdv1-1, pdv2-1, and pdv1 pdv2 (Figures 5C to 5E), suggesting that PDV1 and PDV2 function after FtsZ ring formation, as does ARC5.
Figure 5.
Localization of FtsZ in arc5, pdv1, pdv2, and pdv1 pdv2 Mutants.
Localization of FtsZ in mesophyll cells of young leaves was observed by immunofluorescence microscopy using anti-At FtsZ2-1 antibodies. The strong fluorescence is the GFP signal, and the dim fluorescence is the red autofluorescence of chlorophyll.
(A) Wild type.
(B) to (E) arc5-2 (B), pdv1-1 (C), pdv2-1 (D), and pdv1 pdv2 (E) mutants.
To examine whether ARC5 localization at the division site (Gao et al., 2003; Miyagishima et al., 2003b) (Figures 4G to 4J) is dependent on PDV1 and/or PDV2 and vice versa, we expressed GFP-PDV1 in the arc5-1 mutant and also expressed GFP-ARC5 in pdv1, pdv2, and pdv1 pdv2. Discontinuous localization of GFP-PDV1 was observed at the constricted sites of large chloroplasts in arc5-1 (Figure 6B), similar to its localization in pdv1-1 (Figures 4A to 4F and 6A). This result indicated that ARC5 is not required for the localization of PDV1. Similarly, GFP-ARC5 formed a discontinuous ring structure at the constricted sites of large chloroplasts in pdv1-1, pdv1-2, and pdv2-1, as in the wild type (Figures 4G to 4J and 6C to 6F). By contrast, no specific localization of GFP-ARC5 was detected on chloroplasts in pdv1 pdv2 (Figure 6G), even though a few speckles were observed in the cytosol (not on chloroplasts) in some transformants. Despite the lack of a clearly localized GFP signal, however, immunoblotting showed that GFP-ARC5 is expressed in pdv1 pdv2 (Figure 6H). These results suggest (1) that PDV1 and PDV2 are required for the localization of ARC5 at the division site, (2) that either PDV1 or PDV2 alone is sufficient for ARC5 localization, but (3) that both PDV1 and PDV2 are required for plastid division to occur properly. Together with the FtsZ localization study, these results suggest a hierarchical localization of FtsZ, PDV1 and PDV2, and ARC5 at the division site, in this order.
Figure 6.
Localization of GFP-PDV1 in the arc5 Mutant, and Localization of GFP-ARC5 in pdv1, pdv2, and pdv1 pdv2 Mutants.
(A) to (G) The strong fluorescence is the GFP signal, and the dim fluorescence is the red autofluorescence of chlorophyll. Mesophyll cells of young leaves were observed by fluorescence microscopy. Localization of GFP-PDV1 expressed in pdv1-1 (A) and arc5-1 (B) plants. Localization of GFP-ARC5 expressed in wild-type (C), pdv1-1 (D), pdv1-2 (E), pdv2-1 (F), and pdv1 pdv2 (G) plants. Bar in (G) = 5 μm.
(H) Immunoblotting of extracts from young leaves showing the expression of GFP-ARC5 in pdv1 pdv2 mutant. Blots containing extracts from wild-type (lane 1) and pdv1 pdv2 (lane 2) plants expressing GFP-ARC5 were probed with anti-GFP antibody.
PDV1 Is an Outer Envelope Membrane Protein with Its N-Terminal Region, Including the Coiled-Coil Domain, Exposed to the Cytosol
The lack of cleavage between GFP and PDV1 in plants expressing the GFP-PDV1 fusion protein (Figure 3), as well as the complementation of the pdv1-1 phenotype by the N-terminally tagged GFP-PDV1 (Figure 4), indicated that PDV1 lacks an N-terminal transit peptide. Furthermore, GFP-PDV1 was observed at the plastid division site by fluorescence microscopy (Figure 4), and fractionation experiments showed that PDV1 is an integral membrane protein that fractionates with the low-speed pellet (Figure 3). Together, these results suggested that PDV1 is probably integrated into the chloroplast outer envelope membrane rather than the inner envelope membrane or other small membranous structures around the division site.
To further analyze the localization of PDV1 and to examine its topology, we performed an in vitro chloroplast-import experiment, followed by treatment with thermolysin (Figure 7A). A full-length radiolabeled PDV1 translation product was synthesized in vitro. Control assays were performed with the integral outer envelope protein OEP14 (Li et al., 1991), the integral inner envelope protein Tic110 (Tic110-110N; Lübeck et al., 1997), and the stroma-localized small subunit of ribulose-1,5-bis-phosphate carboxylase/oxygenase (SS). After incubation of the PDV1 translation product with isolated pea (Pisum sativum) chloroplasts, the protein was detected in the chloroplast fraction, as were control proteins (Figure 7A), suggesting that PDV1 was targeted to the chloroplasts. When the chloroplasts were treated with thermolysin, which does not penetrate the outer envelope membrane, PDV1 and OEP14 were digested, whereas the Tic110 and SS mature proteins were protected from the protease (Figure 7A). These results indicate that PDV1 is associated with the outer envelope membrane and suggest that the large portion of PDV1 upstream of the membrane-spanning domain (Figure 2) is exposed to the cytosolic side of the outer envelope. Because we could not detect any smaller fragments of PDV1 after thermolysin treatment even using a 10 to 20% gradient gel and exposing film for an extended period (data not shown), we were not able to determine experimentally whether the short C-terminal region downstream of the transmembrane domain faces the intermembrane space or the cytosol.
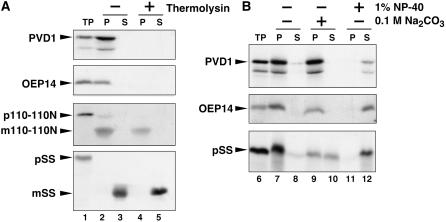
Figure 7.
In Vitro Chloroplast Import, Protease Protection, and Fractionation Assay of PDV1.
35S-labeled PDV1, the integral outer envelope protein OEP14, the precursor of truncated inner envelope protein Tic110-110N (110-110N), and the precursor of the stroma-localized small subunit of ribulose-1,5-bis-phosphate carboxylase/oxygenase (SS) were synthesized in vitro. In vitro–translated proteins were incubated with isolated pea chloroplasts in the presence of 4.0 mM Mg-ATP for 30 min at room temperature. Chloroplasts were recovered by sedimentation through 40% (v/v) Percoll. p and m indicate precursor and mature proteins for 110-110N and SS, respectively.
(A) The recovered intact chloroplasts were incubated without (−; lanes 2 and 3) or with (+; lanes 4 and 5) thermolysin for 30 min at 4°C. Intact chloroplasts were again recovered by centrifugation through 40% (v/v) Percoll and fractionated into total membrane (P; lanes 2 and 4) and soluble (S; lanes 3 and 5) fractions. TP represents 10% of translated product added to a single import assay (lane 1).
(B) The recovered intact chloroplasts were lysed and fractionated into total membrane (P) and soluble (S) fractions (lanes 7 and 8). A portion of the membranes recovered in lane 7 for each import reaction was subsequently extracted with 0.1 M sodium carbonate, pH 11.5 (lanes 9 and 10), or with 1% Nonidet P-40 (lanes 11 and 12) for 1 h on ice. After the treatments, membranes (P) and extractable soluble proteins (S) were recovered and analyzed by SDS-PAGE and fluorography. TP represents 10% of translated product added to a single import assay (lane 6).
To confirm that PDV1 was integrated into the outer envelope in the import experiments, we further treated the chloroplasts with sodium carbonate or Nonidet P-40 (Figure 7B). Neither PDV1 nor OEP14 was extracted by sodium carbonate, but both were solubilized by Nonidet P-40. By contrast, the SS precursor protein, which is peripherally attached to the envelope membrane, was partially extracted by sodium carbonate (Figure 7B). These results indicate that PDV1 was integrated into the outer envelope membrane in our experiments.
The C-Terminal Conserved Gly Residue of PDV1 Is Required for Recognition of the Division Site
Although computational analysis failed to predict any known functional motifs in PDV1 or PDV2 beyond the presence of a coiled-coil domain, the conservation of the C-terminal Gly residue (Figure 2) and the finding that this residue was mutated in pdv1-2 (Figure 1K) suggest that it is critical for PDV1 function. To investigate how the pdv1-2 mutation affects the behavior of PDV1, we introduced the equivalent mutation into GFP-PDV1 and expressed the mutant fusion protein (GFP-PDV1G272D) in both wild-type and pdv1-2 mutant plants. In contrast with GFP-PDV1 expressed in pdv1-1 (Figures 4 and 6), GFP-PDV1G272D did not show any specific fluorescence signals at the chloroplast division sites in either the wild-type or pdv1-2 backgrounds (Figure 8A). Immunoblotting showed that GFP-PDV1G272D was expressed at levels similar to those of GFP-PDV1 (Figure 8B, lanes 1 to 3). Fractionation experiments showed that GFP-PDV1G272D was detected in the low-speed pellet (Figure 8B, lanes 4 to 7 and 12 to 15) and that it was not solubilized by sodium carbonate but was solubilized by Nonidet P-40 (Figure 8B, lanes 8 to 11 and 16 to 19), similar to GFP-PDV1 (Figure 3). In chloroplast import and protease-protection assays, a PDV1G272D in vitro translation product (Figure 8C) behaved identically to the PDV1 translation product (Figure 7); it associated with isolated chloroplasts, was susceptible to thermolysin, fractionated with the chloroplast pellet fraction, and was not solubilized by alkaline treatment but was solubilized by detergent (Figure 8C). These results suggest that GFP-PDV1G272D is integrated into the outer envelope membrane, like GFP-PDV1, but that it cannot localize to the division site. The lack of an obvious fluorescence signal from GFP-PDV1G272D on the chloroplasts is probably attributable to dispersal of the protein on the chloroplast outer membrane.
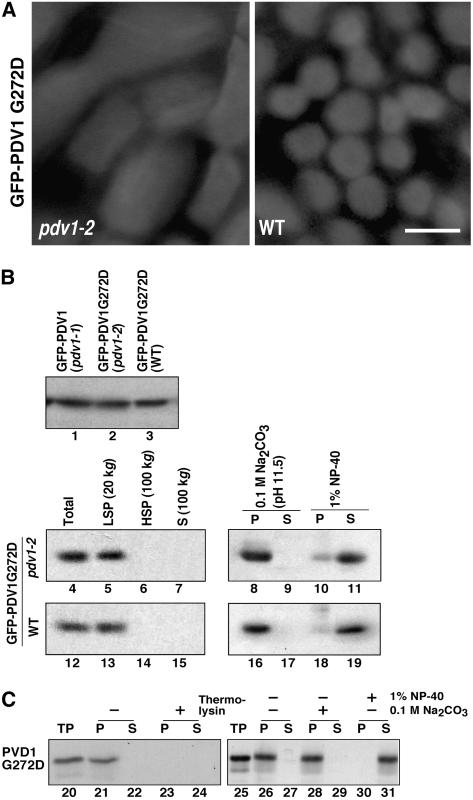
Figure 8.
Effect of the C-Terminal Mutation of PDV1 (pdv1-2) on PDV1 Localization.
(A) GFP-PDV1G272D was expressed in pdv1-2 (left) and wild-type (right) plants. GFP signals in mesophyll cells of young leaves were observed by fluorescence microscopy. Bar = 5 μm.
(B) Expression levels of GFP-PDV1 in pdv1-1 plants (lane 1), which showed ring structures, and of GFP-PDV1G272D in pdv1-2 (lane 2) and wild-type (lane 3) plants were compared. Homogenates of flower buds were analyzed by immunoblotting using anti-GFP antibody. The total homogenates (lanes 4 and 12) were fractionated into the low-speed pellet (LSP; lanes 5 and 13), the high-speed pellet (HSP; lanes 6 and 14), and the supernatant (S; lanes 7 and 15) fractions. The low-speed pellet fraction was treated with 0.1 M sodium carbonate (lanes 8, 9, 16, and 17) or 1% Nonidet P-40 (NP-40; lanes 10, 11, 18, and 19), and then soluble (S; lanes 9, 11, 17, and 19) and insoluble (P; lanes 8, 10, 16, and 18) fractions were separated. Homogenates from pdv1-2 (lanes 4 to 11) and wild-type (lanes 12 to 19) plants expressing GFP-PDV1G272D were fractionated by the same methods as described for Figure 3.
(C) In vitro chloroplast import, protease protection, and fractionation assay of PDV1G272D. 35S-labeled PDV1G272D was synthesized in vitro. The translation product was incubated with isolated chloroplasts, and assays were performed as described for Figure 7. TP represents 10% of translated product added to a single import assay (lanes 20 and 25).
DISCUSSION
PDV1 and PDV2 Function in the Plastid Division Machinery
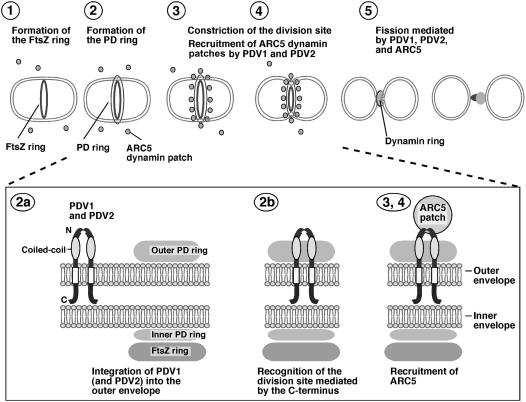
Previous studies established that a plant-specific group of dynamin-related eukaryotic GTPases, defined by ARC5 in Arabidopsis and Cm Dnm2 in the red alga C. merolae, function in the division of chloroplasts (Gao et al., 2003; Miyagishima et al., 2003b). These studies also revealed that host-derived factors, as well as endosymbiont-derived factors, were incorporated into the plastid division machinery during the evolution of photosynthetic eukaryotes. ARC5 and its orthologs redistribute from the cytosol to the surface of the outer envelope membrane at the division site after constriction of the organelle commences, and they are hypothesized to mediate the final scission of the envelope membranes. Here, we have shown that the midplastid localization of ARC5 in Arabidopsis requires the newly identified plastid division proteins PDV1 and PDV2. PDV1 localizes to the outer envelope membrane at the division site with its N-terminal region facing into the cytosol. Although we have not yet determined the localization and topology of PDV2, the similarity between the PDV1 and PDV2 amino acid sequences, along with their partial functional redundancy as shown by analysis of their mutant phenotypes suggest that PDV2 behaves similarly to PDV1. Collectively, our results provide evidence that PDV1 and PDV2 function together in the recruitment of ARC5 from the cytosol to the outer envelope surface at the midplastid division site. A working model summarizing the proposed activity of PDV1 and PDV2 during plastid division is shown in Figure 9.
Figure 9.
Working Model of PDV1 and PDV2 Function during Plastid Division.
The top panel shows an abbreviated model of the plastid division pathway. (For additional details without PDV1 and PDV2, see Miyagishima et al., 2003b; Osteryoung and Nunnari, 2003.) The FtsZ ring, inner plastid–dividing (PD) ring, and outer plastid–dividing ring assemble sequentially (steps 1 and 2) before the onset of constriction (step 3). During constriction, the ARC5 dynamin patches are recruited from the cytosol to the division site by PDV1 and PDV2 (steps 3 and 4) to form a ring structure during a late stage of division (step 5). The bottom panel shows details of proposed PDV1 and PDV2 function. PDV1 (and probably PDV2) is integrated into the outer envelope membrane (step 2a), perhaps interacting via their coiled-coil domains. Localization of PDV1 (and probably PDV2) to the midplastid is mediated at least in part by the conserved Gly at the C terminus, which may be oriented toward the intermembrane space (but see Discussion) (step 2b). PDV1 (and probably PDV2) forms a discontinuous ring structure in the outer envelope membrane; this ring is required for the recruitment of ARC5 dynamin patches to the cytosolic surface of the outer envelope membrane (step 3).
The large chloroplasts in pdv1, pdv2, pdv1 pdv2, and arc5 are frequently constricted and have a dumbbell-shaped appearance, unlike in several other plastid division mutants, in which most of the chloroplasts are round (Pyke, 1999; Osteryoung and Nunnari, 2003). These phenotypic differences suggest that the early and late stages of plastid constriction (or severing) are governed by different biochemical activities and that PDV1, PDV2, and ARC5 are required for the latter. Given the finding that PDV1 and PDV2 mediate ARC5 localization, the arc5-like phenotypes of pdv1, pdv2, and pdv1 pdv2 probably reflect, at least in part, the loss of ARC5 activity at the division site in these mutants.
In contrast with other known division proteins that appear as continuous rings at the plastid division site, including FtsZ (McAndrew et al., 2001; Mori et al., 2001; Vitha et al., 2001; Kuroiwa et al., 2002), ARC6 (Vitha et al., 2003; but see Maple et al., 2005), and ARC3 (Shimada et al., 2004), both ARC5 and its red algal ortholog (Gao et al., 2003; Miyagishima et al., 2003b) and PDV1 exhibit a discontinuous pattern of localization until the final stages of division. The presence of discontinuous PDV1 rings in some of the unconstricted and all of the constricted chloroplasts suggests that these structures form before the onset of constriction (Figure 9). The punctate appearance of the PDV1 ring further suggests that biogenesis of the division complex involves the formation of specialized, discontinuous outer envelope subdomains enriched in PDV1 (and probably PDV2), and the corresponding punctate appearance of the ARC5 ring, along with its dependence on PDV1 and PDV2 for its localization, suggest that the formation of PDV1- and PDV2-enriched subdomains is a prerequisite for ARC5 recruitment to the division site. Such subdomains could also be enriched in specific membrane lipids. Defining the localized composition of the outer and inner envelope membranes at the division site will be important for understanding the organizing principles governing the assembly and dynamic activity of the plastid division complex.
In plastids of pdv1, pdv2, pdv1 pdv2, and arc5 mutants, FtsZ rings assemble at the division site, suggesting that these genes act downstream of FtsZ in the chloroplast division pathway. However, the FtsZ filaments in these mutants coiled around the division site many times, unlike in wild-type chloroplasts, in which only a single FtsZ ring was usually observed, and the region occupied by FtsZ was significantly wider than in wild-type chloroplasts (Figure 5). By contrast, only one discontinuous band of PDV1 was observed at the division sites of arc5 chloroplasts. Similarly, only one or occasionally two bands of ARC5 were observed in pdv1 and pdv2 single mutants. These results indicate that the localization of PDV1 and ARC5 is not directly coupled with that of FtsZ in these mutants. However, it seems likely that one or more adaptor proteins indirectly link and coordinate the localization and activities of the stromal FtsZ ring and the cytosolic ARC5 ring across the two envelope membranes. PDV1 and PDV2 are candidate mediators of this coordination.
The larger N-terminal portion of PDV1 upstream of the predicted transmembrane domain contains a short conserved region and a putative coiled-coil domain. Our results showing that the N-terminal region is exposed to the cytosol suggest that it could interact with ARC5 directly or via other components. In this regard, we have been unable to detect any interaction between PDV1 and ARC5 in yeast two-hybrid assays (data not shown). We are using other methods to investigate the possibility that ARC5, PDV1, and PDV2 interact in a complex.
Mutation of the conserved C-terminal Gly abolished PDV1 localization at the division site but did not affect its localization to the membrane. This result suggests that PDV1 recognizes the division site at least in part through its C-terminal region. One transmembrane domain was predicted for PDV1, which also suggests that the small C-terminal portion (∼5 kD) downstream from the transmembrane domain is in the intermembrane space. If that is the case, PDV1 could convey topological information from the putative C-terminal intermembrane space region to the N-terminal cytosolic region by recognizing the division site (Figure 9). We cannot yet draw firm conclusions about the topology of the PDV1 C-terminal region because we could not detect the small C-terminal fragment after digestion by thermolysin, and the loss of PDV1 function by mutation of the C-terminal Gly indicates that epitope tagging at the C terminus would not yield a functional protein. Further investigation of PDV1 topology will require overcoming the difficulty of tagging the C-terminal region. As indicated above, the similarity of PDV2 to PDV1 in both amino acid sequence and function suggests that its localization and topology are similar to those of PDV1.
Comparison between Plastidic and Mitochondrial Mechanisms for the Recruitment of Dynamin-Related Organelle Division Proteins
The division of mitochondria requires a dynamin-related protein paralogous with ARC5 that assembles into multimeric structures on the surface of the outer mitochondrial membrane to promote fission (reviewed in Miyagishima et al., 2003a; Osteryoung and Nunnari, 2003; Okamoto and Shaw, 2005). The dynamin-related mitochondrial division protein in Saccharomyces cerevisiae, Dnm1p (Bleazard et al., 1999; Sesaki and Jensen, 1999), is recruited from a cytosolic pool to the mitochondrial surface by the WD repeat proteins Mdv1p and Caf4p (Fekkes et al., 2000; Mozdy et al., 2000; Tieu and Nunnari, 2000; Cerveny et al., 2001; Griffin et al., 2005; Bhar et al., 2006; Naylor et al., 2006), paralogous peripheral outer membrane proteins that are partially redundant in their Dnm1p recruitment activity. Early in the yeast mitochondrial division pathway, Mdv1p and Caf4p localize uniformly on the outer membrane via interaction with the integral outer membrane protein Fis1p (Fekkes et al., 2000; Mozdy et al., 2000; Tieu and Nunnari, 2000; Tieu et al., 2002; Cerveny and Jensen, 2003; Griffin et al., 2005; Karren et al., 2005; Bhar et al., 2006; Naylor et al., 2006), and this interaction is required for Dnm1p localization to mitochondria. Thus, Mdv1p and Caf4p function as molecular adaptors between Dnm1p and Fis1p, mediating the Fis1p-dependent recruitment of Dnm1p to the mitochondrial surface. Later in the division pathway, Mdv1p becomes reorganized into division site–localized punctate structures that also contain Fis1p and Dnm1p (Bhar et al., 2006); these puncta represent the active mitochondrial fission complexes.
At present, there is little evidence that plastids and mitochondria use similar mechanisms to recruit dynamin-related fission proteins. Searches based on primary structures have not yielded Fis1p-like proteins in plants or algae that might function in plastid division (although a Fis1p ortholog involved in plant mitochondrial division has been identified [Scott et al., 2006]), and Mdv1p and Caf4p have been found only in fungi (Osteryoung and Nunnari, 2003; Okamoto and Shaw, 2005). Similarly, BLAST searches did not reveal proteins similar to PDV1 and PDV2 in species other than land plants. However, the primary structures of Mdv1p/Caf4p and PDV1/PDV2 are not well conserved among species, suggesting that these proteins have been evolving at faster rates than other proteins. Therefore, searches based on primary structures may not be sufficient to identify possible orthologs or homologs in distantly related organisms. PDV1 and Mdv1p are functionally similar in that they both localize to organelle division sites and play roles in recruiting dynamin-related proteins to the organelle surface. Furthermore, PDV1/PDV2 and Mdv1p/Caf4p (Griffin et al., 2005) have putative central coiled-coil domains, although their other domains are apparently different. These similarities raise the possibility that the regions containing coiled-coil domains in PDV1/PDV2 and Mdv1p/Caf4p have mechanistically related functions in plastid and mitochondrial division, respectively, and that PDV1/PDV2 and Mdv1p/Caf4p may have an evolutionary relationship. Further functional and biochemical characterization of PDV1 and PDV2 and comparison between PDV1/PDV2 and Mdv1p/Caf4p will provide significant insights into the similarities and differences between the plastid and mitochondrial division machineries.
METHODS
Plant Materials and Growth Conditions
The Col-0 ecotype of Arabidopsis thaliana was used as the wild type. The T-DNA insertion line SAIL 71D_11 (arc5-2) was obtained from Syngenta. The T-DNA insertion lines SALK_059656 (pdv2-1) and SAIL_875E10 (pdv2-2) were provided by the ABRC. Seeds were surface-sterilized, sown on Murashige and Skoog plates, and stratified at 4°C for 48 h in the dark before germination. Plants were grown in controlled-environment chambers at a RH of 40% and provided daily with 16 h of light (125 μmol·m−2·s−1) and 8 h of dark at 20°C. Seedlings were transplanted onto soil ∼2 to 3 weeks after germination and were grown in the controlled-environment chambers.
Isolation of pdv1-1 and pdv1-2 Mutants
M2 seeds of Col-0 that had been mutagenized by ethyl methanesulfonate were obtained from Lehle Seeds. The M2 seeds were germinated and grown for 3 weeks on Murashige and Skoog plates. Tips from expanding leaves were put on a glass slide without fixation, covered with a cover slip, and smashed gently. Samples were observed with Nomarski differential interference contrast optics using an Olympus BH-2 microscope (Olympus America).
Among 10,000 plants observed, mesophyll cells of 18 plants showed enlarged chloroplasts. Two allelic recessive mutants, pdv1-1 and pdv1-2, were analyzed further in this study.
Map-Based Cloning of PDV1
The pdv1-1 mutation was mapped with molecular markers based on a cleaved amplified polymorphic sequence (Konieczny and Ausubel, 1993) and simple sequence length polymorphisms (Bell and Ecker, 1994). We used some markers listed on The Arabidopsis Information Resource (TAIR; http://www.Arabidopsis.org); other markers were designed based on polymorphisms listed at TAIR (http://www.Arabidopsis.org/Cereon) in the Monsanto SNP and Ler Sequence Collection. The pdv1-1 homozygous mutant was crossed with Landsberg erecta wild-type plants to generate a mapping population. Analyses using 48 F2 progeny with the pdv1-1 phenotype showed that the mutation is located in a region of ∼0.7 Mb on chromosome 5 (between polymorphisms SGCSNP18686 and SGCSNP220). Using ∼500 F2 plants, we fine-mapped the pdv1-1 mutation to a region of ∼65 kb that includes 12 genes (between polymorphisms CER437355 and SGCSNP18689). Of these 12 genes, 6 were amplified from pdv1-1 and sequenced; a mutation was found in At5g53280. At5g53280 was then amplified from pdv1-2 plants, and a different mutation was found in the same gene (Figure 1K).
DNA Constructs and Plant Transformation
For the pdv1 complementation construct, a genomic fragment containing the annotated PDV1 open reading frame flanked by 0.8 kb at the 5′ end and 0.4 kb at the 3′ end was amplified by PCR with PfuTurbo DNA polymerase (Stratagene) using the primers 5′-AGTTTTTTTACATTTTATCACCGC-3′ and 5′-ACTGTATTTTGTATATTGTTTCCG-3′. The amplified product was cloned into pGEM-T Easy vector (Promega), excised with NotI, and then transferred into pMLBART (Vitha et al., 2001). The final construct was transformed into pdv1-1 and pdv1-2 plants.
To create an N-terminal GFP fusion of PDV1 that is regulated by the PDV1 promoter, two unique restriction sites were added between the PDV1 promoter and the start codon by overlap-extension PCR. We amplified a 0.8-kb 5′ region of PDV1, including the start codon, using primers 5′-AGTTTTTTTACATTTTATCACCGC-3′ and 5′-CATGGTACCCCGGGATCCCATTTCTCCCATTTCATTGACA-3′. A PDV1 open reading frame plus 0.4 kb at the 3′ end was amplified using primers 5′-ATGGGATCCCGGGGTACCATGGAGATCGAAGAAATCGAAG-3′ and 5′-ACTGTATTTTGTATATTGTTTCCG-3′ (BamHI and KpnI sites are underlined, and boldface letters show the overlap). These two amplified fragments were mixed and fused by PCR using primers 5′-GCGGTAGAAGTTATACAATTCCTA-3′ and 5′-TTGTCATCAAAAGTAAAAGGGC-3′. The fused fragment was cloned into pGEM-T Easy. An open reading frame of GFP (S65T [Chiu et al., 1996]) was amplified by primers 5′-CGCGGATCCATGGTGAGCAAG-3′ and 5′-CGGGTACCCTTGTACAGCTCGTCCATGC-3′ (BamHI and KpnI sites, respectively, are underlined), digested with BamHI and KpnI, and cloned between a 5′ flanking region and an open reading frame of PDV1. The resulting PDV1 promoter-GFP-PDV1 fusion was excised with NotI and then transferred into pMLBART. The final construct was transformed into pdv1-1 and arc5-1 plants. (Because the T-DNA insertion in arc5-2 confers resistance to glufosinate, here we used arc5-1.)
To introduce the pdv1-2 mutation (G269D) into the GFP-PDV1 construct, a genomic fragment including the mutation was amplified from pdv1-2 using primers 5′-GTGTTTAGTTAGGGTTTAGATTTACG-3′ and 5′-TTATTACGGTTTAGAGTGATTTGTC-3′. The amplified product was digested with BglII and KpnI and replaced with a wild-type sequence at BglII and KpnI sites in the GFP-PDV1 construct. The final construct was transformed into wild-type and pdv1-2 plants.
Because the previous GFP-ARC5 construct (Gao et al., 2003) yielded a very weak fluorescence signal in transformed plants, we modified the construct. In the previous construct, the ARC5 start codon flanked by the ARC5 promoter and the start codon of GFP were out of frame (promoter-ATGGCGGATCCATG-GFP), potentially reducing the efficiency of translation from the second ATG. Therefore, an additional A was added just before the second ATG to make two in-frame ATG codons. The construct was transformed into wild-type, pdv1-1, pdv1-2, and pdv1 pdv2 plants. To observe GFP-ARC5 in pdv2-1 plants, a transformed pdv1 pdv2 mutant was crossed with a pdv2 mutant, and we observed the next generation (pdv1/PDV1 pdv2/pdv2). The modified construct yielded a stronger fluorescence signal in transformed plants.
All constructs were transferred to Agrobacterium tumefaciens GV3101 and introduced into Arabidopsis plants as described (Vitha et al., 2001). T1 plants were selected by resistance to glufosinate and used for further analyses.
Fractionation and Immunoblot Analyses
Because we could barely detect GFP-PDV1 in leaf extracts by immunoblotting, most likely as a result of its low concentration, we used flower buds for immunoblot analyses. The flower buds were frozen in liquid nitrogen, ground with a pestle, and homogenized in extraction buffer (50 mM Tris, pH 7.6, 2 mM MgCl2, 5 mM EDTA, 100 mM NaCl, 300 mM sucrose, and a protease inhibitor mixture [P2714; Sigma-Aldrich]). The homogenate was filtered through Miracloth (Calbiochem), and the filtrate was used as total extract. The total extract was centrifuged at 20,000g for 20 min at 4°C to sediment the low-speed pellet. The supernatant fraction was centrifuged further at 100,000g for 1 h at 4°C to obtain a high-speed microsomal pellet and a supernatant fraction. For alkaline and detergent treatments, the low-speed pellet was resuspended with either 100 mM sodium carbonate, pH 11.5, or 1% Nonidet P-40 in the extraction buffer. After incubation for 1 h at 4°C, samples were centrifuged at 100,000g for 1 h at 4°C to separate soluble and insoluble fractions. Each fraction obtained from 3 mg of flower buds was suspended in Laemmli sample buffer (50 mM Tris, pH 6.8, 10% glycerol, 2% SDS, 5% 2-mercaptoethanol, and 0.01% bromophenol blue) and then subjected to immunoblot analyses.
To detect GFP-ARC5 by immunoblotting, we ground young expanding leaves with a pestle and directly suspended them in Laemmli sample buffer. Samples of 3 mg from leaves were subjected to immunoblot analyses.
All samples in Laemmli sample buffer were incubated at 65°C for 20 min, separated on 10% SDS-polyacrylamide gels, and transferred onto polyvinylidene difluoride membranes (Immobilon; Millipore). The membrane was blocked for 1 h at room temperature with blocking buffer (5% skim milk in Tris-buffered saline [TBS] plus 0.1% Tween 20) and incubated at room temperature overnight with anti-GFP mouse monoclonal antibody (JL-8; Clontech) diluted at 1:2000. After washing with TBS plus 0.1% Tween 20, the primary antibody was detected by horseradish peroxidase–conjugated goat anti-mouse antibody diluted at 1:10,000. After a 1-h incubation with secondary antibody at room temperature, the blots were washed with TBS plus 0.1% Tween 20 and the signal was detected by SuperSignal West Pico Chemiluminescent Substrate (Pierce Biotechnology).
In Vitro Chloroplast Import Assay
Total RNA was prepared from young leaves using an RNeasy Plant Mini kit (Qiagen). cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen) and oligo(dT) primer. cDNA of PDV1 and PDV1G272D was amplified using primers 5′-ATGGAGATCGAAGAAATCGAAGC-3′ and 5′-CTTAACCACGAGCCATCATTACG-3′ (PDV1) or 5′-CTTAATCACGAGCCATCATTACG-3′ (PDV1G272D) and then cloned into pGEM-T Easy.
PVD1, PVD1G272D, OEP14 (Li et al., 1991), precursors of Tic110-110N (Lübeck et al., 1997), and the small subunit of ribulose-1,5-bis-phosphate carboxylase/oxygenase (Olsen and Keegstra, 1992) were translated and radiolabeled using [35S]Met and the TNT Coupled Reticulocyte Lysate system (Promega) according to the manufacturer's protocol.
Intact chloroplasts were isolated from 8- to 12-d-old pea (Pisum sativum) seedlings and purified over a Percoll gradient as described previously (Bruce et al., 1994). Intact pea chloroplasts were resuspended in the import buffer (330 mM sorbitol and 50 mM HEPES/KOH, pH 8.0) at a concentration of 1 mg chlorophyll/mL. Import reactions were performed as described by Bruce et al. (1994). Import and extraction experiments were performed as described previously by Tranel et al. (1995).
Microscopy
For observation of chloroplast size and number, tips from expanding leaves were cut and fixed with 3.5% glutaraldehyde in water for 1 h at room temperature and then incubated in 0.1 M Na2-EDTA, pH 9.0, for 15 min at 50°C. Samples were analyzed with Nomarski differential interference contrast optics.
Localization of FtsZ in expanding leaves (∼10 mm) was examined by immunofluorescence microscopy using rabbit anti-peptide antibodies recognizing At FtsZ2-1, as described by Vitha et al. (2001).
GFP fluorescence was visualized without fixation in young expanding leaves (∼5 mm long) from ∼3-week-old plants and in flower buds with a Leica DMR A2 microscope (Leica Microsystems).
Accession Numbers
The GenBank accession numbers for the nucleotide sequences (Arabidopsis thaliana Col-0) described in this article are AB252216 (PDV1 genomic), AB252217 (PDV2 genomic), AB252218 (PDV1 cDNA), and AB252219 (PDV2 cDNA). The accession numbers for the sequence data shown in Figure 2 are as follows: Oryza sativa 1 (AP005924), Oryza sativa 2 (AP002835) Oryza sativa 3 (AP004743), Oryza sativa 4 (XM_476440), Physcomitrella patens 1 (BJ172680, BJ977778, BJ969477, and BJ159074 were assembled), Physcomitrella patens 2 (BJ597520), and Physcomitrella patens 3 (BJ952747).
Acknowledgments
We thank the ABRC for providing seeds of pdv2-1 and pdv2-2 and Yasuo Niwa (University of Shizuoka) for providing plasmids encoding an enhanced version of GFP (S65T). We are also grateful to Chieko Saito (RIKEN) and Laura Gilliland for technical suggestions regarding mutant screening and map-based cloning. This study was supported by a Japan Society for the Promotion of Science Postdoctoral Research Fellowship for Research Abroad (to S.-y.M.) and by grants from the National Science Foundation (to K.W.O. and J.E.F).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Katherine W. Osteryoung (osteryou@msu.edu).
References
- Aldridge, C., Maple, J., and Møller, S.G. (2005). The molecular biology of plastid division in higher plants. J. Exp. Bot. 56 1061–1077. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano, T., Yoshioka, Y., Kurei, S., Sakamoto, W., Sodmergen, and Machida, Y. (2004). A mutation of the CRUMPLED LEAF gene that encodes a protein localized in the outer envelope membrane of plastids affects the pattern of cell division, cell differentiation, and plastid division in Arabidopsis. Plant J. 38 448–459. [DOI] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19 137–144. [DOI] [PubMed] [Google Scholar]
- Bhar, D., Karren, M.A., Babst, M., and Shaw, J.M. (2006). Dimeric Dnm1–G385D interacts with Mdv1 on mitochondria and can be stimulated to assemble into fission complexes containing Mdv1 and Fis1. J. Biol. Chem. 281 17312–17320. [DOI] [PubMed] [Google Scholar]
- Bhattacharya, D., Yoon, H.S., and Hackett, J.D. (2004). Photosynthetic eukaryotes unite: Endosymbiosis connects the dots. Bioessays 26 50–60. [DOI] [PubMed] [Google Scholar]
- Bleazard, W., McCaffery, J.M., King, E.J., Bale, S., Mozdy, A., Tieu, Q., Nunnari, J., and Shaw, J.M. (1999). The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffey, S.A., and Lloyd, D. (1988). Division and Segregation of Organelles. (Cambridge, UK: Cambridge University Press).
- Bruce, B.D., Perry, S., Froehlich, J., and Keegstra, K. (1994). In vitro import of protein into chloroplasts. In Plant Molecular Biology Manual, S.B. Gelvin and R.B. Schilperoort, eds (Boston, MA: Kluwer Academic Publishers), pp. 1–15.
- Cavalier-Smith, T. (2004). Only six kingdoms of life. Proc. Biol. Sci. 271 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny, K.L., and Jensen, R.E. (2003). The WD-repeats of Net2p interact with Dnm1p and Fis1p to regulate division of mitochondria. Mol. Biol. Cell 14 4126–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny, K.L., McCaffery, J.M., and Jensen, R.E. (2001). Division of mitochondria requires a novel DMN1-interacting protein, Net2p. Mol. Biol. Cell 12 309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, W., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6 325–330. [DOI] [PubMed] [Google Scholar]
- Colletti, K.S., Tattersall, E.A., Pyke, K.A., Froelich, J.E., Stokes, K.D., and Osteryoung, K.W. (2000). A homologue of the bacterial cell division site-determining factor MinD mediates placement of the chloroplast division apparatus. Curr. Biol. 10 507–516. [DOI] [PubMed] [Google Scholar]
- Fekkes, P., Shepard, K.A., and Yaffe, M.P. (2000). Gag3p, an outer membrane protein required for fission of mitochondrial tubules. J. Cell Biol. 151 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, H., Kadirjan-Kalbach, D., Froehlich, J.E., and Osteryoung, K.W. (2003). ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc. Natl. Acad. Sci. USA 100 4328–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin, E.E., Graumann, J., and Chan, D.C. (2005). The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J. Cell Biol. 170 237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, H. (1986). Double-ring structure around the constricting neck of dividing plastids of Avena sativa. Protoplasma 135 166–172. [Google Scholar]
- Haswell, E.S., and Meyerowitz, E.M. (2006). MscS-like proteins control plastid size and shape in Arabidopsis thaliana. Curr. Biol. 16 1–11. [DOI] [PubMed] [Google Scholar]
- Itoh, R., Fujiwara, M., Nagata, N., and Yoshida, S. (2001). A chloroplast protein homologous to the eubacterial topological specificity factor minE plays a role in chloroplast division. Plant Physiol. 127 1644–1655. [PMC free article] [PubMed] [Google Scholar]
- Karren, M.A., Coonrod, E.M., Anderson, T.K., and Shaw, J.M. (2005). The role of Fis1p-Mdv1p interactions in mitochondrial fission complex assembly. J. Cell Biol. 171 291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4 403–410. [DOI] [PubMed] [Google Scholar]
- Kuroiwa, H., Mori, T., Takahara, M., Miyagishima, S., and Kuroiwa, T. (2002). Chloroplast division machinery as revealed by immunofluorescence and electron microscopy. Planta 215 185–190. [DOI] [PubMed] [Google Scholar]
- Kuroiwa, T., Kuroiwa, H., Sakai, A., Takahashi, H., Toda, K., and Itoh, R. (1998). The division apparatus of plastids and mitochondria. Int. Rev. Cytol. 181 1–41. [DOI] [PubMed] [Google Scholar]
- Li, H.M., Moore, T., and Keegstra, K. (1991). Targeting of proteins to the outer envelope membrane uses a different pathway than transport into chloroplasts. Plant Cell 3 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lübeck, J., Heins, L., and Soll, J. (1997). A nuclear-encoded chloroplastic inner envelope membrane protein uses a soluble sorting intermediate upon import into the organelle. J. Cell Biol. 137 1279–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas, A., van Dyke, M., and Stock, J. (1991). Predicting coiled coils from protein sequences. Science 252 1162–1164. [DOI] [PubMed] [Google Scholar]
- Maple, J., Aldrige, C., and Møller, S.G. (2005). Plastid division is mediated by combinatorial assembly of plastid division proteins. Plant J. 43 811–823. [DOI] [PubMed] [Google Scholar]
- Maple, J., Chua, N.H., and Møller, S.G. (2002). The topological specificity factor AtMinE1 is essential for correct plastid division site placement in Arabidopsis. Plant J. 31 269–277. [DOI] [PubMed] [Google Scholar]
- Maple, J., Fujiwara, M.T., Kitahata, N., Lawson, T., Baker, N.R., Yoshida, S., and Møller, S.G. (2004). GIANT CHLOROPLAST 1 is essential for correct plastid division in Arabidopsis. Curr. Biol. 14 776–781. [DOI] [PubMed] [Google Scholar]
- Margolin, W. (2005). FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell Biol. 6 862–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrison, J.L., Rutherford, S.M., Robertson, E.J., Lister, C., Dean, C., and Leech, R.M. (1999). The distinctive roles of five different ARC genes in the chloroplast division process in Arabidopsis. Plant J. 18 651–662. [DOI] [PubMed] [Google Scholar]
- McAndrew, R.S., Froehlich, J.E., Vitha, S., Stokes, K.D., and Osteryoung, K.W. (2001). Colocalization of plastid division proteins in the chloroplast stromal compartment establishes a new functional relationship between FtsZ1 and FtsZ2 in higher plants. Plant Physiol. 127 1656–1666. [PMC free article] [PubMed] [Google Scholar]
- Mita, T., Kanbe, T., Tanaka, K., and Kuroiwa, T. (1986). A ring structure around the dividing plane of the Cyanidium caldarium chloroplast. Protoplasma 130 211–213. [Google Scholar]
- Miyagishima, S., Itoh, R., Toda, K., Takahashi, H., Kuroiwa, H., and Kuroiwa, T. (1998). Identification of a triple ring structure involved in plastid division in the primitive red alga Cyanidioschyzon merolae. J. Electron Microsc. (Tokyo) 47 269–272. [Google Scholar]
- Miyagishima, S., Nishida, K., and Kuroiwa, T. (2003. a). An evolutionary puzzle: Chloroplast and mitochondrial division rings. Trends Plant Sci. 8 432–438. [DOI] [PubMed] [Google Scholar]
- Miyagishima, S., Nishida, K., Mori, T., Matsuzaki, M., Higashiyama, T., Kuroiwa, H., and Kuroiwa, T. (2003. b). A plant-specific dynamin-related protein forms a ring at the chloroplast division site. Plant Cell 15 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima, S., Wolk, C.P., and Osteryoung, K.W. (2005). Identification of cyanobacterial cell division genes by comparative and mutational analyses. Mol. Microbiol. 56 126–143. [DOI] [PubMed] [Google Scholar]
- Mori, T., Kuroiwa, H., Takahara, M., Miyagishima, S., and Kuroiwa, T. (2001). Visualization of an FtsZ ring in chloroplasts of Lilium longiflorum leaves. Plant Cell Physiol. 42 555–559. [DOI] [PubMed] [Google Scholar]
- Mozdy, A.D., McCaffery, J.M., and Shaw, J.M. (2000). Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor, K., Ingerman, E., Okreglak, V., Marino, M., Hinshaw, J.E., and Nunnari, J. (2006). Mdv1 interacts with assembled dnm1 to promote mitochondrial division. J. Biol. Chem. 281 2177–2183. [DOI] [PubMed] [Google Scholar]
- Okamoto, K., and Shaw, J.M. (2005). Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 39 503–536. [DOI] [PubMed] [Google Scholar]
- Olsen, L.J., and Keegstra, K. (1992). The binding of precursor proteins to chloroplast requires nucleoside triphosphates in the intermembrane space. J. Biol. Chem. 267 433–439. [PubMed] [Google Scholar]
- Osteryoung, K.W., and Nunnari, J. (2003). The division of endosymbiotic organelles. Science 302 1698–1704. [DOI] [PubMed] [Google Scholar]
- Osteryoung, K.W., Stokes, K.D., Rutherford, S.M., Percival, A.L., and Lee, W.Y. (1998). Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell 10 1991–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung, K.W., and Vierling, E. (1995). Conserved cell and organelle division. Nature 376 473–474. [DOI] [PubMed] [Google Scholar]
- Pyke, K.A. (1999). Plastid division and development. Plant Cell 11 549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke, K.A., and Leech, R.M. (1994). A genetic analysis of chloroplast division and expansion in Arabidopsis thaliana. Plant Physiol. 104 201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud, C., Cassier-Chauvat, C., Perennes, C., and Bergounioux, C. (2004). An Arabidopsis homolog of the bacterial cell division inhibitor SulA is involved in plastid division. Plant Cell 16 1801–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud, C., Perennes, C., Reuzeau, C., Catrice, O., Brown, S., and Bergounioux, C. (2005). Cell and plastid division are coordinated through the prereplication factor AtCDT1. Proc. Natl. Acad. Sci. USA 102 8216–8221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, E.J., Rutherford, S.M., and Leech, R.M. (1996). Characterization of chloroplast division using the Arabidopsis mutant arc5. Plant Physiol. 112 149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke, R., Schneider, A., van der Graaff, E., Fischer, K., Catoni, E., Desimone, M., Frommer, W.B., Flugge, U.I., and Kunze, R. (2003). ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, I., Tobin, A.K., and Logan, D.C. (2006). BIGYIN, an orthologue of human and yeast FIS1 genes functions in the control of mitochondrial size and number in Arabidopsis thaliana. J. Exp. Bot. 57 1275–1280. [DOI] [PubMed] [Google Scholar]
- Sesaki, H., and Jensen, R.E. (1999). Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147 699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, H., Koizumi, M., Kuroki, K., Mochizuki, M., Fujimoto, H., Ohta, H., Masuda, T., and Takamiya, K. (2004). ARC3, a chloroplast division factor, is a chimera of prokaryotic FtsZ and part of eukaryotic phosphatidylinositol-4-phosphate 5-kinase. Plant Cell Physiol. 45 960–967. [DOI] [PubMed] [Google Scholar]
- Strepp, R., Scholz, S., Kruse, S., Speth, V., and Reski, R. (1998). Plant molecular gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc. Natl. Acad. Sci. USA 95 4368–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu, Q., and Nunnari, J. (2000). Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 151 353–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu, Q., Okreglak, V., Naylor, K., and Nunnari, J. (2002). The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J. Cell Biol. 158 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranel, P.J., Froehlich, J., Goyal, A., and Keegstra, K. (1995). A component of the chloroplastic protein import apparatus is targeted to the outer envelope membrane via a novel pathway. EMBO J. 14 2436–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha, S., Froehlich, J.E., Koksharova, O., Pyke, K.A., van Erp, H., and Osteryoung, K.W. (2003). ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell 15 1918–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha, S., McAndrew, R.S., and Osteryoung, K.W. (2001). FtsZ ring formation at the chloroplast division site in plants. J. Cell Biol. 153 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, E., Kim, P.S., and Berger, B. (1997). MultiCoil: A program for predicting two- and three-stranded coiled coils. Protein Sci. 6 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]