Abstract
Three genes, BE1, BE2, and BE3, which potentially encode isoforms of starch branching enzymes, have been found in the genome of Arabidopsis thaliana. Although no impact on starch structure was observed in null be1 mutants, modifications in amylopectin structure analogous to those of other branching enzyme II mutants were detected in be2 and be3. No impact on starch content was found in any of the single mutant lines. Moreover, three double mutant combinations were produced (be1 be2, be1 be3, and be2 be3), and the impact of the mutations on starch content and structure was analyzed. Our results suggest that BE1 has no apparent function for the synthesis of starch in the leaves, as both be1 be2 and be1 be3 double mutants display the same phenotype as be2 and be3 separately. However, starch synthesis was abolished in be2 be3, while high levels of α-maltose were assayed in the cytosol. This result indicates that the functions of both BE2 and BE3, which belong to class II starch branching enzymes, are largely redundant in Arabidopsis. Moreover, we demonstrate that maltose accumulation depends on the presence of an active ADP-glucose pyrophosphorylase and that the cytosolic transglucosidase DISPROPORTIONATING ENZYME2, required for maltose metabolization, is specific for β-maltose.
INTRODUCTION
Starch is a polymer of d-glucose that accumulates in the plastids of plants as huge water-insoluble granules. Glucose residues are linked together by α-1,4 and α-1,6 O-glycosidic bonds. The α-1,6 linkages are commonly referred to as the branch points, while the α-1,4 O-glycosidic bonds compose the linear backbone of the polymer. The starch granule is composed of two structurally distinct homopolymers: amylose, which is essentially linear, and amylopectin, which is a moderately branched macromolecule (usually 6% of α-1,6 bonds within the polymer). The asymmetrical distribution of the branch points within amylopectin is responsible for the cluster organization of the molecule that further adopts a semicrystalline structure (Hizukuri, 1986; for review, see Buléon et al., 1998).
The synthesis pathway of amylopectin is rather complex and involves different enzymatic activities usually supported by multiple and genetically distinct forms of enzymes. This pathway includes starch synthases (elongation step), starch branching enzymes (introduction of branches), starch debranching enzymes, and α-1,4-glucanotransferases (maturation of the structure required for further growth and crystallization of the granule) (for review, see Myers et al., 2000; Ball and Morell, 2003).
Starch branching enzymes (SBEs; EC 2.4.1.18; CAZy family GH13) catalyze the formation of the α-1,6 linkages within the polymer. SBEs first cleave a preexisting α-1,4 linkage and transfer the fragment initially positioned at the nonreducing end of the cleaved glucan to an α-1,6 position through an intra- or intermolecular mechanism (for review, see Sivak and Preiss, 1998). The consequence of this reaction is to increase the number of nonreducing ends within the molecule, thus facilitating the elongation by the starch synthases. Two to three genetically independent isoforms of SBEs were observed in most plants (Fisher et al., 1996a; Larsson et al., 1996, 1998; Morell et al., 1997; Mizuno et al., 1992; Nakamura et al., 1992). Depending on their peptide sequence, these SBEs were categorized into two classes named SBEI (or B family) and SBEII (or A family). Some of these forms, essentially maize (Zea mays) and potato (Solanum tuberosum) SBEs, were kinetically characterized after their purification from plant tissues or their heterologous expression in Escherichia coli (Guan and Preiss, 1993; Takeda et al., 1993; Guan et al., 1994; Rydberg et al., 2001). From these studies, it appears that SBEI is essentially active on long linear chains (of amylose type), while SBEII is more active on the shorter chains of amylopectin. Moreover, SBEI preferentially transfers longer chains than SBEII.
Mutant lines defective for SBEI, IIa, and IIb in maize (Stinard et al., 1993; Blauth et al., 2001, 2002; Yao et al., 2004) and rice (Oryza sativa; Nishi et al., 2001; Nakamura, 2002; Satoh et al., 2003) were isolated and analyzed for their ability to synthesize starch. The absence of SBEI in both plants has little or no impact on starch that accumulates in the endosperm, while the absence of SBEIIb leads to the synthesis of a modified amylopectin with a strong decrease in the number of short chains and an increase in the number of long chains. On the other hand, the absence of SBEIIa in maize leads only to the modification of the structure of starch that accumulates in the leaf (transitory starch) without impact on the endosperm starch (storage starch). In Arabidopsis thaliana, two SBEs belonging to class II were cloned (Fisher et al., 1996b), and their expression levels were characterized (Khoshnoodi et al., 1998). The precise function of these two forms of SBEII remains unknown. A third gene putatively encoding an SBE was observed in the nuclear genome of this plant. However, the corresponding protein does not belong to a type I class of SBEs as described in other plants.
The objective of this work was to investigate the function of the three SBEs (BE1, BE2, and BE3) in the leaves of Arabidopsis through the selection and phenotypic characterization of mutants specifically defective in each of the three isoforms. Moreover, we have constructed double mutant combinations defective in two out of the three SBEs. Our results indicate that (1) BE1 has no apparent function in the metabolism of starch in Arabidopsis leaves; (2) BE2 and BE3, which belong to class II of SBEs, are required for the synthesis of normal starch, and their activities are largely redundant; (3) the double mutant line simultaneously defective in both BE2 and BE3 substitutes starch by the accumulation of α-maltose in the cytosol; (4) the latter is generated through degradation of abnormal products of the starch synthesis pathway; and (5) DISPROPORTIONATING ENZYME2 (DPE2) is specific for β-maltose, thereby explaining the rationale of β-amylolysis as the major route of starch degradation in plant leaves.
RESULTS
The Nuclear Genome of Arabidopsis Contains Three Candidate Genes Putatively Encoding SBEs
The three genes putatively encoding SBEs (EC 2.4.1.18; CAZy family GH13) in the Arabidopsis genome are BE1, BE2, and BE3 (Table 1). These genes are expressed in the leaves of Arabidopsis during either short (8 h) or normal (12 h) daylengths (http://www.starchmetnet.org/Datapages/AtBE2/AtBE2Frameset.htm), although the expression of BE1 is very low and rather constant throughout the cycle when compared with those of BE2 and BE3. The transcript level of BE3 increases during the illuminated period to reach a maximum at the transition between day and night. It then regularly falls to a minimum at the end of the dark period. BE2 transcript level is maximal 4 h after light onset and slowly decreases to a minimum near the end of the dark phase.
Table 1.
Description of the Different Arabidopsis Mutant Alleles of SBEs Used in This Work
| Gene | AGI Gene Identificationa | Allele and Line Identification | Position of T-DNAb | Mutant Collection | Genetic Background |
|---|---|---|---|---|---|
| BE1 | At3g20440 | be1-1 (DYK140) | +6461 (intron 20) | Génoplantec | Ws |
| be1-2 (N637880) | +5592 (intron 15) | NASCd | Col-0 | ||
| BE2 | At5g03650 | be2-1 (EFH20) | +5022 (intron 19) | Génoplante | Ws |
| be2-2 (DSA16) | +5487 (exon 22) | Génoplante | Ws | ||
| BE3 | At2g36390 | be3-1 (N548089) | +3262 (exon 10) | NASC | Col-0 |
| be3-2 (EQJ13) | +4485 (exon 15) | Génoplante | Ws |
AGI, Arabidopsis Genome Initiative.
Position of T-DNA is given from the start codon.
Génoplante: http://genoplante-info.infobiogen.fr/FLAGdb/.
Nottingham Arabidopsis Stock Centre: http://nasc.nott.ac.uk/.
The proteins corresponding to the BE1, BE2, and BE3 genes contain a predicted chloroplast-targeting peptide of 49, 61, and 37 amino acids, respectively (ChloroP, http://www.cbs.dtu.dk/services/ChloroP/; Emanuelsson et al., 1999), thereby suggesting that these proteins are likely to be localized within the chloroplast. BE2 and BE3 are two highly conserved proteins with >75% amino acid identity. The relatively well-conserved intron-exon organization in 5′ parts of both BE2 and BE3 genes (Figure 1) suggests that these two genes probably arose from a duplication event. By comparison, the sequence homology between BE1 and both BE2 and BE3 can be considered as low (only 28% and 27% amino acid identity, respectively), suggesting a different ancestral origin of the corresponding gene. Protein sequence comparisons with SBEs from other plant sources indicate that BE2 and BE3 belong to the class A of SBEs, while BE1 cannot be easily classified within plant SBEs. BE1 is not related to the standard plant A- or B-type SBE families but shows more relatedness to the glycogen-branching enzymes from fungi and animals, although the protein sequence similarity is quite low in that case too (<30%). The situation in Arabidopsis is thus different from that observed in other dicots, such as potato, pea (Pisum sativum), and bean (Phaseolus vulgaris), where only one type A of SBE can be found along with one type B of SBE (see dendrogram of branching enzyme families in Supplemental Figure 1 online; for review, see Ball and Morell, 2003).
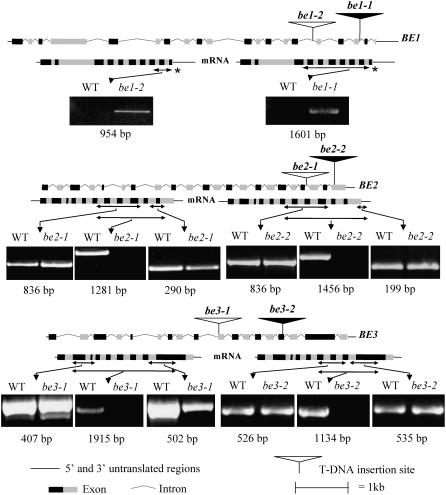
Figure 1.
Molecular Organization of SBE Mutant Alleles and Impact of Insertional Mutations on Transcript Expression.
Both genomic DNA and mRNA schematics are presented for each SBE gene of Arabidopsis with the T-DNA insertion sites. Double-sided arrows indicate mRNA regions amplified by RT-PCR for the different primer pairs designed for each T-DNA insertion (see supplemental data online for primer pair sequences). Agarose gels performed after RT-PCR amplifications are shown below the mRNA structures. * Double-sided arrows are not to scale.
Selection of Insertion Mutations in Genes BE1, BE2, and BE3
Several T-DNA insertion mutant lines for each of the three SBEs were selected from different collections (Génoplante and SALK; Figure 1, Table 1), and the corresponding homozygous mutants were identified for further analysis.
We have investigated the ability of the mutant lines to further express the mRNA corresponding to the SBE genes by RT-PCR. Amplifications were performed on RT products upstream, downstream, and spanning the insertion site of the T-DNA (Figure 1) for both be2 and be3 mutant alleles. The correct amplicons were obtained with the wild-type allele in all cases. However, RT-PCR performed on mutant alleles have shown that no amplification was obtained for the region spanning the insertion site of the T-DNA, although RT-PCR amplicons with the expected size were generated with regions upstream and downstream of the T-DNA insertion site. It is therefore unlikely that these modified transcripts could be functional and lead to the production of active proteins. In the case of be1 mutant alleles, RT-PCR analyses were performed between primers corresponding to the T-DNA and the gene-specific sequences (Figure 1). While an amplicon with the correct expected size was produced in both be1-1 and be1-2 mutants, this amplicon was missing in the wild-type line. The presence of the T-DNA within the transcript is likely to abolish its function in vivo.
We have constructed all three combinations of double mutant lines defective for two out of the three SBEs. The different double mutant combinations were produced with the Wassilewskija (Ws) genetic background and were as follow: be1-1 be2-1, be1-1 be3-2, and be2-1 be3-2. These double mutants were analyzed together with the single mutants for their growth phenotype and for starch accumulation and structure.
Mutations in BE2 and BE3 Abolish Two Distinct SBE Activities
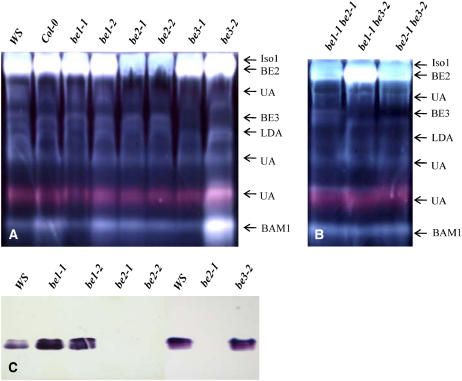
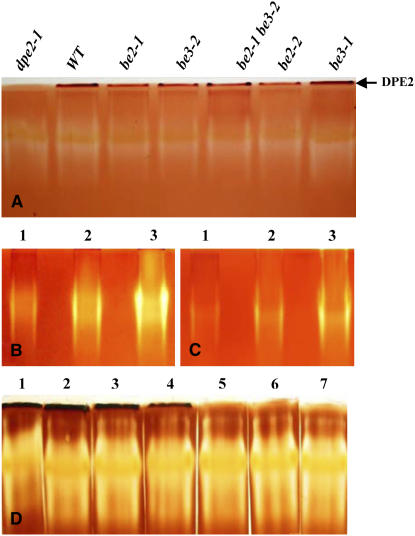
SBE activities in the different mutant backgrounds were assayed by two different approaches: zymograms and in vitro assays. Zymogram analyses were performed in two different ways: either using the ability of branching enzymes to stimulate phosphorylase “a” activity or through the iodine staining of the modified starch structure produced by branching enzymes (see Methods for more details about the different methods used for the characterization of the SBE activities). Only one activity was observed when the phosphorylase “a” stimulation technique was used (Figure 2B). The corresponding band was missing in both be2-1 and be2-2 mutants but was unaffected by the mutations at BE1 and BE3 loci. This result was confirmed by in vitro assay of SBE activities using the phosphorylase “a” stimulation method. In that case, no SBE activity was measured in the be2 mutant background, whereas the SBE activity level was equivalent to that of the wild type in be1 and be3 mutant backgrounds (Table 2). SBE activity was also assayed by an iodine-stained starch containing zymogram (Figure 2A). A clear band in the top part of the gel was systematically missing in all be2 mutant backgrounds, while a band in the middle part of the gel was missing in all be3 mutant backgrounds. The clear band that disappeared in line be2 might be a consequence of the combined action of both Iso1 (Wattebled et al., 2005) and BE2 on the starch contained in the gel. Because both activities were not always fully resolved with this zymogram procedure, the progressive release in the incubation buffer of small malto-oligosaccharides (MOS) is expected as combined products of both enzymes activities (branching enzyme produces small branched glucans that are debranched by the debranching enzyme), thereby leading to a clear band when the gel was stained with iodine. However, when both Iso1 and BE2 were better separated on the gel, the BE2 activity adopts its typical pink stain as expected for a branching enzyme (see Supplemental Figure 2 online). A pink-colored band was also observed for the BE3 activity on the starch-containing gel (see Supplemental Figure 2 online). However, despite several attempts, no activity corresponding to BE1 protein was observed in the conditions used in this work.
Figure 2.
Zymograms of SBE Activities.
(A) and (B) Starch-containing gels. After migration of 100 μg of proteins under native conditions, gels were incubated overnight at room temperature in 50 mM Tris-HCl, pH 7.0, 1 mM DTT, 1 mM CaCl2, and 1 mM MgCl2. Starch-modifying enzymes were observed by staining the gel with lugol solution.
(C) Stimulation of phosphorylase “a” activity. After the migration of 100 μg of proteins under native conditions, the gel was incubated overnight at room temperature in 30 mL of incubation buffer containing glucose-1-phosphate (G1P) at 50 mM and 28 units of rabbit muscle phosphorylase “a.” SBE activities were revealed after soaking the gel in lugol solution. Ws and Col-0 were the wild-type references. UA, unknown activity; LDA, limit dextrinase corresponds to gene At5g04360 as already reported by Wattebled et al. (2005); Iso1, isoamylase activity (ISA1 and ISA2 gene products as described by Wattebled et al. (2005); BAM1 corresponds to gene At4g15210 encoding a β-amylase.
Table 2.
In Vitro Assays of Several Starch-Metabolizing Enzymes Performed with Leaf Crude Extracts of Wild-Type (Ws and Col-0) and Single and Double Mutant Lines Defective for SBE Activities
| SBE | AGPase | AGPase (+ 3-PGA)a | Soluble Starch Synthase | α-Amylase | β-Amylase | d-Enzyme | Pullulanase | Starch-Phosphorylase | |
|---|---|---|---|---|---|---|---|---|---|
| Ws | 107 ± 19 | 3.3 ± 0.2 | 22.9 ± 0.2 | 33.2 ± 0.2 | 55 ± 1 | 147 ± 9 | 63 ± 3 | 13 ± 1 | 5.9 ± 0.1 |
| Col-0 | 107 ± 24 | 3.9 ± 0.2 | 28.7 ± 0.9 | 20.1 ± 0.2 | 37 ± 1 | ND | ND | 8 ± 1 | 2.8 ± 0.1 |
| be1-1 (Ws) | 96 ± 8 | 3.0 ± 0.2 | 25.6 ± 0.7 | 22.0 ± 0.3 | 41 ± 1 | ND | ND | 12 ± 1 | 9.7 ± 0.1 |
| be1-2 (Col-0) | 130 ± 11 | 7.3 ± 0.1 | 34.2 ± 0.3 | 30.9 ± 0.8 | 36 ± 1 | ND | ND | 13 ± 3 | 4.5 ± 0.1 |
| be2-1 (Ws) | 2 ± 1 | 3.9 ± 1.1 | 16.8 ± 1.5 | 50.8 ± 0.5 | 36 ± 1 | ND | ND | 8 ± 2 | 7.8 ± 0.1 |
| be2-2 (Ws) | 1 ± 0 | 4.8 ± 0.1 | 29.9 ± 0.1 | 20.7 ± 0.3 | 52 ± 1 | ND | ND | 8 ± 2 | 6.0 ± 0.1 |
| be3-1 (Col-0) | 82 ± 16 | 3.9 ± 0.3 | 25.6 ± 0.6 | 29.7 ± 0.6 | 74 ± 1 | ND | ND | 13 ± 2 | 5.1 ± 0.1 |
| be3-2 (Ws) | 102 ± 5 | 3.1 ± 0.4 | 21.2 ± 0.6 | 20.8 ± 1.0 | 39 ± 1 | ND | ND | 7 ± 2 | 7.7 ± 0.3 |
| be1-1 be2-1 | 1 ± 0 | 2.9 ± 0.1 | 24.7 ± 0.6 | 26.0 ± 0.8 | ND | ND | ND | 9 ± 1 | 14.0 ± 0.3 |
| be1-1 be3-2 | 51 ± 17 | 3.1 ± 0.3 | 28.1 ± 0.4 | 54.3 ± 1.4 | 55 ± 1 | ND | ND | 10 ± 1 | 5.4 ± 0.1 |
| be2-1 be3-2 | 2 ± 1 | 4.4 ± 0.2 | 27.7 ± 0.4 | 31.0 ± 0.6 | 69 ± 2 | 146 ± 7 | 39 ± 1 | 19 ± 1 | 20.2 ± 0.1 |
Activities are expressed in nmol·min−1·mg−1 of proteins (means ± se; n = 3; independent samples). AGPase = ADP-glucose pyrophosphorylase. ND, not determined.
3 mM of 3-phosphoglyceraldehyde (3-PGA) was added to the incubation buffer.
Other starch metabolizing activities, including ADP-glucose pyrophosphorylase, starch synthases, starch phosphorylases (STPases), α-1,4 glucanotransferases, α- and β-amylases, and pullulanase, were assayed from leaf extracts. None of these activities were significantly affected by the different mutations except for starch phosphorylases. Indeed, in the be2-1 be3-2 double mutant, STPase activity was three to four times higher than in the corresponding wild-type line (Ws) (Table 2). This behavior was confirmed by zymogram analysis for STPase activities where both the cytosolic and the plastidial forms of STPase were increased (see Supplemental Figure 4 online).
Impact of the Mutations on Starch Accumulation Levels and the Growth Phenotype
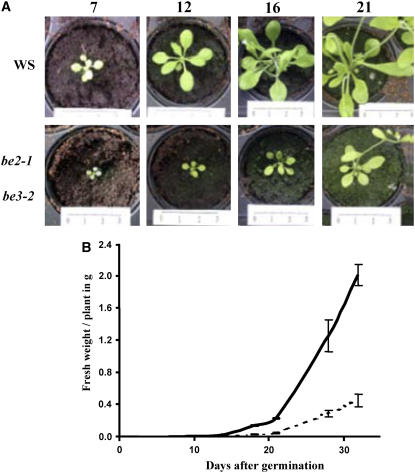
We measured the leaf starch and water-soluble glucan (WSGs) contents at the end of the light period (16 h day/8 h night) in the different single and double mutants and compared them with that of the corresponding wild-type ecotypes (Table 3). We did not detect any significant alteration of the starch content in the single mutants or in both be1-1 be2-1 and be1-1 be3-2 double mutants. However, the be2-1 be3-2 double mutant was free of starch (see Supplemental Figure 6 online). The absence of starch was coupled with the accumulation of very high levels of WSGs (whatever extraction method was used) that were not observed in other lines (Tables 3 and 4). Moreover, this double mutant displayed a lower growth rate, a reduced size of the mature plant, a pale color, and a general wilting of the inflorescence (Figure 3; see Supplemental Figure 5 online). Thirty days after seed germination, the fresh weight of the above-ground organs of the double mutant was only one-fifth of the wild type (Figure 3) under the 16-h-day/8-h-night growth conditions used during this work. The same lower growth rate was observed under the 12-h-day/12-h-night regime, although it was less pronounced. Both dpe2 and mex1 mutants of Arabidopsis that have lost their ability to metabolize maltose display such a dwarf-like phenotype. However, both mutants exhibit a starch excess phenotype and are not affected for polysaccharide biosynthesis. Despite the growth phenotype, the be2-1 be3-2 double mutant was still able to produce siliques and viable seeds after self-pollination, although the flowering rate was low when compared with that of other lines.
Table 3.
Leaf Starch Accumulation in Wild-Type (Ws and Col-0) and SBE Mutant Lines
| Starch Amount in Wild Type (%)a | λmax of Amylopectin (nm) | Amylose Ratio (%) | Branching Level of Amylopectin (%) | WSG Content (mg·g−1 of FW) | |
|---|---|---|---|---|---|
| Ws | 100 | 555 ± 1 (n = 8) | 24 ± 4 (n = 6) | 4.9 (n = 2) | 0.28 ± 0.14 (n = 8) |
| Col-0 | 100 | 547 ± 4 (n = 3) | 21 ± 3 (n = 3) | ND | 0.34 ± 0.29 (n = 3) |
| be1-1 (Ws) | 107 ± 13 (n = 9) | 556 ± 2 (n = 6) | 23 ± 7 (n = 4) | ND | 0.27 ± 0.06 (n = 4) |
| be1-2 (Col-0) | 109 ± 21 (n = 3) | 545 ± 5 (n = 3) | 27 ± 2 (n = 3) | ND | 0.32 (n = 1) |
| be2-1 (Ws) | 86 ± 10 (n = 7) | 564 ± 5 (n = 4) | 28 ± 5 (n = 3) | 4.8 (n = 2) | 0.21 ± 0.09 (n = 3) |
| be2-2 (Ws) | 99 ± 16 (n = 10) | 561 ± 2 (n = 6) | 26 ± 4 (n = 5) | 5.6 (n = 2) | 0.20 ± 0.14 (n = 5) |
| be3-1 (Col-0) | 78 ± 14 (n = 3) | 556 ± 0 (n = 3) | 24 ± 4 (n = 3) | ND | 0.03 ± 0.02 (n = 3) |
| be3-2 (Ws) | 92 ± 12 (n = 10) | 559 ± 2 (n = 7) | 27 ± 6 (n = 5) | 4.9 (n = 1) | 0.17 ± 0.07 (n = 6) |
| be1-1 be2-1 | 122 ± 10 (n = 3) | 557 ± 2 (n = 3) | 29 ± 8 (n = 3) | ND | 0.15 ± 0.06 (n = 3) |
| be1-1 be3-2 | 120 ± 17 (n = 5) | 555 ± 2 (n = 3) | 27 ± 4 (n = 3) | ND | 0.20 ± 0.12 (n = 5) |
| be2-1 be3-2 | Not detected (n = 3) | NA | NA | NA | 11.47 ± 5.48 (n = 4) |
The results are the average of several independents experiments performed on independent samples (means ± se or sd when n = 2). Starch and WSGs were extracted at the end of the illuminated period (16-h-day/8-h-night growth rhythm). The amounts of starch, amylose, and WSG were determined by the amyloglucosidase assays. The λmax and the branching level of amylopectin and the ratio of amylose were measured after purification through size exclusion chromatography on sepharose CL-2B columns. NA, not applicable; ND, not determined.
Average starch contents for Ws (n = 12) and Col-0 (n = 4) are 9.9 ± 3.7 and 15.0 ± 4.6 mg·g−1 of fresh weight (FW), respectively.
Table 4.
Comparison of Starch, WSG, and Sugar Contents in the Wild-Type (Ws) and Mutant Lines
| Line | Starch | WSGa | WSGb | Glucosec | Fructosec | Sucrosec | Glc-1-Pc | Glc-6-Pc |
|---|---|---|---|---|---|---|---|---|
| Wild type (Ws) | 6.7 ± 0.2 | 0.1 ± 0.01 | 0.05 ± 0.04 | 0.03 ± 0.01 | 0.02 ± 0.01 | 0.29 ± 0.06 | 0.004 ± 0.004 | 0.03 ± 0.01 |
| adg1 (Ws) | 1.4 ± 0.1* | Not detected | 0.10 ± 0.01 | 0.06 ± 0.01* | 0.02 ± 0.02 | 0.31 ± 0.05 | 0.011 ± 0.006 | 0.04 ± 0.02 |
| be2-1 be3-2 | Not detected | 12.5 ± 0.03* | 4.53 ± 0.8* | 0.54 ± 0.20* | 0.15 ± 0.10 | 0.29 ± 0.08 | 0.008 ± 0.007 | 0.04 ± 0.02 |
| adg1 be2-1 be3-2 | Not detected | 0.46 ± 0.03 | 0.02 ± 0.01* | 0.15 ± 0.05* | 0.03 ± 0.01 | 0.46 ± 0.09* | 0.011 ± 0.006 | 0.05 ± 0.01 |
Starch, WSG, sugar, and hexose-P contents are expressed in mg·g−1 of FW (means ± se; n = 3 independent samples) and were measured at the end of the light phase on plants cultured under 16-h-light/8-h-dark conditions. * P value < 0.01 (confidence interval of 95%).
Determined after immediate boiling of the sample formerly homogenized in an electric blender in MOPS buffer.
Determined after perchloric acid extraction.
Determined after TCA/diethylether extraction as described in Methods.
Figure 3.
Comparison of Growth Rates of the Wild Type (Ws) and be2-1 be3-2.
(A) Pictures of wild-type and mutant lines were taken periodically after seed germination. Pictures are shown at the same scale, thus allowing direct comparison of wild-type and mutant plants.
(B) Above-ground organs were harvested at 7, 10, 14, 18, 21, 28, and 32 d after germination and immediately weighed. Continuous line = the wild type (Ws); dashed line = be2-1 be3-2 mutant. Vertical bars represent se of three independent measures.
Structural Analysis of Starch and WSG
Whenever possible, we have compared the structures of the starches from the single and double mutants with those of the corresponding wild-type ecotypes. After extraction and purification, the size and morphology of the starch granules were analyzed on ultrathin slices by transmission electron microscopy. The starch granules in be2 and be3 mutants were on average slightly larger than that of the wild type, while no differences were seen for the be1 mutant (data not shown). Wide-angle x-ray diffraction analysis was performed on purified starches from be1-1, be2-1, and be3-2 mutants. No difference was observed when compared with the wild type. All starch samples were of the B crystalline-type and displayed ∼37% level of crystallinity.
Starches purified from the different mutant backgrounds were submitted to size exclusion chromatography on a sepharose CL-2B column to purify amylopectin and measure the amount of amylose. The amount of amylose and the branching level of amylopectin were not significantly different in the mutants when compared with the wild type (Table 3).
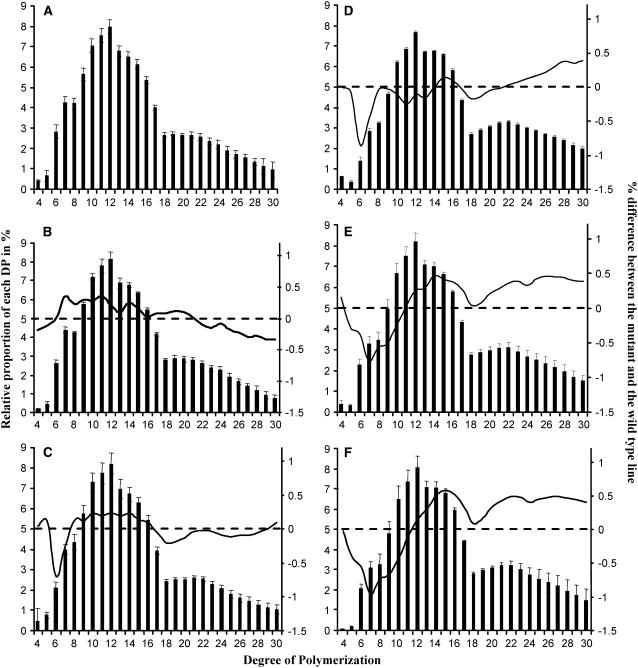
We have established the chain length (CL) distribution profiles of the amylopectin from each single and double mutant. Average profiles produced from several independent analyses are presented in Figure 4. For better clarity, we have combined profiles from mutant lines in both Ws and Columbia (Col-0) genetic backgrounds since individual profiles were the same whatever the genetic background (CL profiles were the same for Ws and Col-0 samples). Both be2 and be3 mutants displayed more or less the same profiles that were slightly different from those of the wild type (Figures 4A, 4C, and 4D). DP 6-7 and DP 5-9 chains were slightly decreased in be2 and be3, respectively, while DP 10-16 chains were slightly increased. No difference was evidenced for be1 in comparison with the wild type (Figure 4B). be1-1 be2-1 (Figure 4E) and be1-1 be3-2 (Figure 4F) double mutants showed the same profiles as the corresponding be2 and be3 single mutants, respectively.
Figure 4.
CL Distribution Profiles of Wild-Type and SBE Mutants.
After purification and enzymatic debranching of amylopectin, the linear glucans were condensed to 8-amino-1,3,6 pyrenetrisulfonic acid (APTS) and subjected to capillary electrophoresis that allows the separation of glucans that differ by only one glucose unit in size. The level of APTS fluorescence is quantitative and indicates the proportion of each glucan in the mix. The diagrams represent the percentage (y axis on the left) of each glucan (x axis) in the total population of glucans that can be detected for each sample. Thin vertical bars correspond to se calculated on several independent analyses for each line (n = 10 for the wild type in [A]; n = 6 for be1 in [B]; n = 7 for be2 in [C]; n = 3 for be1 be2 in [D]; n = 6 for be3 in [E]; n = 3 for be1 be3 in [F]). The continuous line corresponds (y axis on the right) to the difference plot determined for each glucan between the wild type and the mutant line (i.e., mutant profile minus wild-type profile). Dashed lines correspond to 0 level for the y axis on the right (difference plots).
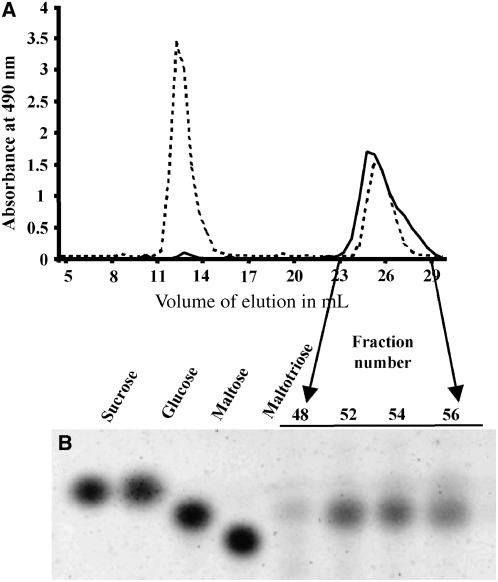
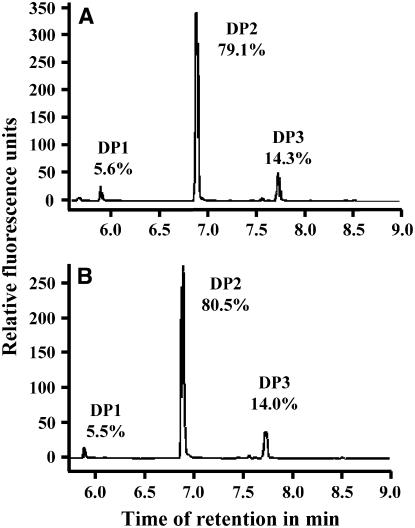
We characterized the structure of the accumulating WSGs in the be2-1 be3-2 mutant. After extraction, the WSGs were submitted to size exclusion chromatography on a TSK HW50 column. The elution profile was compared with that of a mix of glycogen and glucose after the detection of the corresponding sugars by the phenol-sulfuric method (Figure 5A). Only one peak of material that comigrates with glucose was observed for the be2-1 be3-2 double mutant. The fractions corresponding to this peak were analyzed by thin layer chromatography (TLC) (Figure 5B) and fluorophore-assisted capillary electrophoresis (FACE) before and after enzymatic debranching (Figure 6). Our results indicate that WSGs accumulating in the double mutant are composed of very short MOS made of maltose (80%), maltotriose (14%), and glucose (6%). Similar results were obtained whatever the method used for the extraction of WSGs (nonacid or perchloric acid methods; see Supplemental Figures 7 and 8 online).
Figure 5.
Analysis of the WSGs in be2-1 be3-2.
(A) The profile of the WSGs (2 mg) from be2-1 be3-2 (continuous line) was compared with that of a mixture of glycogen (3 mg) and glucose (2 mg) (dashed line) by size exclusion chromatography on a TSK HW50 column. Fractions of 500 μL were collected at a flow rate of 10 mL/h in 10% DMSO. Carbohydrates were detected by the phenol-sulfuric acid method. y axis, absorbance at 490 nm; x axis, elution volume in milliliters.
(B) TLC of fractions 48, 52, 54, and 56.
Figure 6.
FACE Analysis of WSGs That Accumulate in the Double Mutant Line.
After purification, WSGs extracted from be2-1 be3-2 were submitted to FACE analysis before (A) and after (B) enzymatic debranching. x axis, time of retention in minutes; y axis, relative fluorescence level expressed in arbitrary units. DP1, glucose; DP2, maltose; DP3, maltotriose. The numbers under DP1, DP2, and DP3 indicate the relative proportion of each type of glucan in the total content.
The sucrose, glucose, fructose, Glc-6-P, and Glc-1-P levels were determined in the different lines under study after extraction with 16% trichloroacetic acid (TCA) (w/v) in diethylether (Table 4). Sucrose, fructose, Glc-6-P, and Glc-1-P levels were not significantly changed in the double mutant when compared with that of the wild type and other mutants. However, an increase in glucose content was observed in the be2-1 be3-2 mutant as expected from the previous analysis of the composition of the WSGs.
Maltose in be2-1 be3-2 Essentially Accumulates in the Cytosol
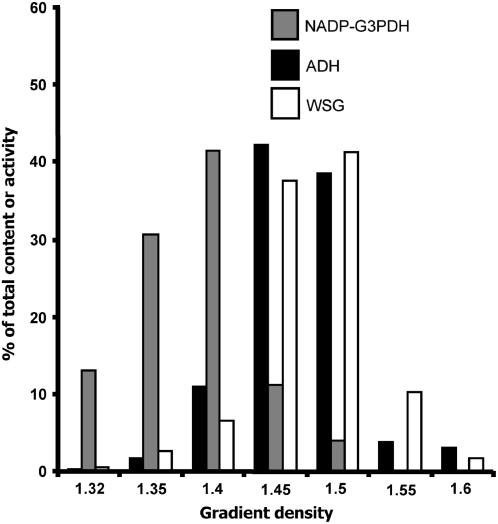
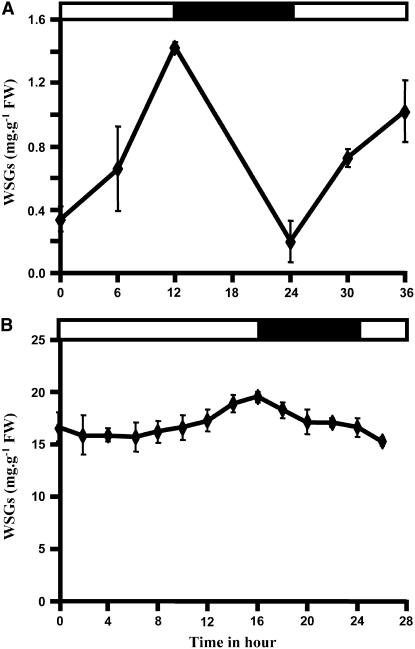
We have performed cell fractionation experiments (nonaqueous method) to determine the intracellular location of the maltose contained in the be2-1 be3-2 mutant. Our results (based on two independent fractionation experiments) indicate that it essentially accumulates in the cytosol (∼80% of WSG is localized in the cytosol) 10 h after the beginning of the 16-h light phase (Figure 7). This result is not surprising since the maltose catabolism is expected to occur in the cytosol after its translocation from the stroma by MEX1, the maltose exporter of the chloroplast envelop. Day/night evolution of MOS content in the be2-1 be3-2 mutant was performed under 12-h-day/12-h-night growth conditions. The plants were left in the dark for 3 d to remove all residual glucans (including starch and MOS) and then transferred to 12-h-day/12-h-night growth rhythms. Leaf samples were harvested 3 d after reilluminating the plants. This experiment shows that MOS accumulates during the day to reach a maximum at the day/night transition and is degraded at night to reach a minimum at the night/day transition (Figure 8A). However, when plants were kept for several days under a 16-h-day/8-h-night rhythm, the MOS concentration was always very high in the double mutant compared with the wild type whatever the time of the cycle (Figure 8B). Nonetheless, an increase was observed during the day, while a decrease was observed during the night. This result suggests that MOS removal is not complete under short nights, leading to an overaccumulation of these molecules after several days of culture in such conditions.
Figure 7.
Fractionation of Plant Leaf Extract to Determine the Intracellular Localization of the MOS That Accumulates in be2-1 be3-2.
The cytosol and the chloroplasts were fractionated by a nonaqueous method after centrifugation on discontinuous density gradient (see Methods for details). WSG content, NADP-G3PDH activity (NADP-glyceraldehyde-3-P dehydrogenase; chloroplastic marker), and ADH activity (alcohol dehydrogenase; cytosolic marker) were assayed on the different density fractions and were expressed as percentages of the total content or activity (y axis) measured in all fractions (x axis). The results presented on this picture are the average of two independent fractionation experiments.
Figure 8.
Diel Variation of MOS Content in be2-1 be3-2.
Total WSGs in the crude extract of be2-1 be3-2 were assayed by the amyloglucosidase assay at different of 12-h-day/12-h-night time points (A) and 16-h-day/8-h-night (B) growth cycles. In (A), the plants were left for 3 d under complete dark before transfer back to a 12-h-light/12-h-night rhythm. WSG content was measured after 3 d under such growth conditions. Values are the means of three independent measures. x axis, time in hours; y axis, total WSG content in mg·g−1 of fresh weight (FW). Thin vertical bars represent the standard error value. Open bar, illuminated period; closed bar, dark period.
Finally, we have determined that α-maltose represents >90% of the total pool of maltose present in the double mutant (the level of α-maltose was determined by the enzymatic method described in Shirokane et al., 2000) at the end of the light period when plants were cultured under a 16-h-light/8-h-dark regime. This test was performed on fresh material immediately after extraction of the MOS to limit the impact of mutarotation in the samples and after immediate boiling to inactivate enzymes. Mutarotation occurs spontaneously when molecules are conserved in solution, although its rate is quite low according to Weise et al. (2005) and based on our own observations. Mutarotation can be defined as the spontaneous conversion of either α- to β-maltose or β- to α-maltose. Indeed, after leaving freshly extracted MOS on melting ice for 1 week, we determined that the β-maltose content had reached 40% of the total maltose, indicating that, in the conditions we used during the purification procedure of the MOS, the mutarotation process is very slow (spontaneous anomerization of maltose may occur at a higher rate at room temperature and is strongly dependent upon the incubation conditions).
We have also determined the maltose anomery at the end of the light period on be2-1 be3-2 plants cultivated under a 12-h-light/12-h-night regime. α-Maltose represents 65% of the total pool of maltose at the end of the light phase in these conditions.
Production of Maltose Is Strongly Reduced in the adg1 Mutant Background
To ensure that the production of maltose in the be2-1 be3-2 double mutant actually occurs through a modified starch pathway, we produced a triple mutant line that is defective for be2, be3, and adg1 (or APS1). The ADG1 locus encodes one of the two small subunits of ADP-glucose pyrophosphorylase. ADP-glucose pyrophosphorylase is responsible for the synthesis of the unique precursor molecule of starch synthesis: ADP-glucose. A mutation at the ADG1 locus leads to a strong reduction of starch accumulation in Arabidopsis leaves as described by Lin et al. (1988) (see Supplemental Figure 6 online). We selected from the Génoplante collection a mutant line of Arabidopsis that has an insertion of a modified T-DNA in the promoter region of the ADG1 gene (line EGZ73 corresponding to flanking sequence tag 511G12). Starch accumulation is not fully abolished in this line but strongly reduced to ∼20% of the wild-type level at the end of the day under 16-h-light/8-h-dark growth conditions (Table 4). In addition, the growth of this mutant line was comparable with that of the wild type in the condition used in this work (see Supplemental Figure 5 online). The presence of the triple mutant adg1 be2-1 be3-2 combination showed a fully restored wild-type growth phenotype when compared with the be2-1 be3-2 double mutant (see Supplemental Figure 5 online). Moreover, a strong reduction of maltose accumulation was observed in the presence of adg1 when compared with the be2-1 be3-2 double mutant (Table 4), suggesting that maltose arises directly from the starch pathway and not from any other route that would be enhanced because of the absence of starch granule formation. This result suggests that the reduced growth of the be2-1 be3-2 double mutant is essentially due to the high level of maltose accumulation in the cells and not because of the absence of starch synthesis under the 16-h-day/8-h-night cycle. High concentrations of maltose might disturb the osmotic balance within the cell and/or might lead to pleiotropic feedback regulation effects on photosynthesis, carbon metabolism, and plant growth.
Transglucosidase DPE2 Requires β-Maltose to Be Active
Weise et al. (2005) previously suggested that DPE2, a cytosolic transglucosidase required for the metabolization of maltose during leaf starch degradation at night (Chia et al., 2004; Lu and Sharkey, 2004), could be specific for β-maltose. The accumulation of α-maltose in the be2-1 be3-2 double mutant provided us with a reliable source that enables us to prove or rule out this hypothesis. We have thus performed zymogram analysis of the transglucosidase activity (Figure 9) of Arabidopsis using either commercially available maltose (50% α- and 50% β-forms) or that purified from the be2-1 be3-2 mutant (90% α- and 10% β-forms). DPE2 activity localized at the very top of the glycogen-containing polyacrylamide gel (Figure 9A) was clearly visible when commercially produced maltose was used as substrate (Figure 9B), even in the be2-1 be3-2 mutant. However, no band was observed when maltose from the be2-1 be3-2 double mutant was used (Figure 9C). This result indicates both that DPE2 is preferentially active on β-maltose and that this enzyme is expressed in the be2-1 be3-2 double mutant (Figure 9A) during the day. To avoid any effect of the cellular extract on the activity of DPE2, the same experiment was performed by incubating DPE2 with maltose freshly made by the degradation of maltoheptaose (DP7) with either β-amylase (from sweet potato [Ipomoea batatas]) or α-amylase (from porcine pancreas). The degradation of DP7 by α- and β-amylases leads to the production of α- and β-maltose, respectively, and maltotriose (see Supplemental Figure 9 online). To this end, a glycogen-containing gel was loaded in each well with 150 μg of proteins of wild-type leaf extract. After migration, gel lanes were separated and incubated in microtubes containing different concentrations of freshly made maltose preparations. DPE2 activity was observed when gel slices were incubated with commercial maltose or with the products of degradation of DP7 by β-amylase (Figure 9D, lanes 1 to 4). However, DPE2 activity was not seen when gel slices were incubated with the products of degradation of DP7 by α-amylase (Figure 9D, lanes 5 to 7). We believe these results demonstrate that DPE2 is specific to β-maltose and cannot use α-maltose as an effective substrate.
Figure 9.
Zymograms of Transglucosidase (DPE2) Activity.
Proteins from leaf extracts (harvested at the middle of the light phase) were loaded onto glycogen-containing polyacrylamide gels and migrated under native conditions for 3 h at 4°C and 15 V·cm−1. After migration, the gels were incubated overnight at room temperature in Tris buffer containing maltose and subsequently stained with iodine solution.
(A) and (B) Gels incubated with 10 mM and 6 mM of commercially available maltose (50% α- and 50% β-forms). In (A), 100 μg of proteins for each sample were loaded onto the gel. dpe2-1: leaf crude extract from a mutant line with T-DNA insertion in gene DPE2 (At2g40840).
(C) Gel incubated with 6 mM maltose purified from be2-1 be3-2 (90% α- and 10% β-forms). In (B) and (C), lanes 1 to 3 correspond to 100, 200, and 400 μg, respectively, of proteins loaded onto the gel.
(D) Small gel pieces incubated with the products of degradation of DP7 by β-amylase (lanes 2 to 4) and α-amylase (lanes 5 to 7) and with 10 mM commercial maltose (lane 1). Wild-type leaf crude extracts were loaded onto the same gel. After migration, the gel was cut into small pieces (corresponding to a zone of the gel where DPE2 is known to be present; i.e., at the border with the stacking gel) that were incubated in microtubes in a final volume of 1.5 mL of buffer. Proteins (150 μg) were loaded onto the gel. Decreasing volumes (100, 50, and 25 μL) of degradation products of DP7 by β- and α-amylases were used in lanes 2 to 4 and in lanes 5 to 7, respectively.
DISCUSSION
In this work, we describe mutant lines of Arabidopsis specifically defective for one out of the three putative SBE genes found in the nuclear genome of the plant (be1, be2, and be3 mutant lines). Moreover, all double mutant combinations (be1-1 be2-1, be1-1 be3-2, and be2-1 be3-2) were produced and their ability to synthesize starch was examined. First, our results suggest that BE1 has no obvious function in the Arabidopsis leaf starch pathway since no modification of the starch phenotype could be recorded when the corresponding gene was disrupted. Moreover, in be2 and be3 mutant backgrounds, the additional mutation at the BE1 locus does not lead to any further changes in the starch phenotype other than those observed in the single be2 and be3 mutants. Finally, despite several attempts, no branching activity corresponding to BE1 was observed, although it is expressed in the leaf of Arabidopsis. The BE1 sequence is undoubtedly related to the SBE family, but its level of similarity to other SBEs (either from Arabidopsis or other plant sources) is low (<40%). Therefore, and at variance with all other vascular plants, Arabidopsis appears to be devoid of B-type SBEs. It remains possible, however, that the function of BE1 might be more specialized and/or restricted to some other parts of the plant. It is also possible that it substitutes for some SBE B elusive functions.
Conversely, the absence of either BE2 or BE3 protein leads to a slight alteration of the starch phenotype essentially characterized by a small but significant modification in the CL distribution profile of amylopectin. DP 5-8 chains are reduced, while DP 9-15 chains are increased in both mutant lines. These results are reminiscent of those previously reported for rice and maize endosperm storage starch, although differences were much more pronounced in the latter cases. Indeed rice mutants (amylose extender) defective for SBEIIb display a striking decrease of DP 5-13 chains and an increase in DP 15-30 chains (Nishi et al., 2001; Tanaka et al., 2004).
Therefore, in Arabidopsis leaves, DP 5-8 glucans may correspond to the main products of the catalytic activities of BE2 and BE3. This range of size matches to that already reported for maize SBEIIa and IIb when incubated in vitro in the presence of amylose (Takeda et al., 1993). Afterward, these DP 5-8 glucans are further elongated by starch synthases to produce longer chains required for the formation of the amylopectin clusters and the linkage between these clusters. SS1 (At5g24300) might represent a good candidate for such elongation, as this enzyme is responsible for the synthesis of the short outer chains of amylopectin (DP 8-12; Delvallé et al., 2005) and is essentially active on short glucans (DP < 10-12; Commuri and Keeling, 2001). SS2 (At3g01180) and SS3 (At1g11720) might also be good candidates for the direct elongation of the glucans produced by SBEs. In maize, SS3 and SBEIIa are suspected to interact physically, as a mutation at the Du1 locus (the structural gene for SS3; Gao et al., 1998) leads to the modification of SBEIIa activity (Boyer and Preiss, 1981). Such an interaction might occur between SS3 and BE2 or BE3 (or both) in the Arabidopsis leaf, leading to the preferential elongation of the end products of SBEs by SS3. This idea is supported by our own observation of SS3 mutants in Chlamydomonas reinhardtii (Fontaine et al., 1993) or Arabidopsis that accumulate a structurally modified amylopectin in which the number of DP 6-7 chains is significantly increased (F. Wattebled, unpublished data). The recent description of a mutant line of Arabidopsis specifically defective for SS3 did not mention such a modification in the amylopectin structure (Zhang et al., 2005), although a slight but significant increase in DP 6-7 chains can be observed in the ss3-2 mutant allele.
The apparent different behavior between BE2 and BE3 with respect to their ability to stimulate phosphorylase “a” might reveal a threshold effect that is specific to this SBE assay. Indeed, the recombinant BE3 expressed in E. coli was clearly observed on a zymogram performed with the phosphorylase “a” stimulation method (see Supplemental Figure 3 online). Moreover, their in vivo functions seem largely (but not fully) redundant, as suggested by the complete absence of starch in the mutant line simultaneously defective for both enzymes. The loss of their activity in the double mutant cannot be overcome by BE1, therefore strengthening our assumption that the latter is not actively involved in starch metabolism in Arabidopsis leaves.
This situation may be related to that described in potato (Safford et al., 1998) where the reduction of SBE activity by 98% in some transgenic lines expressing an antisense sequence of SBE B (class I) does not lead to a dramatic modification of starch structure. The remaining SBE A activity (measured through the phosphorylase stimulation assay) accounts only for a minor part of the total SBE activity in potato tuber but is sufficient to sustain normal starch synthesis. However, reduction of SBE A expression by antisense technology leads to a much bigger modification in starch structure (Jobling et al., 1999) that is somewhat different to the situation observed in Arabidopsis where the contribution of each SBE II (family A) by itself is low, as shown by the marginal phenotype described in each single mutant. This in turn might reveal some basic differences between transitory and storage starches. Only the antisense inhibition of both SBE A and B leads to a strongly altered structure of tuber starch, although starch synthesis was not abolished in these lines and was only reduced by 50% (Schwall et al., 2000). This could be due to the presence of low, albeit significant amounts of residual BE activity.
The origin of maltose accumulation in the double mutant line cannot yet be completely addressed, although we have definitely shown with the analysis of the adg1 be2-1 be3-2 triple mutant that the maltose derives from the starch pathway. The absence of maltose in the starch-free pgm1 mutant (Caspar et al., 1985) and in the very-low-starch ss2 ss3 double mutant (M. James and A. Myers, personal communication) confirms this conclusion. The amount of maltose in the be2-1 be3-2 mutant is 300- to 600-fold higher than that assayed at midcourse of the light phase in the wild type but is more or less equivalent to that measured in dpe2 mutants defective for cytosolic transglucosidase (Chia et al., 2004; Lu and Sharkey, 2004; Weise et al., 2005). The stunted growth phenotype of the double mutant is equally reminiscent of that observed in the dpe2 mutants. The lack of maltose assimilation in the dpe2 mutant can explain why such a high level of maltose accumulates in the cell. Spontaneous mutarotation of maltose could explain why equivalent amounts of both anomers are assayed in the mutant (Weise et al., 2005). In our study, >90% of maltose assayed in the be2-1 be3-2 double mutant was in the α-form. Although no definitive explanation can yet address this difference between our results and those of Weise et al. (2005), several hypotheses may be raised to explain the origin of α-maltose accumulation in the Arabidopsis be2-1 be3-2 double mutant. In this line, despite the absence of SBE activity, the starch synthases could still be able to produce linear α-glucans from ADP-glucose (ADP-glucose pyrophosphorylase and other starch metabolizing enzymes remain unaffected, with the exception of STPases; Table 1). We propose that these linear glucans are rapidly degraded (even during the light phase) by amylases to produce maltose in great majority and some other short MOS (glucose and DP 3 are side products of degradation). After its translocation to the cytosol by the MEX1 maltose exporter (Niittyla et al., 2004; Weise et al., 2004), maltose is degraded by a cytosolic transglucosidase (DPE2; Chia et al., 2004; Lu and Sharkey, 2004) with the help of an acceptor molecule whose nature is not yet known, although the existence of a cytosolic complex soluble heteroglycan was recently reported in Arabidopsis leaves (Fettke et al., 2005a, 2005b). Plastidial β-amylases were shown to interact directly with semicrystalline starch granules and were suggested to be important factors for leaf starch degradation in the potato leaf (Scheidig et al., 2002). However, the situation is obviously different in the be2-1 be3-2 double mutant since no granular starch was observed in this line. The abnormal products of the starch pathway may thus be comparatively more accessible to other enzymes of the starch pathway. In such a modified situation, a plastidial α-amylase might become the most effective enzyme of degradation of those glucans produced by starch synthases in the be2-1 be3-2 line. Since α-amylases exhibit a retaining mechanism (for more details, see http://afmb.cnrs-mrs.fr/CAZY/GH_13.html), the main end product of α-amylases on linear α-1,4–linked glucans is α-maltose, with some other side products like maltotriose and other short MOS (Robyt and French, 1970). α-Maltose is then exported to the cytosol by MEX1 where it will be metabolized by DPE2 after mutarotation. Abnormal accumulation of α-maltose in be2-1 be3-2 may be understood because of the substrate specificity that we now demonstrate for DPE2 (Figures 9B and 9C). Indeed, the latter seems to be significantly active on β-maltose only. β-Maltose is slowly generated by spontaneous mutarotation of α-maltose and is subsequently metabolized by DPE2 while accumulating in the cytosol. The rate of spontaneous mutarotation being quite low (Weise et al., 2005; this work), this could explain why accumulation of α-maltose is observed during the illuminated period (Figure 8). At night (in 12-h-day/12-h-night growth conditions), because de novo synthesis of maltose is turned off, maltose content progressively decreases because of the degradation of the β-form by DPE2. However, under long-day (16 h) growth conditions, maltose cannot be completely degraded since mutarotation is too slow to convert all the α-maltose to β-maltose during the short night. This might explain why maltose progressively accumulates in the leaf cells even at the end of the dark phase when the plants are subjected to a 16-h-day/8-h-night growth regime.
An alternative to this hypothesis is that maltose is produced to the same extent by both α- and β-amylases, leading to the production of both α- and β- maltose. After its transfer to cytosol, β-maltose is rapidly metabolized by DPE2, while α-maltose is only slowly degraded after its spontaneous anomerization to the β-form. Therefore, under long-day/short-night growth conditions, α-maltose concentration gradually increases in the leaf of the plant because it is not a suitable substrate for DPE2.
METHODS
Materials
ADP [U-14C] glucose, [U-14C] glucose-1-phosphate, the CL-2B sepharose column, and Percoll were obtained from Amersham Biosciences. TSK HW50 Toyopearl matrix was purchased from Tosoh Bioscience. ADP-glucose, G1P, and enzymes (α- and β-amylases [from porcine pancreas and sweet potato, respectively], maltose epimerase, and maltose phosphorylase) were from Sigma-Aldrich (unless specified). The starch assay kit was purchased from Enzytec. The fructose and glucose assay kit was obtained from Megazyme.
Arabidopsis Lines, Growth Conditions, and Media
Wild-type (Ws and Col-0) and mutant lines of Arabidopsis thaliana were obtained from the T-DNA mutant collections generated at Génoplante (Unité de Recherche en Génomique Végétale, Institut National de la Recherche Agronomique of Versailles; Bechtold et al., 1993; Bouchez et al., 1993) and the Nottingham Arabidopsis Stock Centre (Alonso et al., 2003). Standard procedures were employed for plant germination and growth. The plants were grown on peat-based compost (seeds were previously incubated at 4°C before sowing) under a 16-h-light/8-h-dark cycle, with temperature ranging from 16°C during the night to 21°C during the day.
RT-PCR Amplifications
Approximately 100 mg of fresh tissue was harvested at the middle of the light phase (see culture conditions described above) for total RNA extraction with the Plant RNeasy kit (Qiagen) following the supplier's instructions. Twenty nanograms of purified total RNA were used to perform RT-PCR amplifications using the One-Step RT-PCR kit (Qiagen). Three different regions surrounding the T-DNA insertion site were targeted for amplification: upstream, downstream, and spanning the T-DNA insertion site (see Supplemental Table 1 online for detailed primer sequences).
Extraction and Purification of Starch
Starch from ∼10 g of leaves was extracted and purified as described by Delvallé et al. (2005).
Extraction of Soluble Carbohydrates (WSGs)
Extraction with Perchloric Acid
All leaves of individuals were harvested at the end of the light period and immediately frozen in liquid nitrogen. Leaves were broken down to rough pieces with a pestle directly in a microtube. Five hundred microliters of 0.7 M perchloric acid was added, and the samples were immediately homogenized with a polytron blender. The samples were then centrifuged for 15 min at 3000g at 4°C. The soluble phase was collected and subsequently neutralized with 2 M KOH, 0.4 M MES, and 0.4 M KCl. The potassium perchlorate precipitate was then removed by centrifugation at 16,000g for 15 min at 4°C. The supernatant was conserved at −80°C before use.
Nonacid Extraction of WSGs
Leaves harvested at the same time as for starch extraction were immediately soaked into liquid nitrogen, broken down to fine powder with pestle and mortar, and placed for 10 min in a boiling water bath to inactivate enzymes. The corresponding samples were then extracted in an ice-cooled buffer (100 mM MOPS, pH 7.2, 5 mM EDTA, 10% [v/v] ethanediol) and further homogenized using a polytron blender. The tubes were always kept on ice during the whole extraction procedure and processed as quickly as possible for analysis to limit the impact of spontaneous mutarotation on maltose anomery. After centrifugation of the homogenate (10 min at 10,000g at 4°C), the supernatant was directly used for the assay of the glucan polymers (amyloglucosidase assay) and the determination of maltose anomery but was lyophilized prior to chromatography by size exclusion on sephadex TSK HW50 matrix (see below).
Determination of Starch and WSG Contents and Spectral Properties of the Iodine-Starch Complex
A full account of λmax (maximal absorbance wavelength of the iodine polysaccharide complex) measure can be found in Delrue et al. (1992). Starch and WSG contents in leaves were determined after extraction (as described above) by the amyloglucosidase assay as described by Delvallé et al. (2005).
Extraction of Soluble Sugars (Glucose, Sucrose, Fructose, and Hexose-P)
Sugars were extracted from 200 to 300 mg of rosette leaves harvested at the end of the light phase and immediately frozen in liquid nitrogen. The leaves were homogenized with an electric polytron blender (the metallic device was replaced by a plastic pestle adapted for 1.5-mL microfuge tubes) in 1 mL of ice-cold 16% TCA (w/v) in diethylether. During the procedure, the samples were kept on ice. EGTA was then added to 5 mM final concentration, and the samples were further homogenized with the polytron blender. Samples were left for 2 h on ice before centrifugation at 16,000g for 5 min at 4°C. The nonaqueous phase was removed, and the aqueous phase was washed four times with diethylether (centrifugation for 5 min at 16,000g at 4°C between each wash). The aqueous phase is transferred to a new tube, neutralized by the addition of 50 mM KOH and 10 mM triethanolamine buffer, and conserved at −80°C.
Determination of Glucose, Sucrose, Fructose, and Hexose-P Contents
Glucose and fructose contents were determined from leaf extract by the use of a specific kit (Megazyme) following the supplier's instructions. Sucrose was assayed as follows: 30 μL of leaf extract was added to 60 μL of 20 mM NaH2PO4, pH 4.5, and 10 units of invertase (Sigma-Aldrich). After 20 min at 55°C, 300 μL of water, 300 μL of triethanolamine buffer, pH 7.6, 1.5 mM NADP, 5 mM ATP, 11 mM MgSO4, 2 units of hexokinase, and 1 unit of G-6-P dehydrogenase (Enzytec) were added to the sample. The reaction was performed at room temperature until absorbance remained stable at 365 nm. The amount of glucose determined previously was deducted from the value obtained in that case. G-6-P content was determined as follow: 30 μL of leaf extract was added to 360 μL of water, and 300 μL of triethanolamine buffer, pH 7.6, 1.5 mM NADP, 5 mM ATP, 11 mM MgSO4, and 1 unit of G-6-P dehydrogenase (Enzytec) were added to the sample. The reaction was performed at room temperature until absorbance remained stable at 365 nm. To measure G-1-P content, 2 units of phosphoglucomutase (Sigma-Aldrich) was added to the previous sample, and absorbance was read at 365 nm at room temperature until it remained stable. G-1-P level was deduced after the subtraction of the amount of G-6-P measured previously.
Separation of Starch Polysaccharides and Water-Soluble Polysaccharides by Size Exclusion Chromatography
Amylopectin and amylose were separated from 1.5 to 2.0 mg of starch by size exclusion chromatography on a CL-2B column as fully described by Delvallé et al. (2005).
Two milligrams of purified and lyophilized WSGs were dissolved in 500 μL of 10% DMSO (v/v) and loaded on a sephadex TSK HW50 column (1 cm i.d. × 50 cm), equilibrated, and eluted with 10% DMSO (v/v). Fractions of 500 μL were collected at a flow rate of 10 mL/h. Carbohydrates in the collected fractions were detected by the phenol-sulfuric acid method. Twenty microliters of 5% phenol were added to 20 μL of sample. After thorough shaking, 100 μL of concentrated sulfuric acid were added, and after gentle shaking, the samples were incubated for 30 min at 80°C. Absorbance was determined at 490 nm and compared with that of a glucose solution of known concentration.
CL Distribution of Amylopectin and WSGs, and Branching Level Determination
Full procedures for the determination of amylopectin CL distribution after enzymatic debranching can be found in Wattebled et al. (2005). CL profile of WSGs was established after FACE using APTS as fluorophore (the protocol for derivatization of the soluble glucans is the same as the one described for amylopectin). Only glucans or sugars with a reducing end can be labeled with APTS.
Branching degree of amylopectin was determined by methylation of glucans as described by Delvallé et al. (2005). Five hundred micrograms of purified amylopectin was used to this end.
TLC
Four microliters of different fractions from the TSK HW50 column (see above) were loaded on silica gel 60 (Merck). Sucrose, glucose, maltose, and maltotriose (2 μg each) were loaded on the sheet as controls. The sheet was developed for 4 h in the following solution: butanol/ethanol/water in ratio 1/1/1 (v/v/v). After migration, the sheet was dried before spraying sulfuric orcinol solution and heating for several minutes at 100°C until sugars appeared as brown bands on the TLC.
Production of Fresh Maltose by α- and β-Amylases
Twenty milligrams of maltoheptaose (Sigma-Aldrich) were incubated for 2 h at 30°C in a final volume of 500 μL with 20 units of α-amylase (from porcine pancreas) or 20 units of β-amylase (from sweet potato). Incubation was extended by 2 h after the addition of an additional 20 units of each enzyme to ensure a complete degradation of DP7. For incubation with α-amylase, the buffer was composed of 25 mM sodium acetate, pH 4.8, 5 mM DTT, and 2 mM CaCl2. For incubation with β-amylase, the buffer was composed of 25 mM sodium acetate, pH 4.8, 5 mM DTT, and 5 mM EDTA. The reaction was stopped by the addition of 500 μL of phenol/chloroform (1/1; v/v). After a thorough mixing, the samples were centrifuged for 10 min at 16,000g at 4°C, and the supernatant was extracted three times with chloroform to remove traces of phenol. The degradation of DP7 to maltose and maltotriose was checked by TLC on silica gel 60 (see above and Supplemental Figure 9 online) and subsequently used at different concentrations for incubation with small pieces of gel containing glycogen and DPE2 activity (Figure 9D; see below for DPE2 zymogram).
Determination of Maltose Anomery
Anomery of maltose was determined using the procedure described by Shirokane et al. (2000). Twelve nanomoles of total purified WSGs were incubated in the following buffer: triethanolamine, pH 7.6, 155 mM NaH2PO4, 1.5 mM NADP, 5 mM ATP, and 11 mM MgSO4 at 30°C in the presence of 2 units of hexokinase and 1 unit of G-6-P dehydrogenase. The reaction was started by the addition of 5 units of maltose phosphorylase alone (to determine the proportion of α-maltose in the sample) or by the addition of both 5 units of maltose phosphorylase and 5 units of maltose epimerase (for α- and β-maltose determination). Absorbance was read at 365 nm in a spectrophotometer (U 2000; Hitachi).
Zymogram Techniques
For a complete description of soluble starch synthase activities, refer to Delvallé et al. (2005).
Starch modifying activities on starch-containing gel (hydrolases and branching enzymes) were tested as follows: 100 μg of leaf extract proteins were loaded onto a native PAGE (7.5% acrylamide) containing potato-soluble starch (Sigma-Aldrich) at 0.3% final concentration and separated for 3 h at 4°C and 15 V·cm−1. The gels were incubated overnight at room temperature in the following buffer: 50 mM Tris-HCl, pH 7.0, 1 mM DTT, 1 mM CaCl2, and 1 mM MgCl2. The hydrolytic activities were revealed by soaking the gel into iodine solution (0.2% I2 [w/v] and 2% KI [w/v]).
For phosphorylase “a” stimulation, 100 μg of proteins from a leaf crude extract were loaded on a native PAGE (6.5% acrylamide). After electrophoresis at 4°C for 2 h and 30 min, the gel was incubated overnight at room temperature in 30 mL of the following buffer: 50 mM HEPES/NaOH, pH 7.0, 10% glycerol (v/v); 50 mM G1P, 2.5 mM AMP, and 28 units of phosphorylase “a” from rabbit muscle. Branching activities were revealed by iodine staining.
For transglucosidase DPE2, 100 μg of proteins from a leaf crude extract were loaded on a native PAGE (7.5% acrylamide) containing 0.3% of rabbit liver glycogen. After migration (under native conditions for 3 h at 4°C at 15 V·cm−1), the gel was preincubated for 1 h in 50 mL of 100 mM Tris-HCl, pH 7.0, 1 mM MgCl2, and 1 mM DTT and incubated overnight at room temperature in the same buffer. Unless otherwise indicated in the text, the maltose concentration was 10 mM. To test for DPE2 activity with degradation products of DP7 by α- or β-amylases (Figure 9D), 150 μg of proteins were loaded onto the gel.
In Vitro Assays of Starch Metabolizing Enzymes
For SBEs, proteins were extracted in 50 mM NaH2PO4, pH 8.0, and 500 mM NaCl. One hundred micrograms of protein were incubated at 30°C for 2 h in 250 μL of the following: 50 mM HEPES/NaOH, pH 7.0, 1 mM AMP, 10% glycerol (v/v), 8 μM [U-14C] G1P (150 mCi/mmol), 50 mM G1P, and 2 units of phosphorylase “a” from rabbit muscle. Reaction was stopped by boiling the sample for 10 min. One hundred microliters from a 100-mg/mL stock solution of glycogen and 1 mL of 75% methanol (v/v)/1% KCl (w/v) were added. The resulting precipitate was then filtered on a glass-fiber filter (Whatmann), rinsed two times with 15 mL of deionized water, two times with 70% ethanol, and dried. The sample was finally counted in a scintillation counter after the addition of 3 mL of counting solution (UltimaGold; Perkin-Elmer).
Soluble starch synthases and ADP-glucose pyrophosphorylase were assayed according to Delvallé et al. (2005).
The α- and β-amylases, pullulanase, α-1,4 glucanotransferase, and starch phosphorylase activities were assayed as described by Delvallé et al. (2005).
Nonaqueous Fractionation of Chloroplasts
The method was previously described (Gerhardt and Heldt, 1984) and modified (Weise et al., 2005). The procedure is briefly described here. Seven to eight grams of fresh leaves were harvested 10 h after the beginning of the 16-h-light period and immediately frozen in liquid nitrogen. Leaves were ground with a mortar and pestle and were continuously kept frozen by the addition of liquid nitrogen. The ground material was then poured into a 50-mL Falcon tube and dried by lyophilization. The powder was resuspended in 20 mL of dry C2Cl4/n-heptane (density = 1.32 g·mL−1) and homogenized by ultrasonication at a power level of 5.5 and 40% duty cycle. The sample was sonicated for 3 min in 10-s periods followed by a pause of 10 s on ice. The homogenate was filtered through two layers of Miracloth (Calbiochem, EMD Biosciences) into a new 50-mL Falcon tube. The retentate was rinsed by 10 mL of dry C2Cl4/n-heptane (density = 1.32 g·mL−1) and then by 10 mL of dry n-heptane. The filtrate was centrifuged for 7 min at 1800g. The supernatant was discarded, and the pellet was resuspended in 4 mL of dry C2Cl4/n-heptane (density = 1.32 g.mL−1) and layered on top of a discontinuous gradient consisting of six density steps, 4 mL each, from 1.60 to 1.35 g·mL−1. It was then centrifuged at 3200g for 1.5 h. The content of the centrifugation was collected in 4-mL fractions. Two volumes of n-heptane were then added to each fraction and then tubes were centrifuged for 10 min at 2200g. The supernatant was discarded, and the pellets were dried in a dessicator for 18 h at room temperature.
Sample Preparation for Assay of Marker Enzyme Activities and MOS Measurement
Two hundred microliters of 50 mM NaH2PO4, pH 8.0, and 500 mM NaCl were added to the dried sample. The suspension was ultrasonicated at a power level of 7 and 70% duty cycle for 30 s in 10-s periods followed by a pause of 20 s on ice. The sample was then centrifuged for 5 min at 10,000g, and the resulting supernatant was used for assaying the marker enzyme activities and the determination of the MOS content.
Assay of Marker Enzyme Activities
The following marker enzymes were assayed at 25°C as described here: for the chloroplastic marker NADP-glyceraldehyde-3-P dehydrogenase, 20 μL of sample was added to 180 μL of the following buffer: 50 mM GlyGly/NaOH, pH 8.2, 80 mM 3-phosphoglyceric acid, 20 mM MgSO4, 10 mM cysteine, 5 mM gluthatione, 0.4 mM NADPH, and 2 units of phosphoglycerat kinase. Oxidation of NADPH was followed in a spectrophotometer at 340 nm for 10 min. The result for each sample is given by the slope of the corresponding curve that is related to the activity rate of NADP-glyceraldehyde-3-P dehydrogenase.
For the cytosolic marker alcohol dehydrogenase, 50 μL of sample was added to 950 μL of 50 mM GlyGly/NaOH, pH 8.7, 2 mM NAD, and 30% (v/v) ethanol. Reduction of NAD was followed at 340 nm for 5 min.
Transmission Electron Microscopy and X-Ray Diffraction Measurements
A full account of these techniques can be found in Delvallé et al. (2005).
Accession Numbers
Arabidopsis Genome Initiative accession numbers for BE1, BE2, and BE3 can be found in Table 1. The Arabidopsis Genome Initiative accession number for ADG1 is At5g48300.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Primer Pairs Used for the Selection of the Mutant Alleles by RT-PCR Amplification.
Supplemental Figure 1. Dendrogram of Branching Enzyme Amino Acid Sequences from Various Sources.
Supplemental Figure 2. Zymogram for Starch Modifying/Degrading Enzymes.
Supplemental Figure 3. Zymogram Analysis of Recombinant BE3 Expressed in E. coli.
Supplemental Figure 4. Zymogram of Starch Phosphorylases.
Supplemental Figure 5. Comparison of Growth Phenotypes.
Supplemental Figure 6. Iodine Staining of Arabidopsis Leaves from Different Lines.
Supplemental Figure 7. Fluorophore-Assisted Capillary Electrophoresis of WSGs Extracted from be2-1 be3-2.
Supplemental Figure 8. Thin Layer Chromatography of WSGs Extracted from be2-1 be3-2.
Supplemental Figure 9. Thin Layer Chromatography of Products Obtained after the Degradation of Maltoheptaose (DP7) by α-Amylase or β-Amylase.
Supplementary Material
Acknowledgments
This work was supported by Génoplante (program Af2001030), the Centre National de la Recherche Scientifique, the Institut National de la Recherche Agronomique, the Région Nord-Pas de Calais, the European Union FEDER (grant ARCir to C.D.), and the Ministère Délégué à la Recherche (Grant JC5145 Action Concertée Incitative Jeunes-Chercheurs). We thank Alan Myers and Martha James (Iowa State University) for sharing unpublished results and for helpful discussions. We are grateful to Frédéric Chirat for his assistance with FACE and Yves Leroy for his assistance with gas chromatography.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Christophe D'Hulst (christophe.dhulst@univ-lille1.fr).
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Ball, S.G., and Morell, M.K. (2003). From bacterial glycogen to starch: Understanding the biogenesis of the plant starch granule. Annu. Rev. Plant Biol. 54 207–233. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Life Sci. 316 1194–1199. [Google Scholar]
- Blauth, S.L., Kim, K.-N., Klucinec, J., Shannon, J.C., Thompson, D., and Guiltinan, M. (2002). Identification of Mutator insertional mutants of starch-branching enzyme (sbe1) in Zea mays L. Plant Mol. Biol. 48 287–297. [DOI] [PubMed] [Google Scholar]
- Blauth, S.L., Yao, Y., Klucinec, J.D., Shannon, J.C., Thompson, D.B., and Guiltinan, M.J. (2001). Identification of Mutator insertional mutants of starch-branching enzyme 2a in corn. Plant Physiol. 125 1396–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchez, D., Camilleri, C., and Caboche, M. (1993). A binary vector based on Basta resistance for in planta transformation of Arabidopsis thaliana. C. R. Acad. Sci. Life Sci. 316 1188–1193. [Google Scholar]
- Boyer, C.D., and Preiss, J. (1981). Evidence for independent genetic control of the multiple forms of maize endosperm branching enzymes and starch synthases. Plant Physiol. 67 1141–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buléon, A., Colonna, P., Planchot, V., and Ball, S. (1998). Starch granules: Structure and biosynthesis. Int. J. Biol. Macromol. 23 85–112. [DOI] [PubMed] [Google Scholar]
- Caspar, T., Huber, S.C., and Somerville, C. (1985). Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana (L.) deficient in chloroplast phosphoglucomutase activity. Plant Physiol. 79 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia, T., Thorneycroft, D., Chapple, A., Messerli, G., Chen, J., Zeeman, S.C., Smith, S.M., and Smith, A.M. (2004). A cytosolic glucosyltransferase is required for conversion of starch to sucrose in Arabidopsis leaves at night. Plant J. 37 853–863. [DOI] [PubMed] [Google Scholar]
- Commuri, P.D., and Keeling, P.L. (2001). Chain-length specificities of maize starch synthase I enzyme: Studies of glucan affinity and catalytic properties. Plant J. 25 475–486. [DOI] [PubMed] [Google Scholar]
- Delrue, B., Fontaine, T., Routier, F., Decq, A., Wieruszeski, J.-M., van den Koornhuyse, N., Maddelein, M.-L., Fournet, B., and Ball, S. (1992). Waxy Chlamydomonas reinhardtii: Monocellular algal mutants defective in amylose biosynthesis and granule-bound starch-synthase activity accumulate a structurally modified amylopectin. J. Bacteriol. 174 3612–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delvallé, D., Dumez, S., Wattebled, F., Roldán, I., Planchot, V., Berbezy, P., Colonna, P., Vyas, D., Chatterjee, M., Ball, S., Mérida, A., and D'Hulst, C. (2005). Soluble starch synthase I: A major determinant for the synthesis of amylopectin in Arabidopsis thaliana leaves. Plant J. 43 398–412. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., and von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fettke, J., Eckermann, N., Tiessen, A., Geigenberger, P., and Steup, M. (2005. a). Identification, subcellular localization and biochemical characterization of water-soluble heteroglycans (SHG) in leaves of Arabidopsis thaliana L.: Distinct SHG reside in the cytosol and in the apoplast. Plant J. 43 568–585. [DOI] [PubMed] [Google Scholar]
- Fettke, J., Poeste, S., Eckermann, N., Tiessen, A., Pauly, M., Geigenberger, P., and Steup, M. (2005. b). Analysis of cytosolic heteroglycans from leaves of transgenic potato (Solanum tuberosum L.) plants that under- or overexpress the Pho2 phosphorylase isozyme. Plant Cell Physiol. 46 1987–2004. [DOI] [PubMed] [Google Scholar]
- Fisher, D.K., Gao, M., Kim, K.-N., Boyer, C.D., and Guiltinan, M.J. (1996. a). Allelic analysis of the maize amylase-extender locus suggests that independent genes encode starch-branching enzymes IIa and IIb. Plant Physiol. 110 611–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, D.K., Gao, M., Kim, K.-N., Boyer, C.D., and Guiltinan, M.J. (1996. b). Two closely related cDNAs encoding starch branching enzyme from Arabidopsis thaliana. Plant Mol. Biol. 30 97–108. [DOI] [PubMed] [Google Scholar]
- Fontaine, T., D'Hulst, C., Maddelein, M.-L., Routier, F., Marianne-Pepin, T., Decq, A., Wieruszeski, J.M., Delrue, B., van den Koornhuyse, N., Bossu, J.P., Fournet, B., and Ball, S.G. (1993). Toward an understanding of the biogenesis of the starch granule. Evidence that Chlamydomonas soluble starch synthase II controls the synthesis of intermediate size glucans of amylopectin. J. Biol. Chem. 268 16223–16230. [PubMed] [Google Scholar]
- Gao, M., Wanat, J., Stinard, P.S., James, M.G., and Myers, A.M. (1998). Characterization of dull1, a maize gene coding for a novel starch synthase. Plant Cell 10 399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt, R., and Heldt, H. (1984). Measurement of subcellular metabolite levels in leaves by fractionation of freeze-stopped material in nonaqueous media. Plant Physiol. 75 542–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, H.-P., Baba, T., and Preiss, J. (1994). Expression of branching enzyme I of maize endosperm in Escherichia coli. Plant Physiol. 104 1449–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, H.-P., and Preiss, J. (1993). Differentiation of the properties of the branching isozymes from maize (Zea mays). Plant Physiol. 102 1269–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hizukuri, S. (1986). Polymodal distribution of the chain lengths of amylopectins, and its significance. Carbohydr. Res. 147 342–347. [Google Scholar]
- Jobling, S.A., Schwall, G.P., Westcott, R.J., Sidebottom, C.M., Debet, M., Gidley, M.J., Jeffcoat, R., and Safford, R. (1999). A minor form of starch branching enzyme in potato (Solanum tuberosum L.) tubers has a major effect on starch structure: Cloning and characterisation of multiple forms of SBE A. Plant J. 18 163–171. [DOI] [PubMed] [Google Scholar]
- Khoshnoodi, J., Larsson, C.-T., Larsson, H., and Rask, L. (1998). Differential accumulation of Arabidopsis thaliana Sbe2.1 and Sbe2.2 transcripts in response to light. Plant Sci. 135 183–193. [Google Scholar]
- Larsson, C.-T., Hofvander, P., Khoshnoodi, J., Ek, B., Rask, L., and Larsson, H. (1996). Three isoforms of starch synthase and two isoforms of branching enzyme are present in potato tuber starch. Plant Sci. 117 9–16. [Google Scholar]
- Larsson, C.-T., Khoshnoodi, J., Ek, B., Rask, L., and Larsson, H. (1998). Molecular cloning and characterization of starch-branching enzyme II from potato. Plant Mol. Biol. 37 505–511. [DOI] [PubMed] [Google Scholar]
- Lin, T.-P., Caspar, T., Somerville, C., and Preiss, J. (1988). Isolation and characterization of a starchless mutant of Arabidopsis thaliana (L.) Heynh lacking ADPglucose pyrophosphorylase activity. Plant Physiol. 86 1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y., and Sharkey, T.D. (2004). The role of amylomaltase in maltose metabolism in the cytosol of photosynthetic cells. Planta 218 466–473. [DOI] [PubMed] [Google Scholar]
- Mizuno, K., Kimura, K., Arai, Y., Kawasaki, T., Shimada, H., and Baba, T. (1992). Starch branching enzymes from immature rice seeds. J. Biochem. (Tokyo) 112 643–651. [DOI] [PubMed] [Google Scholar]
- Morell, M.K., Blennow, A., Kosar-Hashemi, B., and Samuel, M.S. (1997). Differential expression and properties of starch branching enzyme isoforms in developing wheat endosperm. Plant Physiol. 113 201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, A.M., Morell, M.K., James, M.G., and Ball, S.G. (2000). Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiol. 122 989–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, Y. (2002). Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: Rice endosperm as a model tissue. Plant Cell Physiol. 43 718–725. [DOI] [PubMed] [Google Scholar]
- Nakamura, Y., Takeichi, T., Kawaguchi, K., and Yamamouchi, H. (1992). Purification of two forms of starch branching enzyme (Q-enzyme) from developing rice endosperm. Physiol. Plant. 84 329–335. [Google Scholar]
- Nishi, A., Nakamura, Y., Tanaka, N., and Satoh, H. (2001). Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiol. 127 459–472. [PMC free article] [PubMed] [Google Scholar]
- Niittyla, T., Messerli, G., Trevisan, M., Chen, J., Smith, A.M., and Zeeman, S.C. (2004). A previously unknown maltose transporter essential for starch degradation in leaves. Science 303 87–89. [DOI] [PubMed] [Google Scholar]
- Robyt, J.F., and French, D. (1970). The action pattern of porcine pancreatic a-amylase in relationship to the substrate binding site of the enzyme. J. Biol. Chem. 245 3917–3927. [PubMed] [Google Scholar]
- Rydberg, U., Andersson, L., Andersson, R., Aman, P., and Larsson, H. (2001). Comparison of starch branching enzyme I and II from potato. Eur. J. Biochem. 268 6140–6145. [DOI] [PubMed] [Google Scholar]
- Safford, R., Jobling, S.A., Sidebottom, C.M., Westcott, R.J., Cooke, D., Tober, K.J., Strongitharm, B.H., Russell, A.L., and Gidley, M.J. (1998). Consequences of antisense RNA inhibition of starch branching enzyme activity on properties of potato starch. Carbohyd. Polym. 35 155–168. [Google Scholar]
- Satoh, H., Nishi, A., Yamashita, K., Takemoto, Y., Tanaka, Y., Hosaka, Y., Sakurai, A., Fujita, N., and Nakamura, Y. (2003). Starch-branching enzyme I-deficient mutation specifically affects the structure and properties of starch in rice endosperm. Plant Physiol. 133 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidig, A., Fröhlich, A., Schulze, S., Lloyd, J.R., and Kossmann, J. (2002). Downregulation of a chloroplast-targeted β-amylase leads to a starch-excess phenotype in leaves. Plant J. 30 581–591. [DOI] [PubMed] [Google Scholar]
- Schwall, G.P., Safford, R., Westcott, R.J., Jeffcoat, R., Tayal, A., Shi, Y.-C., Gidley, M.J., and Jobling, S.A. (2000). Production of very-high-amylose potato starch by inhibition of SBE A and B. Nat. Biotechnol. 18 551–554. [DOI] [PubMed] [Google Scholar]
- Shirokane, Y., Ichikawa, K., and Suzuki, M. (2000). A novel enzymic determination of maltose. Carbohydr. Res. 329 699–702. [DOI] [PubMed] [Google Scholar]
- Sivak, M.N., and Preiss, J. (1998). Starch: Basic science to biotechnology. In Advances in Food and Nutrition Research, Vol. 41. (San Diego, CA: Academic Press).
- Stinard, P.S., Robertson, D.S., and Schnable, P.S. (1993). Genetic isolation, cloning, and analysis of a mutator-induced, dominant antimorph of the maize amylose extender1 locus. Plant Cell 5 1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, Y., Guan, H.-P., and Preiss, J. (1993). Branching of amylose by branching isoenzymes of maize endosperm. Carbohydr. Res. 240 253–263. [Google Scholar]
- Tanaka, N., Fujita, N., Nishi, A., Satoh, H., Hosaka, Y., Ugaki, M., Kawasaki, S., and Nakamura, Y. (2004). The structure of starch can be manipulated by changing the expression levels of starch branching enzyme IIb in rice endosperm. Plant Biotechnol. J. 2 507–516. [DOI] [PubMed] [Google Scholar]
- Wattebled, F., Dong, Y., Dumez, S., Delvallé, D., Planchot, V., Berbezy, P., Vyas, D., Colonna, P., Chatterjee, M., Ball, S., and D'Hulst, C. (2005). Mutants of Arabidopsis lacking a chloroplastic isoamylase accumulate phytoglycogen and an abnormal form of amylopectin. Plant Physiol. 138 184–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise, S.E., Kim, K.S., Stewart, R.P., and Sharkey, T.D. (2005). beta-Maltose is the metabolically active anomer of maltose during transitory starch degradation. Plant Physiol. 137 756–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise, S.E., Weber, A.P., and Sharkey, T.D. (2004). Maltose is the major form of carbon exported from the chloroplast at night. Planta 218 474–482. [DOI] [PubMed] [Google Scholar]
- Yao, Y., Thompson, D.B., and Guiltinan, M.J. (2004). Maize starch-branching enzyme isoforms and amylopectin structure. In the absence of starch-branching enzyme IIb, the further absence of starch-branching enzyme Ia leads to increased branching. Plant Physiol. 136 3515–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Myers, A.M., and James, M.G. (2005). Mutations affecting starch synthase III in Arabidopsis alter leaf starch structure and increase the rate of starch synthesis. Plant Physiol. 138 663–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.