Abstract
Endoreduplication is a type of cell cycle in which DNA replication continues without cell division. We have isolated several dominant mutants from Arabidopsis thaliana activation tagging lines by flow cytometry. One of the mutants, increased level of polyploidy1-1D (ilp1-1D), showed increased polyploidy in both light- and dark-grown hypocotyls. The corresponding gene of ilp1-1D encodes a protein homologous to the C-terminal region of mammalian GC binding factor. We demonstrate that this protein functions as a transcriptional repressor in vivo. The expression of all members of the CYCLINA2 (CYCA2) family was reduced in an ILP1 overexpressing line, and the mouse (Mus musculus) homolog of ILP1 repressed cyclin A2 expression in mouse NIH3T3 cells. T-DNA insertion mutants of ILP1 showed reduced polyploidy and upregulated all CYCA2 expression. Furthermore, loss of CYCA2;1 expression induces an increase in polyploidy in Arabidopsis. We demonstrate that this protein regulates endoreduplication through control of CYCA2 expression in Arabidopsis.
INTRODUCTION
In plants, many organs are composed of a mixture of cells of different ploidy levels, and this feature is prominent in hypocotyl elongation, leaf expansion, and endosperm development. Polyploidy is caused by endoreduplication, a type of cell cycle where nuclear chromosomal DNA replication occurs without cell division. These polyploid cells are commonly observed in various multicellular organisms, such as insects, mammals, and plants (Joubes and Chevalier, 2000; Edgar and Orr-Weaver, 2001). Polyploid cells are often seen in various developing tissues and correlate with development; hence, polyploidy is thought to be a marker of differentiation (De Veylder et al., 2001). Although endoreduplication can also be observed in several tissues in insects and mammals, this feature is quite characteristic of plant organs, and it distinguishes plant development from that of other organisms. Cells from Arabidopsis thaliana hypocotyls contain as much as 8C (C is a set of the haploid chromosomes) nuclear DNA in light-grown seedlings and as high as 16C in dark-grown seedlings (Gendreau et al., 1997).
The polyploidy levels in hypocotyls are also controlled by phytohormones (Gendreau et al., 1999). constitutively triple response1 (ctr1) is an ethylene signal transduction mutant in which the ethylene signal is constitutively activated and causes a triple response without exogenous ethylene (Kieber et al., 1993). ctr1 has increased polyploidy levels in hypocotyls, having as high as 32C in dark-grown seedlings (Gendreau et al., 1997), indicating that ethylene regulates endoreduplication positively in hypocotyl cells.
Endoreduplication is also involved in the development of the plant's architecture. A trichome consists of a single cell that contains a nucleus of up to 32C (Melaragno et al., 1993). Endoreduplication is also observed in endosperm, and there are several reports of the involvement of cell cycle–related genes in endosperm expansion (Sun et al., 1999; Larkins et al., 2001). Thus, the regulation of endoreduplication plays an important role in plant development and differentiation.
It is reported that some cell cycle regulating factors control endoreduplication. A D-type cyclin gene, CYCLIND3;1 (CYCD3;1), expresses specifically in meristems and developing leaves but not in differentiated tissues in Arabidopsis. When CYCD3;1 was overexpressed, the polyploidy level of the transgenic plants was much reduced and cell size was smaller (Dewitte et al., 2003). This indicates that CYCD3;1 is involved in cell proliferation by inhibiting endoreduplication in plant tissue. An Arabidopsis A-type cyclin gene, CYCA2;1, is expressed in various differentiated cells, such as guard cells (Burssens et al., 2000), where endoreduplication has never occurred (Melaragno et al., 1993). When tobacco (Nicotiana tabacum) CYCA3;2 is overexpressed in Arabidopsis, polyploidy levels are reduced in various tissues (Yu et al., 2003). Loss of Arabidopsis CYCA2;3 function increases polyploidy in mature true leaves (Imai et al., 2006). A-type cyclins could play an important role in regulating endoreduplication in plants.
In addition to cyclins, Arabidopsis E2F, a transcriptional factor regulating the S-phase, is also involved in the control of endoreduplication (De Veylder et al., 2002). Overexpression of E2FA/E2F3 with DPA that encodes for an interacting protein of E2FA causes uncontrolled cell proliferation and upregulation of DNA replication licensing factors including ORC, MCM5, and CDC6. This coexpression of E2FA and DPA also causes increased polyploidy levels, suggesting that E2FA/DPA is crucial for transition from mitosis to the endocycle. This hypothesis is supported by another observation. Ectopic expression of the dominant negative B-type cyclin-dependent kinase CDKB1;1 strongly enhances polyploidy levels in E2FA/DPA overexpressing Arabidopsis plants (Boudolf et al., 2004). This indicates that reduction of mitotic activity regulated by CDKB1;1 is nessesary for the transition from mitosis to the endocycle after stimulation of the S-phase by E2FA/DPA. Atypical E2F genes, DP1-E2F-like (DEL), have been isolated from Arabidopsis. Loss of DEL1 increases polyploidy levels and upregulates expression of DNA replication genes, such as CDT1a (Vlieghe et al., 2005). CDT1a is one of the DNA replication licensing factors involved in the S-phase, and its expression is also controlled by E2F. An extra round of endreduplication is observed in CDT1a overexpressing Arabidopsis plants (Castellano Mdel et al., 2004). These results indicate that regulation of the S-phase is important for endoreduplication.
To understand the mechanism of endoreduplication, we set up a strategy to screen systematically for endoreduplication mutants. Using flow cytometric analysis and activation tagging mutant lines, we isolated several dominant mutants that had increased ploidy levels. They were categorized into two groups according to their dependence on light. Group 1 has increased polyploidy both in light- and dark-grown seedlings. Group 2 has increased polyploidy only under dark conditions.
We discuss here one mutant belonging to Group 1, the corresponding gene encoding a nuclear protein that has homology with the C-terminal region of the GC binding factor (GCF) protein of human (Homo sapiens) (Kageyama and Pastan, 1989). We demonstrate that this protein regulates endoreduplication and represses the expression of Cyclin A2 in Arabidopsis and mouse (Mus musculus) cell lines.
RESULTS
Classification of Endoreduplication Mutants
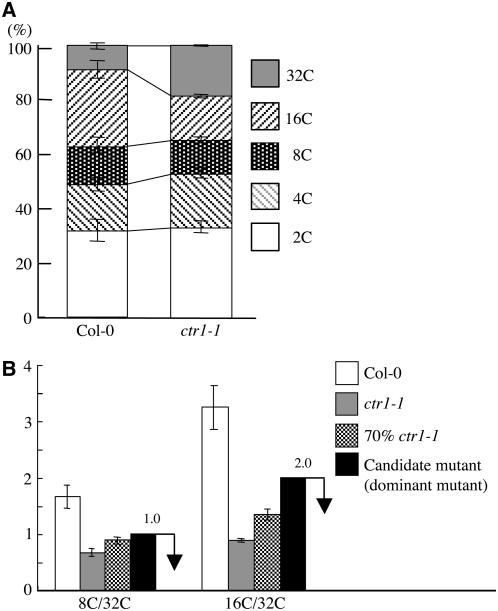
To understand the mechanism of polyploidy control in plants, we used a strategy based on the screening of gain-of-function mutants by flow cytometry. We used dark-grown Arabidopsis seedlings and measured the ploidy levels of hypocotyl cells. To define the screening conditions, we used the ethylene signal transduction mutant ctr1-1 as a positive control (Gendreau et al., 1997). The 32C peak was significantly increased in dark-grown ctr1-1 hypocotyls, although other ploidy peaks, including 2C, 4C, 8C, and 16C, also appeared in ctr1-1 as they did in the wild type (Columbia-0 [Col-0]) but at different ratios (Figure 1A) (Gendreau et al., 1997). Under our screening conditions with sucrose in the growth plates, we could observe cells of as high as 32C in dark-grown hypocotyls. We calculated the relative ratios of both 8C/32C and 16C/32C of ctr1-1 and the wild type. We mixed ctr1-1 seedlings with the wild type at a ratio of 7 to 3 instead of 3 to 1 to represent the appearance of a dominant polyploid mutation to account for heterozygous siblings. We calculated the values of 8C/32C and 16C/32C and set screening criteria of <1.0 for 8C/32C and 2.0 for 16C/ 32C (Figure 1B). We used these values for the isolation of dominant polyploid mutants.
Figure 1.
Categories Used for Dominant Polyploid Mutant Screening.
(A) Relative ratio of each cell ploidy of the wild type (Col-0) and ctr1-1. Approximately 5000 nuclei were counted in the wild type and ctr1-1 mutant.
(B) Ratio of 8C/32C and 16C/32C of dark-grown seedlings of the wild type (Col-0), ctr1-1, and a mixture of Col-0 and ctr1-1 at a ratio of 3 to 7. Black bars indicate the categories used for mutant screening.
For each ploidy measurement, at least 20 seedlings were used, and it was replicated three times. Error bars indicate standard deviation.
We used ∼20 seedlings for each T2 activation tagging line. In our screening, it is difficult to isolate recessive mutations since mutants will appear in only one-quarter of the T2 seeds, and at this frequency any difference in ploidy levels will be buried in wild-type patterns. For gain-of-function or dominant mutations, the mutant siblings appear in three-quarters of the population and can be monitored by our flow cytometric assay. Dark-grown seedlings were used for this assay because they were easy to harvest and conditions were reproducible.
We isolated six dominant mutants from 4500 independent activation tagging lines (Ichikawa et al., 2003; Nakazawa et al., 2003; http://rarge.gsc.riken.jp/activationtag/top.php). These mutants had an increased number of polyploid cells and showed a high 32C polyploidy peak in the hypocotyls of dark-grown seedlings like the ctr1-1 mutant.
These six mutants could be classified into two groups according to hypocotyl length, root length, and dependency on light. We named them Group 1 and Group 2 (Table 1). Three mutants belonging to Group 1 showed increased ploidy levels in both dark- and light-grown seedlings compared with the wild type. They had longer roots, but the hypocotyl length was almost the same as the wild type. In Group 2, three mutants had increased polyploidy only under dark conditions, and they had almost the same ploidy levels as the wild type in the light. This indicated that the Group 2 phenotype is light dependent. They also had longer hypocotyls in dark-grown seedlings.
Table 1.
Two Groups of Polyploidy Mutants
| Ploidy (D, Hyp.) | Ploidy (WL, Cot.) | Hypocotyl Length (D) | Root Length (D) | Other Features | |
|---|---|---|---|---|---|
| Group 1 | + | + | − | + | Large cotyledons and thick hypocotyls |
| Group 2 | + | − | + | + |
(D, hyp.), hypocotyls of dark-grown seedlings; (WL, cot.), cotyledons of white light–grown seedlings; (D), dark-grown seedlings; +, increased; −, no difference from Col-0.
ilp1-1D Increased Polyploidy Levels in Both Light and Dark Conditions
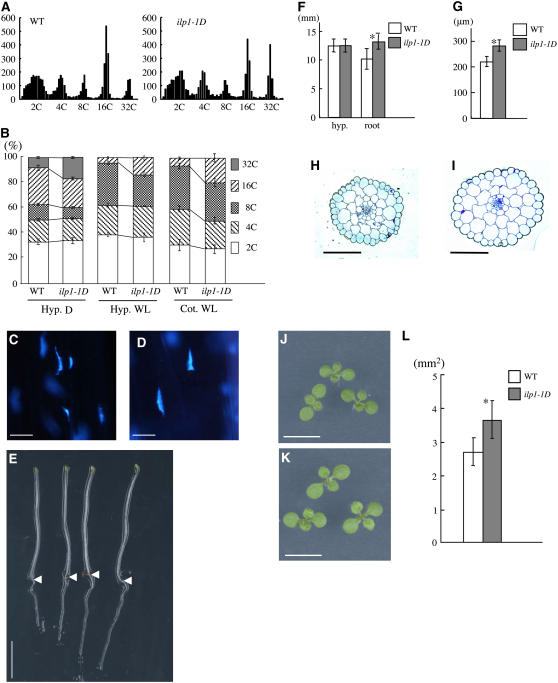
We characterized a dominant mutant Z010521 that we designated as increased level of polyploidy1-1D (ilp1-1D). The hypocotyls of dark-grown seedlings contained cells with ploidy levels as high as 32C in our assay (Figure 2A). Homozygous ilp1-1D also contained cells with levels as high as 32C, but the ratio of the 32C peak was greater compared with the wild type (isogenic siblings without a T-DNA insertion) in darkness (Figure 2A). This result was more apparent when the total cell numbers for each ploidy level were compared (Figure 2B). In dark-grown hypocotyls, the ratio of 32C cells was significantly increased in ilp1-1D compared with the wild type. In ilp1-1D, the values of 8C/32C and 16C/32C were 0.44 and 1.32, respectively, indicating that the mutant falls into our category. This result showed that the degree of endoreduplication was increased in this mutant. We measured increased nuclear volume by staining dark-grown hypocotyls with 4′,6-diamidino-2-phenylindole (DAPI). ilp1-1D seedlings had much enlarged nuclei compared with the wild type (Figures 2C and 2D).
Figure 2.
Ploidy Levels Were Highly Increased in Dark- and Light-Grown ilp1-1D Seedlings.
(A) Histograms of ploidy levels of hypocotyl cells of 7-d-old dark-grown seedlings. Left panel, wild type; right panel, homozygous ilp1-1D; x axis, nuclear ploidy; y axis, cell count. Approximately 5000 nuclei were counted in the wild type and ilp1-1D.
(B) Relative ratio of each cell ploidy of dark- and light-grown wild type and ilp1-1D. At least 20 seedlings were used for ploidy analysis, and it was replicated three times. Hyp. D, hypocotyl cells of dark-grown seedlings; Hyp. WL, hypocotyl cells of light-grown seedlings; Cot. WL, cotyledon cells of light-grown seedlings. Approximately 3000 to 5000 nuclei were counted in the wild type and ilp1-1D.
(C) and (D) DAPI staining of nuclei of the lower part of the hypocotyl of the wild type (C) and ilp1-1D (D).
(E) Morphology of 7-d-old dark-grown seedlings of the wild type (two left seedlings) and ilp1-1D (two right seedlings). Arrowheads indicate the junction of hypocotyl and root.
(F) Hypocotyl and root length of 7-d-old dark-grown wild-type and ilp1-1D seedlings.
(G) Diameter of hypocotyls of 7-d-old dark-grown wild-type and ilp1-1D seedlings.
(H) and (I) Transverse sections of dark-grown hypocotyls of the wild type (H) and ilp1-1D (I).
(J) and (K) Cotyledons of 7-d-old light-grown seedlings of the wild type (J) and ilp1-1D (K).
(L) Cotyledonal areas of 7-d-old light-grown wild-type and ilp1-1D seedlings.
At least 20 seedlings were measured in (F), (G), and (L). Bars in (B), (F), (G), and (L) indicate standard deviation. Bars = 10 μm in (C) and (D), 5 mm in (E), (J), and (K), and 100 μm in (H) and (I). Student's t test: * 0.001 > P versus the wild type in (F), (G), and (L).
In light conditions, wild-type hypocotyls contained cells of up to 16C in our assay, and the ratio of 16C cells was increased in ilp1-1D (Figure 2B). Cotyledons of light-grown ilp1-1D also had an increased number of 16C cells similar to the hypocotyl cells (Figure 2B).
Cell Volume Was Increased in ilp1-1D
We compared the ilp1-1D phenotype with the wild type in light and in darkness. The ilp1-1D homozygous lines did not show differences in hypocotyl length compared with the wild type when grown in the dark (Figures 2E and 2F). Instead of elongating, ilp1-1D hypocotyls became thicker than the wild type, indicating cells increased their volume along their horizontal axis (Figure 2G). We examined the cells in the hypocotyls by making transverse sections. We found that the cortical and endodermal cells of ilp1-1D had increased in diameter, resulting in thicker hypocotyls compared with the wild type (Figures 2H and 2I). There was almost no difference in the number of cells comprising the cortex and endodermis. These results indicated that the rise in the ploidy level in ilp1-1D increased the diameter of hypocotyl cells, resulting in an increase in cell volume. In addition to these hypocotyl phenotypes, we observed an increase in primary root length (Figures 2E and 2F).
In light-grown seedlings, the ilp1-1D homozygous mutant showed much expanded cotyledons compared with the wild type (Figures 2J to 2L). We examined the number of cells along the major and minor axes of the cotyledons. There was no difference in cell number between ilp1-1D and the wild type, indicating that the large cotyledon size of the mutant was caused by an increase in individual cell size and not by an increase in cell number (see Supplemental Figure 1A online). Adult ilp1-1D plants were almost the same height as the wild type (see Supplemental Figure 1B online).
ILP1 Encodes a Nuclear Protein Homologous to the C-Terminal Domain of GCF
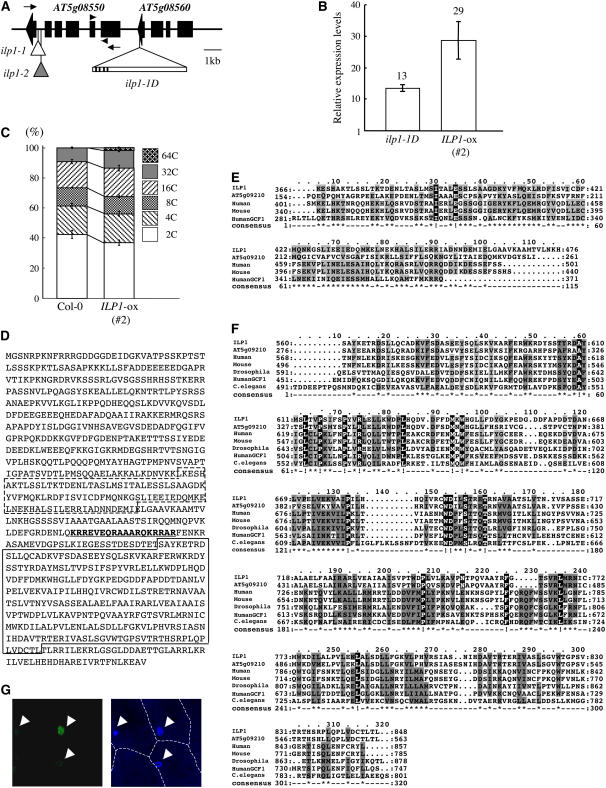
The activation T-DNA contains the hygromycin resistance gene as the selection marker. We examined T2 progenies of ilp1-1D heterozygous plants and found that ∼70% of the progeny showed hygromycin resistance, suggesting that there was only one T-DNA in the genome. All the hygromycin-resistant plants showed increased ploidy levels in the T3 generation. These results strongly suggest that the activation-tagged T-DNA was responsible for the increased polyploidy phenotype. The T-DNA flanking sequence was isolated by plasmid rescue. After sequencing, we found that the T-DNA was inserted in the coding region of AT5g08560 (Figure 3A). The distances between the putative start codons and the right border of the T-DNA were ∼1 kb for AT5g08550 and 7.4 kb for AT5g08560 (Figure 3A). We examined expression of AT5g08550 in a heterozygote of ilp1-1D and the wild type by real-time PCR and found that in ilp1-1D it was ∼13 times higher than in the wild type (Figure 3B). To determine whether the insertion in AT5g08560 caused increased polyploidy, we examined a T-DNA insertion line from the SALK T-DNA collections (SALK_095495) (Alonso et al., 2003). The T-DNA was inserted in the first exon in AT5g08560 (data not shown). This line did not show altered polyploidy (data not shown). These results indicated that AT5g08550 was a candidate for the gene responsible for the activation phenotype of ilp1-1D.
Figure 3.
Cloning and Characterization of the ILP1 Gene.
(A) T-DNA insertion sites in ilp1-1D. The triangle with bar indicates the activation-tagging T-DNA insertion site in ilp1-1D. Black lines on bar indicate the four copies of the CaMV 35S enhancer near the right border. Small white and gray triangles indicate the T-DNA insertion sites of ilp1-1 (SALK_030650) and ilp1-2 (SALK_135563), respectively. Arrowheads indicate primer positions for real-time PCR in Figures 3B and 6C and Supplemental Figure 2B online, and arrows indicate primer positions for semiquantitative RT-PCR in Figure 4B.
(B) Real-time PCR analysis showing expression of AT5g08550 (ILP1) in the wild type (Col-0), ilp1-1D, and ILP1-ox. Relative expression levels: expression levels of the ILP1 genes in ilp1-1D and AT5g08550 (ILP1) overexpressing line (#2) (ILP1-ox) relative to the wild type. Error bars indicate standard deviation.
(C) Relative ratio of each cell ploidy of dark-grown wild type (Col-0) and ILP1-ox (#2). Approximately 5000 nuclei were counted in the wild type and ILP1-ox.
(D) Amino acid sequence of ILP1 protein. Box with dashed line indicates motif 1, and box with solid line indicates motif 2. Underlined bold letters indicate putative NLS.
(E) Alignment of ILP1 motif 1 and its homologs. The ILP1 motif 1 was aligned with similar regions of other proteins: Arabidopsis, human, mouse, and human GCF1. The amino acid identity and similarity between the ILP1 motif 1 and its homologs are 38 and 42% for Arabidopsis, 27 and 48% for human, 27 and 48% for mouse, and 28 and 52% for human GCF1.
(F) Alignment of ILP1 motif 2 and its homologs. Motif 2 was aligned with similar regions of other proteins; Arabidopsis, human, mouse, Drosophila, human GCF1, and C. elegans. All alignments were performed using ClustalW and Mac Boxshade software. The amino acid identity and similarity between ILP1 motif 2 and its homologs are 72 and 77% for Arabidopsis, 27 and 45% for human, 27 and 44% for mouse, 28 and 48% for Drosophila, 22 and 43% for human GCF1, and 25 and 44% for C. elegans.
For (E) and (F), gray letters indicate functionally conserved amino acid residues in at least three members. White letters with black background indicate conserved amino acid residues in all members.
(G) Localization of ILP1:GFP. Left panel indicates fluorescence of ILP1:GFP. Right panel is a DAPI-stained nuclear image. Arrowheads indicate nuclei. The experiment was repeated three times.
To confirm this, transgenic plants overexpressing the AT5g08550 cDNA isolated by RT-PCR under the control of the cauliflower mosaic virus 35S (CaMV 35S) promoter were generated. Eight out of 15 lines showed significantly increased ploidy levels in the T2 generation recapitulating the ilp1-1D phenotype. AT5g08550 was highly expressed in these lines (Figure 3B). We examined the ploidy of dark-grown seedlings of a homozygous line. The relative ratio of the 32C peak was increased, and cells as high as 64C were observed (Figure 3C). Transgenic lines (#1 and #3) that did not show polyploidy phenotypes had almost the same ILP1 expression level as the wild type (see Supplemental Figures 2A and 2B online). AT5g08550 overexpression also reproduced the other phenotypes of ilp1-1D, such as enlarged cotyledons, thick hypocotyls, and elongation of primary roots (see Supplemental Figures 2C to 2H online). However, there were almost no differences in adult plant height and seed size compared with the wild type. These results strongly suggest that AT5g08550 is the corresponding gene for the ilp1-1D mutation. We designated AT5g08550 as ILP1.
The ILP1 gene encodes for a protein of 908 amino acid residues. We searched for ILP1 homologs in the protein database using the BLASTP program to identify conserved motifs. This search revealed that ILP1 had similarity to the C-terminal region of the GCF of human and other species (Figures 3D to 3F). The GCF protein was first isolated as a transcriptional repressor that bound to GC-rich sequence in the promoter region of the epidermal growth factor receptor (EGFR), β-actin, and calcium-dependent protease genes (Kageyama and Pastan, 1989). However, the first reported GCF cDNA clone was a chimeric gene, and the N terminus of the protein bound to a GC-rich region; its C-terminal region was from another cDNA with unknown function (Reed et al., 1998; Takimoto et al., 1999). To prevent confusion, we will call the gene containing this DNA binding domain authentic GCF and the gene encoding the C-terminal region CTILP1 (for C-terminal region of ILP1). ILP1 shows similarity with CTILP1. CTILP1 has homologs in mouse, Drosophila melanogaster, and Caenorhabditis elegans (Figures 3E and 3F). ILP1 has a homologous gene in the Arabidopsis genome (AT5g09210) (Figures 3E and 3F). We found two conserved motifs in ILP1 and other CTILP1 proteins. Motif 1 is at residues 371 to 465 of ILP1 (Figures 3D and 3E), and motif 2 is at residues 571 to 852 (Figures 3D and 3F). These two motifs are well conserved in CTILP1s of various species. Motif 2 in particular is well conserved, but motif 1 cannot be found in proteins of Drosophila and C. elegans. Significant similarity was not found in the N terminus region of CTILP1 proteins. We could not obtain any predicted structures of these two motifs using 3D-PSSM (Kelley et al., 2000).
In these two conserved regions, we found a putative nuclear localization signal (NLS) using the PSORT program (Nakai and Horton, 1999). This sequence is at residues 522 to 539 of ILP1 and is rich in Arg residues, a typical bipartite NLS (Figure 3D). The presence of this putative NLS motif suggests that ILP1 is a nuclear protein. To confirm this prediction, ILP1 was expressed as a fusion protein with the N-terminal region of the green fluorescent protein (GFP) under the control of the CaMV 35S promoter (ILP1:GFP). We examined localization in onion (Allium cepa) epidermal cells using biolistic bombardment. The ILP1:GFP fusion protein was detected in the nucleus, indicating that ILP1 is a nuclear protein (Figure 3G).
The T-DNA Insertional Mutation of ILP1 Causes Reduction of Polyploidy and Inhibition of Hypocotyl Elongation
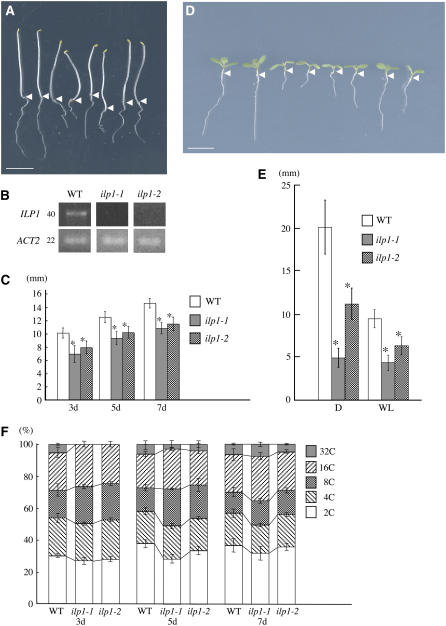
We isolated two T-DNA insertional mutants from the SALK T-DNA insertion lines (Alonso et al., 2003). Both mutants have T-DNA insertions in different positions in the 5th intron of ILP1 (Figure 3A, indicated by small triangles). Expression of the ILP1 gene in both these mutants was examined, and none was detected in lines homozygous for the T-DNA (Figure 4B), although a primer set (arrowheads in Figure 3A) specific for the region did amplify a PCR product (data not shown). This indicates that these mutants lacked full-length transcripts rather than having null mutations. We designated these homozygous mutants as ilp1-1 (SALK_030650) and ilp1-2 (SALK_135563), respectively. Both ilp1-1 and ilp1-2 had shorter hypocotyls and roots compared with their wild-type siblings in darkness (Figures 4A, 4C, and 4E). In the light, they both showed shorter hypocotyls and smaller cotyledons compared with the wild type, and there was inhibition of root elongation (Figures 4D and 4E). ilp1-1 that has the T-DNA insertion closer to the splicing acceptor site of the 5th intron exhibited a more severe morphological phenotype than ilp1-2. To examine complementation of these lines, ilp1-1 and ilp1-2 were crossed to each other. The F1 plants also showed short hypocotyls and roots compared with the wild type both in light and darkness (Figures 4A and 4D). This result indicated that these lines were allelic, and loss of ILP1 caused the short hypocotyl phenotype. To address the relationship between polyploidy and ILP1 function, we examined the ploidy levels of homozygous ilp1-1 and ilp1-2 in the dark. We observed a reduction in the number of 32C cells in both ilp1-1 and ilp1-2 in 3-d-old seedling hypocotyl cells (Figure 4F). To examine the relationship between hypocotyl length and ploidy in ilp1-1 and ilp1-2, these mutants were analyzed at different stages of seedling development. ilp1-1 and ilp1-2 had shorter hypocotyls compared with the wild type at all stages of development in darkness (Figure 4C). However, reduced ploidy levels of hypocotyl cells were recovered to those of the wild type at day 7 after imbibition, indicating that reduced polyploidy is not the consequence of short hypocotyl length.
Figure 4.
Loss of ILP1 Function Decreases Polyploidy Levels.
(A) Morphology of dark-grown wild type, ilp1-1, and ilp1-2. Seedlings were grown for 5 d. Each pair of seedlings from left to right is wild type, ilp1-1, heterozygotes of ilp1-1 and ilp1-2, and ilp1-2, respectively. Isogenic wild-type siblings of ilp1-1 were used as the wild type. The same result was obtained from wild-type siblings of ilp1-2. Arrowheads indicate the junction of hypocotyl and root.
(B) Semiquantitative RT-PCR for the expression of ILP1. Number on the left indicates cycles of PCR. ACT2 was used as a control.
(C) Hypocotyl length of 3-, 5-, and 7-d-old dark-grown wild-type, ilp1-1, and ilp1-2 seedlings.
(D) Seven-day-old light-grown seedlings of the wild type, ilp1-1, and ilp1-2. Alignment of seedlings is same as in (A). Arrowheads indicate the junction of hypocotyl and root.
(E) Root length of 7-d-old dark- and light-grown wild type, ilp1-1, and ilp1-2. D, darkness; WL, white light.
(F) Relative ratio of each cell ploidy of 3-, 5-, and 7-d-old dark-grown wild-type, ilp1-1, and ilp1-2 homozygotes. Approximately 3000 nuclei were counted in the wild type, ilp1-1, and ilp1-2.
Bars in (A) and (D) = 5 mm. Error bars in (C), (E), and (F) indicate standard deviation. Student's t test: * 0.001 > P versus the wild type in (C) and (E). At least 20 seedlings were measured in (C) and (E).
ILP1 Functions as a Transcriptional Repressor
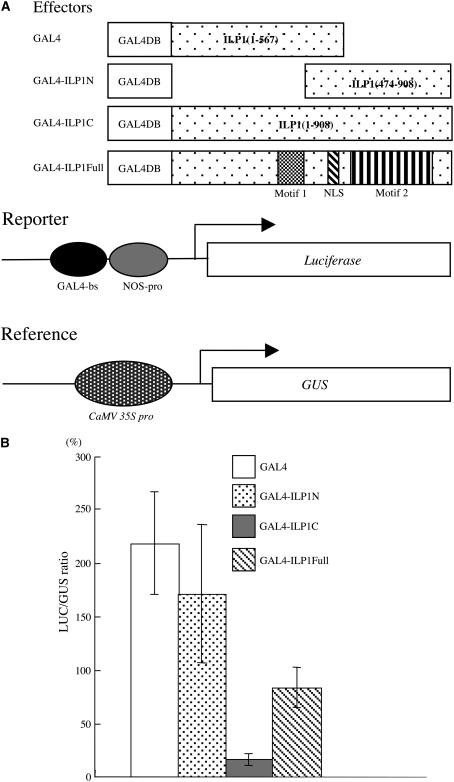
It has been reported that the chimeric GCF first identified functioned as a transcriptional repressor (Kageyama and Pastan, 1989). The N-terminal portion of this protein (GCF) has homology with GCF2 and has DNA binding activity (Reed et al., 1998). ILP1 has homology with CTILP1s, but they have not been examined in detail in mammalian cells. To understand the function of ILP1, we performed an in vivo transcriptional assay (Yamamoto and Deng, 1998). ILP1 cDNA was fused to the C-terminal region of the GAL4 DNA binding domain (GAL4-ILP1Full). This chimeric plasmid was introduced into tobacco leaf cells by biolistic bombardment along with a luciferase (LUC) reporter plasmid containing the GAL4 binding sequence in the promoter region (Figure 5A). The reporter plasmid was prepared from an Escherichia coli strain that lacked DNA methylase to ensure it was demethylated. This would enhance basal activity in case there was any transcriptional repression. When GAL4-ILP1Full was used, we observed a reduction in reporter activity (Figure 5B). ILP1 has two conserved motifs. We expressed the part of the ILP1 protein that contains one of these motifs with an NLS. GAL4-ILP1N is a chimera containing the GAL4 DNA binding domain with the N-terminal region of ILP1 (residues 1 to 567) (Figure 5A). This chimera contains motif 1, and the NLS and did not show the strong repression as observed with the GAL4-ILP1Full protein (Figure 5B). However, when the C-terminal region of ILP1 (residues 474 to 908) that contains motif 2 was used, we observed much stronger repression of LUC reporter activity (Figures 5A and 5B). These results indicated that ILP1 functions as a transcriptional repressor in vivo and that motif 2 is responsible for this repressor activity.
Figure 5.
ILP1 Functions as a Transcriptional Repressor in Plant Cells.
(A) The constructs used for the in vivo transcription assay. GAL4-ILP1N, GAL4 DNA binding domain (GAL4DB) is fused to the N-terminal region of ILP1 (residues 1 to 567); GAL4-ILP1C, GAL4DB is fused to the C-terminal region of ILP (residues 474 to 908); GAL4ILP1Full, GAL4DB is fused to the full-length ILP1. The reporter plasmid contains a GAL4 binding site and 0.2 kb of the nopaline synthase promoter (NOS-pro) upstream of the LUC reporter gene. The reference plasmid serves to monitor the transformation efficiency by β-glucuronidase (GUS) expression controlled by a constitutive CaMV 35S promoter.
(B) In vivo transcription assay in tobacco leaves. LUC/GUS ratio: LUC expression (reporter) was normalized with GUS expression (reference). Error bars indicate standard error. The experiment was repeated five times.
ILP1 Regulates CYCA2 Expression in Arabidopsis
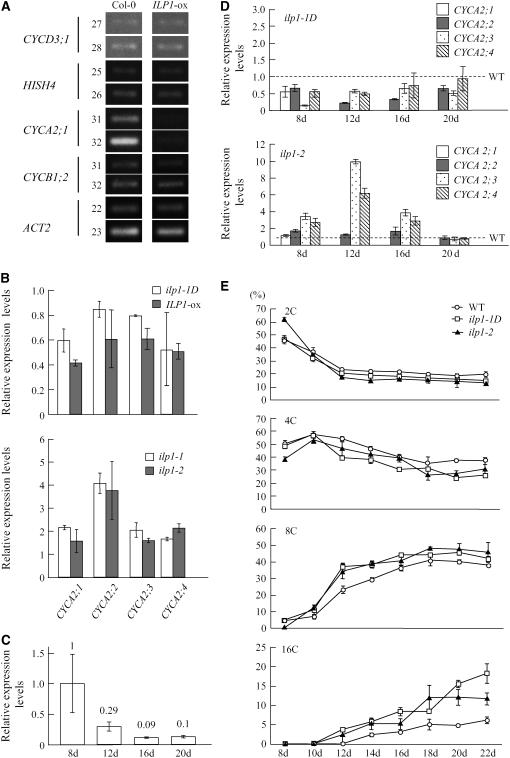
Endoreduplication is a type of cell cycle, and switching to this cycle from the regular mitotic cell cycles may involve different cell cycle–related gene expression. We examined the expression of some cell cycle–related genes that express at specific phases of the mitotic cell cycle. We used CYCD3;1 for G1-phase-specific genes (Riou-Khamlichi et al., 2000), E2FB/E2F1 and E2FD/DEL2 for G1/S-phase-specific genes, CDC6 and HistonH4 (HISH4) for S-phase-specific genes (Mariconti et al., 2002), CYCA2;1 for S/G2-phase-specific genes, and CYCB1;2 for G2/M-phase-specific genes (Shaul et al., 1996). Expression of these genes was analyzed by semiquantitative RT-PCR. An ILP1 overexpressing line (ILP1-ox, line #2, Figure 3B) that showed increased polyploidy was used, and expression of these cell cycle–related genes in dark-grown seedlings was examined. There were no differences in expression of CYCD3;1, E2FB, E2FD/DEL2, CDC6, HISH4, and CYCB1;2 between the wild type (Col-0) and the ILP1 overexpressing line (Figure 6A; E2FB, E2FD/DEL2, and CDC6 in Supplemental Figure 3A online). However, expression of CYCA2;1 was significantly reduced in the transgenic line compared with the wild type (Figure 6A). CYCA2;1 is part of a gene family, and there are four CYCA2 members in the Arabidopsis genome (Vandepoele et al., 2002). We examined the expression of the CYCA2 genes in ILP1-ox and ilp1-1D more precisely using real-time PCR. We observed reduced expression of all of the CYCA2 members (Figure 6B); in particular, expression of CYCA2;1 in the ILP1-ox line was reduced to ∼40% of that of the wild type. Examination of expression of the CYCA2 genes in the ILP1 insertion mutants revealed that both ilp1-1 and ilp1-2 showed an increase in all of them (Figure 6B).
Figure 6.
ILP1 Represses CYCA2 Expression in Arabidopsis.
(A) Semiquantitative RT-PCR analysis of cell cycle–specific genes. CYCD3;1, HISH4, CYCA2;1, and CYCB1;2 are G1-, S-, G2-, and M-phase-specific markers, respectively. ACT2 was used as a control. Numbers on left side indicate cycles of PCR.
(B) Real-time PCR analysis of CYCA2 gene family members. Expression levels of the CYCA2 family genes were normalized with ACT2 expression. Relative expression levels: expression levels of the CYCA2 genes in each mutant line and an ILP1-ox line relative to the wild type. RNA was isolated from 7-d-old dark-grown hypocotyls of ilp1-1D and ILP1-ox (top panel) and from 3-d-old dark-grown hypocotyls of ilp1-1 and ilp1-2 (bottom panel). The experiment was repeated four times.
(C) Real-time PCR analysis of ILP1 in the wild type (Col-0). The numbers indicate ILP1 expression relative to day 8. The experiment was repeated four times.
(D) Real-time PCR analysis of CYCA2 gene family members in first leaves of ilp1-1D and ilp1-2 at four developmental stages. Expression levels of the CYCA2 family genes were normalized with ACT2 expression. Relative expression levels: expression levels of the CYCA2 genes in each mutant line relative to the wild type. CYCA2;1 expression was not detected in the wild type and ilp1-1D after day 12. The experiment was repeated four times.
(E) Ploidy distribution patterns of first leaves of the wild type, ilp1-1D, and ilp1-2 at several developmental stages. The fraction of each ploidy was plotted as wild type (open circle), ilp1-1D (open square), and ilp1-2 (closed triangle). Isogenic wild-type siblings of ilp1-1D were used as the wild type. The same result was obtained from wild-type siblings of ilp1-2. The experiment was repeated three times.
Error bars in (B) to (E) indicate standard deviation.
We investigated expression of ILP1 during leaf development. Expression was gradually reduced in accordance with first leaf development, and at day 20 after imbibition it reduced to one-tenth of the day 8 level when the first leaves are in the proliferating phase (Vlieghe et al., 2005) (Figure 6C). We also investigated expression of the CYCA2 genes in comparison with the wild type during leaf development (Figure 6D). All the CYCA2 genes had high expression at day 8, and this was gradually reduced as in ILP1 (data not shown) (Imai et al., 2006). In ilp1-1D, expression of all CYCA2 genes was reduced compared with the wild type (Figure 6D). However, expression of all the CYCA2 genes was increased in ilp1-2 compared with the wild type, and relatively high expression was observed in CYCA2;3 and CYCA2;4 at day 12 (Figure 6D). We could not detect CYCA2;1 expression after day 8 in the wild type, ilp1-1D, and ilp1-2.
Observation of polyploidy levels during leaf development revealed a reduction in the 2C fraction and an increase in the 8C and 16C fractions in the wild type, ilp1-1D, and ilp1-2, although there were apparent differences between them. ilp1-1D gradually increases in the 8C and 16C fractions compared with the wild type (Figure 6E) after day 10. At day 22, the fraction of 16C cells was increased to 18% in ilp1-1D compared with 7% in the wild type (Figure 6E). The 2C fraction was >60%, and the 8C and 16C fractions were not detected in ilp1-2 at day 8 (Figure 6E). However, the 8C and 16C fractions were increased in ilp1-2 as in ilp1-1D after day 10 (Figure 6E).
Mouse ILP1 Regulates Cyclin A2 Gene Expression in Mammalian Cells
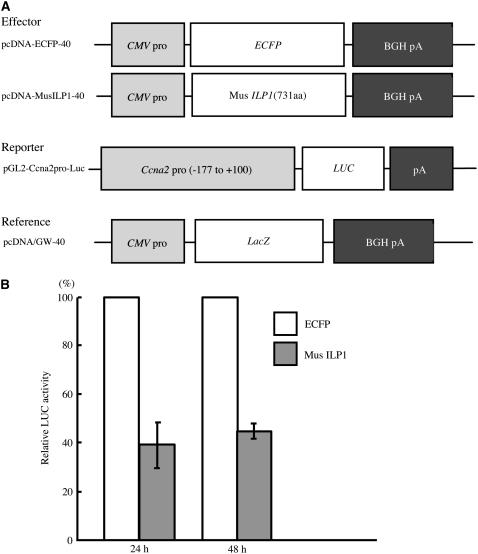
To understand whether the reduction of cyclin A2 expression is also observed in mammalian cells, a cotransfection assay was performed using mouse NIH3T3 cells. Mouse ILP1 homolog cDNA (Figures 3E and 3F) was isolated by RT-PCR and cloned into an expression plasmid containing the Cytomegalovirus promoter (Figure 7A). This cDNA was cotransfected into NIH3T3 cells with a mouse cyclin A2 (Ccna2) promoter-LUC reporter by lipofection. We used a Ccna2 promoter that contained −177 to +100 of the transcriptional start site (Huet et al., 1996). This region shows conservation between mouse and human cyclin A2 promoters. As an internal standard for this assay, we used the β-galactosidase (LacZ) gene. As shown in Figure 7B, we observed a reduction in reporter activity in the cells transfected with the mouse ILP1 gene both 24 and 48 h after transfection.
Figure 7.
ILP1 Represses Ccna2 Expression in Mouse NIH3T3 Cells.
(A) The constructs used for the in vivo transcription assay in mouse NIH3T3 cells. pcDNA-ECFP-40 contains the enhanced cyan fluorescent protein (ECFP) gene and was used as a control, and pcDNA-MusILP1-40 contains the mouse ILP1 cDNA (731 amino acids). The reporter plasmid consists of the Ccna2 promoter region (−177 to +100 bp of the transcriptional start site) fused to the LUC gene. The reference plasmid serves to monitor the transfection efficiency by β-galactosidase (LacZ) expression. CMV pro, Cytomegalovirus promoter; BGH pA, bovine growth hormone polyadenylation site.
(B) In vivo transcription assay in mouse NIH3T3 cells. LUC activity was normalized with β-galactosidase activity. Relative LUC activity: LUC activity of mouse ILP1 relative to enhanced cyan fluorescent protein. Activities were measured 24 and 48 h after transfection. Error bars indicate standard deviation. The experiment was repeated four times.
Reduction of CYCA2;1 Expression Induces Increase in Polyploidy
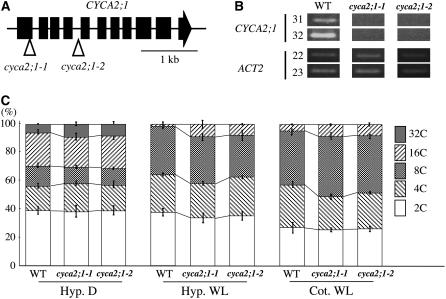
Of the CYCA2 family, CYCA2;1 has been extensively studied, and this gene's expression is reported to be specific to the S/G2-phase (Shaul et al., 1996). To test whether reduction of CYCA2;1 expression is related to endoreduplication, we examined the ploidy levels of CYCA2;1 T-DNA insertion mutants obtained from the SALK T-DNA collection (Alonso et al., 2003). Two independent T-DNA insertion lines were examined. In insertion line 1 (cyca2;1-1), the T-DNA was in the 1st exon (SALK_121077), and in insertion line 2 (cyca2;1-2), it was in the 4th intron (SALK_136750) (Figure 8A). RT-PCR analysis suggested these two lines were null (Figure 8B). Both cyca2;1-1 and cyca2;1-2 homozygous lines showed almost no morphological differences compared with the wild type at the adult stage. Dark-grown seedling morphology was also the same as the wild type. When we observed the ploidy levels in these T-DNA insertion lines, both showed an increased ratio of 32C cells compared with the wild type in dark-grown hypocotyls (Figure 8C). In the hypocotyls of light-grown seedlings, we observed an increase in levels of 16C cells (Figure 8C). We also examined ploidy levels in light-grown cotyledonal cells. Although the size of the cotyledons was not changed compared with the wild type, the 16C fraction was increased in both cyca2;1-1 and cyca2;1-2 (Figure 8C). These data indicate that loss of CYCA2;1 expression induces an increase in polyploidy.
Figure 8.
Loss of CYCA2;1 Reduced Polyploidy Level.
(A) Loci of T-DNA insertions in CYCA2;1. Triangles indicate insertion sites of T-DNAs of cyca2;1-1 (SALK_121077) and cyca2;1-2 (SALK_136750).
(B) Semiquantitative RT-PCR analysis of CYCA2;1. Numbers on left side indicate cycles of PCR.
(C) Relative ratio of each cell ploidy of dark- and light-grown wild-type, cyca2;1-1, and cyca2;1-2 homozygotes. Hyp. D, hypocotyl cells of dark-grown seedlings; Hyp. WL, hypocotyl cells of light-grown seedlings; Cot. WL, cotyledon cells of dark-grown seedlings. Isogenic wild-type siblings of cyca2;1-1 were used as the wild type. The same result was obtained from wild-type siblings of cyca2;1-2. Approximatley 3000 nuclei were counted in the wild type, cyca2;1-1, and cyca2;1-2. Error bars indicate standard deviation.
DISCUSSION
ILP1 Acts as an Enhancer for Endoreduplication
ilp1-1D has increased polyploidy both in hypocotyls and cotyledons, and the relative fractions of the highest polyploidy cells were increased (Figures 2A and 2B). However, two ILP1 insertion mutants (ilp1-1 and ilp1-2) had a reduction in the number of cells with the highest polyploidy early in dark-grown hypocotyl growth (Figure 4F). These results indicate that ILP1 enhances the endocycle in endoreduplicating cells in seedlings. Although no clear connection between polyploidy and cell volume has been reported, we observed thicker hypocotyls in ilp1-1D dark-grown seedlings (Figures 2G to 2I) and shorter hypocotyls in the ILP1 insertion mutants (Figures 4A, 4C, and 4D). The number of cells comprising the hypocotyl cell files in ilp1-1D and the ILP1 insertion mutants had not changed. We concluded that ILP1 affects cell volume through an increase in polyploidy levels. The ILP1 insertion mutants have short hypocotyls at day 7, and polyploidy levels of ilp1-1 and ilp1-2 were recovered to that of the wild type at this time (Figures 4A and 4F). Reduced polyploidy levels at day 3 in ilp1-1 and ilp1-2 affect the mature hypocotyl length (Figure 4C). This may be due to the presence of an ILP1 homolog (AT5g09210) or some unknown compensation mechanism for the control of endoreduplication.
ILP1-ox plants had much higher ILP1 expression compared with ilp1-1D (Figure 3B). They had as high as 64C cells in their hypocotyls, indicating that higher expression of ILP1 causes much enhancement of polyploidy compared with ilp1-1D (Figure 3C). This observation indicated that increased polyploidy level is caused by overexpression of the ILP1 gene.
ILP1 Controls Endoreduplication through Regulation of CYCA2 Transcription
We found ILP1 functions as a transcriptional repressor both in plants and mammals, and it specifically represses CYCA2 genes. We could not observe any differences in the expression of CYCA1;1, a gene that is also expressed in the S/G2-phase (see Supplemental Figure 3B online). In Medicago sativa, cycA2;2 expression was observed in dividing cells but not in endoreduplicating cells (Roudier et al., 2003). Tobacco CYCA3;2 resulted in impaired cell differentiation and endoreduplication when overexpressed in Arabidopsis (Yu et al., 2003). It was reported that Arabidopsis CYCA2;1 is expressed not only in meristems but also in differentiated cells, such as guard cells, where endoreduplication is strictly inhibited (Burssens et al., 2000). In Drosophila, it has been reported that Cyclin A is one of the key components of chromosomal DNA replication that prevents reinitiation of DNA replication, and overexpression of Drosophila cyclin A caused a reduction in polyploidy levels and inhibition of the endocycle (Hayashi and Yamaguchi, 1999). These observations suggested that the CYCA2 genes' primary role is the control of endoreduplication, and this control is brought about by the absence or a reduction in the CYCA2s. It is possible that ILP1 controls the degree of the endocycle by repressing the expression of the CYCA2 genes. However, the ILP1 insertion mutants caused several morphological changes, including short hypocotyls and roots, small cotyledons, and upregulation of CYCA2 genes. Strong induction of CYCA2;3 using an estrogen-inducible system did not completely mimic the ILP1 insertion mutant phenotypes (Imai et al., 2006). This result indicates that ILP1 controls several genes involved in endocycle control and that CYCA2 genes are one of the targets of ILP1.
Two T-DNA insertional mutants of CYCA2;1 were examined, and both had increased polyploidy levels in dark-grown hypocotyl cells. This increase was also observed in cotyledon cells in light-grown seedlings (Figure 8C). However, increased cell volume was not observed in CYCA2;1 mutants. This may be explained if the CYCA2 members play overlapping roles in endoreduplication to ensure maintenance of this cell cycle that is so important for development. This assumption was supported by the observation that increased polyploidy levels in CYCA2;1 mutants were lower compared with ilp1-1D and ILP1-ox. Since ILP1 does not bind to the CYCA2;1 promoter in in vitro assays and CYCA2 genes also have tissue specificity, ILP1 may regulate CYCA2 gene expression by interacting with tissue-specific transcription factors or by controlling upstream genes that regulate their expression.
ILP1 Controls First-Leaf Polyploidy
Leaf development is another good model in which to analyze polyploidy control. At different developmental stages, true leaves are made up of different proportions of dividing and endoreduplicating cells. Cell division is frequently observed during the proliferation stage in 8-d-old first leaves (Boudolf et al., 2004; Vlieghe et al., 2005). ILP1 expression is high at day 8 and gradually decreases during leaf development (Figure 6C). At day 8, first leaves contain many proliferating cells and many endoreduplicating cells, such as 4C. As the leaf develops, these endoreduplicating cells continue endoreduplication and a proportion of the 4C cells become 8C cells. Again, as the leaf develops, a proportion of the 8C cells become 16C cells (Figure 6E). Reduced ILP1 expression is correlated with the proportion of cells endoreduplicating (Figures 6C and 6E). Arabidospsis CYCA2;3 is expressed during the termination period of endoreduplication in trichomes, and loss of CYCA2;3 results in increased polyploidy in mature leaves (Imai et al., 2006). Reduced ILP1 expression during leaf development causes an increase in expression of the CYCA2 genes, especially of CYCA2;3 to suppress the endocycle, resulting in maturation of the leaves (Figures 6C and 6E). In ilp1-1D, the proportion of higher polyploidy cells was increased compared with the wild type after day 12 when the leaves start to expand. After day 20, the first leaves enter the maturation stage and ilp1-1D shows an increase in the highest cell fraction at this stage compared with the wild type, and the expression of CYCA2s was repressed in ilp1-1D (Figures 6D and 6E). Increased ILP1 in ilp1-1D could inhibit the termination of endoreduplication by reducing the activities of the CYCA2s that are required for leaf maturation.
ilp1-2 has reduced polyploidy profiles at day 8 (Figure 6E). Contrary to expectation, the proportions of 8C and 16C cells in ilp1-2 were increased to the levels of ilp1-1D after day 12, although CYCA2 expression was much increased (Figures 6D and 6E). A similar tendency of recovery of polyploidy patterns was observed in 7-d-old hypocotyls of ilp1-1 and ilp1-2 (Figure 4F). Endoreduplication, subsequent cell elongation, and expansion are important for plant development. Loss of one of the components of the endocycle may be compensated for during hypocotyl and leaf development by other mechanisms. One of these mechanisms may be posttranslational regulation of Cyclin A. Fizzy-related protein (FZR) is a substrate-specific activator of the anaphase promoting complex and is reported to be a negative regulator of Cyclin A in Drosophila. Loss of fzr causes ectopic Cyclin A accumulation and inhibits endoreduplication in the salivary gland (Sigrist and Lehner, 1997). CCS52 of Medicago is a plant homolog of FZR, and it degrades mitotic cyclin in fission yeast. Antisense expression of ccs52 also reduces endoreduplication in Medicago (Cebolla et al., 1999). Therefore, the effect of increased CYCA2 expression on polyploidy in ilp1-2 could be compensated for by Arabidopsis CCS52. Control of endoreduplication through CYCA2/Cyclin A may be regulated both at the transcriptional level by ILP1 and at the protein level by CCS52.
Possible Mechanism by Which ILP1 Represses cyclin A2 Expression
The mouse Ccna2 promoter used for the transfection assay is only 277 nucleotides in length and is conserved between mouse and human. There are at least four cis-elements in this region: the cAMP response element, nuclear factor-1/nuclear factor-Y, a cell cycle responsive element (CCRE), and a cell cycle gene homology region (CHR). When this region was fused to the LUC reporter gene, it was shown to function as a proliferation-regulated promoter in mouse NIH3T3 cells, and this reporter expression is negatively regulated by the arrest of cell proliferation (Huet et al., 1996). A mutation in the CCRE sequence causes relief of transcriptional repression and results in a constitutively active promoter (Huet et al., 1996). Brahma/SNF2α (Brm) is a catalytic subunit of the SWI/SNF complex, and it was found to occupy the cyclin A promoter on cell cycle arrest (Dahiya et al., 2001; Kadam and Emerson, 2003). Ccna2 is constitutively expressed in Brm-deficient cells and is repressed through CCRE and CHR by Brm (Coisy et al., 2004). These observations in combination with our results using NIH3T3 cells may indicate that CCRE and CHR are the target sequences by which Mus ILP1 controls Ccna2 expression. Arabidopsis CYCA2;1 promoter sequences (nucleotides −860 to +160 or −1900 to +160) were used in a gel shift assay. However, ILP1 produced by in vitro translation did not bind to CYCA2;1 promoter fragments (data not shown). We concluded that ILP1 functions as a repressor that regulates CYCA2 genes in Arabidopsis and Ccna2 in mouse NIH3T3 cells (Figure 7), although indirectly. We speculate that ILP1 regulates CYCA2/Ccna2 expression in conjunction with Brm. It will be interesting to know whether common regulatory elements also exist in plants.
ILP1 Maintains Potential for Endoreduplication
It has been reported that overexpression of Arabidopsis CDT1 increases polyploidy levels in first leaves (Castellano Mdel et al., 2004). CDT1 is known to be an essential licensing factor and is suppressed by phosphorylation by Cyclin A and CDKs in human cells (Sugimoto et al., 2004). Overexpression of ILP1 also increases polyploidy levels in first leaves. Although more data are required, ILP1 may control endoreduplication through regulation of CYCA2 in first leaves, and CDT1 is involved in this regulation. However, overexpression of E2FA-DPA upregulates S-phase-specific genes, including some licensing factors, such as ORC, MCM, and CDC6. Overexpression of E2FA-DPA increases polyploidy levels and at the same time increases cell number in seedlings compared with the wild type, indicating that extra endoreduplication and cell division occur simultaneously (De Veylder et al., 2002). These phenotypes have not appeared in ILP1-ox. Therefore, the E2FA-DPA–dependent polyploidy control mechanism may be different from that of ILP1. E2FA-DPA may promote endoreduplication through initiation of the endocycle, whereas ILP1 maintains the potential of endoreduplication by controlling the CYCA2s.
METHODS
Plant Material and Growth Conditions
All plants were grown on germination medium (GM) plates (Valvekens et al., 1988) containing 10 mg/mL of sucrose with or without antibiotics. Plants were grown in a temperature-controlled incubation room at 22°C under white light conditions (15 W/m2 for light-grown cotyledons or 5 W/m2 for light-grown hypocotyls) or in complete darkness. SALK T-DNA insertion mutants were backcrossed with Col-0 twice to purify mutations for physiological experiments.
Ploidy Analysis
Nuclei were extracted and stained with CyStain UV precise P (Partec) following the manufacturer's protocol. Flow cytometric analysis was performed by a Ploidy Analyzer (Partec).
Generation of ILP1 Overexpression Transgenic Lines
ILP1 cDNA spanning the full coding region was amplified by PCR from the Super Script Arabidopsis thaliana cDNA library (Invitrogen) using primers ILP1-F (5′-GGGGTACCATGGGAAGTAACCGTCCTAAG-3′) and ILP1-R (5′-ACGCGTCGACTCAAACTGCCTCCTTAAGATT-3′). The ILP1 cDNA obtained was digested with KpnI and SalI and cloned into the yy45 vector, a derivative of pPZPY122 (Yamamoto et al., 2001). ILP1 cDNA was cloned between the CaMV 35S promoter and the nopaline synthase terminator of yy45 (yy45:ILP1). Agrobacterium tumefaciens (strain GV3101) was transformed with yy45:ILP1 by electroporation, and transformants were selected on Luria-Bertani medium containing 70 μg/mL of chloramphenicol. Arabidopsis wild type (Col-0) was transformed by the floral dipping method (Clough and Bent, 1998). Transgenic seedlings were selected on GM plates containing 50 μg/L of kanamycin and 100 μg/L of cefotaxime.
Intracellular Protein Localization
For ILP1:GFP, GFP was amplified from yy217 using GFPn-F (5′-TCTAGAGGATCCCCCGGGGGTACCGTCGACATGGCAATGAGTAAAGGAGAA-3′) and GFPn-R (5′-CGAGCTCTTATTTGTAAAGTTCATC-3′) primers. The GFP fragment was digested with XbaI and SacI and cloned into yy45 (yy45GFPn). ILP1 cDNA was amplified using ILP1-F and ILP-R2-SAL (5′-ACGCGTCGACAACTGCCTCCTTAAGATTG-3′) primers from the Super Script Arabidopsis cDNA library and cloned into the KpnI and SalI sites of yy45GFPn to generate ILP1:GFP. Onion (Allium cepa) epidermal cells were peeled off and placed on GM plates. The ILP1:GFP construct was loaded onto gold particles (1-μm diameter) following the manufacturer's protocol. Particles were delivered into the onion epidermal cells using the Biolistic PDS-1000/He system (Bio-Rad). The bombardment parameters were as follows: rapture disk bursting pressure, 600 psi; distance to target tissue, 9 cm. GFP fluorescence was observed with a BX60 microscope (Olympus) 18 to 36 h after bombardment.
Semiquantitative RT-PCR and Real-Time PCR Analysis
Semiquantitative RT-PCR analysis was performed as described previously (Kimura et al., 2001). Seeds were sown on GM plates containing sucrose, and plates were treated at 5°C for 5 d and incubated at 22°C under white light for 3 d. Seedlings were harvested, and total RNA was isolated as described previously (Yoshizumi et al., 1999). The primer sets used for amplification of the CYCA2;1, CYCB1;2, and CYCD3;1 cDNAs were as described by Richard et al. (2001); primers for amplifying E2FB, E2FD/DEL2, and HISH4 cDNAs were as described by Mariconti et al. (2002); primers for amplification of the ACT2 cDNA were as described by Himanen et al. (2002); primers for amplifying CDC6 were CDC6F (5′-CTGTTTCAATCCCTAAGACC-3′) and CDC6R (5′-CAGACACTGTTTCAGGACAA-3′). For the expression studies of CYCA2;1 in the CYCA2;1 T-DNA insertion mutants and ILP1 in ilp1-1D and ILP1-ox, the following sets of primers were used: CYCA2;1, CycA2;1-F (5′-GGACTAGTGAGCTCGCACACTAATGCGAAGAAAG-3′) and CycA2;1-R (5′-CCGCTCGAGTCTAGAGCAGATGCATCTAAAGATTC-3′).
Real-time PCR was performed according to the protocol of Mx3000P (Stratagene). Total RNA was isolated from seedlings as described above using TRIzol (Invitrogen) and used as a template to synthesize first-strand cDNA using a SuperScript first-strand synthesis system following the manufacturer's protocol (Invitrogen). The PCR used SYBR Green Realtime PCR Master Mix (TOYOBO) and was analyzed by the Mx3000P multiplex quantitative PCR system (Stratagene). To investigate expression levels of the ILP1 gene and the CYCA2 gene family, the following sets of primers were used: ILP1, ILP1realF (5′-AGCTTGCCAAGAAGGCATTG-3′) and ILP1realR (5′-TCATCAACGACGCAGTCAGA-3′); CYCA2;1, CycA2;1-F (5′-CGCTTCAGCGGTTTTCTTAG-3′) and CycA2;1-R (5′-ATCCTCCATTGCAAGTACCG-3′); CYCA2;2, CycA2;2-F (5′-TGTATGTGTTGGCCGTAATG-3′) and CycA2;2-R (5′-TGGTGTCTCTTGCATGCTTA-3′); CYCA2;3, CycA2;3-F (5′-CTCTATGCCCCTGAAATCCA-3′) and CycA2;3-R (5′-ACCTCCACAAGCAATCAAC-3′); CYCA2:4, CycA2;4-F (5′-CAAAGCCTCCGATCTCAAAG-3′) and CycA2;4-R (5′-CTTGTCCGGTAGCTCTCCAG-3′); CYCA1:1, CycA1;1-F (5′-CGATGACGAAGAAACGAGCA-3′) and CycA1;1-R (5′-TGGCATTAACGCAAACACTTG-3′); and ACT2, Act2-F (5′-CTGGATCGGTGGTTCCATTC-3′) and Act2-R (5′-CCTGGACCTGCCTCATCATAC-3′).
Light Microscopy
Plant material was fixed in 4% paraformaldehyde in a buffer containing 20 mM sodium cacodylate, pH 7.0, for 24 h at 4°C, dehydrated through an ethanol series, and then embedded in Technovit 7100 resin (Kulzer and Co.). The 2.5-μm-thick sections were cut with a glass knife on an ultramicrotome, placed on cover slips, and dried. They were stained with 1% toluidine blue O in 0.1 M phosphate buffer saline, pH 7.0, for 30 s and then washed in distilled water for 10 s. The samples were observed with an Olympus IX70 microscope.
In Vivo Transcription Assay
The −150 to +5 region of the nopaline synthase promoter was amplified by PCR from pMA560 (Ma et al., 1988) using the following primers: 5′-GGGGGATCCGCGGGTTTCTGGAGTTTAATG-3′ and 5′-CCTCTAGAGACTCTAATTGGATACCGAGG-3′. The amplified fragment was digested with BamHI and XbaI and cloned into the BamHI-XbaI site of yy76 (Yamamoto and Deng, 1998), leaving a second BamHI site between the XbaI site and GUS in the resultant clone. This clone, yy78, was digested with BamHI and HindIII and cloned into the BamHI-HindIII site of pBIL221 (Nakamura et al., 2002) to give yy97. The yy97 plasmid was prepared from the GM2163 (Dam−/Dcm−) strain for this assay. To make effector plasmids, various-length ILP1 cDNAs were amplified using ILP1-F and ILP1-R primers for GAL-ILP1Full, ILP1-F and ILP1-No 2-R (5′-GGGGTACCTTAGGATCCGTCACTCTCATCAGTGCT-3′) primers for GAL4-ILP1N, and ILP1-No 5-F (5′-GCTCTAGAGGATCCATGACAGTTCTAAACAAACAT-3′) and ILP1-R primers for GAL4-ILP1C. The cDNAs obtained were digested with KpnI and SalI and cloned into the KpnI-SalI site of yy64 (Yamamoto and Deng, 1998). Biolistic bombardment was performed as described above into tobacco leaves (Nicotiana tabacum cv SR1). LUC activity was measured using a Lumat LB9507 luminometer (Perkin-Elmer).
Cell Culture and Transfection
Mouse (Mus musculus) NIH3T3 cells were cultured in DMEM medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen). For transfection, ∼2.0 × 105 NIH3T3 cells were seeded in each well of a 12-well titer plate. After incubation in a CO2 incubator (5% CO2) for 2 d, transfection was performed with Lipofectamine 2000 (Invitrogen). After 24 and 48 h of transfection, LUC activity was measured according to the manufacturer's protocol (Promega) using a TD-20/20 luminometer (Promega). The mouse ILP1 gene was amplified from total RNA prepared from NIH3T3 cells using the following primer set: mouseILP1 forward (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGCCACCATGGACATGGAGAGCGAGAAGG-3′) and mouseILP1 reverse (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCCTATTTTCCTTCAATCAGAGACTT-3′). It was confirmed by sequencing. The PCR fragment was cloned into pcDNA-DEST40 using the Gateway cloning system (Invitrogen).
Accession Numbers
The cDNA sequence for ILP1 was submitted to DDBJ under accession number AB253763. Sequence data of ILP1 homologs from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Arabidopsis, AT5g09210; human, AAK68721; mouse, AAK68725; human GCF1, AAA35598; Drosophila, AAF54074; and C. elegans, NP492341.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Morphological Phenotype of ilp1-1D.
Supplemental Figure 2. Morphological Phenotype in Dark- and Light-Grown Seedlings of ILP1-ox.
Supplemental Figure 3. Cell Cycle–Related Gene Expression in ILP1-ox.
Supplementary Material
Acknowledgments
We thank T. Hirayama for helpful discussions and for providing ctr1-1 seeds, R. Motohashi and F. Myoga for providing tobacco leaves, T. Wada, T. Kurata, Y. Yamauchi, Y. Kuroki, J. Aoki, and T. Gohara for technical help, Y. Ichikawa and R. Nakazawa for DNA sequencing (Bioarchitect Research Group, RIKEN), and A. Enju for sequencing. We thank J.M. Blanchard for providing the pGL2-cycApro-luc. We also thank the ABRC and the Salk Institute Genomic Analysis Laboratory for providing the T-DNA insertion mutant lines and the RIKEN Bio-Resource Center for providing NIH3T3 cells. This work was performed during functional analysis of activation tagging lines. These lines were generated as material for functional genomics using Arabidopsis at the RIKEN Genomic Sciences Center. T.Y. is a recipient of a Special Postdoctoral Researcher Fund of RIKEN and is also supported by a Grant-in-Aid for Young Scientists (B) from the Ministry of Education, Culture, Sports, and Technology of Japan (14740452).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Minami Matsui (minami@postman.riken.go.jp).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Boudolf, V., Vlieghe, K., Beemster, G.T., Magyar, Z., Torres Acosta, J.A., Maes, S., Van Der Schueren, E., Inze, D., and De Veylder, L. (2004). The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16 2683–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burssens, S., de Almeida Engler, J., Beeckman, T., Richard, C., Shaul, O., Ferreira, P., Van Montagu, M., and Inzé, D. (2000). Developmental expression of the Arabidopsis thaliana CycA2;1 gene. Planta 211 623–631. [DOI] [PubMed] [Google Scholar]
- Castellano Mdel, M., Boniotti, M.B., Caro, E., Schnittger, A., and Gutierrez, C. (2004). DNA replication licensing affects cell proliferation or endoreplication in a cell type-specific manner. Plant Cell 16 2380–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebolla, A., Vinardell, J., Kiss, E., Olah, B., Roudier, F., Kondorosi, A., and Kondorosi, E. (1999). The mitotic inhibitor ccs52 is required for endoreduplication and ploidy-dependent cell enlargement in plants. EMBO J. 18 4476–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Coisy, M., Roure, V., Ribot, M., Philips, A., Muchardt, C., Blanchard, J.M., and Dantonel, J.C. (2004). Cyclin A repression in quiescent cells is associated with chromatin remodeling of its promoter and requires brahma/SNF2alpha. Mol. Cell 15 43–56. [DOI] [PubMed] [Google Scholar]
- Dahiya, A., Wong, S., Gonzalo, S., Gavin, M., and Dean, D.C. (2001). Linking the Rb and polycomb pathways. Mol. Cell 3 557–569. [DOI] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T., de Almeida Engler, J., Ormenese, S., Maes, S., Naudts, M., Van Der Schueren, E., Jacqmard, A., Engler, G., and Inzé, D. (2002). Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J. 21 1360–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T., Krols, L., Terras, F., Landrieu, I., van der Schueren, E., Maes, S., Naudts, M., and Inzé, D. (2001). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13 1653–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte, W., Riou-Khamlichi, C., Scofield, S., Healy, J.M., Jacqmard, A., Kilby, N.J., and Murray, J.A.H. (2003). Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, B.A., and Orr-Weaver, T.L. (2001). Endoreplication cell cycles: More for less. Cell 105 297–306. [DOI] [PubMed] [Google Scholar]
- Gendreau, E., Orbovic, V., Höfte, H., and Traas, J. (1999). Gibberellin and ethylene control endoreduplication levels in the Arabidopsis thaliana hypocotyl. Planta 209 513–516. [DOI] [PubMed] [Google Scholar]
- Gendreau, E., Traas, J., Desnos, T., Grandjean, O., Caboche, M., and Höfte, H. (1997). Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 114 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, S., and Yamaguchi, M. (1999). Kinase-independent activity of Cdc2/cyclin A prevents the S phase in the Drosophila cell cycle. Genes Cells 4 111–122. [DOI] [PubMed] [Google Scholar]
- Himanen, K., Boucheron, E., Vanneste, S., de Almeida Engler, J., Inzé, D., and Beeckman, T. (2002). Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14 2339–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet, X., Rech, J., Plet, A., Vié, A., and Blanchard, J.M. (1996). Cyclin A expression is under negative transcriptional control during the cell cycle. Mol. Cell. Biol. 16 3789–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa, T., et al. (2003). Sequence database of 1172 T-DNA insertion sites in Arabidopsis activation-tagging lines that showed phenotypes in T1 generation. Plant J. 36 421–429. [DOI] [PubMed] [Google Scholar]
- Imai, K.K., Ohashi, Y., Tsuge, T., Yoshizumi, T., Matsui, M., Oka, A., and Aoyama, T. (2006). The A-type cyclin CYCA2;3 is a key regulator of ploidy levels in Arabidopsis Endoreduplication. Plant Cell 18 382–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubes, J., and Chevalier, C. (2000). Endoreduplication in higher plants. Plant Mol. Biol. 43 735–745. [DOI] [PubMed] [Google Scholar]
- Kadam, S., and Emerson, B.M. (2003). Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol. Cell 11 377–389. [DOI] [PubMed] [Google Scholar]
- Kageyama, R., and Pastan, I. (1989). Molecular cloning and characterization of a human DNA binding factor that represses transcription. Cell 59 815–825. [DOI] [PubMed] [Google Scholar]
- Kelley, L.A., MacCallum, R.M., and Sternberg, M.J.E. (2000). Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299 499–520. [DOI] [PubMed] [Google Scholar]
- Kieber, J.J., Rothenberg, M., Roman, G., Feldmann, K.A., and Ecker, J.R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72 427–441. [DOI] [PubMed] [Google Scholar]
- Kimura, M., Yoshizumi, T., Manabe, K., Yamamoto, Y.Y., and Matsui, M. (2001). Arabidopsis transcriptional regulation by light stress via hydrogen peroxide-dependent and -independent pathways. Genes Cells 6 607–617. [DOI] [PubMed] [Google Scholar]
- Larkins, B.A., Dilkes, B.P., Dante, R.A., Coelho, C.M., Woo, Y.M., and Liu, Y. (2001). Investigating the hows and whys of DNA endoreduplication. J. Exp. Bot. 52 183–192. [PubMed] [Google Scholar]
- Ma, J., Przibilla, E., Hu, J., Bogorad, L., and Ptashne, M. (1988). Yeast activators stimulate plant gene expression. Nature 334 631–633. [DOI] [PubMed] [Google Scholar]
- Mariconti, L., Pellegrini, B., Cantoni, R., Stevens, R., Bergounioux, C., Cella, R., and Albani, D. (2002). The E2F family of transcription factors from Arabidopsis thaliana. Novel and conserved components of the retinoblastoma/E2F pathway in plants. J. Biol. Chem. 277 9911–9919. [DOI] [PubMed] [Google Scholar]
- Melaragno, J.E., Mehrotra, B., and Coleman, A.W. (1993). Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5 1661–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai, K., and Horton, P. (1999). PSORT: A program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24 34–36. [DOI] [PubMed] [Google Scholar]
- Nakamura, M., Tsunoda, T., and Obokata, J. (2002). Photosynthesis nuclear genes generally lack TATA-boxes: A tobacco photosystem I gene responds to light through an initiator. Plant J. 29 1–10. [DOI] [PubMed] [Google Scholar]
- Nakazawa, M., Ichikawa, T., Ishikawa, A., Kobayashi, H., Tsuhara, Y., Kawashima, M., Suzuki, K., Muto, S., and Matsui, M. (2003). Activation tagging, a novel tool to dissect the functions of a gene family. Plant J. 34 741–750. [DOI] [PubMed] [Google Scholar]
- Reed, A.L., Yamazaki, H., Kaufman, J.D., Rubinstein, Y., Murphy, B., and Johnson, A.C. (1998). Molecular cloning and characterization of a transcription regulator with homology to GC-binding factor. J. Biol. Chem. 273 21594–21602. [DOI] [PubMed] [Google Scholar]
- Richard, C., Granier, C., Inzé, D., and De Veylder, L. (2001). Analysis of cell division parameters and cell cycle gene expression during the cultivation of Arabidopsis thaliana cell suspensions. J. Exp. Bot. 52 1625–1633. [PubMed] [Google Scholar]
- Riou-Khamlichi, C., Menges, M., Healy, J.M., and Murray, J.A.H. (2000). Sugar control of the plant cell cycle: Differential regulation of Arabidopsis D-type cyclin gene expression. Mol. Cell. Biol. 20 4513–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier, F., Fedorova, E., Lebris, M., Lecomte, P., Gyorgyey, J., Vaubert, D., Horvath, G., Abad, P., Kondorosi, A., and Kondorosi, E. (2003). The Medicago species A2-type cyclin is auxin regulated and involved in meristem formation but dispensable for endoreduplication-associated developmental programs. Plant Physiol. 131 1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul, O., Mironov, V., Burssens, S., Van Montagu, M., and Inzé, D. (1996). Two Arabidopsis cyclin promoters mediate distinctive transcriptional oscillation in synchronized tobacco BY-2 cells. Proc. Natl. Acad. Sci. USA 93 4868–4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist, S.J., and Lehner, C.F. (1997). Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell 90 671–681. [DOI] [PubMed] [Google Scholar]
- Sugimoto, N., Tatsumi, Y., Tsurumi, T., Matsukage, A., Kiyono, T., Nishitani, H., and Fujita, M. (2004). Cdt1 phosphorylation by cyclin A-dependent kinases negatively regulates its function without affecting geminin binding. J. Biol. Chem. 279 19691–19697. [DOI] [PubMed] [Google Scholar]
- Sun, Y., Flannigan, B.A., and Setter, T.L. (1999). Regulation of endoreduplication in maize (Zea mays L.) endosperm. Isolation of a novel B1-type cyclin and its quantitative analysis. Plant Mol. Biol. 41 245–258. [DOI] [PubMed] [Google Scholar]
- Takimoto, M., Mao, P., Wei, G., Yamazaki, H., Miura, T., Johnson, A.C., and Kuzumaki, N. (1999). Molecular analysis of the GCF gene identifies revisions to the cDNA and amino acid sequences. Biochim. Biophys. Acta 1447 125–131. [DOI] [PubMed] [Google Scholar]
- Valvekens, D., Van Montagu, M., and Van Lijsebettens, M. (1988). Agrobacterium tumefaciens-mediated transformation of Arabidopsis thalinana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA 85 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele, K., Raes, J., De Veylder, L., Rouze, P., Rombauts, S., and Inzé, D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe, K., Boudolf, V., Beemster, G.T., Maes, S., Magyar, Z., Atanassova, A., de Almeida Engler, J., De Groodt, R., Inzé, D., and De Veylder, L. (2005). The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Curr. Biol. 15 59–63. [DOI] [PubMed] [Google Scholar]
- Yamamoto, Y.Y., and Deng, X.W. (1998). A new vector set for GAL4-dependent transactivation assay in plants. Plant Biotechnol. 15 217–220. [Google Scholar]
- Yamamoto, Y.Y., Deng, X.W., and Matsui, M. (2001). CIP4, a new COP1 target, is a nucleus-localized positive regulator of Arabidopsis photomorphogenesis. Plant Cell 13 399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizumi, T., Nagata, N., Shimada, H., and Matsui, M. (1999). An Arabidopsis cell cycle -dependent kinase-related gene, CDC2b, plays a role in regulating seedling growth in darkness. Plant Cell 11 1883–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y., Steinmetz, S., Meyer, D., Brown, S., and Shen, W.H. (2003). The tobacco A-type Cyclin, Nicta;CYCA3;2, at the nexus of cell division and differentiation. Plant Cell 15 2763–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.