A system is a network of mutually dependent and thus interconnected components comprising a unified whole. Every system exhibits emergent behavior, a unique property possessed only by the whole system and not shared to any great degree by the individual components on their own. Systems biology is currently undergoing enormous expansion, but there seems little awareness of either the history of systems biology or the behavior of systems that make them exciting to study. The aim of this article is to expand on both themes. Original references that pioneered changes in perception of systems structure and behavior are indicated, and a few modern developments are briefly referenced to indicate progress. The essay focuses on systems biology prior to the age of genomics and large-scale biology, with the intent of giving modern systems biologists a sense of the extensive foundations of the field.
The understanding of systems has had enormous impact on what are loosely regarded as human sciences, including economics, sociology, psychology, and medicine. Systems biology has generated revolutions in ecology, population biology, and evolutionary studies and is slowly making inroads into biochemistry, development, genetics, and whole-plant biology. But it is only very recently that molecular biology has adopted a systems approach. The enormous growth in genomics now makes this possible. Currently, this is an age of systems, and systems structure and behavior should form the core of all student biology courses. All biological systems are effectively systems within systems, as indicated by Jacob above. Understanding the complexity of biological systems represents the greatest intellectual and experimental challenge yet faced by any biologist.
This article is structured as follows. First, consideration is given to how systems approaches developed and what, in turn, they replaced or refined. The hierarchical structure of systems is then explained and the possibility of a definition examined. Since systems are composed of interlinked components, the connections and communication within the various parts of the hierarchy are outlined, and the article finishes with some of the less-understood, and sometimes counterintuitive, aspects of systems behavior.
AN UNDERSTANDING OF SYSTEMS INITIATED A PARADIGM CHANGE IN THE EARLY 20th CENTURY
Two important concepts underpinned investigative biology by the end of the 19th century, both of which had their roots in the 17th century.
The first is identified with René Descartes (1596–1650). He formulated the notion that complex situations can be analyzed by reducing them to manageable pieces, examining each in turn, and reassembling the whole from the behavior of the pieces. Descartes' reductionism as it is now known was formulated when biology as a subject was nonexistent. The main scientific input was to physics and mathematics. Newton's success in mathematically describing planetary movements and characterizing gravity were powerful influences toward the easy belief that reductionism would provide all the necessary answers. Reductionist investigations still form an important component of present-day plant biology and lead to the simple assumption that higher levels in a biological hierarchy can easily be understood from the behavior of the lower levels.
Mechanistic biology also had its roots in the 17th century and developed as a result of the same powerful influences that saw reductionism to the fore. The evident success of the development of physics and more particularly the construction of simple clockworks were crucial contributions (Toulmin and Goodfield, 1965). Not only did the ease of disassembly and reassembly of clockwork predispose early scientific thinking toward Cartesian reductionist approaches, but with time carried the implication that everything, including organisms, was based on simple clockwork-like, easily understood, deterministic principles. Mechanistic biology found its ultimate expression in a famous text by Jacques Loeb (1912), an early and productive plant biologist. His book reflected the common view of the time and was based on his mechanistic attitudes toward the simple response of seedlings to light and gravity. All biological behavior, he concluded, was predetermined, forced, and identical between all individuals of a particular species; organisms were thus merely complex machines. Although environmental transduction mechanisms were completely unknown, Loeb assumed they must be rigid, invariant, physico-chemical mechanisms like the cogs in a clock.
Limitations of the Reductionist Mechanistic Approach
Reaction against these predictive mechanistic and reductionist attitudes began among a few biologists (e.g., Smuts, 1926; Woodger, 1929; Weiss, 1940; von Bertallanfy, 1950a, 1950b) in the early part of the 20th century. The objections to reductionism were twofold.
First, the ancient Greek physician Aristotle (384–322 B.C.) had stated that “the whole is something over and above its parts and not just the sum of them all” (Aristotle, 1946). Aristotelean views had dominated science up to the 17th century but had disappeared with the development of experimental physics and later biology. This emphasis on whole behavior as being basically indivisible was now resuscitated. Jan Smuts (1870–1950), naturalist/philosopher and twice Prime Minister of South Africa, coined the commonly used term holism. Whole systems such as cells, tissues, organisms, and populations were proposed to have unique (emergent) properties. It was impossible to try and reassemble the behavior of the whole from the properties of the individual components, and new technologies were necessary to define and understand the behavior of systems.
Second, it was apparent from simple investigations on the brain and animal development that the structure of an entire system actually orchestrated and constrained the behavior of the component parts. Reductionist mechanistic investigations would miss the vital element of orchestration. These early texts (and subsequently many others) began to change the simple description of organism characteristics, such as growth, respiration, excretion, reproduction, and irritability, which Loeb (1912) had emphasized. Instead, a new language of life developed in which complexity, organization, uniqueness, emergence, holism, unpredictability, openness, interconnectedness, teleonomy, disequilibrium, and evolution became more dominant terms.
Although reductionism and holism are often posed in opposition to each other, they can be reconciled. There is a need to understand how organisms are put together (reductionism) just as in turn there is a need to understand why they are put together in the way that they are (systems; holism). Both lines of approach are productive and answer different questions. The study of biological systems does, however, require an understanding of control and design structures, elements of structural stability, resilience, and robustness, which are not easily constructed from mechanistic information. Better understanding will follow from computer modeling of biological complexity.
Williams' Systems Revolution and the Defeat of Loeb's Mechanistic Approach
The first experimental attack on Loeb's mechanistic approach was launched by Weiss (1925) in his PhD dissertation. He observed that changes in light and gravity on insect behavior led to an identical final behavioral response among all individuals, but each individual approached this ultimate response by a unique behavioral route. Whatever the mechanism involved, it possessed neither the characteristics of clockwork nor Loeb's precise mechanistic sequence. A current view would see the behavior as resulting from negative feedback toward a predetermined set point. The individual trajectories toward the set point result from the known variation in molecular constituents present in each individual (see further discussion of negative feedback below).
Measurements of the growth trajectories of young rhizomes, seedlings, roots, and hypocotyls in response to light or gravity are similarly individually variable (Bennet-Clerk and Ball, 1951; Rich and Smith, 1986; Ishikawa et al., 1991). These data thus contradict the very foundations on which Loeb had formulated his belief of mechanistic behavior. While eventually the ultimate response to gravity and light of most individual seedlings is fairly predictable, the growth trajectory to reach that new position is individually unique in time, space, or both. These individual responses are not more widely recognized because measurements are usually expressed as a population average, which is then assumed to apply to all individuals.
An acceleration of systems understanding came with the publication of the first ground-breaking text compiling molecular, physiological, and anatomical individuality in animals (Williams, 1956), which has been described as a revolution (Elsasser, 1987). First published 50 years ago, it has recently been reissued—the mark of a classic. Several tables illustrate the enormous variation, often 20- to 50-fold, in numerous biochemical, hormonal, and physiological parameters and in organ size between normal, healthy, fertile human individuals. Substantial variation in protein composition between individual cells has also been observed (van Roon et al., 1989; Ko et al., 1990; Yui et al., 2006). Thus, each individual organism has a unique and distinctive biochemical and hormonal pattern, which is based upon anatomical and physiological potentialities and the intricate balance that exists between each, a fundamental property of systems, which, unlike machines, can tolerate such variation.
What little quantitative biochemical and hormonal data are available in plants would suggest similar degrees of chemical variation. For example, measurements of mineral and vitamin content of the same species can vary 10- to 20-fold, ethylene content may vary 100-fold between individual apple fruits at the same stage of ripening, and individual poppy seed production can vary up to one millionfold (Goodall and Gregory, 1947; White, 1979; Trewavas, 1999). A compendium of molecular variation in plants would further demonstrate this property of plants and also establish that they are robust systems.
Machines can only function properly with exacting specification of their constituents; their error tolerance is extremely low. Living cells and organisms are clearly not machines, not even complex ones, as Loeb (1912) proposed. Since Williams' data were constructed from mammals within a normal range of phenotype, robust compensation mechanisms must be present that avoid turning such huge molecular variation into equivalent phenotypic variation. For example, low levels of hormones can be compensated for by increasing hormone sensitivity; individuals with small stomachs may eat more frequently, etc. Compensation of this kind is a systems characteristic wherein the whole orchestrates the behavior of the parts.
Williams' (1956) introductory chapter marked the end of strictly mechanistic beliefs. He showed clearly that the average individual, the statistical mean, is not a biological reality. Every individual in a wild population is a variant in certain characteristics and exhibits some individual responses in response to environmental perturbation. Clausen et al. (1948; and references within) made similar observations in plants in a series of classic publications describing the cloning of individual plants and their growth in three very different environmental circumstances. Responses were measured in numerous phenotypic characteristics. These data have now been expressed as a norm of reaction (Gupta and Lewontin, 1982). Each individual exhibited a unique norm of reaction and all crossed over each other with no apparent segregation (Schlichting and Pigliucci, 1998).
THE STRUCTURE OF BIOLOGICAL SYSTEMS: HIERARCHIES OF ORGANIZATION WITH EMERGENT PROPERTIES
The properties of systems are the result of two important characteristics: systems have a hierarchical structure, and the structure is held together by numerous linkages to construct very complex networks.
Recognition That Systems Are a Hierarchy of Organization
Woodger (1929) in an early influential text pointed to organization as a critical property of living systems. He emphasized that most higher organisms start their life cycle as single cells. During development, multiple cells appear in typical diversity, typical spatial distribution, and typical temporal order. The cells are subordinated to this developmental order, and their freedom of behavior is restricted by it. Isolate individual plant cells from each other and, once released from organizational constraints, their behavior changes. Organization is thus a supracellular property and represents a level of control higher than that of any individual cell. Likewise, tissues are organized complexes of cells; isolate any individual tissue from a plant and its behavior changes relative to those remaining on an intact plant. The later recognition that many systems were constructed from hierarchies of organization thus represented an important advance in understanding (Boulding, 1953; Leake, 1969; Whyte, 1969; Pattee, 1973).
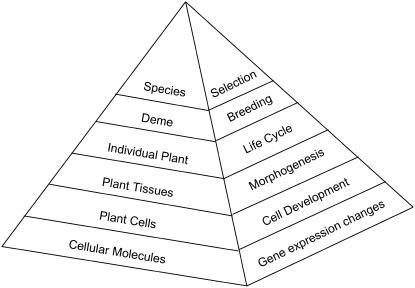
Figure 1 illustrates a familiar but oversimplified biological hierarchy. Each level in the hierarchy above that of molecules is an emergent property resulting from the very complex interactions between the constituents of the lower level. Each level in turn contains numerous recognizable subsystems: some simple, some complex, but each presenting emergent properties that can also be arranged in a hierarchy of organization.
Figure 1.
Biological Systems Arranged in a Hierarchy of Increasing Organizational Complexity.
Each lower level through numerous and complex interactions of its constituents creates the level above as an emergent property. On the left is a static compositional hierarchy and on the right a dynamic developmental and evolutionary hierarchy incorporating time dependence of change.
In cells, for instance, aggregation of subunits of multienzyme complexes or calcium/calmodulin with dependent enzymes creates the simple emergent property of novel enzyme activity. Tubulin or actin polymerization in the test tube creates the emergent behavior of isolated microtubules or filaments. By contrast, the organized cellular behavior of cyclins and other numerous regulatory proteins underpins the emergent property of the cell cycle (Kohn, 1999; Strogatz, 2001). This process, like circadian phenomena or growth, results from an integrated, organized collective of complex feedback controls, protein phosphorylation regulation, second messenger distribution, structural interactions, organelle interactions, and other as yet uncovered control mechanisms. This level of organization is much more complex than that of individual enzymes, and how this emergent property is constructed is the province of systems biology. Systems construction of embryological development points the way (Davidson et al., 2002).
Evolutionary Implications of Systems Definitions
Weiss (1973) in his final scientific contribution attempted to define the critical characteristics of biological systems using the (then) recent recognition of their hierarchical structure. Two interconnected points were made. (1) All systems express much greater variation at lower levels than at higher levels of organization. Information such as that from Williams (1956) established this point for molecules, tissues, and phenotype. (2) The output of individual pathways is more ordered within a system than would be expected from random operation of those pathways outside the system. Molecular behaviors are thus subject to restraints on their behavior and are orchestrated by the higher level property that emerges.
This definition was substantially enlarged by Bateson (1972). He pointed to the intricate relationship and ability of all organisms to deal with a variable environmental contingency and to the two-way interaction between genes and environment that had earlier been established by Schmalhausen (1949), Waddington (1953, 1957), and later by Rendel (1967). In other words, the real system extends outwards from the individual organism, encompassing its environmental and biological parameters.
This unusual systems perspective has had a powerful influence on the understanding of evolutionary mechanisms. The intricate behavior elicited from an organism by its complex variable environment came to be regarded as the real focus of selective forces in nature (Gupta and Lewontin, 1982; Levins and Lewontin, 1985; Vrba and Gould, 1986). On this systems basis, the whole life cycle may be the object of natural selection (McNamara and Houston, 1996; Schlichting and Pigliucci, 1998). A striking early contrast thus palpably emerged between a later, reductionist alternative that emphasizes the gene as the object of selection. Gould (2002) provides a devastating critique of the gene selection hypothesis from a strong systems perspective.
von Bertallanfy's General Systems Theory
Once the hierarchical structure of systems was clarified, an important generalization was suggested by von Bertallanfy (1950b, 1968). He argued that all systems shared the common property of being composed of interlinked components, in which case they might share similarities in detailed structure and control design. This prophetic and imaginative general systems theory has recently received striking (if partial) support with the recognition that a common stable systems structure is represented by hubs and connectors. Hubs are connected to many components (connectors), each of which is linked only to a few other components (Barabasi, 2002; Buchanan, 2002a, 2002b). Currently, numerous economic, industrial, linguistic, political, managerial, information theory (IT), cellular, ecological, and psychological systems are believed to possess this kind of structure, including protein networks inside cells (Forrester, 1961, 1969; Beer, 1965, 1972; Bateson, 1972; Pattee, 1973; Ravasz et al., 2002; Cohen et al., 2003). This is an oversimplification of general systems theory and is merely used here to illustrate how diverse networks might share topological features.
Whether the control properties of one system provide critical insights into the behavior of others has been productively investigated many times. Three examples, among many, are original investigations of individual plant or ecosystem behavior using economic analogies (Bloom et al., 1985; Herendeen, 1991); parallels between social insect foraging and plant foraging (Lopez et al., 1994); and the term superorganism used to describe the organic and biological nature of human organization and the frequently made parallel with living organisms (Beer, 1972; Bloom, 1995; Pech and Oakely, 2005).
COMMUNICATION AND CONTROL WITHIN SYSTEMS
Biological systems, like all systems, are composed of networks of interdependent components that integrate the system into a unified whole. Linkages are demonstrated by modifying the level of one component and observing the communicated effects on others. Furthermore, the strength (sensitivity) of the linkage can be ascertained by measuring the extent of the response. The various forms of communication that operate within the hierarchy of a system are therefore essential to understanding overall systems behavior.
The development of IT was a major advance by communication engineers (Shannon and Weaver, 1949) and was much discussed by early systems proponents. IT has found only little use in biology because it uses statistical uncertainty and binary bits for communicated information that is unrelated to biological information. Communicated information in cells and organisms is usually context-dependent and user-specific. Plants, for example, use phytochrome to gain information about the light environment and a transduction path that is, in part, specific to activated phytochrome.
Interactions between Different System Levels: Upward and Downward Causation
Polyani (1968) first clarified the relationship between levels in a hierarchy. He showed that adjacent levels mutually constrain but do not determine each other and emphasized that the upper level harnesses the constituents of the lower level to carry out behaviors that they would not perform on their own (echoed in Weiss, 1973; see above). Several examples were explained in detail. The simplest was a linguistic hierarchy (letters, words, phrases, sentences, paragraphs, etc.) arranged in terms of perception of increasing complexity. Consider specifically a sentence (higher level) and its component words (level below). The meaning of a sentence is its emergent property, and the meaning constrains the words that can be employed in any sentence. The meaning provides an overall top-down constraint on the choice of words and harnesses them to serve the function of the higher level. But in turn, the words constrain what meanings can be constructed. How this works in biological practice can be illustrated by several examples.
Crick (1966) stated that modern biology aimed to explain everything in terms of physics and chemistry, a strongly reductionist position. Any arrangement of organic bases in DNA is compatible with the laws of physics and chemistry. However, only in the cell are certain crucial nucleotide sequences constrained to act as a code. This systems perspective marks where biology departs from chemistry and physics.
Mutations in DNA can have an effect on the higher emergent levels of cell, phenotype, deme, and even species (Figure 1) by what is commonly called upward causation. In reverse, the expression of any mutation (or gene) is constrained by its genetic background issuing, in this case, from the higher level of the cell. Campbell (1974) was the first to describe these higher-level constraints as downward causation. Demes are composed of small groups of interbreeding (and thus interconnected) individuals; an emergent property is generated via a complex of reproductive linkages. Species populations in turn are constructed from a shifting balance of demes with different proliferative capacities (Wright, 1931, 1982). A mutant phenotype will likely experience a different fitness (reproductive yield) in one kind of deme compared with another (Gould, 2002). Individual fitness is thus constrained by all the other interconnected deme members. At every level of the biological hierarchy (Figure 1), influence goes in both directions: upwards and downwards.
Downward causation may have a more direct influence than simple constraint, although this is more contentious (Andersen et al., 2000). In one example, Cairns et al. (1988) observed that the majority of bacterial lac operon mutations seemed to appear only in the presence of the selection agent. An example in plants is the observation that environmental effects of different mineral balances on flax branching morphology (and tobacco) can last for 10 to 12 generations (Durrant, 1962, 1971; Hill, 1965). In response to stress, both the plant phenotype and, astonishingly, the genotype seem to have changed (Ries et al., 2000; Kovalchuk et al., 2003; Henikoff, 2005). Even specific mutations seem capable of repair (Lolle et al., 2005). These and other examples (Jablonka and Lamb, 1995; Bjedov et al., 2003) suggest that environmental signals, mediated through the cellular system, can in some circumstances instigate or repair mutations in DNA with some degree of specificity, providing evidence of direct influence resulting from downward causation. Environmental influence through control of DNA methylation and transposon activity are two intriguing possible mechanisms among many (McClintock, 1985; Jablonka and Lamb, 1995; Cullis, 2005). New mechanisms of downward causation cry out to be uncovered, and there are some exciting research times ahead.
Control Design in Systems: Communication by Negative Feedback and Homeostasis
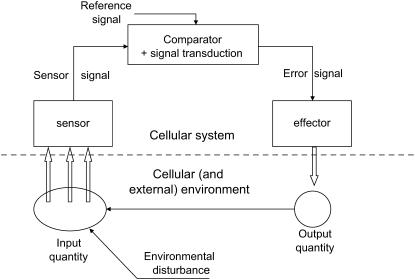
Negative feedback is one of the most important control elements found in biological systems (Figure 2). Information about the actual performance of a set of reactions (or behavior) is monitored and fed back to an earlier stage, enabling a reduction in the difference between a desired or optimal outcome and the actual performance. Negative feedback stabilizes outputs and enables biological systems to operate with resilience (Powers, 1973). There are probably hundreds, even thousands, of such control loops in biological systems operating inside cells and between cells, tissues, and even populations.
Figure 2.
Basic Control Design of a System Manipulating Many Aspects of Plant Behavior and Incorporating Negative Feedback.
The primary element involves an assessment of output by a feedback loop using a comparator that assesses current input against a predetermined reference signal. In a simple metabolic system or pathway, the end product uses the first enzyme in the sequence as the reference signal and controls metabolic flux through inhibition. In more complex cellular and tissue systems, the reference signal is currently unknown. Note that in growth and tissue development there are feedback loops operating continuously because growth in turn continually alters the external environment that feeds back into the system.
Environmental variety can threaten internal stability. Transcription/translation and replication mechanisms are likely to be surrounded by negative feedback and fidelity-reading mechanisms to block or undo damage. Heat shock controls and DNA repair are familiar examples. Environmental variety is also transduced into beneficial behavioral changes. However, continual monitoring of a shifting environment requires that transduction systems, such as cytosolic calcium concentrations and associated molecules, are also subject to immediate feedback regulation to maintain surveillance.
Recognition of feedback controls was early illustrated by the remarkable stability of numerous blood properties in mammals in the face of enormous environmental change (Bernard, 1885; Cannon, 1932). Cannon (1932) termed the process homeostasis, and his book was extremely influential for early systems proponents. Weiner's (1948) revolutionary text initiating cybernetics and raising the profile of negative feedback owed much to Cannon.
Although Cannon (1932) dealt mainly with blood composition, he also included hunger and thirst. For example, the experience of hunger leads to a search for food and eventual satisfaction by eating. There are recognizable feedback qualities here and an internal set point indicating satisfaction. Analogous drives exist in many plants that compensate for low availability of light energy, minerals, and water by redirection of tissue growth and compensatory changes in leaf and root area to improve nutrient absorption. Some internal plant reference signals (Figure 2) must be present and enmeshed with the stage of development. A search for their identity could be of value because the level of stored resources strongly relates to seed number and potential fitness (Thomas and Stoddart, 1980). An internal turgor reference point might explain plant thirst, but the situation is less clear for light and minerals.
The first examples of molecular feedback were detected in bacteria in which the final products (Ile and CTP) inhibited an earlier enzyme in the synthetic sequence (Umbarger, 1956; Yates and Pardee, 1956). All the early characterized enzymes that responded in this fashion were found to be multimeric enzymes that bind a regulatory molecule (often the end product of the metabolic sequence) at distinct sites removed from the catalytic site, causing conformational changes that affect catalysis. Such enzymes were termed allosteric (Monod et al., 1963), and they are found in abundance in plant cells. In plants, numerous examples of feedback have been described, such as in Lys biosynthesis, nitrogen and carbohydrate metabolism, gibberellin biosynthesis, and shoot meristem activity (Galili, 1995; Martin et al., 1996; Lejay et al., 1999; Schoof et al., 2000). If there is a delay in feedback, then long-term oscillations may be generated, as observed in plant growth or even perhaps cytosolic calcium (Bose, 1924; McAinsh et al., 1995).
Control Designs for Feed-Forward Activities
Environmental signals can lead to feed-forward mechanisms operating within individual organisms and cells. These mechanisms are necessarily different from negative feedback mechanisms that are concerned with stability rather than change. Feed-forward is activated in individual cells but only after receipt of information by the whole plant. One of the most prominent cellular feed-forward mechanisms, change in protein phosphorylation, serves to illustrate some facets of control. The presence of a thousand protein kinases and numerous protein phosphatases in the plant genome and potentially one-quarter to one-third of cellular proteins as protein substrates indicates its cellular importance. Because protein phosphorylation is fast, protein kinase regulation enables the rapid creation or destruction of network linkages or simple modifications of linkage strength. But in every case, the net result is specifically to redirect the flow of information through the complex of cellular and metabolic networks and change the expression of many genes.
Protein kinases and phosphatases were identified first in animals (Cori and Green, 1943; Burnett and Kennedy, 1954) and only later in plants (Trewavas, 1972; Keates and Trewavas, 1974). Second messengers are small molecules released into the cytoplasm or the plane of a cellular membrane as a result of receptor occupation, and they relay information to downstream signal transduction components, triggering a series of molecular interactions that alter the physiologic state of the cell. The detection of the first second messenger kinase (cyclic 3′5′AMP-dependent protein kinase) in the animal kingdom highlighted the crucial importance of phosphorylation as a major cellular control mechanism (Kuo and Greengard, 1969). The first plant second messenger, Ca2+-dependent kinase, was identified a decade later (Hetherington and Trewavas, 1982).
Attempts to understand the control of feed-forward mechanisms that enable robust changes in systems behavior earlier had led to the following three basic principles, and protein phosphorylation mechanisms represent the biological versions of these control specifications.
Draper's Control Timing
This control design followed from studies on inertial guidance. To control a given system, the control elements must be able to effect changes in the system in times that are less than one-tenth of the characteristic time of change of the parts of the system that are to be controlled (Draper, 1960, 1981). A change in gene expression normally takes minutes, and second messenger and phosphorylation control take seconds. A simple example explains this idea: if a car steering wheel required a minute to turn, traveling at 30 mph down a winding road would soon result in a crash.
Law of Requisite Variety
This law was deduced from first principles and states that there must be an equivalent variety in the controls as there is environmental variety to be controlled. In biological systems, both internal and external environmental signals need to be transduced by individual cells, and the plethora of protein kinases offers some of the complexity of control potential needed. Ashby (1956) shows that this principle is intuitively obvious: photographing 20 subjects that require different exposures and distance requires a camera capable of at least 20 different settings if all the negatives are to be brought to a uniform density and sharpness.
Changing Systems Structure and Function
Because systems lack single, overall, limiting factors, the manipulation of many linkages is essential to systems change (Churchman, 1971; Beer 1972). Innovation systems theory is the most recent description of how changes in multiple linkages involved in feed-forward mechanisms propel human systems in a certain direction (Nelson, 1993). A plethora of protein kinases and kinase substrates enables complex alterations in many metabolic and signaling steps.
Communication within the Plant System: Transmission Accuracy Problems
IT specifies a sender, receiver, and a relatively noise-free channel of communication for accurate information transmission. Whole plants present three difficulties that complicate the accurate transmission of information between the major tissues: (1) the transmission length varies according to the stage of development, (2) there is uncertainty in the homeostatic control of the major conducting tissues, and (3) outside environmental variation (notably temperature) is a daily occurrence.
Polar auxin transport involves shoot synthesis with potentially transmissible effects on root development (Keeble et al., 1930). In a file of cells, each upper cell secretes auxin through the wall to lower cells, but this channel is noisy because of potential sideways diffusion of auxin while in transit in the wall. Temperature variation also influences secretion. A simple relay mechanism represents a possible resolution to these noisy channels. For example, Dictyostelium amoebae aggregate in an oscillatory fashion by secreting cyclic AMP from nucleation sites. Each cell perceives the message and its direction and resynthesises the cyclic AMP and secretes it from the base of the cell together with phosphodiesterase to limit the diffusion period and improve polar aggregation. This provides a signaling mechanism that overcomes noise. By analogy, polar auxin transport could involve resynthesis of the auxin signal on perception, subsequent vesiculation, and then secretion to the basipetal cell. Some evidence supports this suggestion. Habituation is a phenomenon whereby processes dependent on external auxin application such as cell division switch to become auxin-independent. Auxin-induced auxin biosynthesis is the believed basis of habituation (Gautheret, 1950). The presence of an auxin oxidase, either in vesicles or secreted to the wall, might reduce lateral movement and false signaling. Waves of auxin movement have been observed for many years, analogous to the oscillations expressed by aggregating Dictyostelia (Hertel and Flory, 1968; Shen-Miller, 1973).
Fidelity of signal transmission through noisy channels can be improved by feedback, indicating that the signal has been received. In plants, simple feedback loops using both sugars and nitrogenous compounds are obvious candidates to provide information loops enabling some balance between root and shoot development to be maintained (Forde, 2002; Sachs, 2005). An early example can be found in the source-sink relationship in germinating barley seedlings. The growing embryo secretes a gibberellin signal that helps mobilize amylase secretion from the aleurone into the endosperm. Normally this sugar is used by the growing embryo, but if the embryo stops growing, sugar accumulation elevates the osmotic pressure, inhibiting further embryo-initiated, gibberellin-dependent amylase production (experiments conducted by Pfeffer and Hansteen circa 1926; cited in von Bertallanfy, 1968, p. 143). Thus, the gibberellin signal is controlled later in the circuitry by direct feedback onto the aleurone.
SPECIFIC SYSTEMS PROPERTIES AND BEHAVIOR
Supraorganism Design Structures: Marginal Value and Game Theory
Using a simple mathematical analysis, Simon (1956) showed that if organism behavior was random, then the chances of survival effectively would be zero. Organisms instead use cues in their environment that enable nonrandom selection of specific adaptive paths, leading with high probability to a need-satisfying food or reproductive point. Schutzenberger (1945) and Russell (1946) specifically identified such behavior as goal seeking, highlighted in early systems discussion as a holistic property. Higher plants, for instance, can forage for resources by exploration, grow up along gradients of resources (light, water, and minerals), and proliferate when rich resource patches are located, changing phenotype (Sutherland and Stillman, 1988; de Kroon and Hutchings, 1995). The goal is optimal fitness.
But fitness is relative and represents a complex systems property structured by other species members and based on a mixture of cooperation and competition. Seed number is, in part, related to accumulated reserves of sugars and minerals. The conflict that arises from competition for limited resources and an internal drive in the individual toward optimizing fitness necessitates the introduction of particular tactics for resource collection.
Marginal value foraging tactics, first characterized by Charnov (1976), optimize foraging to ensure that the ratio of energy expenditure to energy gain is maximized. Such foraging tactics now familiar in animal foraging have also been reported in plants (Kelly, 1990). Behavioral interactions between individuals are also described by game theory, first outlined by Von Neumann and Morganstern (1947) and expanded substantially by Maynard-Smith (1982). Gersani et al. (2001) first tested game theory predictions of selfish behavior by plants competing actively for resources. It was observed that root proliferation was greatly enhanced to steal nutrients from neighboring competitors, thus fulfilling the theoretical predictions. Many other games played by plants against competitors (and against nature) have been summarized recently by de Jong and Klinkhamer (2005).
Systems Stability and Response to Perturbation
An inbuilt tendency of any system is stability. Systems have a kind of informational entropy that carries them to a more probable state of optimal stability (Beer, 1965). The natural tendency of subsystems toward individual stability or resilience is overridden by more complex interactions, communication, and feedback controls within the whole system. Such behavior emphasizes the difficulty of investigation by the reductionist approach of holding invariant the behavior of internal and external components with which subsystems are systemically interacting in real life.
Early systems proponents emphasized that organisms were open systems that were maintained by a continual (steady state) flow of energy and matter (Hill, 1930; Burton, 1939; Denbigh et al., 1948; von Bertallanfy, 1950a; Denbigh, 1951). Simple model systems were constructed, but unfortunately they incorporated an unrecognized negative feedback. When input was changed, the output of these simple systems reached a new steady state by three kinds of behavior: simple hyperbolic change, initial overshoot, or more surprisingly, undershoot. Whichever trajectory was adopted depended simply on the initial conditions, that is, the state of the system at the time. All three responses were observed by different individual roots responding to a gravitational stimulus (Ishikawa et al., 1991), and they have been observed in other system perturbations, such as changes in respiratory rates.
Systems Output Control
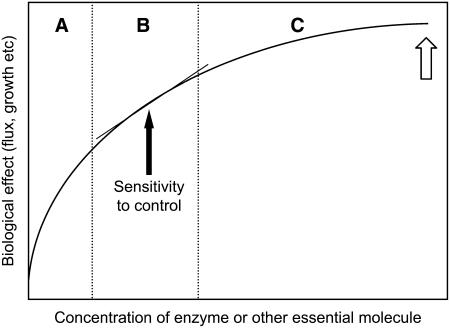
Metabolic pathways are very simple systems. A group of enzymes (components) are linked together by substrates and products, and the pathway has a measurable output (flux). Present technology enables precise manipulation of the levels of particular enzymes in many pathways that has led to great advances in understanding system behavior. Kacser and Burns (1973) developed unambiguous methods for measuring the control exerted by any particular enzyme. The simplest form of control is to measure the sensitivity with which the output responds to a progressive reduction in a particular enzyme. Figure 3 summarizes the response curves found for numerous enzymes (and for other important regulatory molecules). The typical position found for virtually all enzymes investigated is indicated by the open arrow. In other words, in vivo, enzymes typically exist at concentrations that are neither rate-limiting nor exerting substantive control. A valuable review of many of these measurements can be found in Fell (1997).
Figure 3.
Typical Dose–Response Curve of the Concentration of Necessary Molecules against Response.
The graph indicates the variation in response against concentration either of an enzyme or other essential molecule to the output of a metabolic pathway or a complex system process, such as growth or development. With increasing concentration, the curve results from increasing constraints by other linked molecules. Region A is typically occupied by systems in which mutation or inhibition has been used to lower the amount of the essential molecule. Region B is more typical of molecules whose amount influences the control characteristics of the system. Region C is typical of many essential molecules that have little direct influence on the control behavior of the system but by mutation or inhibition can be moved into region A. The sensitivity to control is the tangent to the curve and is indicated only for region B. The sensitivity is much higher or lower in regions A and C, respectively. The open arrow marks the typical position of an enzyme in a metabolic pathway.
Control is usually shared among all the enzymes in the sequence to differing degrees and to some enzymes outside the direct sequence. Traditional ideas of rate-limiting steps in which all control was posited in one enzyme have not been supported experimentally, and this simple notion has been largely discarded. Only by increasing the activity of virtually all enzymes in the sequence can overall flux through the pathway be substantially increased (Neiderberger et al., 1992). The manipulation of systems behavior can only be sensibly accomplished by modifying many steps, a theme echoed by Waddington (1977) in considering more complex systems. Trewavas (1986, 1987) outlines application of these approaches to plant growth control.
However, detailed control analysis of photosynthesis, respiration, and other pathways clearly indicated that the control values of each enzyme changed enormously as internal and external environmental situations were altered (Groen et al., 1982; Hafner et al., 1990; Stitt et al., 1991). The sensitive steps that can be used to change a system change in turn with environmental circumstances, thus limiting the value of control theory to predict possible control points. This also recalls the earlier deductions of Bateson (1972) relating systems behavior to its environmental context.
CONCLUSIONS
Systems approaches enable plant scientists to understand the structural stability of plants, their control and design structure, and how these lead to robust and resilient behavior. These capabilities are the result of a complex biological system in which control operates at many different levels (Figure 1). Complexity is a serious biological problem, and it is likely that biological systems are the most complex known. Increasingly, scientists are going to have to depend on computational biologists to construct models that can then be tested back in laboratory conditions. However, as indicated here, laboratory conditions are only one environmental circumstance among many in which plant systems develop. In 10 years, my own estimate is that plant molecular research groups will be half modelers and half wet investigators producing new data for modelers. While mechanistic approaches will still be a valuable first step, their relevance will diminish as the need for understanding the construction of system design modules increases. However, models can only provide the basis for how a system might operate. Wet science will remain an absolute requirement for testing and refining the models. As evolution underpins the linkages and control of systems, and evolution rarely works in a linear or simple progression, the initial, simple models will usually require significant refinement. This essay is mainly historical and has not dealt with the present developing views on system behavior in yeast and bacteria, tractable organisms in which ready advances will be made and which will provide clues for investigative approaches and modeling for plants. While Arabidopsis will still be the choice for plant systems analysis, it is only a pioneer plant, and eventually a need for investigating the systems strategy and tactics in other plants and in other environments will emerge.
The critical points established in the past indicate that control of systems behavior is shared; control mechanisms with a system meet constraints from other parts of the system, accounting for the typical hyperbolic curve shown in Figure 3. Systems are hierarchical structures in which influence extends in both directions in ways that are only partly understood. Emergent properties, which are the result of complex interactions and controls at many places in the system, remain an outstanding problem, but advanced modeling will reveal more information about some of the more complex situations. Now is the time to incorporate computational modeling as much as the centrifuge, thermocycler, and other laboratory tools into plant science research programs and not merely to manipulate language as done here.
Acknowledgments
I am indebted to Peter Corning, who has so frequently and patiently explained many aspects of systems behavior to me over many years.
References
- Andersen, P.B., Emmeche, C., Finnemann, N.O., and Christiansen, P.V. (2000). Downward Causation. (Aarhus, Denmark: Aarhus University Press).
- Aristotle (1946). The Politics (translated). E. Barker. (Oxford, UK: Oxford University Press).
- Ashby, W.R. (1956). Introduction to Cybernetics. (New York: John Wiley & Sons).
- Barabasi, A.L. (2002). Linked: The New Science of Networks. (Cambridge, MA: Perseus Publishing).
- Bateson, G. (1972). Steps to an Ecology of Mind. (New York: Ballantine).
- Beer, S. (1965). The world, the flesh and the metal. The prerogatives of systems. Nature 205 223–231. [DOI] [PubMed] [Google Scholar]
- Beer, S. (1972). Brain of the Firm. (London: Allen Lane).
- Bennet-Clerk, T.A., and Ball, N. (1951). The diageotropic behaviour of rhizomes. J. Exp. Bot. 2 169–203. [Google Scholar]
- Bernard, C. (1885). Leçons sur la Phénonèmes de la Vie Communs aux Animaux et aux Vegetaux, 2nd ed, Vol. 1. (Paris: J.B. Balliere).
- Bjedov, I., Tenaillon, O., Gerard, B., Souza, V., Denamur, E., Radman, M., Taddei, F., and Matic, I. (2003). Stress-induced mutagensis in bacteria. Science 300 1404–1409. [DOI] [PubMed] [Google Scholar]
- Bloom, A.J., Chapin, F.S., and Mooney, H.A. (1985). Resource limitation in plants: An economic analogy. Annu. Rev. Ecol. Syst. 16 363–392. [Google Scholar]
- Bloom, H. (1995). The Lucifer Principle: A Scientific Expedition into the Forces of History. (New York: Atlantic Monthly Press).
- Bose, J.C. (1924). Plant Response as a means of Physiological Investigation. (London: Longmans).
- Boulding, K.E. (1953). The Organisational Revolution. (New York: Harper & Row).
- Buchanan, M. (2002. a). Small World, Uncovering Natures Hidden Networks. (London: Wiedenfield and Nicolson).
- Buchanan, M. (2002. b). Nexus. Small World and the Groundbreaking Science of Networks. (New York: Norton).
- Burnett, G., and Kennedy, E.P. (1954). The enzymatic phosphorylation of proteins. J. Biol. Chem. 211 969–981. [PubMed] [Google Scholar]
- Burton, A.C. (1939). The properties of the steady state compared to those of equilibrium as shown in characteristic biological behaviour. J. Cell. Comp. Physiol. 14 327–349. [Google Scholar]
- Cairns, J., Overbaugh, J., and Miller, S. (1988). The origin of mutants. Nature 335 142–145. [DOI] [PubMed] [Google Scholar]
- Campbell, D.T. (1974). Downward causation in hierarchically organised biological systems. In Studies in the Philosophy of Biology, F.J. Ayala and T. Dobzhansky, eds (London: Macmillan), pp. 139–163.
- Cannon, W.B. (1932). The Wisdom of the Body. (New York: Norton).
- Charnov, E.L. (1976). Optimal foraging, the marginal value theorem. Theor. Popul. Biol. 9 129–136. [DOI] [PubMed] [Google Scholar]
- Churchman, C.W. (1971). The Design of Inquiring Systems. (New York: Basic Books).
- Clausen, J., Keck, D., and Hershey, W.M. (1948). Experimental studies on the nature of plant species, III. Environmental responses of climatic races of Achillea, Publication 581. (Washington, DC: Carnegie Institute of Washington).
- Cohen, J.E., Johnson, T., and Carpenter, S.R. (2003). Biological community description using the food web, species abundance and body size. Proc. Natl. Acad. Sci. USA 100 1781–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cori, G.T., and Green, A.A. (1943). Crystalline muscle phosphorylase. II. Prosthetic group. J. Biol. Chem. 151 31–43. [Google Scholar]
- Crick, F.C. (1966). Of Molecules and Men. (Seattle, WA: University of Washington Press).
- Cullis, C.A. (2005). Mechanisms and control of rapid genomic changes in flax. Ann. Bot. (Lond.) 95 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, E.H., et al. (2002). A genomic regulatory network for development. Science 295 1669–1678. [DOI] [PubMed] [Google Scholar]
- de Jong, T.J., and Klinkhamer, P.G.L. (2005). Evolutionary Ecology of Plant Reproductive Strategies. (Cambridge, UK: Cambridge University Press).
- de Kroon, H., and Hutchings, M.J. (1995). Morphological plasticity in clonal plants: The foraging concept reconsidered. J. Ecol. 83 143–152. [Google Scholar]
- Denbigh, K.G. (1951). The Thermodynamics of the Steady State. (London: Methuen).
- Denbigh, K.G., Hicks, M., and Page, F.M. (1948). The kinetics of open reaction systems. Trans. Faraday Soc. 44 479–494. [Google Scholar]
- Draper, C.S. (1960). The inertial gyro: An example of basic and applied research. Am. Sci. 48 9–19. [Google Scholar]
- Draper, C.S. (1981). Origins of inertial guidance. J. Guid. Control Dyn. 4 449–463. [Google Scholar]
- Durrant, A. (1962). Induction, reversion and epitrophism of Flax genotypes. Nature 196 1302–1304. [Google Scholar]
- Durrant, A. (1971). The induction and growth of flax genotrophs. Heredity 27 277–298. [Google Scholar]
- Elsasser, W. (1987). Reflections on a Theory of Organisms. (Quebec, Canada: Orbis).
- Fell, D. (1997). Understanding the Control of Metabolism. (London: Portland Press).
- Forde, B.G. (2002). Local and long-range signaling pathways regulating plant responses to nitrate. Annu. Rev. Plant Biol. 53 202–235. [DOI] [PubMed] [Google Scholar]
- Forrester, J.W. (1961). Industrial Dynamics. (Cambridge, MA: MIT Press).
- Forrester, J.W. (1969). Principles of Systems. (Cambridge, MA: Wright-Allen Press).
- Galili, G. (1995). Regulation of lysine and threonine synthesis. Plant Cell 7 899–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautheret, R.J. (1950). Remarques sur les baisins nutritifs des culture de tissues de tissues vegetaux. C. R. Soc. Biol. 144 173–174. [PubMed] [Google Scholar]
- Gersani, M., Brown, J.S., O'Brien, E.E., Maina, G.M., and Abramsky, Z. (2001). Tragedy of the commons as a result of root competition. J. Ecol. 89 660–669. [Google Scholar]
- Goodall, D.W., and Gregory, F.G. (1947). Chemical composition of plants as an index of their nutritional status. In Imperial Bureau of Horticultures and Plantation Crops, Technical Communication 17. (Aberystwyth, Wales: IAB Central Branch).
- Gould, S.J. (2002). The Structure of Evolutionary Theory. (Cambridge, MA: Harvard University Press).
- Groen, A.K., Wanders, R.J.A., Westerhoff, H.V., van der Meer, R., and Tager, J.M. (1982). Quantification of the contribution of various steps to the control of mitochondrial respiration. J. Biol. Chem. 257 2754–2757. [PubMed] [Google Scholar]
- Gupta, A.P., and Lewontin, R.C. (1982). A study of reaction norms in natural populations of D. pseudoobscura. Evolution 36 934–948. [DOI] [PubMed] [Google Scholar]
- Hafner, R.P., Brown, G.C., and Brand, M.D. (1990). Flux control coefficients change dramatically with respiration rate. Eur. J. Biochem. 188 313–319. [DOI] [PubMed] [Google Scholar]
- Henikoff, S. (2005). Rapid changes in plant genomes. Plant Cell 17 2852–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herendeen, R. (1991). Do economic-like principles predict ecosystem behaviour under changing resource constraints? In Theoretical Studies of Ecosystems: the Network Perspective, M. Higashi and T.P. Burns, eds (Cambridge, UK: Cambridge University Press), pp. 261–287.
- Hertel, R., and Flory, R. (1968). Auxin movement in corn coleoptiles. Planta 82 123–144. [DOI] [PubMed] [Google Scholar]
- Hetherington, A.M., and Trewavas, A.J. (1982). Calcium dependent protein kinase in pea shoot membranes. FEBS Lett. 145 67–71. [Google Scholar]
- Hill, A.V. (1930). Membrane phenomena in living matter: Equilibrium or steady state. Trans. Faraday Soc. 26 667–678. [Google Scholar]
- Hill, J. (1965). Environmental induction of heritable changes in Nicotiana rustica. Nature 207 732–734. [Google Scholar]
- Ishikawa, H., Hasenstein, K.H., and Evans, M.L. (1991). Computer based video digitizer analysis of surface extension in maize roots. Planta 183 381–390. [DOI] [PubMed] [Google Scholar]
- Jablonka, E., and Lamb, M.J. (1995). Epigenetic Inheritance in Evolution. The Lamarckian Dimension. (Oxford, UK: Oxford University Press).
- Jacob, F. (1974). The Logic of Living Systems. (London: Allen Lane).
- Kacser, H., and Burns, J.A. (1973). The control of flux. In Rate Control of Biological Processes, D.D. Davies, ed (Cambridge, UK: Cambridge University Press), pp. 65–104.
- Keates, R.A.B., and Trewavas, A.J. (1974). Protein kinase activity associated with ribosomes of peas and Lemna. Plant Physiol. 54 95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeble, F., Nelosson, M.G., and Snow, R. (1930). The integration of plant behaviour. II. The influence of the shoot on the growth of roots in seedlings. Proc. R. Soc. Lond. Ser. B 106 182–188. [Google Scholar]
- Kelly, C.L. (1990). Plant foraging: A marginal value model and coiling response in Cuscuta subinclusa. Ecology 71 1916–1925. [Google Scholar]
- Ko, M., Nakauchi, H., and Takahashi, N. (1990). The dose dependence of glucocorticoid inducible gene expression results from changes in the number of transcriptionally active templates. EMBO J. 9 2835–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn, K.W. (1999). Molecular interaction map of the mammalian cell cycle and DNA repair system. Mol. Biol. Cell 10 2705–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk, I., Kovalchuk, O., Kalk, V., Boyko, V., Filkowsi, J., Heinlein, M., and Hohn, B. (2003). Pathogen-induced systemic plant signal triggers DNA rearrangements. Nature 423 760–762. [DOI] [PubMed] [Google Scholar]
- Kuo, J.F., and Greengard, P. (1969). Cyclic nucleotide dependent protein kinase. IV. Widespread occurrence of 3′5′cyclic nucleotide dependent protein kinase in various tissues and phyla of the animal kingdom. Proc. Natl. Acad. Sci. USA 64 1349–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leake, C.D. (1969). Historical aspects of the concept of organisational levels of living material. In Hierarchical Structures, L.L. Whyte, A.G. Wilson, and D. Wilson, eds (New York: Elsevier), pp. 147–161.
- Lejay, L., Tillard, P., Lepetit, M., Olive, F.D., Filleur, S., Daniel-Vedele, F., and Gojon, A. (1999). Molecular and functional regulation of two NO3 uptake systems by N and C-status of Arabidopsis plants. Plant J. 18 509–519. [DOI] [PubMed] [Google Scholar]
- Levins, R., and Lewontin, R.C. (1985). The Dialectical Biologist. (Cambridge, MA: Harvard University Press).
- Loeb, J. (1912). The Mechanistic Conception of Life (reprinted 1964). (Cambridge, MA: Belknap Press).
- Lolle, S.J., Victor, J.L., Young, J.M., and Pruitt, R.E. (2005). Genome-wide non-mendelian inheritance of extra-genomic information in Arabidopsis. Nature 434 505–509. [DOI] [PubMed] [Google Scholar]
- Lopez, F., Serrano, J.M., and Acosta, F.J. (1994). Parallels between the foraging strategies of ants and plants. Trends Ecol. Evol. 4 150–153. [DOI] [PubMed] [Google Scholar]
- Martin, D.N., Proebsting, W.M., Parks, T.D., Dougherty, W.G., Lange, T., Lewis, M.J., Gaskin, P., and Hedden, P. (1996). Feedback regulation of gibberellin biosynthesis and gene expression in Pisum sativum. Planta 200 159–166. [DOI] [PubMed] [Google Scholar]
- Maynard-Smith, L. (1982). Evolution and the Theory of Games. (Cambridge, UK: Cambridge University Press).
- McAinsh, M.R., Webb, A.A.R., Taylor, J.E., and Hetherington, A.M. (1995). Stimulus-induced oscillations in guard cell cytosolic free calcium. Plant Cell 7 1207–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B. (1985). Significance of the responses of the genome to challenge. Science 226 792–801. [DOI] [PubMed] [Google Scholar]
- McNamara, J.M., and Houston, A.I. (1996). State dependent life histories. Nature 380 215–221. [DOI] [PubMed] [Google Scholar]
- Monod, J., Changeux, J.-P., and Jacob, F. (1963). Allosteric proteins and cellular control systems. J. Mol. Biol. 6 306–329. [DOI] [PubMed] [Google Scholar]
- Neiderberger, P., Prasad, R., Miozzari, G., and Kacser, H. (1992). A strategy for increasing an in vivo flux by genetic manipulations. The tryptophan system of yeast. Biochem. J. 287 473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, R.R. (1993). National Innovation Systems: A Comparative Analysis. (Oxford, UK: Oxford University Press).
- Pattee, H.H. (1973). Hierarchy theory: The challenge of complex systems. (New York: Brazilier).
- Pech, R.J., and Oakely, K.E. (2005). Hormesis: An evolutionary “predict and prepare” survival mechanism. Leadership Organ. Develop. J. 26 673–687. [Google Scholar]
- Polyani, M. (1968). Life's irreducible structure. Science 160 1308–1311. [DOI] [PubMed] [Google Scholar]
- Powers, W.T. (1973). Feedback: Beyond behaviourism. Science 179 351–356. [DOI] [PubMed] [Google Scholar]
- Ravasz, E., Somera, A.L., Mongru, D.A., Olivai, Z.N., and Barabasi, A.L. (2002). Hierarchical organisation of modularity in metabolic networks. Science 297 1551–1556. [DOI] [PubMed] [Google Scholar]
- Rendel, J.M. (1967). Canalisation and Gene Control. (London: Academic Press).
- Rich, T.S.G., and Smith, H. (1986). Comparison of lag times in plant physiology. Plant Cell Environ. 9 707–709. [Google Scholar]
- Ries, G., Heller, W., Puchta, H., Sandemann, H., Seidlitz, H.K., and Hohn, B. (2000). Elevated UV-B radiation reduces genome stability in plants. Nature 406 98–101. [DOI] [PubMed] [Google Scholar]
- Russell, E.S. (1946). The Directiveness of Organic Activities. (Cambridge, UK: Cambridge University Press).
- Sachs, T. (2005). Auxin's role as an example of the mechanism of shoot-root relations. Plant Soil 268 13–19. [Google Scholar]
- Schlichting, C.D., and Pigliucci, M. (1998). Phenotypic Evolution: A Reaction Norm Perspective. (Sunderland, MA: Sinauer Associates).
- Schmalhausen, I.I. (1949). Factors of Evolution: The Theory of Stabilising Selection. (Philadelphia: Blakeston).
- Schoof, H., Lenhard, M., Haeckeer, A., Mayer, K.F.X., Jurgens, G., and Laux, T. (2000). The stem cell regulation of Arabidopsis shoot meristem is maintained by a regulatory loop between the CLAVATA and Wuschel genes. Cell 100 635–644. [DOI] [PubMed] [Google Scholar]
- Schutzenberger, M.P. (1945). A tentative classification of goal seeking behaviours. J. Ment. Sci. 100 97–102. [PubMed] [Google Scholar]
- Shannon, C.E., and Weaver, W. (1949). The Mathematical Theory of Communication. (Urbana, IL: University of Illinois Press).
- Shen-Miller, J. (1973). Rhythmicity in the basipetal transport of indoleacetic acid through coleoptiles. Plant Physiol. 51 615–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, H.A. (1956). Rational choice and the structure of the environment. Psychol. Rev. 63 129–138. [DOI] [PubMed] [Google Scholar]
- Smuts, J.C. (1926). Holism and Evolution. (New York: Viking Press).
- Stitt, M., Quick, W.P., Schurr, U., Schulze, E.D., Rodernel, S.R., and Bogorad, L. (1991). Decreased ribulose-1,5-biphosphate carboxylase-oxygenase in transgenic tobacco transformed with antisense RBCS. 2. Flux-control coefficients for photosynthesis in varying light, CO2 and air humidity. Planta 183 555–566. [DOI] [PubMed] [Google Scholar]
- Strogatz, S.H. (2001). Exploring complex networks. Nature 410 268–276. [DOI] [PubMed] [Google Scholar]
- Sutherland, W.I., and Stillman, R.A. (1988). The foraging tactics of plants. Oikos 52 239–244. [Google Scholar]
- Thomas, H., and Stoddart, J.L. (1980). Leaf senescence. Annu. Rev. Plant Physiol. 31 83–111. [Google Scholar]
- Toulmin, S.E., and Goodfield, J. (1965). The Discovery of Time. (London: Hutchinson).
- Trewavas, A.J. (1972). The phosphorylation of ribosomal protein in Lemna minor. Plant Physiol. 51 760–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewavas, A.J. (1986). Understanding the control of development and the role of growth substances. Aust. J. Plant Physiol. 13 447–457. [Google Scholar]
- Trewavas, A.J. (1987). Sensitivity and sensory adaptation in growth substance responses. In Hormone Action in Plant Development, C.V. Hoad, J.R. Lenton, M.B. Jackson, and R.K. Atkin, eds (London: Butterworths), pp. 19–39.
- Trewavas, A.J. (1999). The importance of individuality. In Plant Responses to Environmental Stresses, H.R. Lerner, ed (New York: Marcel Dekker), pp. 27–43.
- Umbarger, H.E. (1956). Evidence for a negative feedback mechanism in the biosynthesis of leucine. Science 123 848. [DOI] [PubMed] [Google Scholar]
- van Roon, M.A., Aten, J.A., Vanoven, C.H., Charles, R., and Lamers, W.H. (1989). The initiation of hepatocyte gene expression within embryonic hepatocytes is a stochastic event. Dev. Biol. 136 508–516. [DOI] [PubMed] [Google Scholar]
- von Bertallanfy, L. (1950. a). The theory of open systems in physics and biology. Science 111 23–29. [DOI] [PubMed] [Google Scholar]
- von Bertallanfy, L. (1950. b). An outline of general systems theory. Br. J. Philos. Sci. 1 139–164. [Google Scholar]
- von Bertallanfy, L. (1968). General System Theory. (New York: Brazillier).
- Von Neumann, J., and Morganstern, O. (1947). Theory of Games and Economic Behaviour. (Princeton, NJ: Princeton University Press).
- Vrba, E., and Gould, S.J. (1986). The hierarchical expansion of sorting and selection: Sorting and selection cannot be equated. Paleobiology 10 146–171. [Google Scholar]
- Waddington, C.H. (1953). The genetic assimilation of an acquired character. Evolution 7 118–126. [Google Scholar]
- Waddington, C.H. (1957). The Strategy of the Genes. (New York: Macmillan).
- Waddington, C.H. (1977). Tools for Thought. (London: Jonathan Cape).
- Weiner, N. (1948). Cybernetics or Control and Communication in the Animal and Machine. (Boston: MIT Press).
- Weiss, P. (1925). Animal behaviour as system reaction: The orientation towards light and gravity in the resting postures of butterflies. Biologia Generalis 1 167–248. [Google Scholar]
- Weiss, P. (1940). The problem of cell individuality. Am. Nat. 74 34–46. [Google Scholar]
- Weiss, P. (1973). The Science of Life. (New York: Futura Publishing).
- White, J. (1979). The plant as a metapopulation. Ann. Rev. Ecol. Syst. 10 109–145. [Google Scholar]
- Whyte, L.L. (1969). Structural hierarchies: A challenging class of physical and biological problems. In Hierarchical Structures, L.L. Whyte, A.G. Wilson, and D. Wilson, eds (New York: Elsevier), pp. 3–17.
- Williams, R.J. (1956). Biochemical Individuality. The Key for the Genetotrophic Concept. (New York: John Wiley & Sons).
- Woodger, J.H. (1929). Biological Principles. (London: Kegan Paul Trench & Trubner).
- Wright, S. (1931). Evolution in Mendelian populations. Genetics 16 97–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S. (1982). Character change, speciation and the higher taxa. Evolution 36 427–443. [DOI] [PubMed] [Google Scholar]
- Yates, R.A., and Pardee, A.B. (1956). Control of pyrimidine biosynthesis in Escherichia coli by a feedback mechanism. J. Biol. Chem. 221 757–770. [PubMed] [Google Scholar]
- Yui, J., Xiao, J., Ren, X., Lao, K., and Xie, S. (2006). Probing gene expression in live cells, one protein molecule at a time. Science 311 1600–1603. [DOI] [PubMed] [Google Scholar]