Abstract
In contrast with the model Escherichia coli Clp protease, the ATP-dependent Clp protease in higher plants has a remarkably diverse proteolytic core consisting of multiple ClpP and ClpR paralogs, presumably arranged within a dual heptameric ring structure. Using antisense lines for the nucleus-encoded ClpP subunit, ClpP6, we show that the Arabidopsis thaliana Clp protease is vital for chloroplast development and function. Repression of ClpP6 produced a proportional decrease in the Clp proteolytic core, causing a chlorotic phenotype in young leaves that lessened upon maturity. Structural analysis of the proteolytic core revealed two distinct subcomplexes that likely correspond to single heptameric rings, one containing the ClpP1 and ClpR1-4 proteins, the other containing ClpP3-6. Proteomic analysis revealed several stromal proteins more abundant in clpP6 antisense lines, suggesting that some are substrates for the Clp protease. A proteolytic assay developed for intact chloroplasts identified potential substrates for the stromal Clp protease in higher plants, most of which were more abundant in young Arabidopsis leaves, consistent with the severity of the chlorotic phenotype observed in the clpP6 antisense lines. The identified substrates all function in more general housekeeping roles such as plastid protein synthesis, folding, and quality control, rather than in metabolic activities such as photosynthesis.
INTRODUCTION
Proteolytic enzymes play an essential role in all living organisms. Basic cellular functions such as growth and differentiation require a continuous rate of protein turnover facilitated by specific proteases. This constant degradation of polypeptides performs a range of functions. Eventually, most proteins lose activity as a result of their involvement in various cellular activities. Such damaged polypeptides can routinely arise from translational errors, loss of structural integrity (e.g., misfolding/denaturation), and chemical modifications such as oxidation. Left unchecked, these nonfunctional proteins may eventually accumulate and begin to impair associated processes. The role for proteases in removing these inactive polypeptides is particularly important during periods of stress, when the potential for protein damage is considerably greater. In addition to these housekeeping duties, proteases also perform other essential functions, including the recycling of amino acids and the regulation of key enzymes and regulatory proteins (Vierstra, 1993).
Because of their fundamental importance, many different types of proteases are located throughout the eukaryotic cell. In higher plants, the best-defined proteases are those that require ATP, such as the cytosolic 26S proteasome (Smalle and Vierstra, 2004). Chloroplasts, consistent with their endosymbiotic origin, contain various proteases of bacterial ancestry (Adam et al., 2005; Clarke et al., 2005). One of the first identified was the ATP-dependent Clp protease localized primarily in the stroma (Shanklin et al., 1995). The model for the Ser-type Clp protease has long been the one in Escherichia coli, which consists of a proteolytic core flanked on one or both sides by a HSP100 molecular chaperone. The proteolytic core is a barrel-shaped structure consisting of a single ClpP subunit arranged in two-tiered heptameric rings, with the catalytic active site housed within the central cavity (Wang et al., 1997). Adjacent to the entrance of the proteolytic core is a single homogeneous hexameric ring of either ClpA or ClpX (Grimaud et al., 1998), two members of the HSP100 family of molecular chaperones. The HSP100 partners confer substrate specificity to the Clp protease and unfold the protein substrate in an energy-dependent manner (Kim et al., 2001). Denatured substrates are then translocated into the proteolytic core and rapidly degraded to small peptide fragments that later diffuse out of the complex (Ortega et al., 2002; Kang et al., 2004).
Members of the Clp protein family are by far more diverse and numerous in higher plants than in any other group of organisms. The model species Arabidopsis thaliana has at least 23 individual Clp proteins (Adam et al., 2001; Peltier et al., 2004; Clarke et al., 2005), 10 of which are HSP100 chaperones (ClpB1-4, ClpC1-2, ClpD, and ClpX1-3), six of which are paralogs of the proteolytic subunit ClpP (ClpP1-6), and four of which are paralogs of a ClpP-like subunit (ClpR) that apparently lacks the catalytic triad (ClpR1-4). Arabidopsis also possesses two unique Clp proteins with sequence similarity to the N-terminal domain of HSP100 proteins. These proteins have been designated ClpS1-2 (Peltier et al., 2004), but they should not be confused with the functionally distinct bacterial ClpS, for which the ortholog in Arabidopsis has been termed ClpT (Peltier et al., 2004). Most Clp proteins in Arabidopsis are located in the chloroplast stroma (ClpB3, ClpC1-2, ClpD, ClpP1, ClpP3-6, ClpR1-4, ClpS1-2, and ClpT) and all are nucleus-encoded, except for the plastomic ClpP1 (Nakabayashi et al., 1999; Peltier et al., 2001; Zheng et al., 2002). The bulk of chloroplast Clp proteins associate into a single 325- to 350-kD proteolytic core complex consisting of the ClpP1, ClpP3-6, and ClpR1-4 paralogs along with the two novel ClpS1-2 proteins (Peltier et al., 2001, 2004). This core complex presumably forms a double heptameric ring structure analogous to the E. coli ClpP proteolytic core, although to date little is known about the organization of the constituent ClpP/R/S proteins within this oligomeric structure.
Despite recent progress, essentially nothing is known concerning the specific function of the Clp protease within plant chloroplasts. It has been assumed that Clp functions as a stromal housekeeping protease, but the evidence for such a role remains sparse. Genetic studies have revealed that, in general, all chloroplast ClpP paralogs are necessary for chloroplast development and thereby plant viability. The plastomic ClpP1 protein is vital for shoot development and cell viability in tobacco (Nicotiana tabacum) (Shikanai et al., 2001; Kuroda and Maliga, 2003), whereas no viable Arabidopsis mutants for any of the nucleus-encoded ClpP paralogs have yet been reported, despite extensive screening of all available T-DNA insertion lines (Clarke et al., 2005). Key to elucidating the precise role of the chloroplast Clp protease is the identification of targeted protein substrates, none of which has yet been found in higher plants. In this study, we use antisense technology to repress the level of one subunit (i.e., ClpP6) of the Clp proteolytic core in Arabidopsis. We show that the loss of ClpP6 causes pleiotropic changes, the most prominent of which are leaf variegation, reduced growth and photosynthetic rates, and impaired chloroplast development. More importantly, we demonstrate that the Clp proteolytic core consists of smaller subcomplexes with a defined ClpP/R protein composition, and using the clpP6 antisense lines, we identify native protein substrates for the chloroplast Clp protease in higher plants.
RESULTS
Screening and Phenotypic Appearance of Arabidopsis clpP6 Antisense Lines
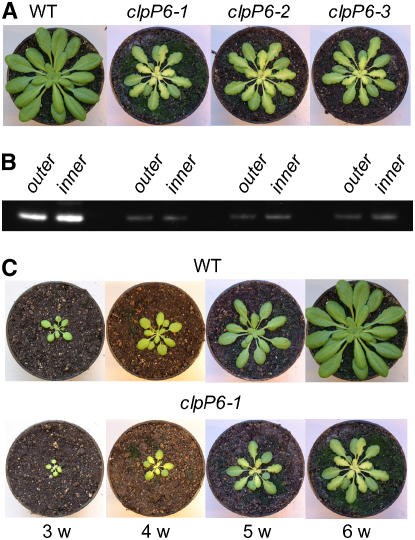
Antisense clpP6 lines were made by transforming wild-type Arabidopsis with Agrobacterium tumefaciens containing the antisense clpP6 plasmid. After immunologically screening >100 individual positive transformants from several independent lines, ∼20% were found to have significant decreases in ClpP6 protein content. It was only within these transformants that a phenotypic change was observed, the severity of which correlated to the level of ClpP6 repression. The main characteristics of the phenotype were slow growth and leaf chlorosis. For more detailed analysis, three lines were selected based on their phenotypic similarity and level of ClpP6 antisense repression. When grown under the standard conditions of 150 μmol·m−2·s−1 illumination for an 8-h day, all three antisense lines exhibited leaf chlorosis, a chlorosis that was not observed in the cotyledons. The extent of chlorosis was more severe in the midvein region, giving the leaf a variegated yellow-heart appearance. These affected leaves also had a wrinkled appearance along the blade edges. During growth, however, the severity of chlorosis gradually decreased until eventually expanded leaves in the outer whorl appeared green, like those in wild-type plants. This was most evident in 6- to 8-week-old antisense plants, in which the younger inner whorl leaves exhibited the chlorotic phenotype and the older, more developed leaves did not (Figure 1A). Despite this change in phenotype, the level of repression of ClpP6 protein (i.e., 80 to 90%) in these antisense lines was relatively constant within the inner and outer leaves (Figure 1B). Interestingly, the chlorotic phenotype of the young developing leaves only slowed the growth of the antisense lines by ∼1 week relative to the wild type (Figure 1C). Because of this size difference, however, all further comparisons between wild-type Arabidopsis and the clpP6 antisense lines were performed on plants of the same developmental size rather than age.
Figure 1.
Antisense Repression of clpP6 Produces a Chlorotic Phenotype.
(A) Comparison between 6-week-old wild-type Arabidopsis and three independent clpP6 antisense lines (clpP6-1, clpP6-2, and clpP6-3). Plants were grown under the same standard conditions of 23/18°C day/night temperatures, 8-h photoperiod with ∼150 μmol·m−2·s−1 light, and 60 to 65% RH.
(B) Shown underneath each photograph in (A) is the level of ClpP6 protein in each plant as determined by immunoblotting. Because of the more extensive chlorosis within the inner leaves of the antisense lines, the level of ClpP6 protein was examined in both inner and outer rosettes of all plants. Samples were prepared from total leaf extracts and separated by one-dimensional PAGE on the basis of equal protein content.
(C) Comparison of growth rate and phenotype development in a representative clpP6 antisense line (clpP6-1) relative to the wild type over a 3- to 6-week (w) period.
Inner Leaves of clpP6 Antisense Lines Have Impaired Photosynthesis
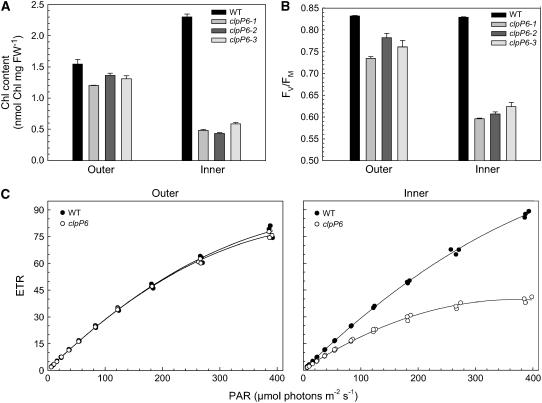
Because of the phenotypic difference between the leaf whorls in the three clpP6 antisense lines, both the outer and inner leaves were compared with those of the wild type. For wild-type plants, the younger inner leaves had a significantly higher chlorophyll content (2.302 ± 0.043 nmol/mg; n = 3) than the older outer leaves (1.544 ± 0.073 nmol/mg; n = 3) (Figure 2A). By contrast, the inner leaves of the three clpP6 antisense lines had considerably less chlorophyll (0.501 ± 0.040 nmol/mg; n = 9) than the corresponding wild-type leaves, consistent with their chlorotic appearance. Again, consistent with their appearance, the more developed outer leaves of the antisense lines had recovered much of the chlorophyll content (1.293 ± 0.052 nmol/mg; n = 9), reaching a level similar to that in the outer wild-type leaves (Figure 2A). Photosynthetic parameters also showed a similar trend between the inner and outer leaves in clpP6 antisense lines relative to the wild type. Measurements of the photochemical efficiency of photosystem II (PSII) (FV/FM) showed no significant difference between the inner (0.829 ± 0.001; n = 3) and outer (0.832 ± 0.001; n = 3) leaves from the wild type. By contrast, FV/FM values in the chlorotic inner leaves of the clpP6 antisense lines showed significant inhibition of PSII photochemical efficiency (0.609 ± 0.009; n = 9), which eventually recovered in the green outer leaves (0.759 ± 0.016; n = 9). Electron transport rates also showed a dramatic decrease in both photosynthetic efficiency (i.e., quantum yield as determined by the initial slope of the light-response curve) and capacity (as measured at the highest irradiance) in the inner leaves of the clpP6 antisense lines (Figure 2C). Again, this inhibition of photosynthesis recovered in the outer leaves of the antisense lines to levels comparable to those in the wild-type outer leaves.
Figure 2.
Antisense Repression of clpP6 Reduces Chlorophyll Content and Photosynthetic Performance in Inner Leaves.
(A) Chlorophyll (Chl) content in inner and outer leaves of wild-type Arabidopsis and clpP6 antisense lines (clpP6-1 to -3). Values shown are averages ± se (n = 3). FW, fresh weight.
(B) Photochemical efficiency of PSII in inner and outer leaves of wild-type Arabidopsis and clpP6 antisense lines (clpP6-1 to -3). Values shown are averages ± se (n = 3).
(C) Photosynthetic electron transport (ETR) rates in inner and outer leaves of wild-type Arabidopsis and clpP6 antisense lines (clpP6-1 to -3). Electron transport rate as determined by chlorophyll fluorescence was measured at different PAR levels between 0 and 400 μmol·m−2·s−1. Values shown are from three independent wild-type plants and one plant from each clpP6 antisense line (clpP6-1 to -3).
In all panels, plants were compared at the same developmental stage.
Inner Leaves of clpP6 Antisense Lines Have Impaired Chloroplast Development
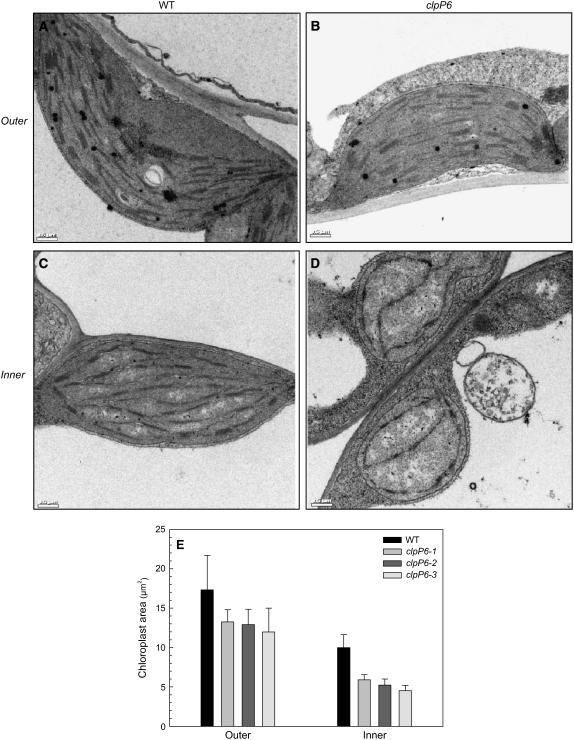
Given their chlorotic phenotype, leaves from the clpP6 antisense lines were examined anatomically in terms of chloroplast ultrastructure using transmission electron microscopy. For this analysis, samples from both the outer and inner leaves of wild-type Arabidopsis and clpP6 antisense lines were used. Wild-type chloroplasts in both types of leaves had the expected morphology and well-structured thylakoid membrane organization (Figures 3A and 3C), although the inner chloroplasts were only half the size of those in the outer leaves (Figure 3E). Chloroplasts within the outer leaves of the clpP6 antisense lines were smaller than those in the wild-type outer leaves but still retained the wild-type morphology and thylakoid membrane structure (Figure 3B). By contrast, however, chloroplasts in the inner leaves of the clpP6 antisense lines showed dramatic changes, with a poorly developed thylakoid membrane network and reduced overall membrane content (Figure 3D). Chloroplasts in these leaves were also only half the size of those in the outer leaves and even smaller than those in the wild type. Morphologically, these chloroplasts had a spherical appearance reminiscent of immature proplastids instead of the more ovoid shape typical of normally differentiated wild-type chloroplasts.
Figure 3.
Microscopic Analysis of Chloroplast Structure in Wild-Type Arabidopsis and clpP6 Antisense Lines.
(A) to (D) Electron micrographs of inner ([C] and [D]) and outer ([A] and [B]) leaves of wild-type Arabidopsis ([A] and [C]) and clpP6 antisense lines ([B] and [D]). Plants were compared at the same developmental stage (5 weeks for the wild type, 6 weeks for clpP6), with all micrographs being representative of chloroplasts from three individual wild-type plants and one plant each from the three clpP6 antisense lines. Bars = 0.5 μm.
(E) Chloroplast size as determined by area in outer and inner leaves of wild-type Arabidopsis and clpP6 antisense lines (clpP6-1 to -3). All values shown are averages ± sd (n = 6).
Reduced Levels of Photosynthetic Proteins in Inner Leaves of clpP6 Antisense Lines
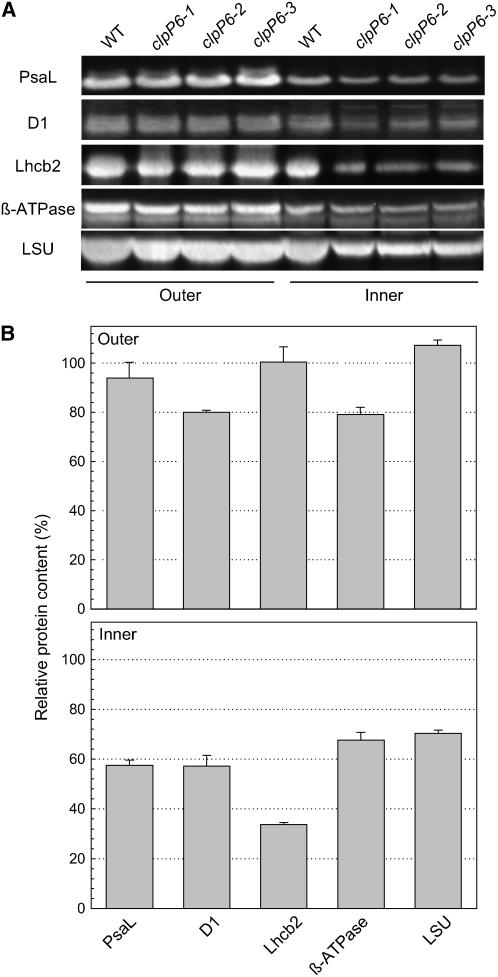
Because of the changes in photosynthetic performance, chlorophyll content, and chloroplast ultrastructure in the clpP6 antisense lines, we next analyzed the relative amounts of photosynthetic protein complexes using antibodies specific to the following marker proteins: PsaL for PSI; D1 and Lhcb2 for the PSII reaction center and outer antennae, respectively; β-subunit for ATPase; and large subunit (LSU) for ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) (Figure 4A). Again, the inner and outer leaves of both wild-type and clpP6 plants were analyzed. As can be seen in Figure 4, there was no significant change in PSI, PSII outer antennae, or Rubisco contents in the outer leaves of the clpP6 antisense lines relative to the wild type, whereas a small decrease (20%) was observed in the levels of ATPase and PSII reaction centers (Figure 4B). By contrast, the levels of PSI, PSII, ATPase, and Rubisco all decreased significantly (30 to 40%) in the chlorotic inner leaves of the clpP6 antisense lines relative to the wild type (Figures 4A and 4B). Lhcb2 content decreased even more markedly (∼70%), consistent with the extent of chlorophyll loss in the inner leaves of the clpP6 antisense lines.
Figure 4.
Reduced Levels of Photosynthetic Protein Complexes in Inner Leaves of the clpP6 Antisense Lines.
(A) Amounts of marker proteins for different photosynthetic protein complexes were determined by immunoblotting in outer and inner leaves of 5-week-old wild-type Arabidopsis and 6-week-old clpP6 antisense lines (clpP6-1 to -3). Total leaf proteins were separated by one-dimensional PAGE on the basis of equal protein content. Antibodies were used to detect specific marker proteins for each photosynthetic protein complex: PsaL for PSI; D1 and Lhcb2 for PSII; β-subunit for ATPase; and LSU for Rubisco.
(B) Quantification of the amount of each photosynthetic marker protein in the outer and inner leaves of clpP6 antisense lines (clpP6-1 to -3) relative to the wild type. Values shown are averages ± se (n = 3), with the wild-type values set to 100%.
Changes in Clp Protein Content in the clpP6 Antisense Lines
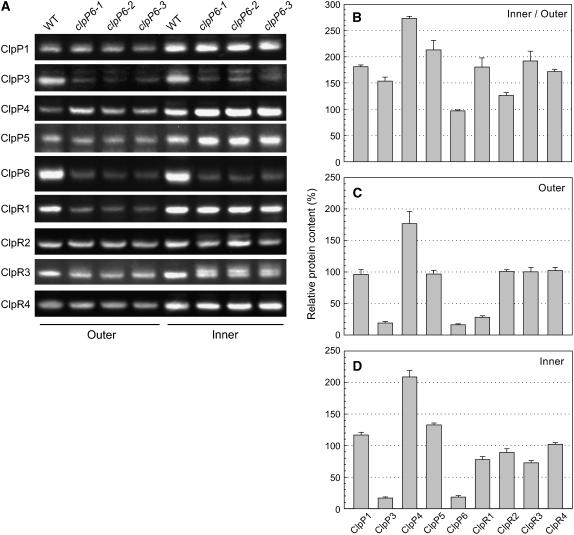
Given that all of the chloroplast ClpP and ClpR proteins associate together into a single proteolytic core complex, we next investigated whether the reduced ClpP6 content in the antisense lines affected the levels of the other ClpP/R proteins. For this analysis, whole cell proteins were extracted from both inner and outer leaves of wild-type Arabidopsis and clpP6 antisense lines, and the amount of each ClpP/R protein was determined by immunoblotting using specific antibodies (Figure 5A). With the exception of ClpP6, which showed no significant change, all chloroplast-localized ClpP/R proteins were more abundant in the inner leaves of wild-type plants than in the outer leaves (Figures 5A and 5B). The extent of this increase varied considerably among the different proteins, ranging from only a 25% increase for ClpR2 to a 2.5- to 3-fold increase for ClpP4. When comparing the wild type with the clpP6 antisense lines, considerable variation also occurred in the level of each ClpP/R protein. In the outer leaves, no significant change occurred to the amount of ClpP1, -P5, -R2, -R3, or -R4 in the antisense lines, whereas ClpP4 content increased almost twofold. Interestingly, the levels of ClpP3 and ClpR1 decreased in the antisense lines to levels similar to that of ClpP6 (Figures 5A and 5C). A similar overall pattern was also observed in the inner leaves of the clpP6 antisense lines, with little variation in the amounts of ClpP1, ClpP5, and ClpR2-4 proteins, a significant increase in ClpP4 content (twofold), and an 80% loss of ClpP3 (Figures 5A and 5D). The only major difference from the outer leaves was a less extensive decrease in the amount of ClpR1 (i.e., 20%) in the inner leaves of the antisense lines.
Figure 5.
Changing Levels of Chloroplast Clp Proteins in the clpP6 Antisense Lines.
(A) Relative amounts of ClpP and ClpR proteins in the inner and outer leaves of wild-type Arabidopsis and clpP6 antisense lines (clpP6-1 to -3). Total cell extracts were isolated from 5-week-old wild-type Arabidopsis and 6-week-old clpP6 antisense lines and separated by one-dimensional PAGE on the basis of equal protein content. The amount of each chloroplast ClpP and ClpR paralog was determined by immunoblotting with specific polyclonal antibodies.
(B) to (D) Quantification of the relative amounts of each chloroplast ClpP and ClpR paralog in wild-type Arabidopsis and clpP6 antisense lines: wild-type inner leaves relative to outer leaves (B); clpP6 antisense outer leaves (C); and clpP6 antisense inner leaves (D). Values shown are averages ± se (n = 3), with the wild-type values for outer and inner leaves ([C] and [D]) set to 100%.
Given that the level of ClpP3 decreased in both inner and outer antisense leaves to the same extent as ClpP6, we checked for possible cross repression of the clpP3 transcript by the clpP6 antisense construct. Using RT-PCR, no significant decrease in the amount of clpP3 mRNA was detected in the clpP6 antisense lines relative to the wild type (data not shown), indicating that the loss of ClpP3 protein was attributable to a specific downregulation and not to interference of its mRNA by the clpP6 antisense transcript.
The Chloroplast Clp Proteolytic Core Comprises Two Distinct Subcomplexes
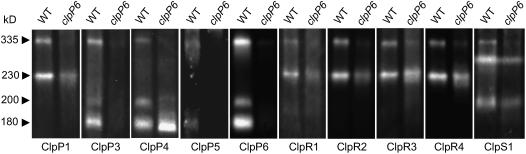
Because of the variation in the amount of each chloroplast ClpP/R protein in the clpP6 antisense lines, we next investigated how this would affect the Clp proteolytic core complex. For this analysis, stromal proteins were isolated from young leaves of wild-type Arabidopsis and clpP6 antisense lines and separated by native PAGE, and then each Clp protein was detected by immunoblotting using specific antibodies. As shown in Figure 6, all of the ClpP/R antibodies detected a 335-kD protein complex in the wild type matching the Clp proteolytic core complex, as described previously (Peltier et al., 2004). This 335-kD complex decreased by 80 to 90% in the clpP6 antisense lines, matching the level of antisense repression of ClpP6 observed in these plants (Figure 1B) and suggesting that ClpP6 is necessary for the formation of the Clp proteolytic core complex.
Figure 6.
Clp Protein Complexes in Wild-Type Arabidopsis and clpP6 Antisense Lines.
Clp protein complexes in isolated stromal fractions from 3-week-old wild-type Arabidopsis and 4-week-old clpP6 antisense lines were separated by native PAGE on the basis of equal protein content. The different Clp protein complexes were visualized by immunoblotting using specific antibodies for Arabidopsis ClpP1, ClpP3-6, ClpR1-4, and ClpS1 as indicated below each panel. The size of each Clp protein complex is indicated at left.
In addition to the 335-kD core complex, smaller Clp protein complexes were also detected in the wild-type stromal fractions. The first of these was a 230-kD complex containing only ClpP1 and ClpR1-4, whereas two additional complexes of 180 and 200 kD were also found containing the remaining ClpP3-6 proteins (Figure 6). Given that the chloroplast Clp core likely consists of two heptameric rings, the 230-kD subcomplex containing ClpP1 and ClpR1-4 would correspond to one such ring, whereas the 180-kD complex with ClpP3-6 would match the other. To determine the identity of the 200-kD complex, we next examined whether this larger ClpP3-6 protein complex might contain one of the two other Clp proteins (i.e., ClpS1 and ClpS2) associated with the Clp proteolytic core. For this, we attempted to prepare antibodies specific for either ClpS1 or ClpS2 using synthetic peptides. Unfortunately, only the ClpS1-specific peptide produced a strong immune response. Using these antibodies, ClpS1 was indeed detected in the 200-kD ClpP3-6 subcomplex in addition to the 335-kD proteolytic core, as expected (Figure 6). Interestingly, a third protein complex containing ClpS1 was also detected, a complex of 260 kD containing no ClpP/R protein nor any of the chloroplast HSP100 proteins ClpC or ClpD (data not shown). ClpS2 was also detected in this 260-kD complex, but the signal was relatively weak as a result of the low antigenicity of the antibodies (data not shown). When comparing the subcomplexes in the clpP6 antisense lines with those in the wild type, the ClpP6-containing subcomplexes of 180 and 200 kD were essentially absent (5 to 15% remaining), again consistent with the degree of ClpP6 antisense repression. By contrast, the ClpP1 and ClpR1-4 subcomplex of 230 kD only decreased by ∼25% in the antisense lines relative to the wild type; a similar small decrease was also observed for the 260-kD complex containing ClpS1 (Figure 6). Surprisingly, a new complex of 175 kD was also observed in the antisense lines containing just the ClpP4 protein. The amount of this novel complex matched the increased level of monomeric ClpP4 protein observed previously in the antisense lines (Figure 5). This finding suggests that, in the absence of the other nucleus-encoded ClpP paralogs (i.e., ClpP3, -5, and -6), the excess ClpP4 protein can form a homogeneous 175-kD complex, but one that cannot complex further to form a functional Clp proteolytic core.
Identification of Potential Protein Substrates for the Chloroplast Clp Protease
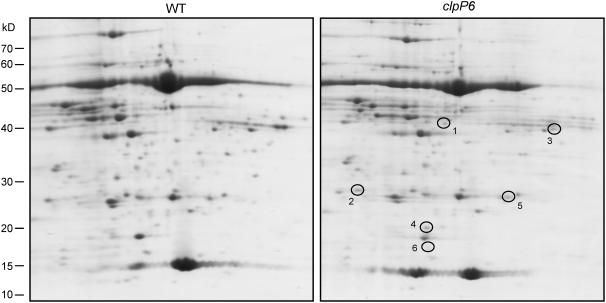
One of the key characteristics of the chloroplast Clp protease in higher plants yet to be addressed is the identity of its specific protein substrates. Based on the rationale that any such substrate would be expected to accumulate when the amount of Clp protease is considerably reduced, we first checked for significant changes in stromal protein content in the clpP6 antisense lines relative to the wild type. For proteins of >60 kD, one-dimensional SDS-PAGE was used because of their better resolution characteristics, whereas two-dimensional PAGE was used to resolve the more complex mixture of stromal proteins of <60 kD. In this analysis, no large molecular mass protein was detected in the stroma of clpP6 antisense chloroplasts greater than threefold more abundant than that in wild-type chloroplasts (data not shown). Of the smaller proteins, five were found to accumulate in the antisense lines, of which only two were also detectable in the wild type by Coomassie blue staining (Figure 7, Table 1). For the other three proteins, silver staining was used to quantify their abundance in the clpP6 antisense lines relative to the wild type. During the silver stain analysis, an additional sixth protein spot was found that accumulated in the clpP6 antisense lines. The six proteins were identified as the Calvin cycle enzymes fructose bisphosphate aldolase and ribose 5-phosphate isomerase, a putative RNA binding protein, uracil phosphoribosyl-transferase (UPRT), a cyclophilin peptidyl-prolyl cis-trans isomerase (PPIase), and a nucleoside diphosphate (NDP) kinase.
Figure 7.
Changes in Stromal Protein Composition in Wild-Type Arabidopsis and clpP6 Antisense Lines.
Isolated stromal proteins from wild-type Arabidopsis and clpP6 antisense lines were separated by two-dimensional PAGE and visualized by Coomassie blue staining. Those proteins consistently more abundant in the clpP6 antisense lines relative to the wild type are circled and numbered. Shown are representative results from three replicate assays for each line. The identity of each numbered protein is detailed in Table 1.
Table 1.
Identification of Stromal Proteins Accumulating in the Arabidopsis clpP6 Antisense Lines
| No. | Identity | Gel Mass (kD)a | Protein Identifierb | Protein Scorec | Peptides Matchedd | Folde |
|---|---|---|---|---|---|---|
| 1 | Fructose bisphosphate aldolase | 42 | At4g38970 | 92 | 9 | >5 |
| 2 | Putative ribose 5-phosphate isomerase | 28 | At3g04790 | 107 | 10 | 5 |
| 3 | Putative RNA binding protein | 41 | At1g09340 | 129 | 11 | >5 |
| 4 | Uracil phosphoribosyl-transferase | 20 | At3g53900 | 456 | 21 | 10 |
| 5 | Peptidyl-prolyl cis-trans isomerase | 27 | At3g62030 | 619 | 130 | >4 |
| 6 | Nucleoside diphosphate kinasef | 18 | At5g63310 | 169 | 7 | 4 |
Identification by MALDI-TOF MS and HPLC-MS/MS of those proteins most abundant in the clpP6 antisense lines relative to the wild type. Each numbered protein corresponds to the same numbered protein circled in Figure 7.
Molecular mass calculated from gel size markers.
Protein identification as Arabidopsis Genome Initiative gene code.
Protein scores > 66 are significant (P < 0.05).
Peptide match at mass tolerance of ±100 ppm, allowing a maximum of one missed cleavage.
Fold upregulation based on quantification of stained gel spots.
Detected with silver staining.
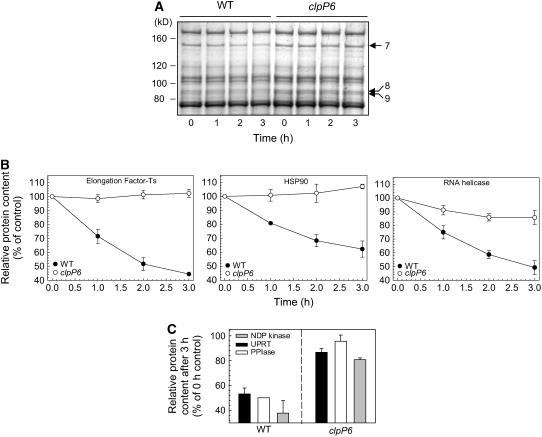
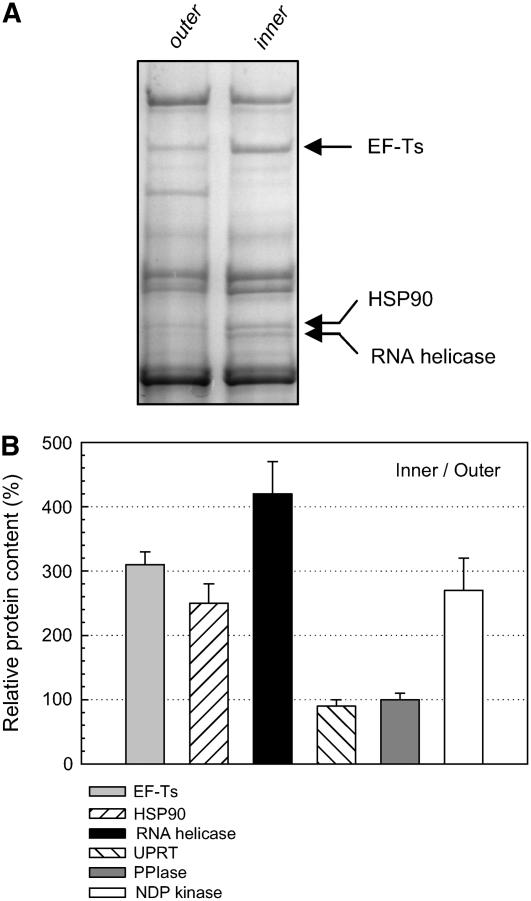
To better identify potential substrates for the Clp protease in Arabidopsis, we next developed an in vivo chloroplast degradation assay to compare the stability of different stromal proteins in wild-type Arabidopsis and clpP6 antisense lines. For this assay, intact chloroplasts were isolated from wild-type and clpP6 plants. Equal numbers and concentrations of intact chloroplasts were then incubated in the presence of ATP at 25°C and constant irradiance for 3 h, with samples taken at 1-h intervals. For each sample, chloroplasts were separated into stromal and thylakoid membrane protein fractions and then resolved by both one-dimensional and two-dimensional PAGE. Degradation rates were determined by simply quantifying the amount of each stained protein during the time course of the assay. In the case of plastid-encoded proteins, assays were performed with the addition of lincomycin to inhibit chloroplast protein synthesis. One-dimensional gels were used to resolve the degradation of larger proteins at all four time points (0, 1, 2, and 3 h), whereas two-dimensional gels were used to resolve the smaller proteins at the 0-h control and 3-h time points. For the larger polypeptides, three potential substrates were identified by their slower degradation rates in the clpP6 antisense lines: elongation factor containing Ts motifs (EF-Ts), the molecular chaperone HSP90, and an RNA helicase (Figures 8A and 8B, Table 2). All three substrate proteins were more abundant in the clpP6 antisense lines than in the wild type before the degradation assay, but only by 30 to 60% (Table 2), which was why they were not identified earlier (Figure 7, Table 1). Another three substrates of low molecular mass were also found: UPRT, PPIase, and NDP kinase (Figure 8C). These three additional putative substrates decreased by 50 to 60% in wild-type chloroplasts by the end of the 3-h assay, whereas the degradation was only 5 to 20% in chloroplasts from the clpP6 antisense lines. It should be noted that in the absence of ATP and light, no significant degradation of the identified protein substrates was observed in wild-type chloroplasts.
Figure 8.
Identification of Protein Substrates for the Chloroplast Clp Protease.
(A) An equal number of intact chloroplasts from wild-type Arabidopsis and the clpP6 antisense lines were incubated for 3 h in the presence of light and ATP. At 1-h intervals, aliquots were taken and chloroplasts were ruptured. After fractionation, stromal proteins were separated by SDS-PAGE and visualized by Coomassie blue staining. Those proteins showing significant degradation over the 3-h time course in the wild type but not in the clpP6 antisense lines were identified by matrix-assisted laser-desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) (see Table 2 for details). Shown are results from a representative degradation assay, with the three putative protein substrates indicated by arrows at right.
(B) Degradation profiles for the high molecular mass substrate proteins EF-Ts, HSP90, and RNA helicase in intact chloroplasts from wild-type Arabidopsis and the clpP6 antisense lines. Values shown are averages ± se (n = 3) plotted as a percentage of the amount of each protein at time 0, which was set to 100%.
(C) Extent of degradation for the three small molecular mass protein substrates in intact chloroplasts of wild-type Arabidopsis and the clpP6 antisense lines over the 3-h time course. After fractionation, stromal proteins were separated by two-dimensional PAGE and quantified after either Coomassie blue or silver staining. The relative amount of each protein remaining after 3 h is shown as an average + se (n = 3) plotted as a percentage of the time-0 value, which was set to 100%.
Table 2.
Identity of Three Putative Protein Substrates for the Chloroplast Clp Protease as Determined by MALDI-TOF MS
| No. | Identity | Gel Mass(kD)a | Protein Identifierb | Protein Scorec | Peptides Matchedd | Folde |
|---|---|---|---|---|---|---|
| 7 | EF-Ts | 150 | At4g29060 | 123 | 13 | 1.3 |
| 8 | HSP90 | 87 | At2g04030 | 222 | 20 | 1.4 |
| 9 | RNA helicase | 85 | At5g26742 | 169 | 15 | 1.6 |
Each numbered protein corresponds to the same numbered protein indicated in Figure 8A.
Molecular mass calculated from gel size markers.
Protein identification as Arabidopsis Genome Initiative gene code.
Protein scores > 66 are significant (P < 0.05).
Peptide match at mass tolerance of ±100 ppm, allowing a maximum of one missed cleavage.
Fold upregulation based on quantification of stained gel spots.
Most Potential Substrates for Clp Protease Are More Abundant in Younger Leaves
Because the chlorotic phenotype exhibited by the clpP6 antisense lines was more severe in the younger inner leaves than in the more mature outer leaves, we next examined the relative abundance of each identified protein substrate in these two types of leaves in the wild type. For the larger protein substrates, all three were 2.5- to 4-fold more abundant in the inner wild-type leaves (Figures 9A and 9B). Similarly, the amount of one of the smaller substrates, NDP kinase, was also higher in the inner leaves (2.5 fold; Figure 9B), whereas the levels of the other two (i.e., URPT and PPIase) were identical between the two ages of leaves. This finding suggests that most of the putative stromal substrates for the chloroplast Clp protease are more abundant, and thus likely more functionally important in younger leaves, correlating with the severity of the chlorotic phenotype in the clpP6 antisense lines.
Figure 9.
Abundance of Putative Substrates for the Chloroplast Clp Protease during Leaf Development.
(A) Isolated stromal proteins from outer and inner wild-type leaves were analyzed by SDS-PAGE based on equal protein content. Arrows indicate the three high molecular mass substrates for the Clp protease.
(B) Abundance of the six putative Clp protein substrates in the inner leaves of wild-type Arabidopsis relative to that in the outer leaves. Shown are averages + se (n = 3), with the level of each protein in the outer leaves set to 100%.
DISCUSSION
In this study, we have shown that the chloroplastic Clp proteolytic core, which is essential for chloroplast development in Arabidopsis, comprises two distinct subcomplexes and have revealed several potential protein substrates. Because all putative T-DNA insertion mutants of the five nucleus-encoded chloroplast-localized ClpP proteins have to date proven to be embryo-lethal, the use of antisense lines specific for individual ClpP paralogs has been invaluable in studying the composition of the Clp proteolytic core and its activity. In the case of clpP6, all three antisense lines exhibited the same phenotype, with young chlorotic inner leaves that gradually became greener during development, so that the older outer leaves appeared similar to those in wild-type plants. Chlorosis was also more severe within the midvein region of the affected leaves, and the leaf edges were wrinkled in appearance. Interestingly, antisense inhibition of clpP6 produces a similar but less severe chlorotic phenotype to the antisense repression of clpP4, despite both sets of transgenic lines having similar levels of protein repression (i.e., 80 to 90%). Despite this difference in severity, the chlorotic phenotype of the clpP4 antisense lines lessened upon maturation, as did that in the clpP6 antisense lines (Zheng et al., 2006), consistent with the fact that both ClpP paralogs function within the same proteolytic core (Peltier et al., 2001, 2004).
The severity of the chlorotic phenotype in young leaves of the clpP6 antisense lines suggests that the Clp proteolytic activity in chloroplasts is most important during early leaf development. Not only do the affected younger leaves have significantly lower chlorophyll contents, they also exhibit impaired photosynthetic activity and reduced amounts of the major photosynthetic protein complexes. More dramatic was the compromised chloroplast ultrastructure within the chlorotic clpP6 antisense leaves. Not only were the plastids half the size of wild-type plastids, but they contained no well-defined thylakoid membrane network. Morphologically, the plastids from the affected antisense leaves appeared more like proplastids, suggesting that stromal Clp proteolytic activity plays a critical role in chloroplast biogenesis. Interestingly, a similar involvement in chloroplast development also has been proposed for the thylakoid membrane–bound FtsH protease. Both the VAR1 (FtsH5) and VAR2 (FtsH2) mutants in Arabidopsis exhibit a more severe variegated phenotype in young leaves compared with the fully expanded mature leaves (Chen et al., 1999; Sakamoto et al., 2002; Zaltsman et al., 2005).
It is well known that the model Clp proteolytic core in E. coli consists of a single ClpP subunit organized as two opposing heptameric rings of ∼300 kD (Wang et al., 1997). A single Clp proteolytic core also exists in the chloroplast stroma, but its composition is remarkably complex compared with any other known Clp protease. The stromal Clp proteolytic core consists of five ClpP and four ClpR paralogs (Peltier et al., 2001), presumably arranged as dual heptameric rings like the bacterial ClpP core. Moreover, the stromal Clp core also contains two novel Clp proteins, ClpS1 and ClpS2, that together with the ClpP and ClpR paralogs form a proteolytic core complex of 320 to 350 kD (Peltier et al., 2004). Using our suite of Clp-specific antibodies, we were able to confirm the existence of a single stromal proteolytic core complex (335 kD) containing all of the chloroplastic ClpP/R/S proteins. Furthermore, we revealed that the core complex consists of two distinct subcomplexes, one of 230 kD containing ClpP1 and ClpR1-4, the other of 180 kD containing ClpP3-6. Given the likely oligomeric structure of the core complex, each of these subcomplexes would represent one of the two heptameric rings. The fact that the combined sizes of the two rings is larger than that of the core complex suggests a degree of spatial overlap when the two rings associate. Previous estimations for the composition of the chloroplastic Clp proteolytic core have proposed a stoichiometry of P1, R2, R4, (R1+R3+P3), P4, P5, P6 = 1:1:2:(3):3:3:1 based on Coomassie blue staining, taking into account protein size and amino acid composition (Peltier et al., 2004). Such a stoichiometry, however, is now inconsistent with the paralog composition of each subcomplex as described in this study. As a consequence, we now propose a new stoichiometry based on changing the least certain assumptions from the previous estimation (i.e., the relative amounts of ClpR1, -R3, and -P5). Based on a 14-mer conformation, the ClpP1/ClpR1-4 heptamer would have a stoichiometry of P1, (R1+R3), R2, R4 = 1:(3):1:2, and the ClpP3-6 heptamer would have P3, P4, P5, P6 = 1:3:2:1.
In addition to the ClpP and ClpR paralogs, the proteolytic core complex has been proposed to also contain a single subunit each of ClpS1 and ClpS2 (Peltier et al., 2004). The ClpS structure has been modeled previously using a PSSM three-dimensional threading tool with the N-terminal E. coli ClpA domain as a template (Peltier et al., 2004). The proteins were predicted to dock to the Clp proteolytic core close to the axial access pores. They are presumed to function either competitively with the HSP100 chaperone partner (i.e., ClpC and/or ClpD) for available binding sites to the Clp proteolytic core or as a wedge to keep the axial opening of the core complex free for degradation products to diffuse out. In this study, we show that ClpS1 is present in a 200-kD subcomplex containing the ClpP3-6 paralogs, consistent with a single ClpS1 protein bound to the 180-kD ClpP3-6 heptameric ring. Neither ClpS1 nor ClpS2, however, was found attached to the ClpP1/R1-4 ring, consistent with the observation of only one subcomplex containing the ClpP1/ClpR1-4 paralogs. Apart from its expected association with the 335-kD proteolytic core, ClpS2 was only detected in the 260-kD complex also containing ClpS1 but no other known Clp protein. We are now pursuing the identity of this 260-kD complex and how it affects the association of both ClpS1 and ClpS2 with the Clp proteolytic core.
The amount of the Clp proteolytic core in the clpP6 antisense lines decreased by 80 to 90% relative to the wild type, matching the level of the least abundant ClpP/R subunit (which was ClpP6). This correlation suggests that the formation of a functionally active Clp protease relies on the correct stoichiometry of each ClpP and ClpR subunit within the proteolytic core. Interestingly, the strong repression in ClpP6 protein content caused a proportional decrease in the ClpP3-6 subcomplex (80 to 90%) but a much less severe decrease in the ClpP1/R1-4 subcomplex (∼25%), suggesting that the loss of ClpP6 causes far greater instability of its associated subcomplex. Within the ClpP3-6 subcomplex, repression of ClpP6 caused a proportional downregulation of ClpP3, suggesting that the two paralogs interact directly within the subcomplex. By contrast, the ClpP5 subunit remained relatively unchanged in the clpP6 antisense lines, whereas ClpP4 was markedly upregulated, likely as a compensatory response to the loss of both ClpP3 and ClpP6. Indeed, the increased level of ClpP4 formed a homogeneous ClpP4 subcomplex slightly smaller than the native ClpP3-6 subcomplex. Despite this, the ClpP4 self-complex could not associate with the ClpP1/R1-4 subcomplex to form an intact Clp proteolytic core and thus could not compensate for the loss of ClpP6. In an earlier study, antisense repression of clpP4 was also shown to cause a downregulation of associated ClpP paralogs but no significant change in the subunits of the ClpP1/R1-4 subcomplex (Zheng et al., 2006). In this case, both ClpP3 and ClpP5 are downregulated to the same extent as ClpP4 (∼80%), whereas ClpP6 is downregulated to a lesser extent (∼30%) (Zheng et al., 2006). Given that ClpP4 is the most abundant subunit within the ClpP3-6 subcomplex based on current predictions, it is not surprising that its loss causes greater instability of associated subunits compared with the less abundant ClpP6 protein.
One of the remaining challenges within the field of chloroplast proteases is the identification of native protein substrates, and this study has identified such substrates for the chloroplast Clp protease in higher plants. Only in the green alga Chlamydomonas reinhardtii has a putative substrate for the chloroplast Clp protease been found previously, that being the cytochrome b6/f complex during nitrogen starvation (Majeran et al., 2000). To identify possible protein substrates for the plant chloroplast Clp protease, we developed an in vivo proteolytic assay using intact chloroplasts isolated from Arabidopsis. By comparing wild-type plants with the clpP6 antisense lines, those chloroplast proteins whose degradation rate was inhibited by the near absence of the Clp proteolytic core could be readily detected. After examining a wide range of chloroplast proteins based on size, localization (thylakoid membrane or stroma), and genetic origin (plastomic or nuclear), six soluble nucleus-encoded polypeptides were eventually identified as putative substrates: EF-Ts, HSP90, RNA helicase, UPRT, PPIase, and NDP kinase. Interestingly, the functions of all of these identified substrates are related more to chloroplast homeostasis than to any specific metabolic pathway, such as photosynthesis. EF-Ts functions as a nucleotide-exchange factor for the elongation factor Tu. It does not specifically bind to the ribosome but is necessary for the dissociation of GDP from elongation factor Tu, which enables the continued elongation of nascent polypeptides during protein synthesis (Kristensen et al., 2002). HSP90 proteins are abundant and highly conserved molecular chaperones involved in a wide range of specific functions. They are often involved in regulating the structure and activity of key regulatory proteins as well as in cooperating with other chaperone systems, such as HSP70 and PPIase, in the correct folding of nascent polypeptides (Sangster and Queitsch, 2005). Indeed, PPIase was another putative substrate for the chloroplast Clp protease. It performs a critical role in protein folding by catalyzing the rapid isomerization of prolyl bonds from the cis to the trans configuration, a process that is essential for the correct folding and maturation of many polypeptides (Romano et al., 2005). RNA helicases are equally important enzymes required for many homeostatic processes, such as transcription and mRNA processing, ribosome biogenesis, initiation of translation, and RNA catabolism (Rocak and Linder, 2004). UPRT performs a similar housekeeping function in the salvaging of the pyrimidine base uracil, an important component in the recycling processes that provide sufficient nucleobases for the continued synthesis of nucleic acids (Zrenner et al., 2006). Finally, NDP kinases also perform a critical housekeeping role involving the generation of nucleoside triphosphates, thereby regulating the available nucleotide pool (Hasunuma et al., 2003).
Given the obvious importance of such proteins to chloroplast biogenesis and maintenance, it is conceivable that impairment of their degradation rate by reduced Clp proteolytic activity could explain the severe phenotype exhibited by the clpP6 antisense lines, especially in the early stages of leaf growth. The nature of the functions of these putative substrates is also consistent with the Clp protease being present in plastids other than chloroplasts. Not only is the Clp proteolytic core present in root plastids (Peltier et al., 2004), it is also conserved in plastids of the nonphotosynthetic parasitic plant Epifagus (Wolfe et al., 1992) and in Apicomplexan parasites such as Plasmodium falciparum (Roos et al., 2002). The types of protein substrates for the chloroplast Clp protease also might be linked to why the plastomic clpP1 gene is coexpressed with two ribosomal proteins in most higher plants (Clarke et al., 1994). As would be expected for substrates whose degradation was impaired, most of the identified proteins accumulated in the clpP6 antisense lines relative to the wild type. The three high molecular mass substrates were also more abundant in an Arabidopsis clpC1 mutant, in which the level of the likely chaperone partner to the Clp proteolytic core, ClpC, was reduced by ∼65% (Sjögren et al., 2004). Moreover, most of the identified protein substrates were more abundant in younger leaves of wild-type plants than in mature leaves, probably because the developing chloroplasts in younger leaves are more metabolically active, with higher rates of transcription, translation, and protein turnover. This would explain, at least in part, why the antisense repression of clpP6 and the subsequent loss of Clp proteolytic activity severely affected chloroplast development and function in younger leaves, and why this phenotype lessened during leaf maturation. Overall, it appears that the stromal Clp protease plays an integral role in chloroplast development and homeostasis via the degradation of key regulatory proteins and enzymes. To address this housekeeping function in more detail, we are continuing to search for additional protein substrates to identify the different chloroplast processes affected by Clp proteolytic activity in higher plants.
METHODS
Plant Growth Conditions
Seeds for Arabidopsis thaliana wild type (ecotype Columbia-0) and clpP6 antisense lines were sown in a Perlite/soil mix (1:5) after vernalization at 4°C for at least 48 h to break dormancy. All plants were grown individually in pots or as lawns under the following standard conditions: 8-h photoperiod with white light (∼150 μmol·m−2·s−1), 23/18°C day/night temperatures, and 65% RH.
Antisense Construct and Arabidopsis Transformation
The clpP6 antisense construct was prepared in the plant expression vector pSJ10 (Ganeteg et al., 2001). As for the clpP4 antisense lines described previously (Zheng et al., 2006), the full-length Arabidopsis clpP6 gene was cloned into the pSJ10 vector between the 35S cauliflower mosaic virus promoter and the nopaline synthase polyadenylation signal. The correct orientation of the clpP6 insert was confirmed by DNA sequencing. The verified clpP6 antisense construct was transformed into the Agrobacterium tumefaciens strain C58rifR/EHA105, which was cultured according to standard procedures (Sambrook et al., 1989). The floral-dip method (Clough and Bent, 1998; Strand et al., 2000) was used to transform wild-type Arabidopsis with Agrobacterium containing the clpP6 antisense construct. Transformed seeds were selected in the T1 generation on Murashige and Skoog plates with 50 μg/mL kanamycin. Kanamycin-resistant plants were screened immunologically for reduced ClpP6 protein content to verify the degree of antisense inhibition. Lines with significant loss of ClpP6 were again screened at the T2 stage, with plants from the T3 generation used for all subsequent experimental work.
Chlorophyll and Photosynthetic Measurements
Arabidopsis leaf discs or small leaves from 5- and 6-week-old wild-type and clpP6 plants, respectively, were weighed and then incubated in dimethylformaldehyde for several hours in darkness at room temperature, followed by a further overnight incubation at 4°C. Absorbance of the resulting extract was measured at 663 and 646 nm using a UV-2401PC spectrophotometer (Shimadzu), and the total chlorophyll content was calculated as described previously (Porra et al., 1989). The photochemical efficiency of PSII (FV/FM) and the photosynthetic electron transport rate were measured in 8- to 9-week-old outer and inner leaves by chlorophyll fluorescence using a pulse-amplitude-modulated fluorometer (PAM-2000; Heinz-Waltz) according to the manufacturer's recommendations. Electron transport rate values were modified according to Evans (1996) because of the different chlorophyll concentrations in wild-type Arabidopsis and clpP6 antisense lines.
Light and Electron Microscopy
Outer and inner leaves of equal length from 5- and 6-week-old wild-type and clpP6 plants were fixed in fixate in 1.5% glutaraldehyde, 1.5% paraformaldehyde, 0.15 M sucrose, 2 mM CaCl2, and 0.05 M cacodylate buffer, pH 7.0, at 4°C, with additional fixation done according to Soikkeli (1980). Triple replicates for the inner and outer leaves from the wild type and three clpP6 antisense lines were prepared for light microscopy as described by Sutinen (1987). Cross sections of whole leaf (40× magnification) and palisade and spongy cell sections (100× magnification) were digitally photographed. Digital images were analyzed with Adobe Photoshop version 6.0 (Adobe Systems). Thin sections for transmission electron microscopy were prepared from the same leaf samples used for light microscopy. Palisade and spongy cells of one leaf per treatment were photographed with the transmission electron microscope (JEM EX; JEOL) at 5000× magnification. The negatives were scanned, and digital images were analyzed using Adobe Photoshop version 6.0.
Production of ClpS-Specific Antibodies
Polyclonal antibodies specific to Arabidopsis ClpS1 and ClpS2 were generated using synthetic peptides corresponding to unique amino acid sequences within each protein: ClpS1, VEKSMNEDVDLSFKKQGQ; ClpS2, NLHIEAYDHRLEPGNRPG. The peptides were conjugated to BSA and then injected into rabbits intramuscularly and subcutaneously (AgriSera).
Immunoblotting of Leaf Protein Extracts
Total cell proteins were isolated from outer and inner leaves of wild-type Arabidopsis and clpP6 antisense lines at the same developmental stage (5 and 6 weeks old, respectively) by grinding in liquid nitrogen in prechilled mortars. The frozen powder (∼50 mg) was transferred to precooled microfuge tubes, and NuPAGE lithium dodecyl sulfate sample buffer (Invitrogen) was added to a final concentration of 0.1 mg/mL. Samples were mixed, incubated at 75°C for 5 min, and then centrifuged at 20,000g for 10 min at 4°C to separate soluble proteins. To determine protein concentration, an aliquot of sample was precipitated in 80% acetone to remove chlorophyll and DTT and resolubilized in NuPAGE sample buffer without DTT. Protein concentrations were determined using a bicinchoninic acid (BCA) protein assay according to the manufacturer's protocol (Pierce Chemical). Samples for PAGE were loaded with equal protein content (15 μg) on precast NuPAGE gels (Invitrogen): 3 to 8% Tris-acetate gels for proteins of ≥70 kD, 12% Bis-Tris gels using MOPS buffer for proteins of 40 to 70 kD, and 12% Bis-Tris gels using MES buffer for proteins of <40 kD. Proteins separated electrophoretically were transferred to nitrocellulose using an Xcell blotting apparatus (Invitrogen). Arabidopsis Clp proteins were detected using specific polyclonal antibodies as described previously (Zheng et al., 2002; Sjögren et al., 2004) or as detailed above. Antibodies specific for other chloroplast proteins were obtained as gifts: Lhcb2, PsaL, and β-ATPase from S. Jansson (Umeå University), and D1 and Rubisco LSU from G. Öquist (Umeå University). Primary antibodies were detected using enhanced chemiluminescence peroxidase-labeled anti-rabbit secondary antibody (Amersham Pharmacia) and visualized by enhanced chemiluminescence (ECL Advance Western Blotting detection kit; Amersham Pharmacia). Chemiluminescent signals were detected and quantified using the ChemiGenius2 imaging system (Syngene) and associated software.
Protein Complex Separation by Native PAGE
Chloroplasts were isolated from lawns of wild-type and clpP6 plants at the same developmental stage (3 and 4 weeks old, respectively) according to Aronsson and Jarvis (2002). Isolated chloroplasts were ruptured by hypotonic shock in buffer A (4 mM MgCl2 and 10 mM MOPS, pH 7.6) on ice for 10 min. Stroma were separated from thylakoid membranes by centrifugation (20,000g for 10 min at 4°C). Stromal proteins were concentrated (Microsep 10K omega biomolecular separation kit; Pall), and protein concentrations were determined using the BCA protein assay according to the manufacturer's protocol (Pierce Chemical). Protein samples from wild-type Arabidopsis and clpP6 antisense lines (60 μg) were loaded on 4 to 13% polyacrylamide Tris-borate gels (45 mM Tris and 45 mM boric acid, pH 8.3) and electrophoresed at 20 mA for 16 h, conditions in which the molecular mass standards had reached their pore limitation for more accurate size determination of native protein complexes (Clarke and Critchley, 1992). Molecular mass standards were ferritin (440-kD monomer, 880-kD dimer), urease (272-kD trimer), and BSA (66-kD monomer, 132-kD dimer). After native PAGE, proteins were transferred to nitrocellulose using a discontinuous buffer system with the Trans-Blot Semi-Dry electrophoretic transfer cell (Bio-Rad). Different Clp protein complexes was detected using polyclonal antibodies specific for each Clp protein described above and visualized by chemiluminescence (ECL Advance Western Blotting detection kit; Amersham Pharmacia) using the ChemiGenius2 imaging system (Syngene).
Two-Dimensional PAGE
Stromal protein fractions were obtained as described above and purified further using the ReadyPrep 2D Cleanup Kit (Bio-Rad) according to the manufacturer's recommendations. Purified protein (i.e., 200 to 400 μg) was resuspended in isoelectric focusing (IEF) buffer (8 M urea, 2 M thiourea, 50 mM DTT, 4% CHAPS, 0.2% Bio-Lyte pH 5-8 ampholytes, and 0.0002% bromophenol blue) and loaded on 17-cm IEF ReadyStrips (IPG nonlinear pH 5 to 8; Bio-Rad). Strips were rehydrated for 12 h at 25°C in a Protean IEF cell (Bio-Rad), followed by IEF using the preset rapid ΔV program. After focusing, strips were immediately equilibrated for SDS-PAGE in DTT buffer (6 M urea, 2.5% SDS, 20% glycerol, 2% DTT, 0.002% bromophenol blue, and 50 mM Tris-HCl, pH 8.8) for 15 min followed by a 10-min incubation in iodoacetamide buffer (6 M urea, 2.5% SDS, 20% glycerol, 2.5% iodoacetamide, 0.002% bromophenol blue, and 50 mM Tris-HCl, pH 8.8). IEF strips were then placed on 8 to 16% acrylamide gradient SDS-PAGE gels (Tris-Gly) and run overnight at 12°C in a Protean II xi system (Bio-Rad). Gels were electrophoresed in simultaneous sets of four to reduce variation, with triplicate gels run for each treatment. Gels were stained with colloidal Coomassie Brilliant Blue G 250 (Sigma-Aldrich) and analyzed using specialized software (2D Phoretix; Nonlinear Dynamics). Spots were selected for mass spectrometric identification based on significant differences in the three replicates between sample pairs; identification was done with MALDI-TOF MS or HPLC-MS/MS (Finnigan LTQ-FT; Thermo Electron) at the SweGene Proteomics Center (Göteborg University).
Protein Degradation Assay
Lawns of wild-type and clpP6 plants at the same developmental age (3 and 4 weeks, respectively) were blended in homogenization buffer (0.33 M sorbitol, 10 mM EDTA, 5 mM MgCl2, 10 mM NaHCO3, and 20 mM HEPES/KOH, pH 8) using an Ultra-Turrax (Janke and Kunkel). Homogenate was filtered through a double layer of prewetted Miracloth (Calbiochem) and then pelleted by centrifugation at 1300g for 8 min. The crude chloroplast pellet was carefully resuspended in a small volume of homogenization buffer, loaded on a two-step 40/70% Percoll gradient in resuspension buffer (0.33 M sorbitol, 5 mM MgCl2, 10 mM NaHCO3, and 20 mM HEPES/NaOH, pH 8), and centrifuged at 1500g for 10 min. Intact chloroplasts were recovered from the interface of the Percoll cushion and diluted in 5 volumes of resuspension buffer. After centrifugation at 1000g for 5 min, intact chloroplasts were carefully resuspended in a small volume of resuspension buffer and counted by phase-contrast microscopy (Olympus BX50) using a hemocytometer. The chloroplast suspension was then diluted in additional resuspension buffer to 1.5 × 106 chloroplasts/μL, at a final concentration of 5 mM Mg-ATP, and 2.5 mM phosphocreatine and 50 mg/mL creatine phosphokinase as an ATP regeneration system (a control assay was also included without the addition of ATP and the regeneration system). For the degradation assay, an equal number of intact chloroplasts were incubated for 0 to 3 h in ∼60 μmol·m−2·s−1 light at 25°C, with samples taken every hour (counting after the 3-h incubation showed no significant difference in the number of intact chloroplasts between wild-type Arabidopsis and clpP6 antisense lines). Each chloroplast sample was ruptured in 5 volumes of rupture buffer (10 mM MgCl2 and 20 mM HEPES/NaOH, pH 7.6) and then frozen in liquid nitrogen. Thawed samples were centrifuged at 20,000g for 10 min to remove thylakoid membranes, with the stromal supernatant transferred to a new tube. Stromal protein concentration was determined using the BCA protein assay described above. Stromal protein samples were separated by one-dimensional denaturing PAGE (precast 3 to 8% polyacrylamide Tris-acetate gels; Invitrogen) for proteins of ≥60 kD and by two-dimensional PAGE for proteins of <60 kD. Gels were stained with either Coomassie Brilliant Blue G 250 or silver (Plus One silver staining kit; GE Healthcare). All proteins identified as potential substrates for the Clp protease were quantified using the ChemiGenius2 imaging system (Syngene). Proteins exhibiting >30% degradation in wild-type chloroplasts during the time course of the assay were identified by MALDI-TOF MS and HPLC-MS/MS (SweGene Proteomics Center, Göteborg University).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: β-ATPase, ATCG00480; ClpP1, ATCG00670; ClpP3, At1g66670; ClpP4, At5g45390; ClpP5, At1g02560; ClpP6, At1g11750; ClpR1, At1g49970; ClpR2, At1g12410; ClpR3, At1g09130; ClpR4, At4g17040; ClpS1, At4g25370; ClpS2, At4g12060; ClpC1, At5g50920; D1, ATCG00020; EF-Ts, At4g29060; FtsH5, At5g42270; FtsH2, At2g30950; HSP90, At2g04030; Lhcb2, At2g05100; LSU, ATCG00490; NDP kinase, At5g63310; PPIase, At3g62030; PsaL, At4g12800; RNA helicase, At5g26742; and UPRT, At3g53900.
Acknowledgments
This work was supported by grants to A.K.C. from the Swedish Research Council for Environment, Agricultural Science, and Spatial Planning and the Swedish Foundation for International Cooperation in Research and Higher Education. T.M.S. acknowledges the Natural Sciences and Engineering Research Council of Canada for an overseas postgraduate scholarship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Adrian K. Clarke (adrian.clarke@botany.gu.se).
References
- Adam, Z., Adamska, I., Nakabayashi, K., Ostersetzer, O., Haussuhl, K., Manuell, A., Zheng, B., Vallon, O., Rodermel, S.R., Shinozaki, K., and Clarke, A.K. (2001). Chloroplast and mitochondrial proteases in Arabidopsis: A proposed nomenclature. Plant Physiol. 125 1912–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam, Z., Zaltsman, A., Sinvany-Villalobo, G., and Sakamoto, W. (2005). FtsH proteases in chloroplasts and cyanobacteria. Physiol. Plant. 123 386–390. [Google Scholar]
- Aronsson, H., and Jarvis, P. (2002). A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett. 529 215–220. [DOI] [PubMed] [Google Scholar]
- Chen, M., Jensen, M., and Rodermel, S. (1999). The yellow variegated mutant of Arabidopsis is plastid autonomous and delayed in chloroplast development. J. Hered. 90 207–214. [DOI] [PubMed] [Google Scholar]
- Clarke, A.K., and Critchley, C. (1992). The identification of a heat shock protein complex in chloroplasts of barley leaves. Plant Physiol. 100 2081–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, A.K., Gustafsson, P., and Lidholm, J.Å. (1994). Identification and expression of the chloroplast clpP gene in the conifer Pinus contorta. Plant Mol. Biol. 26 851–862. [DOI] [PubMed] [Google Scholar]
- Clarke, A.K., MacDonald, T.M., and Sjögren, L.L.E. (2005). The ATP-dependent Clp protease in chloroplasts of higher plants. Physiol. Plant. 123 406–412. [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Evans, J.R. (1996). Developmental constraint on photosynthesis: Effects of light and nutrition. In Photosynthesis and the Environment, N.R. Baker, ed (Dordrecht, The Netherlands: Kluwer), pp. 281–304.
- Ganeteg, U., Strand, Å., Gustafsson, P., and Jansson, S. (2001). The properties of the chlorophyll a/b-binding proteins Lhca2 and Lhca3 studied in vivo using antisense inhibition. Plant Physiol. 127 150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud, R., Kessel, M., Beuron, F., and Stevens, A.C. (1998). Enzymatic and structural similarities between the Escherichia coli ATP-dependent proteases, ClpXP and ClpAP. J. Biol. Chem. 273 12476–12481. [DOI] [PubMed] [Google Scholar]
- Hasunuma, K., Yabe, N., Yoshida, Y., Ogura, Y., and Hamada, T. (2003). Putative functions of nucleoside diphosphate kinase in plants and fungi. J. Bioenerg. Biomembr. 35 57–65. [DOI] [PubMed] [Google Scholar]
- Kang, S.G., Maurizi, M.R., Thompson, M., Mueser, T., and Ahvazi, B. (2004). Crystallography and mutagenesis point to an essential role for the N-terminus of human mitochondrial ClpP. J. Struct. Biol. 148 338–352. [DOI] [PubMed] [Google Scholar]
- Kim, Y.I., Levchenko, I., Fraczkowska, K., Woodruff, R.V., Sauer, R.T., and Baker, T.A. (2001). Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nat. Struct. Biol. 8 230–233. [DOI] [PubMed] [Google Scholar]
- Kristensen, O., Laurberg, M., Liljas, A., and Selmer, M. (2002). Is tRNA binding or tRNA mimicry mandatory for translation factors? Curr. Protein Pept. Sci. 3 133–144. [DOI] [PubMed] [Google Scholar]
- Kuroda, H., and Maliga, P. (2003). The plastid clpP1 protease gene is essential for plant development. Nature 425 86–89. [DOI] [PubMed] [Google Scholar]
- Majeran, W., Wollman, F.A., and Vallon, O. (2000). Evidence for a role of ClpP in the degradation of the chloroplast cytochrome b(6)f complex. Plant Cell 12 137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi, K., Ito, M., Kiosue, T., Shinozaki, K., and Watanabe, A. (1999). Identification of clp genes expressed in senescing Arabidopsis leaves. Plant Cell Physiol. 40 504–514. [DOI] [PubMed] [Google Scholar]
- Ortega, J., Lee, H.S., Maurizi, M.R., and Steven, A.C. (2002). Alternating translocation of protein substrates from both ends of ClpXP protease. EMBO J. 21 4938–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier, J., Ripoll, D.R., Friso, G., Rudella, A., Cai, Y., Ytterberg, J., Giacomelli, L., Pillardy, P., and van Wijk, K.J. (2004). Clp protease complexes from photosynthetic and non-photosynthetic plastids and mitochondria of plants, their predicted three-dimensional structures, and functional implications. J. Biol. Chem. 279 4768–4781. [DOI] [PubMed] [Google Scholar]
- Peltier, J.B., Ytterberg, J., Liberles, D.A., Roepstorff, P., and van Wijk, K.J. (2001). Identification of a 350 kDa ClpP and protease complex with 10 different Clp isoforms in chloroplasts of Arabidopsis thaliana. J. Biol. Chem. 276 16318–16327. [DOI] [PubMed] [Google Scholar]
- Porra, R.J., Thompson, W.A., and Kriedemann, P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975 384–394. [Google Scholar]
- Rocak, S., and Linder, L. (2004). DEAD-box proteins: The driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 5 232–241. [DOI] [PubMed] [Google Scholar]
- Romano, P., Gray, J., Horton, P., and Luan, S. (2005). Plant immunophilins: Functional versatility beyond protein maturation. New Phytol. 166 753–769. [DOI] [PubMed] [Google Scholar]
- Roos, D.S., Crawford, M.J., Donald, R.G.K., Fraunholz, M., Harb, O.S., He, C.Y., Kissinger, J.C., Shaw, M.K., and Striepen, B. (2002). Mining the Plasmodium genome database to define organellar function: What does the apicoplast do? Philos. Trans. R. Soc. Lond. B Biol. Sci. 357 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, W., Tamura, T., Hanba-Tomita, Y., Murata, M., Sodmergen (2002). The VAR1 locus of Arabidopsis encodes a chloroplastic FtsH and is responsible for leaf variegation in the mutant alleles. Gene Cell 7 769–780. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sangster, T.A., and Queitsch, C. (2005). The HSP90 chaperone complex, an emerging force in plant development and phenotypic plasticity. Curr. Opin. Plant Biol. 8 86–92. [DOI] [PubMed] [Google Scholar]
- Shanklin, J., Dewitt, N.D., and Flanagan, J.M. (1995). The stroma of higher plant plastids contain ClpP and ClpC, functional homologs of Escherichia coli ClpP and ClpA: An archetypal two-component ATP-dependent protease. Plant Cell 7 1713–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai, T., Shimizu, K., Ueda, K., Nishimura, Y., Kuroiwa, T., and Hashimoto, T. (2001). The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant Cell Physiol. 42 264–273. [DOI] [PubMed] [Google Scholar]
- Sjögren, L.L.E., MacDonald, T.M., Sutinen, S., and Clarke, A.K. (2004). Inactivation of the clpC1 gene encoding a chloroplast Hsp100 molecular chaperone causes growth retardation, leaf chlorosis, lower photosynthetic activity, and a specific reduction in photosystem content. Plant Physiol. 136 4114–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle, J., and Vierstra, R.D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55 555–590. [DOI] [PubMed] [Google Scholar]
- Soikkeli, S. (1980). Ultrastructure of the mesophyll in Scots pine and Norway spruce: Seasonal variation and molarity of fixative buffer. Protoplasma 103 241–252. [Google Scholar]
- Strand, Å., Zrenner, R., Trevanion, S., Stitt, M., Gustafsson, P., and Gardeström, P. (2000). Decreased expression of two key enzymes in the sucrose biosynthesis pathway, cytosolic fructose-1,6-bisphosphate and sucrose phosphate synthase, has remarkably different consequences for photosynthetic carbon metabolism in transgenic Arabidopsis thaliana. Plant J. 23 759–770. [DOI] [PubMed] [Google Scholar]
- Sutinen, S. (1987). Cytology of Norway spruce needles. I. Changes during ageing. Eur. J. Forest Pathol. 17 65–73. [Google Scholar]
- Vierstra, R.D. (1993). Proteolysis in plants: Mechanisms and functions. Plant Mol. Biol. 32 275–302. [DOI] [PubMed] [Google Scholar]
- Wang, J., Hartling, J.A., and Flanagan, J.M. (1997). The structure of ClpP at 2.3 Å resolution suggests a model for ATP-dependent proteolysis. Cell 91 447–456. [DOI] [PubMed] [Google Scholar]
- Wolfe, K.H., Morden, C.W., and Palmer, J.D. (1992). Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc. Natl. Acad. Sci. USA 89 10648–10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman, A., Feder, A., and Adam, Z. (2005). Developmental and light effects on the accumulation of FtsH protease in Arabidopsis chloroplasts—Implications for thylakoid formation and photosystem II maintenance. Plant J. 42 609–617. [DOI] [PubMed] [Google Scholar]
- Zheng, B., Halperin, T., Hruskova-Heidingsfeldova, O., Adam, Z., and Clarke, A.K. (2002). Characterization of chloroplast Clp proteins in Arabidopsis: Localization, tissue specificity and stress responses. Physiol. Plant. 114 92–101. [DOI] [PubMed] [Google Scholar]
- Zheng, B., MacDonald, T.M., Sutinen, S., Hurry, V., and Clarke, A.K. (May 17, 2006). A nuclear-encoded ClpP subunit of the chloroplast ATP-dependent Clp protease is essential for early development in Arabidopsis thaliana. Planta http://dx.doi.org/10.1007/s00425-006-0292-2. [DOI] [PubMed]
- Zrenner, R., Stitt, M., Sonnewald, U., and Boldt, R. (2006). Pyrimidine and purine biosynthesis and degradation in plants. Annu. Rev. Plant Biol. 57 805–836. [DOI] [PubMed] [Google Scholar]