The circadian clock of the honey bee is implicated in ecologically relevant complex behaviors. These include time sensing, time-compensated sun-compass navigation, and social behaviors such as coordination of activity, dance language communication, and division of labor. The molecular underpinnings of the bee circadian clock are largely unknown. We show that clock gene structure and expression pattern in the honey bee are more similar to the mouse than to Drosophila. The honey bee genome does not encode an ortholog of Drosophila Timeless (Tim1), has only the mammalian type Cryptochrome (Cry-m), and has a single ortholog for each of the other canonical “clock genes.” In foragers that typically have strong circadian rhythms, brain mRNA levels of amCry, but not amTim as in Drosophila, consistently oscillate with strong amplitude and a phase similar to amPeriod (amPer) under both light-dark and constant darkness illumination regimes. In contrast to Drosophila, the honey bee amCYC protein contains a transactivation domain and its brain transcript levels oscillate at virtually an anti-phase to amPer, as it does in the mouse. Phylogenetic analyses indicate that the basal insect lineage had both the mammalian and Drosophila types of Cry and Tim. Our results suggest that during evolution, Drosophila diverged from the ancestral insect clock and specialized in using a set of clock gene orthologs that was lost by both mammals and bees, which in turn converged and specialized in the other set. These findings illustrate a previously unappreciated diversity of insect clockwork and raise critical questions concerning the evolution and functional significance of species-specific variation in molecular clockwork.
Circadian clocks enable organisms to anticipate predictable environmental changes, schedule activities for an advantageous time during the day, and coordinate internal processes among themselves and with the environment. The molecular bases for rhythm generation in organisms as diverse as cyanobacteria, plants, fruit flies, and mammals consist of interlocked autoregulatory transcriptional/translational feedback loops with positive and negative elements (Young and Kay 2001; Bell-Pedersen et al. 2005). The molecular control of circadian rhythms in animals is best known for the fruit fly Drosophila melanogaster, and is based largely on the analysis of mutations and molecular manipulations in “clock genes.” The current model for rhythm generation in central pacemaker cells of Drosophila involves interactions among six transcription factors—Period (dPER), Timeless (dTIM1), Clock (dCLK), Cycle (dCYC), Par Domain Protein 1 (dPDP1), and Vrille (dVRI); the kinases Double-Time (dDBT),Shaggy (dSGG), and Casein Kinase 2 (dCK2); the Protein Phosphatase 2a (dPP2a); and the protein degradation protein Super-numerary Limbs (dSLMB) (Hardin 2004, 2005; Bell-Pedersen et al. 2005). The protein products of dClk and dCyc (dCLK and dCYC, respectively) interact and form a complex that binds E-box elements (CACGTG) in regulatory sequences of the dPer and dTim1 promoter regions to activate their transcription. The mRNA transcripts of these genes accumulate in the cytoplasm of pacemaker cells, where they are translated into proteins. The protein products of dPer (dPER) and dTim1 (dTIM1) accumulate during the night, eventually entering the nucleus and binding to the dCLK/dCYC complex. The binding of dPER (and perhaps dTIM1) to the dCLK/dCYC complex interferes with dCLK/dCYC binding to the E-box and results in a cessation of transcriptional activity (Lee et al. 1999). This creates a negative feedback loop with dPER and dTIM1 inhibiting their own transcription. Degradation of dTIM1 in the late night renders dPER unstable and leads to its degradation later in the morning. These events release the inhibition from dCLK/dCYC activity and enable a new round of dPer, dTim1, dVri, and dPdp1 transcription. The negative feedback loop is fine tuned by the action of dCRY, the kinases, and the phosphatases (for recent reviews, see Hardin 2004, 2005). dCRY allows the period and phase of the clock to adjust to changing photo-periods. On exposure to light, dCRY is thought to associate with dTIM1 to promote its rapid degradation via a proteasome-dependent pathway (Ceriani et al. 1999; Rosato et al. 2001; Busza et al. 2004; Dissel et al. 2004). The dCLK/dCYC complex is involved in a second autoregulatory loop in the fly's pacemaker, controlling the cycling levels of dCLK. dClk mRNA is produced with a circadian rhythm but cycles in antiphase to dPer, and dTim1. The dCLK/dCYC complex binds to E-boxes on the pro-motors of dVri and dPdp1 and activates their transcription. After translation, dVRI and dPDP1 proteins feed back to negatively or positively (respectively) regulate dClk expression (Cyran et al. 2003; Glossop et al. 2003).
Comparison of the mechanism for rhythm generation in vertebrates and flies shows that there is a high degree of conservation not only in the general design and function of the clockwork but also in that similar principal clock genes are involved in these two models for a circadian molecular circuit. Despite this conservation, some of these genes appear to take on a different function in the clocks of Drosophila and mouse (see Dunlap 1999; Edery 2000; Rosato and Kyriacou 2001; Young and Kay 2001; Panda et al. 2002; Stanewsky 2003; Bell-Pedersen et al. 2005). Mammals have two paralogs for CRY and three for PER. In contrast to Drosophila, mCRYs are indispensable components of the mouse central pacemaker (van der Horst et al. 1999; Vitaterna et al. 1999). mCRYs interact with mPERs and are essential for their translocation to the nucleus and the inhibition of mCLK/mBMAL transcriptional activity (BMAL is the mammalian ortholog of Drosophila’s CYC). An additional key difference from Drosophila is that the activity of vertebrate CRY does not appear to depend on light (Griffin Jr. et al. 1999; Kume et al. 1999; Froy et al. 2002). Thus, mammalian CRY functions in the negative limb of the clock, with a similar function to that of dTIM1 in Drosophila pacemaker cells. It is important to note, however, that although mammalian and Drosophila CRY proteins certainly differ in both their structure and biochemical activity (Froy et al. 2002; Busza et al. 2004; Green 2004; Partch et al. 2005; Zhu et al. 2005; Chaves et al. 2006), dCRY is implicated in the regulation of clock gene expression and rhythm generation in peripheral clocks (Ivanchenko et al. 2001; Krishnan et al. 2001; Levine et al. 2002; Collins et al. 2006), and mammalian CRYs are thought to be involved in circadian light response (Cashmore 2003; Sancar 2003). Mammals do not have a true ortholog of Drosophila TIM1 but rather have an ortholog of Drosophila Timeout (dTIM2), a gene with no known function in the fly's clock. Mammalian TIM is expressed in the suprachiasmatic nucleus (SCN), the site of the mammalian pacemaker, but there is an ongoing debate concerning its role, if any, in central rhythm generation (Zylka et al. 1998; Takumi et al. 1999; Field et al. 2000; Gotter at al. 2000; Barnes et al. 2003). mTIM is apparently not involved in photic input to the mammalian clock because its levels are not affected even by a strong resetting light pulse; light resetting instead appears to be mediated by rapid modulation of mPER1 and mPER2 levels (Field et al. 2000). In both mammals and Drosophila, CLK and BMAL/CYC function as positive elements in the interlocked feedback loops. In Drosophila dCLK contains a transactivation domain, and the amounts of its products oscillate in virtual antiphase to those of Per. In contrast in mammals, a transactivation domain is found on BMAL1 that also oscillates in antiphase to Per. The amounts of the products of dCyc in Drosophila and Clk in mammals essentially do not vary during the day.
With this evidence for conservation between flies and mice, it is tempting to assume that the clocks of other insects are similar to the fly model. However, several lines of evidence from studies with insects, including the honey bee, are not easily reconciled with the Drosophila model (see Sauman and Reppert 1996; Wise et al. 2002; Bloch et al. 2003; Sehadova et al. 2003; Zavodska et al. 2003). These inconsistencies may imply that not all insect clockworks are similar to Drosophila. But a detailed description of the molecular biology of the clock is only available for a few insects other than Drosophila (e.g., Sauman and Reppert 1996; Chang et al. 2003; Zhu et al. 2005). Thus, the degree of species-specific variation in the clockwork, as well as its functional significance, is mostly unknown.
The honey bee is an attractive model organism for molecular analysis of circadian rhythms because its clock system is implicated in a set of complex behaviors such as time memory, sun compass navigation, and dance language communication (for review, see von Frisch 1967; Moore-Ede et al. 1983). Another level of complexity emerges from studies showing remarkable plasticity in the circadian rhythms of bees that is at least in part modulated by social factors and therefore may contribute to the temporal organization of their societies. Bees socially synchronize their clocks (Moritz and Kryger 1994), have an endogenous ontogeny of circadian rhythms (Moore 2001), and switch between activity with or without circadian rhythms according to their task (Moore et al. 1998; Bloch and Robinson 2001).
We searched the honey bee genome, cloned, and characterized putative Apis homologs of Per, Tim2, Cry, Cyc, Clk, Vri, and Pdp1. We describe the brain expression profile during the day for the first five genes. Both our bioinformatic and expression analyses point to significant inconsistencies with the Drosophila model and surprising similarities to the mammalian model. We also provide phylogenetic analyses with new data from additional insect genomes that suggest that mammalian and Drosophila clocks evolved from ancestors that probably had both Tim1 and Tim2 and both the Drosophila and mammalian type Cry. These results pose profound challenges to our understanding of the evolution of specific clock genes and the circadian clock as a whole.
Results
Identification and cloning of putative clock genes in the honey bee
We identified homologs for most of the known Drosophila clock genes in the honey bee genome sequence (Supplemental Table S1), one of which, amPer, was already known (Toma et al. 2000), and the genome sequence reveals no additional Per homologs. We PCR cloned and obtained the 5′ and 3′ ends on cDNA samples obtained from brain transcripts of these genes (see Methods). We deduced the amino acid residues from the cloned cDNA and genomic sequences and used these for further analyses of protein structure and phylogenetic relationships. The honey bee genome encodes a single homolog of each of the canonical animal clock genes. There are orthologs for Clk, Cyc, Pdp1, Vri and Per from Drosophila and other insects. In contrast, the honey bee genome does not encode true orthologs of Drosophila Cry and Tim1 genes. Rather, there are only orthologs of Drosophila Timeout (Tim2) and mammalian-type Cry (Cry-m, see below).
Phylogenetic relationships
Clock and Cycle/Bmal
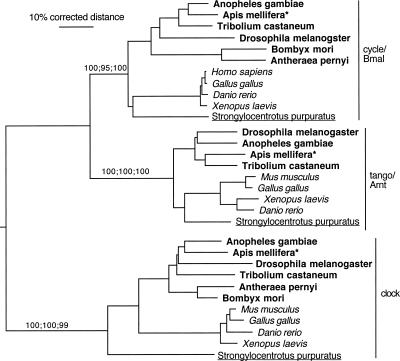
Figure 1 shows the phylogenetic relationships between the clock proteins CLK and CYC/BMAL, and the related protein lineage Tango/Arnt. Note that the species relationships in the phylogenetic tree do not always exactly fit the known taxonomic relationships between the species, as expected for trees using single relatively short proteins. Nevertheless, the agreement with common taxonomy is typically good. For example, the sea urchin (Strongylocentrotus purpuratus) proteins usually cluster near the base of the vertebrates. There is good support for the orthology of each of these three proteins (Tango/Arnt, Clock, and Cycle/Bmal) in vertebrates and insects. Importantly, the honey bee proteins cluster unambiguously with orthologous proteins from other insects.
Figure 1.
Phylogenetic relationships of the Cycle/Bmal, Tango/Arnt, and Clock protein family. We used the Clock protein as an outgroup to root the tree based on its divergence from the Cycle/Bmal and Tango/Arnt sister proteins. Support levels are shown only for the main protein lineages, which are separated slightly vertically for visual clarity (percentage of trees showing a branch in distance and parsimony bootstrapping, followed by percentage of maximum likelihood quartet puzzling steps). Distinct font styles are used to highlight major taxonomic lineages. Bold for insects, italics for vertebrates, underline for the sea urchin, and plain for all the others. The honey bee Apis mellifera is highlighted with an asterisk.
Vrille and Pdp1
The honey bee orthologs amVRI and amPDP1 clearly cluster with those of other insects and are distinct from related proteins of vertebrates (Supplemental Fig. 1).
Timeless/Timeout
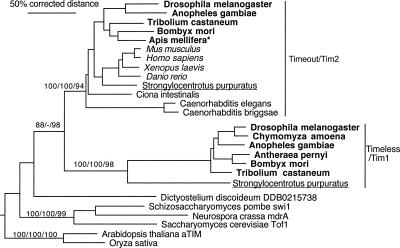
Phylogenetic analysis for the Timeless-Timeout family (Fig. 2) suggests that these two paralogous protein lineages evolved from gene duplication in early animals; a single convincing ortholog is available from plants, fungi, and the social amoeba Dictyostelium (Eichinger et al. 2005). The canonical Timeless/TIM1 protein of D. melanogaster that is missing from the honey bee is present in all other available insect genomes. Because Daphnia, the only sequenced crustacean genome (H.M. Robertson, unpubl.), the basal flour beetle Tribolium castaneum, and the basal dipteran mosquito Anopheles gambiae have TIM1, it must have been lost from the honey bee. The antiquity of TIM1 is demonstrated by the presence of an ortholog in the sea urchin, S. purpuratus (underlined in Fig. 2). This relatively rapidly evolving TIM1 lineage was apparently also lost from most deuterostomes, including chordates, and from the Caenorhabditis nematodes. In contrast, the Timeout (TIM2) protein, often confusingly called “Timeless” in the vertebrate literature (this issue of orthology or paralogy of insect and vertebrate Timeless/Timeout proteins is also discussed in Benna et al. 2000; Gotter et al. 2000), is present throughout the available deuterostomes, including in the unpublished sea urchin genome and in all available insect genomes. Thus, by losing TIM1 the honey bee has “converged” with vertebrates that have only the Timeout/TIM2 ortholog.
Figure 2.
Phylogenetic relationships of the Timeless and Timeout proteins. We used the plant TIMELESS protein as an outgroup to root the tree. For additional details, see legend to Figure 1.
Cryptochromes
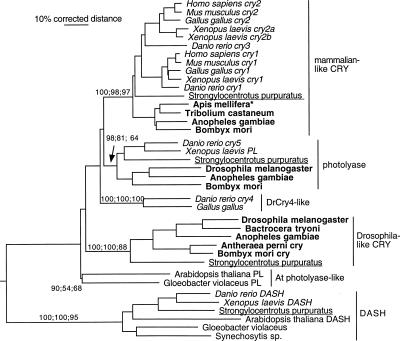
Our analysis (Fig. 3) suggests that there are at least three, and most likely four, closely related lineages of Photolyases/Cryptochromes in animals, presumably resulting from a series of gene duplications in early animals given the presence of a photolyase lineage in plants and bacteria. Rooting the analysis with the recently described DASH Photolyase/Cryptochrome protein (Daiyasu et al. 2004) that is present in some bacteria, plants, sea urchin, and basal vertebrates, but not insects or mammals, shows these relationships particularly well (Fig. 3).
Figure 3.
Phylogenetic relationships of animal Cryptochrome and related Photolyase proteins. We used the CRY-DASH protein family as an outgroup to root the tree (based on its position in a larger tree that was rooted with another distantly related photolyase protein lineage that is present in insects and vertebrates) (data not shown). We termed the mammalian-like CRY proteins “CRY-m” and the Drosophila-like proteins “CRY-d.” For additional details, see legend to Figure 1.
Animal CRY proteins are clearly divided into two distinct clusters. One cluster contains the Drosophila-type CRY (that we termed “CRY-d”; see Discussion), and the other includes all the vertebrate CRYs (that we termed “CRY-m”; see Discussion). CRY-d is present in other flies and moths (Sauman et al. 2005; Zhu et al. 2005) but is absent from the honey bee and appears to be missing from Tribolium (based on the current stage of its genome sequencing). The presence of this type of CRY in most insects and in the sea urchin genome confirms that this is an ancient animal protein lineage that was lost independently from honey bees and (perhaps) Tribolium beetles. The CRY-m protein (mammalian-type), in addition to being duplicated at least once again in vertebrates, is also an old lineage, being present not only in the sea urchin but also in insects such as the bee, Bombyx, Anopheles, and Tribolium, except D. melanogaster. Examination of the D. pseudobscura genome (Richards et al. 2005) and the 10 newly available Drosophila genomes representing the entire genus (see Robertson 2005; http://flybase.bio.indiana.edu/blast/) shows that this loss occurred before the origin of the genus, but its presence in the mosquito A. gambiae indicates it occurred sometime after the split of the suborders Nematocera and Brachycera ∼250 Myr ago (Gaunt and Miles 2002).
The other two lineages of this Photolyase/Cryptochrome set are a Photolyase in basal vertebrates, the sea urchin, and some insects that has been independently lost from mammals (Kato et al. 1994), bee, and Tribolium and a lineage represented by the zebrafish Danio rerio CRY4 protein (Daiyasu et al. 2004). This lineage is also present in the chicken, but not the sea urchin, insects, or mammals, and presumably was again lost independently in these lineages. It becomes evident from this phylogenetic analysis that this lineage of Photolyase/Cryptochrome proteins is replete with both ancient and recent duplications and ancient and recent independent gene losses. By losing CRY-d the honey bee has converged with vertebrates that only have the CRY-m proteins.
Putative functional domains and motifs on honey bee clock genes
amClock and amCycle
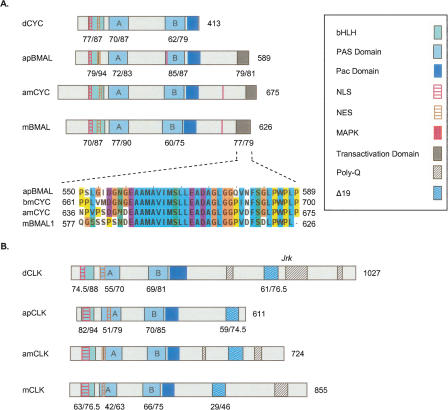
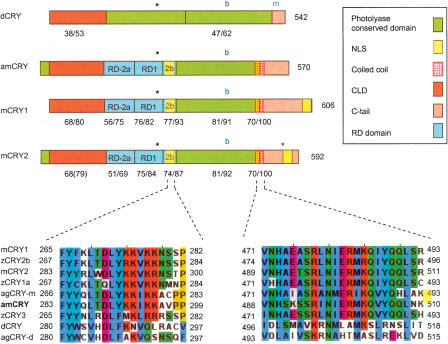
CLK and CYC/BMAL are PAS-bHLH transcription factors. We termed the honey bee ortholog of CYC/BMAL “amCYCLE” based on its definite clustering with dCYC in the phylogenetic analysis (Fig. 1). amCYC protein contains highly conserved PAS-A, PAS-B, Pac, and bHLH; domains; and sequences identical (100%) to the putative N-terminal NLS, and NES signals of other CYC/BMAL proteins (Fig. 4A; Chang et al. 2003; Hirayama and Sassone-Corsi 2005). The amCYC protein is significantly longer than dCYC (675 compared with 413 amino acids) and comparable in size to BMAL proteins of other animals. Importantly, the C-terminal end of amCYC (termed “BCTR” by Chang et al. 2003) is highly conserved (85% identity, 87% similarity) with those of the mouse and A. pernyi, in which this domain has potent transcriptional activity in vitro (Takahata et al. 2000; Chang et al. 2003). Thus, amCYC is phylogenetically related to dCYC but shares important structural similarities with BMAL proteins.
Figure 4.
Schematic presentation of putative functional domains and motifs on Cycle/Bmal and Clock proteins from the honey bee Apis mellifera, the fruit fly Drosophila melanogaster, the giant silk moth Anthereae pernyi, and the mouse Mus musculus. See legend for domain and motif identity and text for additional details on each domain. Numbers below domains indicate identity/similarity with corresponding sequences on the honey bee ortholog. The numbers at the end of each diagram indicate protein size (number of amino acid residues). (A) Cycle/Bmal proteins. Inset shows a CLUSTALW multiple sequence alignment of a putative domain that is thought to be necessary for apBMAL and mBMAL transcriptional activity (see text for details). bmCYC is the CYC ortholog from the moth Bombyx mori. Alignments were generated with CLUSTALW and colored with JalView according to the default CLUSTALX convention. (B) Clock proteins.
CLK proteins are characterized by a highly conserved N-terminal with PAS-A, PAS-B, PAC, bHLH, NLS, and NES domains and a more variable and typically glutamine-rich (Q-rich) C-terminal. All the N-terminal domains are highly conserved in amCLK and are organized in the same order as in CLK proteins of other insects and mammals (Fig. 4B; Chang et al. 2003; Hirayama and Sassone-Corsi 2005). amCLK is significantly shorter than dCLK (724 compared to 1027 amino acids) and does not contain the part on dCLK downstream of the Jrk mutation. This part of the protein contains two poly-glutamine (poly-Q) repeats and is thought to be essential for dCLK transcriptional activity (Allada et al. 1998; Darlington et al. 1998). Nevertheless, amCLK contains two poly-Q sequences (following the method of Chang et al. 2003, who defined poly-Q as a sequence in which ≥ 60% amino acid residues are glutamine) on amino acids 477–486 and 644–688. The C-terminal sequence corresponding to exon 19 on mCLK, which is implicated in transcriptional activity of mCLK but is dispensable in apCLK (King et al. 1997; Takahata et al. 2000; Chang et al. 2003), is also conserved in the honey bee as well as in other insect CLK proteins (Fig. 4B). The C-terminal poly-Q repeats of amCLK are comparable to those of the mouse, in which in vitro experiments with various cell lines suggest that it is not the main transactivation domain but rather plays a regulatory and/or structural role helping BMAL display its transcriptional activity (Takahata et al. 2000). Taken together, the sequence analyses of amCYC and amCLK suggest that the transactivation domain is located on the honey bee's ortholog of BMAL (amCYC), as in mammals, and not on CLK as in Drosophila.
amVrille and amPAR Domain Protein 1
VRI and PDP1 are basic zipper transcription factors that are characterized by a basic region leucine zipper and a highly conserved DNA binding domain. The DNA binding domain of dVRI is essential for binding to sequence elements within dClk promoter region in vitro and probably mediates dVRI repression of dClk expression in vivo (Glossop et al. 2003). Importantly, the DNA binding domain of amVRI is 100% identical to that of dVRI and 86% identical to that of mE4BP4, the mammalian homolog of VRI (Supplemental Fig. 2A) (Cowell et al. 1992; Glossop et al. 2003). Although amVRI is relatively G/S rich (44 of the last 194 amino acid residues), it does not contain a true G/S-rich domain comparable to that of dVRI (Cyran et al. 2003).
PDP1 and related proteins are characterized by a PAR (Prolin and Acidic amino acid Rich) and TDA (a putative transactivation domain) domains that are conserved in amPDP1 (Supplemental Fig. 2B; Lin et al. 1997). The N-terminal of amPDP1 is Q-rich and contains two poly-Q stretches, but not an alanine-rich region (Cyran et al. 2003). The DNA binding domain of amPDP1 is 94%–100% identical to those of other PDP1 proteins (Supple-mental Fig. 2B; Lin et al. 1997). The DNA binding domains of amVRI and amPDP1 are very similar (11/14 identity, 13/14 similarity) (Supplemental Fig. 2C), suggesting that they can bind similar DNA sequences (Cyran et al. 2003). In sum, functional domains on amVRI and amPDP1 are highly conserved with those on orthologs of Drosophila and other insects.
amTimeout
amTIM2 does not contain sequences similar to the two PER binding sites of dTIM1; it also does not contain the NLS or the C terminus cytoplasmic leading domain (CLD) that are implicated in dTIM1 subcellular localization (Supplemental Fig. 3; Gekakis et al. 1995; Saez and Young 1996). By use of the threading server 3D-PPSM (Kelley et al. 2000), we found two helical Armadillo (Arm)/HEAT repeats similar to those found on dTIM1 and dTIM2 (Vodovar et al. 2002; Perry 2005). The first is located on the N-terminal between amino acids 7–506 (90% certainty, E-value = 0.095; note that for 3D-PPSM E < 0.5 is considered significant), and the second between amino acids 570–954 (70% certainty, E-value = 0.32). Residues 32–499, which correspond well with the first domain, were also recognized as an Arm repeat (score = 0.8; E-value = 9 × E10−4) by the threading server GenTHREADER (Jones 1999; McGuffin and Jones 2003). In a 3D-PSSM analysis of the mouse mTIM2, we discovered two ARM repeats on residues 9–471 (90% certainty, e = 0.07) and 493–942 (70% of certainty; e = 0.31). The threading server GenTHREADER predicts one continuous Armadillo/HEAT on residues 19–849 of this protein (score = 0.81; E-value = 6 × E10−4). A search with PredictNLS algorithm identified an RKKQKSR sequence on residues 1300–1307 of the amTIM2 and a similar motif PKKVQKR on residues 998–1004 on dTIM2 as putative NLS sequences. We also found a second putative NLS on dTIM2 amino acids 1357–1373 (RKKPAKVDGEPAKRRRL) and three predicted NLSs on mTIM2 (residues 316–323, 526–537, 940–945) (Supplemental Fig. 3). In sum, our analyses suggest that amTIM2 does not function as does dTIM1 in the negative limb of the Drosophila brain clock because it does not contain domains and motifs that are thought to be important for dTIM1 biochemical activity.
amCryptochrome
CRY proteins have two main domains, an N-terminal conserved domain, and a carboxy-terminal “tail” that is intrinsically unstructured and varies considerably in length and primary amino acid sequence (Sancar 2003; Green 2004; Partch et al. 2005). Both domains are important for mammalian CRY ability to inhibit CLK/BMAL driven transcription (Chaves et al. 2006). It appears that the N-terminal photolyase conserved domain of dCRY is necessary for its transcriptional repressing activity in Drosophila peripheral clocks (Busza et al. 2004; Collins et al. 2006), but specific dCRY domains involved in this function have not yet been defined. Hirayama et al. (2003) generated chimeras between the transcription repressing zCRY1a and the non-repressing zCRY3 proteins of the Zebrafish and identified three regions (RD1, RD2a, and RD2b), and a putative nuclearlocalization signal (NLS) within the RD-2b region that are necessary for mammalian-type CRY nuclear localization and subsequent repression of CLK/BMAL1 transcription. We found that the overallsimilarity between amCRY and mammalian-type CRYs includes these three functional domains (51%–77% identity,69%–93% similarity in primary aminoacid sequence) (Fig. 5) that are not conserved in dCRY. Such high amino acid identity suggests that these domains have a similar three-dimensional structure (Xu et al. 2000). The correspondingamCRY sequence is identical in twelve out of the first fourteen amino acids to the putative RD2b NLS motif. This motifis highly conserved among CRY-m proteins of insects and vertebrates but notin CRY-d proteins such as dCRY, agCRY-d, and the non-repressing Zebra fish zCRY3 (Fig. 5, left lower alignment; Hirayama et al. 2003; Chaves et al. 2006). Much of the variation in light dependent CRY function between Drosophila, mammals, and plants is attributed to the C-terminal (Green 2004; Partch et al. 2005). The C-tail of amCRY shares no similarity with the C-terminal domain of dCRY. The initial part of amCRY's C-tail contains a coiled-coil region thought to be involved in PER and BMAL binding (Chaves et al. 2006). This coiled-coil domain is conserved in CRY-m proteins but not in CRY-d proteins (Fig. 5, right lower alignment). The C-tail of amCRY does not contain a second putative NLS found on the C-tails of mCRY2 and xCRY2b (Zhu et al. 2003; Sakakida et al. 2005; Chaves et al. 2006). These analyses show that amCRY does not contain sequences thought to be necessary for dCRY photoreceptor function. On the other hand, domains implicated in the transcription repressive function of mammalian-type CRY proteins are highly conserved.
Figure 5.
Schematic presentation of putative functional domains and motifs in Cryptochrome proteins from mouse (mCRY1 and mCRY2), honey bee (amCRY), and fruit fly (dCRY). See legend for domain/motif identity and text for additional details on each domain. Phosphorylation sites for MAPK (Sanada et al. 2004) are marked with asterisks. The blue “b” and “m” mark the location of cryb (D401N mutation) and crym (truncation of the last 19 residues in Drosophila) mutations in dCRY. Low panels show multiple sequence alignments of a putative nuclear localization signal (NLS) in the RD2b domain (left) and the coiled-coil region (right). Aligned are mammalian-type (mouse, mCRY1 and mCRY2; Zebrafise, zCRY1–3; mosquito, agCRY-m; honey bee, amCRY) and Drosophila-type (Drosophila, dCRY; mosquito, agCRY-d) CRY proteins. zCRY3 is a mammalian-type CRY protein but has no transcription repressing function in vitro according to the method of Hirayama et al. (2003).
Expression of putative clock genes in the honey bee brain
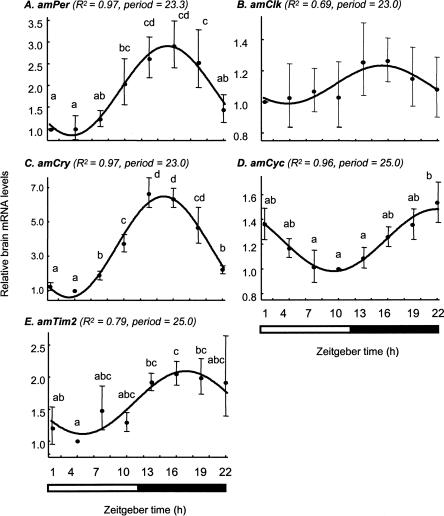
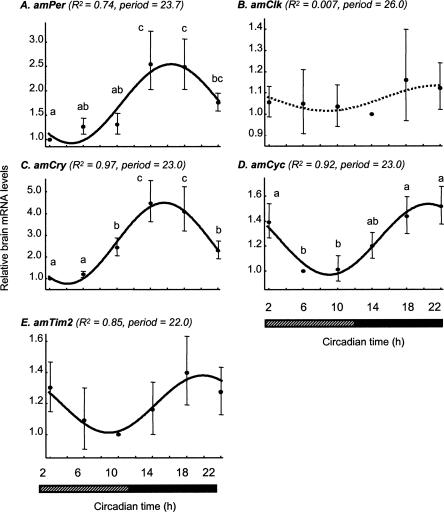
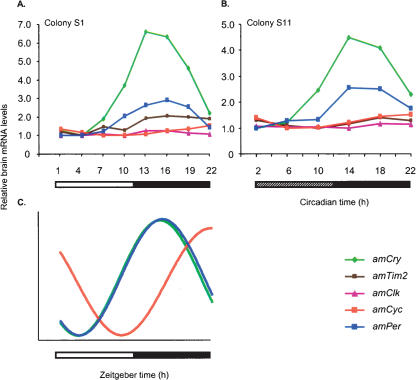
The temporal expression pattern of putative honey bee clock genes was similar in light:dark (LD) and constant darkness (DD) illumination regimes (with the exception of amTim, see below). Brain amPer mRNA levels vary over time with a peak at night that was 2.4- to 3.2-fold (three independent experiments, each with bees from a different source colony) higher than the daily trough, and an excellent fit with a cosinus model with a period of 23–25 h (Fig. 6A). We found similar oscillations in three experiments in which we collected bees in DD, although the peak/trough ratio was somewhat smaller and ANOVA analyses were not statistically significant (1.8–2.5) (Figs. 7A, 8). These findings are consistent with previous analyses of brain amPer mRNA levels in foragers (Toma et al. 2000; Bloch et al. 2001, 2004). There were also consistent and robust oscillations in amCry mRNA levels under both LD and DD illumination regimes (Figs. 6C, 7C). Peak/trough ratio was 3.6–6.6 (ANOVA, P < 0.001 in all six experiments), higher than that of amPer. Most of the variation (>85%) in amCry transcript abundance over time is explained by a cosinus model with a cycle of ∼24 h (adjusted R2 > 0.85, besides colony S8 for which R2 = 0.58) and has a similar phase as amPer in both LD and DD illumination regimes. amPER and amCRY transcript levels were low during the day (or subjective day for bees collected in DD), started to increase in the evening, and reached their maxima at night (Figs. 6–8). Brain amCyc mRNA levels also appear to cycle, but with relatively small amplitude (peak/trough ratio = 1.4–1.9; Figures 6D, 7D, 8). We nevertheless believe that these weak oscillations represent true circadian influence on amCyc expression. Our claim is based on the following observations: (1) the variation over time in amCyc mRNA levels was statistically significant in three out of six experiments (ANOVA, P < 0.01); (2) there was a 12 h difference between peak and trough levels, and the general pattern of transcript variation over time fits well with a cosinus model (adjusted R2 = 0.36–0.96, with the exception of colony S4); and (3) the pattern of amCyc variation over time was consistent in all six experiments (in both LD and DD illumination regimes). The oscillations in amCyc mRNA levels were in an almost anti-phase to those of amPer and amCry; peak levels occur at late night or early morning; levels gradually decreased during the day and reached a trough at late day or early night (Figs. 6–8). In contrast to amCyc, brain amClk mRNA levels did not vary with time (ANOVA, P > 0.25; peak/trough ratio = 1.16–1.59; in all six experiments). The time of peak and trough were typically not 12 h apart, and the overall variation in transcript abundance over time did not fit a cosinus model (Figs. 6B, 7B, 8; adjusted R2 < 0.01, with the exception of colony S1 for which adjusted R2 = 0.69; thus, Figure 6B that shows data from colony S1 is somewhat nonrepresentative), independent of the illumination regime. The temporal pattern of brain amTim expression was the most variable, partly due to differences between DD and LD illumination regimes. In two of three experiments in LD illumination regime (with bees from colonies S1 and S8) (see Fig. 6E), there was a significant variation in amTim RNA levels over time (ANOVA, P < 0.05; peak/trough ratio = 2.05, 1.86, respectively) and a good fit with a cosinus model (adjusted R2 = 0.79, 0.81, respectively), but not in the third colony (bees from colony S4; ANOVA, P > 0.3; peak/trough ratio = 1.6; no significant fit with a cosinus model). In contrast, there was no significant time effect on brain amTim RNA levels in three experiments in DD illumination regimes (ANOVA, P > 0.51). However, despite the small peak/trough ratio (1.4–1.8, in experiments with colonies S11, S12, and H1), there was a good fit with a cosinus model in the experiments with colonies S11 and S12 (adjusted R2 = 0.85, 0.74, respectively) and a weak fit in the last experiment (colony H1, adjusted R2 = 0.28). The phase of the apparent oscillations in brain amTim mRNA levels was similar to amCry and amPer in three experiments in which bees were collected in LD (bees from colonies S1, S4, and S8), but was similar to amCyc in two other experiments in which bees were collected in DD (bees from colonies S11 and H1). In one experiment in DD, the apparent oscillation in amTim mRNA abundance was in a phase intermediate between those of amPer and amCyc (bees from colony S12). In sum, our expression analyses revealed consistent, robust, and similar oscillations for the transcripts of amCry and amPer; consistent but weak oscillations in amCyc; and no oscillations in amClk. The pattern of amTim mRNA abundance over time was difficult to interpret because it was less consistent and appeared different in LD and DD illumination regimes.
Figure 6.
Brain transcript abundance over time in foragers entrained and collected in LD illumination regime. The plots show the correlation between average (±SE) relative mRNA levels for each time point (filled circles and bars) and a cosinus model with a cycle of 23–25 h (continuous line) for bees from colony S1. (A) amPeriod; (B) amClock; (C) amCrypto-chrome; (D) AmCycle; (E) AmTimeout. In parentheses are the adjusted R2 and period of the cosinus model for each gene. Time points with different letters are significantly different (ANOVA, P < 0.05; LSD post hoc test, P < 0.05). In two additional experiments, each with bees from a different, independent colony, we obtained similar results (see text for details). The bars at the bottom of plots indicate the illumination regime during sample collection. Black bar indicates dark; open bar, light. Sample size = 6.
Figure 7.
Brain transcript abundance over time in foragers entrained in LD and collected in DD illumination regime. Details of plots as in Figure 6, but show correlations with a cosinus model with a cycle of 22–26 h for bees from colony S11. (A) amPeriod; (B) amClock; (C) amCryptochrome; (D) AmCycle; (E) AmTimeout. Dashed regression line indicates that a co-sinus model accounts for <1% of the variation over time in amClk mRNA levels. We obtained similar results in two additional experiments, each with bees from a different, independent colony. The bars at the bottom of plots indicate illumination regime during sample collection. Black bar indicates dark; striped bar, subjective day. Sample size = 6.
Figure 8.
The relationships between brain transcript abundance over time for five putative honey bee clock genes in foragers from free-flying colonies. Brain mRNA levels for all genes were measured from the same RNA sample. (A) Foragers entrained and collected in light: dark illumination regime (LD). Colony S1, n = 6 bees/time point (same data as in Fig. 6). (B) Foragers entrained in LD and collected in constant darkness. Colony S11, n = 6 bees/time point (same data as in Fig. 7). (C) Schematic representation of the oscillations of clock genes in the honey bee brain in LD and DD illumination regimes. The phase of mRNA cycling is shown for amPer,amCry, and amCyc for which there is a strong correlation with a cosinus model with about a 24-h cycle. The phase of amCyc transcript is almost in antiphase to that of amPer and amCry. For clarity, the model does not include amClk that does not oscillate and amTim for which the pattern of mRNA variation over time was not consistent across experiments. Amplitudes for the various genes are not to scale. For additional details, see Figures 6 and 7.
Discussion
Our findings on the honey bee clock are not consistent with the Drosophila brain pacemaker, the current model for insect clocks. We found however, remarkable similarities to the mouse clock, the mammalian model. First, honey bees have only the mammalian-type orthologs of CRY and TIM, two proteins that are thought to function differently in central pacemakers of mammals and Drosophila. Second, the amino acid sequence of amCRY and amTIM lack domains and motifs thought to be essential for dTIM1 and dCRY function in the Drosophila clock. This suggests that amTIM2 and amCRY cannot function in exactly the same way as dTIM1 and dCRY in the brain pacemaker of Drosophila. amCRY is similar in structure to CRY-m (mammalian-type) proteins of insects and vertebrates and includes domains and motifs implicated in the transcription repressive function of these proteins. Third, the transactivation domain of the CLK/CYC (BMAL) complex appears to be on amCYC, as in mammals, and not on amCLK, as in Drosophila. Fourth, the temporal expression patterns of amCry, amTim2, amClk, and amCyc are not consistent with the Drosophila model but are similar to the patterns in the mouse. This suggests that products of these genes behave more like the mouse equivalents than Drosophila. Our phylogenetic analyses shed new light on the evolution of animal clocks and suggest that the higher similarity of honey bees to mammals than to Drosophila is due to both divergence of Drosophila from the typical insect clock and molecular convergence in the clocks of bees and mammals.
By using phylogenetic analyses with data from newly available genomes of insects and basal animal taxa, we address the question of orthology or paralogy of the mammalian-and Drosophila-type CRY and TIM proteins. Regarding the Timeless/Timeout protein family, we confirm and extend earlier suggestions that Timeless and Timeout are paralogous and not orthologous proteins (Benna et al. 2000; Gotter et al. 2000, Chang et al. 2003). Our finding of a TIM1 orthologous in the sea urchin confirms the antiquity of this lineage. Our analysis of Cryptochromes (Fig. 3) indicates that animals have two types of CRY proteins, a mammalian-type and a Drosophila-type. Importantly, we show that many insects have a mammalian-type CRY, consistent with the recent report by Zhu et al. (2005). Our analysis clearly shows that the mammalian-type and Drosophila-type CRYs are paralogous proteins with an ancient origin. The mammalian-type CRY is found in the basal insect T. castaneum genome sequence and in the basal deuterostome sea urchin sequence. This evidence for the ancient origin of mammalian-type CRY may imply that circadian photoreception was not necessarily primeval relative to core clock function in animal clocks as is commonly assumed (see Cashmore et al. 1999; Ivanchenko et al. 2001; Cashmore 2003; Gehring and Rosbash 2003; Sancar 2003; Busza et al. 2004, Tauber et al. 2004). It is also possible that a dual circadian function—photoreception and core clock activity—is the ancient role of CRY in animal clocks (Cashmore 2003; Sancar 2003).
The finding that insects have both the Drosophila-type and the mammalian-type cryptochromes calls for new terminology that will clearly distinguish between these two paralog proteins. Zhu et al. (2005) termed the Drosophila-type cryptochromes of insects “CRY1” and the mammalian-type proteins “CRY2.” But this terminology is similar to the numerical labeling of the vertebrate paralogs that are all in the mammalian-like CRY branch (Fig. 3). We therefore propose to term the Drosophila-type crypotochromes of insects “CRY-d” and the mammalian-type crypotochromes of insects“CRY-m.”
The evidence that the honey bee has only orthologs to CRY-m and Tim2 raises the issue of whether the products of amCry and amTim2 also behave similarly to those of mammals. In Drosophila, dCRY has both photoreceptor and core clock functions that appear to vary between tissues (Ivanchenko et al. 2001; Krishnan et al. 2001; Levine et al. 2002; Collins et al. 2006). In the brain's central pacemaker, dCRY is the primary circa-dian photoreceptor and is not necessary for rhythm generation. Its light-dependent function requires both the conserved photolyase homology domain (N-terminal) and the highly variable C-terminal (C-tail) (Rosato et al. 2001; Froy et al. 2002; Busza et al. 2004; Dissel et al. 2004). The current model states that light activated changes in the C-tail conformation expose the core domain and allow it to interact with TIM (and perhaps PER) (Busza et al. 2004). Our sequence analysis indicates that the C-tail is the most divergent domain between amCRY and dCRY. This finding, together with the overall low similarity between amCRY and dCRY, provides no support for the hypothesis that the two proteins function in the same way. In contrast, in mammals and other vertebrates, CRY forms a complex with PER, enters the nucleus, and inhibits CLK/BMAL1 transcriptional activity. We found that the overall high similarity between amCRY and vertebrate CRYs includes significant conservations in domains and motifs implicated in mammalian-type CRY nuclear localization and interaction with PER, CLK, and BMAL1 (Kobayashi et al. 2000; Hirayama et al. 2003; Chaves et al. 2006). The hypothesis that CRY-m proteins of insects behave similar to mammalian CRY proteins is further supported by recent in vitro analyses in Drosophila Schneider 2 cells (S2). Zhu et al. (2005) showed that CRY-m proteins from the mosquito A. gambiae and the monarch butterfly Danaus plexippus, as well as mCRY1, repressed transcriptional activity but were not degraded by 6-h light pulse. In contrast, dCRY and CRY-d proteins of these same insects (termed CRY1 by the investigators) were degraded by light but have no transcriptional repressive activity. The hypothesis that CRY-m proteins of insects are potent transcriptional repressors may explain how the feedback loop is closed in insects such as the honey bee (Bloch et al. 2003) in which amPER immunoreactivity is cytoplasmatic throughout the day (Zhu et al. 2005).
If indeed the honey bee amCRY functions in the clock as mCRY1/2 do in mammals and bees have lost the CRY-d circadian photoreceptor type protein, how do bees entrain their circadian rhythms to daylight cycles? One possibility is that amCRY has a dual function, a circadian photoreceptor and a core clock component, as has been suggested for mammalian CRY (Cashmore 2003; Sancar 2003) and was recently shown for Drosophila (Collins et al. 2006; see also Hall 2000; Rosato and Kyriacou 2001). It is also possible that the honey bee clock relies on other photopigments such as opsins. One highly appealing candidate is the insect Pteropsin protein, which is orthologous to the vertebrate visual and pineal opsins but was lost from Drosophila (Velarde et al. 2005). Pteropsin is a nonvisual opsin that is expressed in the honey bee in a cluster of cells in an optic lobe area that is commonly implicated in coordinating insect clocks (Helfrich-Forster et al. 1998). Cells expressing the putative clock gene Pigment Dispersing Factor (PDF) are located in the same area in the bee brain, which sets the stage for anatomical interactions between pteropsin and PDF-expressing clock cells (Bloch et al. 2003; Zavodska et al. 2003).
In our phylogenetic analysis, honey bee amCLK and amCYC cluster together with orthologs of other insects (Fig. 1), but amCYC contains a highly conserved transactivation domain at the very C-terminal end that is absent from dCYC (Rutila et al. 1998; Takahata et al. 2000; Chang et al. 2003). Honey bee amCLK is much shorter than dCLK and lacks the part of the C-terminal that is implicated in dCLK transcriptional activity (Allada et al. 1998; Darlington et al. 1998). Thus, the structure of amCYC and amCLK suggests that the honey bee is different from Drosophila but similar to the mouse and A. pernyi in which the main trans-activation domain of the CLK/BMAL complex is in the C-terminal end of BMAL.
The hypothesis that the molecular clockwork in the bee is similar to Drosophila further predicts similarity in the temporal pattern of gene expression. The phase relationships between oscillating clock gene transcripts are influenced by the molecular organization of the transcriptional/translational feedback loops in pacemaker cells. Although additional regulatory mechanisms, as well as post-transcriptional processes, may influence clock gene transcript levels, products of genes that are transcribed and translated together typically oscillate with a similar phase. Similarly, transcription factors are likely to cycle in a different, often almost opposite, phase to that of the genes they transcribe (Reppert and Weaver 2001; Williams and Sehgal 2001; Glossop et al. 2003). This premise is generally supported by the available data. For example, in insects that have both Tim1 and the Cry-d, such as the fly Sarcophaga crassipalpis (Goto and Denlinger 2002) and the moths Bombyx mori (Iwai et al. 2006) and Antheraea pernyi (Sauman and Reppert 1996), Tim1 and Per mRNA oscillate with a similar phase. The situation is more complex in vertebrates that have multiple paralogs for CRY and PER, but still there is evidence that Cry products oscillate with strong amplitude and a phase similar to that of Per in the mouse and other vertebrates that have only the mouse type Cry and Tim2 (see Yoshimura et al. 2000; Avivi et al. 2002, 2004; Fu et al. 2002). The temporal pattern of gene expression in the honey bee brain is strikingly distinct from Drosophila. In the honey bee, amPer and amCry mRNA levels oscillate strongly with a similar phase, whereas in Drosophila they are almost in anti-phase (Emery et al. 1998; Glossop et al. 2003). In the honey bee, amTim2 oscillates with a low amplitude at best, has an inconsistent pattern in LD and DD illumination regimes, and does not cycle with a phase similar to amPer in DD illumination regime (Figs. 7, 8); in Drosophila brain clock, dTim1 mRNA abundance oscillates with strong amplitude and a phase similar to dPer (Williams and Sehgal 2001). Additional inconsistencies with the Drosophila model emerge from the temporal expression pattern of amClk and amCyc. In the honey bee, amClk mRNA levels did not vary over time, but amCyc appears to oscillate with low amplitude and an almost anti-phase to amPer and amCry. This pattern is distinct from Drosophila in which dClk mRNA levels oscillate in virtual antiphase to dPer (and dTim) and dCyc mRNA levels do not vary with time. Overall, the temporal pattern of clock gene expression in the honey bee brain is very similar to the mouse and other mammals (Reppert and Weaver 2001).
An important question raised by our findings is how the honey bee ended up with a clock more similar to mammals than to a fly. We suggest, based on our phylogenetic analyses, that two evolutionary mechanisms contributed to this outcome. The first is that the Drosophila clock profoundly diverged from that of the ancestral insect. The second is an evolutionary convergence of the molecular clocks of honey bees and mammals. Basal animal lineages such as Daphnia (“water fleas”) and Strongylocentrotus (sea urchins) have both CRY-d and CRY-m proteins that persist in Anopheles (mosquito) and Bombyx (silk moth), and perhaps others. The sea urchin, water fleas, flies, flour beetles, and the silk moth B. mori also have both types of TIM. Our new findings that amCYC and a sea urchin ortholog (data not shown) contain a conserved transactivation domain supports and extends the analysis of Chang et al. (2003), suggesting that a BMAL with a C-terminal transactivation domain is the ancestral situation in animals. The transition from a transactivation domain on BMAL as found in most insects to a transactivation domain on CLK as is the case in Drosophila was perhaps facilitated by having trans-activation domains on both CLK in BMAL as currently appears to be the situation in the basal dipteran A. gambiae. However, CLK containing poly-Q repeats might also be an ancient situation because it was found in mammals, bees, and the prawn Macro-brachium rosenbergii (gi|61353791, with a stretch of 148 Qs interrupted by only nine other amino acids). Based on our findings we suggest that Drosophila diverged from most other insects in that (1) it lost CRY-m, (2) it specialized on TIM1 in the negative limb of the feedback loop, and (3) it has a transactivation domain on the C-terminal of CLK instead of the C-terminal end of BMAL. Thus, while Drosophila has been a remarkable and pioneering model system for the study of circadian rhythms, major parts of their molecular clockworks appear to be somewhat idiosyncratic and evolutionarily derived. The evidence for the convergence of honey bees with mammals is the loss of Cry-d and Tim1 from the honey bee genome and the apparent specialization in using CRY-m and TIM2. The completeness of the honey bee genome assembly makes it unlikely that a large gene such as timeless, and even a relatively small gene such as Cry-d, would be missing from the current assemblies (see The Honey Bee Genome Sequencing Consortium 2006). The honey bee is novel among insects in losing both CRY-d and TIM1.
Our characterization of the canonical clock genes sets the stage for molecular and biochemical analysis of complex behaviors such as social synchronization, sun compass navigation, and time memory in honey bees. We reveal a previously unappreciated molecular variety in insect clocks and remarkable similarities between bees and mammals. These findings stimulate new and exciting questions concerning the origin of animal clocks, and the evolution and functional significance of species-specific variation in the molecular clockwork.
Methods
Identification and cloning of putative clock genes in the honey bee
We searched the honey bee genome assemblies, culminating in Assembly 4.0 (v20060310; http://hgsc.bcm.tmc.edu/projects/honeybee/), for homologs for the following Drosophila and mouse “clock” genes: Period (Per), Timeless (Tim), cryptochrome (cry), Clock (Clk), Cycle (Cyc = Bmal1), Vrille (Vri), and Par Domain Protein 1 (Pdp1). We used standard algorithms to identify exons and introns on the genomic sequence of each gene and designed primers for an initial PCR amplification. We used these primers to PCR amplify (Expand High Fidelity PCR System, Roche; Bio-X-act, Accuzyme, and BioTaq DNA Polymerases from BioLine) cDNA templates that we reverse transcribed (with BioScript reverse transcription kit, BioLine) from RNA that we purified from bee brains (with Invisorb Spin Tissue RNA Mini Kit, Invitek; RNeasy Midi Kit, Qiagen; Oligotex mRNA Mini Kit, Qiagen). To obtain the ends of the 3′ and 5′ coding sequences for each gene of interest, we performed rapid amplification of cDNA ends (RACE; FirstChoice RLM-RACE Kit, Ambion). The 5′ RACE for Cycle was problematic in that multiple bands were obtained and indicated several possible alternative transcription and translation starts. We employed one of these instead of the official honey bee gene model for this protein, GB11309, which may nevertheless reflect yet another alternative N terminus for this protein. Because the 3′ RACE product of amCry did not include a stop codon, we deduced the protein C-terminal end (amino acids 535–570) bioinformatically from the corresponding genomic sequence. PCR products were sequenced directly after gel extraction and purification (PCR clean-up Gel extraction, Machrey-Nagel; MinElute Gel Extraction Kit, Qiagen) or following cloning into the pGEM-T Vector System (Promega; DH5a competent cells).
Phylogenetic analyses
We searched public genomic DNA databases at NCBI (Benson et al. 2002), FlyBase (Drysdale et al. 2005), Bombyx mori genome (Mita et al. 2004; Xia et al. 2004), BeeBase (http://racerx00.tamu.edu/bee_resources.html), BeetleBase (Brown et al. 2003), and the Baylor College of Medicine Human Genome Sequencing Center (http://www.hgsc.bcm.tmc.edu/projects/) for genes encoding homologs of known circadian rhythm proteins using TBLASTN (Altschul et al. 1997). Gene models were built manually in the PAUP editor (Swofford 2001) using the BLAST output and the expected exon/intron structures from D. melanogaster and mammalian genes as guides, and the Neural Network Splice Predictor program at the Berkeley Drosophila Genome Project to locate likely intron splice sites (http://www.fruitfly.org/seq_tools/splice.html). Protein alignments using CLUSTALX (Jeanmougin et al. 1998) were used to indicate instances of unusual gene structure. For the sea urchin S. purpuratus, the recently available NCBI RefSeq protein sequences were in some cases employed in place of the manually assembled ones.
For phylogenetic analyses, the irregularly long and poorly aligning N and C termini of the Cycle/Tango/Clock proteins and the highly divergent long C terminus of the Timeless/Timeout proteins were removed from the alignment in the final PAUP file. Amino acid distances calculated between each pair of proteins were corrected for multiple amino acid changes using TREE-PUZZLE v5.0 (Schmidt et al. 2002), with its maximum likelihood model, the BLOSUM62 amino acid exchange matrix, and uniform rates based on the actual sequences. Phylogenetic trees were constructed by using neighbor-joining followed by a heuristic search for better trees using tree-bisection-reconnection branch-swapping in PAUP* v4.0b10 (Swofford 2001). Bootstrap analysis was performed by using 1000 neighbor-joining replications with uncorrected distances and 1000 replications of maximum parsimony using full heuristic searches in PAUP*, as well as 10,000 quartet puzzling maximum likelihood steps with the BLOSUM62 matrix in TREE-PUZZLE.
Putative functional domains and motifs on honey bee clock genes
We used the SMART server (Schultz et al. 1998; Letunic et al. 2004) to demarcate sequences of motifs and domains available in its database. These sequences were confirmed with the NCBI (Marchler-Bauer et al. 2005), Pfam (Bateman et al. 2004), and ProSite (Hulo et al. 2006) databases. We relied on Hirayama and Sassone-Corsi (2005), Chang et al. (2003), and references cited therein to define additional putative clock functional domains. Additional domains and motifs were delineated based on relevant literature in which their biochemical function was defined (see Results). We used the EBI Global Alignment program (Rice et al. 2000) to demarcate domains, motifs, and post-translational modifications on honey bee orthologs and other focal proteins, and to determine the degree of amino acid residue identity/similarity between corresponding amino acid sequences on different proteins. We defined a domain as conserved only if identity >40% compared with the original sequence defined for this domain (Xu et al. 2000). For multiple sequence alignments, we used the CLUSTALX/CLUSTALW algorithm (Higgins et al. 1994). We used the Columbia University NLS-Predict algorithm (http://cubic.bioc.columbia.edu/cgi/var/nair/resonline.pl) to search for putative nuclear localization signals.
The expression of putative clock genes in the honey bee brain
Source colonies from which we obtained bees for our experiments were maintained in the field at a bee research facility at the Givat-Ram campus of the Hebrew University of Jerusalem, Jerusalem, Israel, according to standard commercial techniques. These bees were derived from a mixture of European races of Apis mellifera typical to this region. Each source colony was headed by a queen instrumentally inseminated with semen from a single drone (besides colony H1, which was headed by a naturally mated queen). Single-drone insemination helps to reduce genetic variability among bees within each experiment (average coefficient of relatedness = 0.75 due to haplodiploidy) (Page and Laidlaw 1988).
We established experimental colonies with three cohorts (500–1200 bees per cohort) of bees: 1-d-old bees, nurses, and foragers and their mother queen (all from the same source colony). We housed the colonies in a two-frame observation hive with transparent glass walls. One frame contained pollen and honey, and the second was empty for the queen to lay eggs. Foragers and nurses were identified according to standard criteria (see Moore et al. 1998; Bloch and Robinson 2001). To obtain 1-d-old bees, we removed honeycomb frames containing pupae (sealed in cells) from source colonies in the field and immediately transferred them to a light-proof container that was then placed inside a dark incubator in (32 ± 0.5°C, 55 ± 5% relative humidity) from which we collected emerging bees 0–24 h after pupal eclosion.
The observation hive was placed in an environmental chamber (29 ± 1°C; 50 ± 5% relative humidity) and connected to the outside by a clear plastic tube (length = 0.6m, diameter = 3 cm). For the analysis of brain clock gene mRNA abundance, we collected foragers because bees performing this task are known to have strong circadian rhythms in behavior and physiology (Bloch et al. 2001; Moore 2001). In each experiment, we paint-marked a few hundred foragers that we identified in the daytime so they could be collected during times when foragers are inactive (e.g., at night). We entrained the experimental colonies for 3–6 d in a 12-h light/12-h dark (LD) illumination regime. Fluorescent lights (100 ± 30 lum/square foot) were positioned to ensure even lighting of the colonies, minimizing the possibility that bees could avoid light by hiding in dark corners of the hive. In the first set of experiments, we collected foragers on the seventh day in LD. We collected bees for mRNA analysis (N = 15–20 bees/time point) every 3 h directly into liquid N2, and stored them at −80°C until brain dissection (not all bees collected were analyzed for mRNA levels). We repeated this experiment three times, with bees from source colonies S1, S4, and S8. Circadian rhythmicity and entrainment for a sample of foragers were confirmed at the end of each trial by an analysis of locomotor activity for 7 d in DD (Bloch et al. 2006). In the second set of experiments, we similarly entrained the bees but collected the foragers in a DD instead of an LD illumination regime. On the night of the seventh day, we detached the tube leading to the outside, placed it inside a transparent box inside the environmental chamber (in the first experiment, we only closed the tube opening), and switched the illumination regime to DD. During day eight, we collected samples of foragers every 4 h according to the procedure used in the LD experiments. We performed all collections, manipulations, and observations during the dark phase under dim red light that bees cannot see (von Frisch, 1967). We repeated this experiment three times, with bees from source colonies S11, S12, and H1.
RNA analysis
We removed and freeze-dried bee heads and dissected the brains on a frozen dissecting dish in dry ice; the tissue remained frozen during the entire procedure. We removed compound eyes, ocelli, hypopharyngeal glands, and any other glandular tissues during dissection. Because clock gene mRNA levels are expressed on a per brain basis, we discarded all brains in which pieces of tissue were lost and analyzed only intact brains. We stored each brain individually at –80°C until mRNA quantification.
We measured mRNA levels with real-time quantitative RTPCR using an ABI Prism 7000 appliance (Winer et al. 1999). To measure amPer mRNA levels, we established and validated a multiplex PCR reaction in which amPer and amEF-1α are amplified in the same reaction tube. Total brain RNA was isolated (Invisorb Spin Tissue RNA Mini Kit, Invitek), treated with DNAse (RQ1 RNase-Free DNase, Promega) and reverse-transcribed in 20–25μL 1 × RT buffer + 2.5 U/μL Reverse Transcriptase (BioScript, Bio-Line), 4 mM deoxy NTPs mixture (Fermentas), 25 ng/μL random hexamers (Invitrogen), and 1 U/μL RNase inhibitor (RiboLock Ribonuclease Inhibitor, Fermentas). RNA and Random hexamers were incubated for 5 min at 70°C and immediately transferred to ice. Reverse transcription was carried out for 10 min at 25°C, for 60 min at 42°C, for 10 min at 70°C, and then incubated at 4°C. Amplification reactions (20–25μL) contained 1 × TaqMan Universal PCR Master Mix (ABI Applied Biosystems), 0.1 μM of each primer, 0.2 μM TaqMan probe, and 20–24 ng cDNA (control samples had no reverse transcriptase). Amplification thermal profile was for 2 min at 50°C, for 10 min at 95°C, (for 15 sec at 95°C, for 1 min at 60°C) × 40 cycles. We excluded outliers (SD among triplicates >0.3) from our analyses.
We measured levels of amCry, amTim, amClk, and amCyc with the SYBR green dye protocol. Amplification reaction (20 μL) was similar to the above but contained SYBR green master mix (ABI Applied Biosystems) instead of TaqMan Universal PCR Master Mix and did not contain oligo probes. Each cDNA sample was analyzed in triplicate. PCR reactions for all focal genes and amEF-1α were loaded on the same 96-well analysis plate. For each gene we optimized primer concentration and confirmed that efficiency was similar to that of amEF-1α according to ABI User Bulletin 2 (see primer sequence and concentration in Table S2 in the Supplemental material). To prevent amplification of genomic DNA, we designed the PCR primers to span over an exon–exon boundary. Therefore, in these protocols there was no need to perform a DNase treatment. All clock genes and EF1α levels were measured from the same cDNA sample, which was obtained with the same RNA used to produce cDNA for Per measurements.
We quantified clock gene levels as in Bloch et al. (2001, 2004), using the 2−ΔΔCt method and amEF-1α as control gene for normalization (ABI User Bulletin 2) (see also Winer et al. 1999). Measurements with dot blots, Northern blots, and real-time RTPCR indicated that levels of amEF-1α did not vary with age, task or time of day (Toma et al. 2000; Bloch et al. 2001, 2004). For statistical analyses, we used ΔΔCt values. We used relative mRNA levels for the correlations with a cosinus model.
Acknowledgments
We thank Ravid Sachar for help with the bees, Kim Walden for technical assistance, and the Baylor College of Medicine Human Genome Sequencing Center for making the Apis mellifera, Tribolium castaneum, and Strongylocentrotus purpuratus genome assemblies publicly available before publication. Financial support was provided by the Israel-US Binational Science Foundation (BSF) grant 2003-151 (to G.B. and H.M.R.), NIH grant AI56081 (to H.M.R.), and Israel Science Foundation (ISF, grant number 606/02, to G.B.).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5094806.
References
- Allada R., White N.E., So W.V., Hall J.C., Rosbash M., White N.E., So W.V., Hall J.C., Rosbash M., So W.V., Hall J.C., Rosbash M., Hall J.C., Rosbash M., Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. . Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang Z., Miller W., Lipman D.J., Miller W., Lipman D.J., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avivi A., Oster H., Joel A., Beiles A., Albrecht U., Nevo E., Oster H., Joel A., Beiles A., Albrecht U., Nevo E., Joel A., Beiles A., Albrecht U., Nevo E., Beiles A., Albrecht U., Nevo E., Albrecht U., Nevo E., Nevo E. Circadian genes in a blind subterranean mammal II: Conservation and uniqueness of the three Period homologs in the blind subterranean mole rat,Spalax ehrenbergi superspecies. . Proc. Natl. Acad. Sci. 2002;99:11718–11723. doi: 10.1073/pnas.182423299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avivi A., Oster H., Joel A., Beiles A., Albrecht U., Nevo E., Oster H., Joel A., Beiles A., Albrecht U., Nevo E., Joel A., Beiles A., Albrecht U., Nevo E., Beiles A., Albrecht U., Nevo E., Albrecht U., Nevo E., Nevo E. Circadian genes in a blind subterranean mammal III: Molecular cloning and circadian regulation of cryptochrome genes in the blind subterranean mole rat,Spalax ehrenbergi superspecies. . J. Biol. Rhythms. 2004;19:22–34. doi: 10.1177/0748730403260622. [DOI] [PubMed] [Google Scholar]
- Barnes J.W., Tischkau S.A., Barnes J.A., Mitchell J.W., Burgoon P.W., Hickok J.R., Gillette M.U., Tischkau S.A., Barnes J.A., Mitchell J.W., Burgoon P.W., Hickok J.R., Gillette M.U., Barnes J.A., Mitchell J.W., Burgoon P.W., Hickok J.R., Gillette M.U., Mitchell J.W., Burgoon P.W., Hickok J.R., Gillette M.U., Burgoon P.W., Hickok J.R., Gillette M.U., Hickok J.R., Gillette M.U., Gillette M.U. Requirement of mammalian Timeless for circadian rhythmicity. Science. 2003;302:439–442. doi: 10.1126/science.1086593. [DOI] [PubMed] [Google Scholar]
- Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L.L., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L.L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L.L., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L.L., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L.L., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L.L., Khanna A., Marshall M., Moxon S., Sonnhammer E.L.L., Marshall M., Moxon S., Sonnhammer E.L.L., Moxon S., Sonnhammer E.L.L., Sonnhammer E.L.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Pedersen D., Cassone V.M., Earnest D.J., Golden S.S., Hardin P.E., Thomas T.L., Zoran M.J., Cassone V.M., Earnest D.J., Golden S.S., Hardin P.E., Thomas T.L., Zoran M.J., Earnest D.J., Golden S.S., Hardin P.E., Thomas T.L., Zoran M.J., Golden S.S., Hardin P.E., Thomas T.L., Zoran M.J., Hardin P.E., Thomas T.L., Zoran M.J., Thomas T.L., Zoran M.J., Zoran M.J. Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat. Rev. Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benna C., Scannapieco P., Piccin A., Sandrelli F., Zordan M., Rosato E., Kyriacou C.P., Valle G., Costa R., Scannapieco P., Piccin A., Sandrelli F., Zordan M., Rosato E., Kyriacou C.P., Valle G., Costa R., Piccin A., Sandrelli F., Zordan M., Rosato E., Kyriacou C.P., Valle G., Costa R., Sandrelli F., Zordan M., Rosato E., Kyriacou C.P., Valle G., Costa R., Zordan M., Rosato E., Kyriacou C.P., Valle G., Costa R., Rosato E., Kyriacou C.P., Valle G., Costa R., Kyriacou C.P., Valle G., Costa R., Valle G., Costa R., Costa R. A second timeless gene in Drosophila shares greater sequence similarity with mammalian. tim. Curr. Biol. 2000;10:R512–R513. doi: 10.1016/s0960-9822(00)00594-7. [DOI] [PubMed] [Google Scholar]
- Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Rapp B.A., Wheeler D.L., Karsch-Mizrachi I., Lipman D.J., Ostell J., Rapp B.A., Wheeler D.L., Lipman D.J., Ostell J., Rapp B.A., Wheeler D.L., Ostell J., Rapp B.A., Wheeler D.L., Rapp B.A., Wheeler D.L., Wheeler D.L. GenBank. Nucleic Acids Res. 2002;30:17–20. doi: 10.1093/nar/30.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch G., Robinson G.E., Robinson G.E. Reversal of honeybee behavioural rhythms. Nature. 2001;410:1048. doi: 10.1038/35074183. [DOI] [PubMed] [Google Scholar]
- Bloch G., Toma D.P., Robinson G.E., Toma D.P., Robinson G.E., Robinson G.E. Behavioral rhythmicity, age, division of labor and period expression in the honey bee brain. J. Biol. Rhythms. 2001;16:444–456. doi: 10.1177/074873001129002123. [DOI] [PubMed] [Google Scholar]
- Bloch G., Solomon S.M., Robinson G.E., Fahrbach S.E., Solomon S.M., Robinson G.E., Fahrbach S.E., Robinson G.E., Fahrbach S.E., Fahrbach S.E. Patterns of PERIOD and pigment-dispersing hormone immunoreactivity in the brain of the European honeybee (Apis mellifera): Age-and time-related plasticity. J. Comp. Neurol. 2003;464:269–284. doi: 10.1002/cne.10778. [DOI] [PubMed] [Google Scholar]
- Bloch G., Rubinstein C.D., Robinson G.E., Rubinstein C.D., Robinson G.E., Robinson G.E. Period expression in the honey bee brain is developmentally regulated and not affected by light, flight experience, or colony type. Insect Biochem. Mol. Biol. 2004;34:879–891. doi: 10.1016/j.ibmb.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Bloch G., Shemesh Y., Robinson G.E., Shemesh Y., Robinson G.E., Robinson G.E. Seasonal and task-related variation in free running activity rhythms in honey bees (Apis mellifera) Insectes Soc. 2006;53:115–118. [Google Scholar]
- Brown S.J., Denell R.E., Beeman R.W., Denell R.E., Beeman R.W., Beeman R.W. Beetling around the genome. Genet. Res. 2003;82:155–161. doi: 10.1017/s0016672303006451. [DOI] [PubMed] [Google Scholar]
- Busza A., Emery-Le M., Rosbash M., Emery P., Emery-Le M., Rosbash M., Emery P., Rosbash M., Emery P., Emery P. Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science. 2004;304:1503–1506. doi: 10.1126/science.1096973. [DOI] [PubMed] [Google Scholar]
- Cashmore A.R. Cryptochromes: Enabling plants and animals to determine circadian time. Cell. 2003;114:537–543. [PubMed] [Google Scholar]
- Cashmore A.R., Jarillo J.A., Wu Y.J., Liu D., Jarillo J.A., Wu Y.J., Liu D., Wu Y.J., Liu D., Liu D. Cryptochromes: Blue light receptors for plants and animals. Science. 1999;284:760–765. doi: 10.1126/science.284.5415.760. [DOI] [PubMed] [Google Scholar]
- Ceriani M.F., Darlington T.K., Staknis D., Mas P., Petti A.A., Weitz C., Kay S.A., Darlington T.K., Staknis D., Mas P., Petti A.A., Weitz C., Kay S.A., Staknis D., Mas P., Petti A.A., Weitz C., Kay S.A., Mas P., Petti A.A., Weitz C., Kay S.A., Petti A.A., Weitz C., Kay S.A., Weitz C., Kay S.A., Kay S.A. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science. 1999;285:553–556. doi: 10.1126/science.285.5427.553. [DOI] [PubMed] [Google Scholar]
- Chang D.C., McWatters H.G., Williams J.A., Gotter A.L., Levine J.D., Reppert S.M., McWatters H.G., Williams J.A., Gotter A.L., Levine J.D., Reppert S.M., Williams J.A., Gotter A.L., Levine J.D., Reppert S.M., Gotter A.L., Levine J.D., Reppert S.M., Levine J.D., Reppert S.M., Reppert S.M. Constructing a feedback loop with circadian clock molecules from the Silkmoth,Antheraea pernyi. . J. Biol. Chem. 2003;278:38149–38158. doi: 10.1074/jbc.M306937200. [DOI] [PubMed] [Google Scholar]
- Chaves I., Yagita K., Barnhoorn S., Okamura H., van der Horst G.T., Tamanini F., Yagita K., Barnhoorn S., Okamura H., van der Horst G.T., Tamanini F., Barnhoorn S., Okamura H., van der Horst G.T., Tamanini F., Okamura H., van der Horst G.T., Tamanini F., van der Horst G.T., Tamanini F., Tamanini F. Functional evolution of thePhotolyase/Cryptochrome protein family: Importance of the C terminus of mammalian CRY1 for circadian core oscillator performance. Mol. Cell. Biol. 2006;26:1743–1753. doi: 10.1128/MCB.26.5.1743-1753.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins B., Mazzoni E.O., Stanewsky R., Blau J., Mazzoni E.O., Stanewsky R., Blau J., Stanewsky R., Blau J., Blau J. Drosophila CRYPTOCHROME is a circadian transcriptional repressor. Curr. Biol. 2006;16:441–449. doi: 10.1016/j.cub.2006.01.034. [DOI] [PubMed] [Google Scholar]
- Cowell I.G., Skinner A., Hurst H.C., Skinner A., Hurst H.C., Hurst H.C. Transcriptional repression by a novel member of the bZIP family of transcription factors. Mol. Cell. Biol. 1992;12:3070–3077. doi: 10.1128/mcb.12.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyran S.A., Buchsbaum A.M., Reddy K.L., Lin M.C., Glossop N.R.J., Hardin P.E., Young M.W., Storti R.V., Blau J., Buchsbaum A.M., Reddy K.L., Lin M.C., Glossop N.R.J., Hardin P.E., Young M.W., Storti R.V., Blau J., Reddy K.L., Lin M.C., Glossop N.R.J., Hardin P.E., Young M.W., Storti R.V., Blau J., Lin M.C., Glossop N.R.J., Hardin P.E., Young M.W., Storti R.V., Blau J., Glossop N.R.J., Hardin P.E., Young M.W., Storti R.V., Blau J., Hardin P.E., Young M.W., Storti R.V., Blau J., Young M.W., Storti R.V., Blau J., Storti R.V., Blau J., Blau J. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–341. doi: 10.1016/s0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- Daiyasu H., Ishikawa T., Kuma K., Iwai S., Todo T., Toh H., Ishikawa T., Kuma K., Iwai S., Todo T., Toh H., Kuma K., Iwai S., Todo T., Toh H., Iwai S., Todo T., Toh H., Todo T., Toh H., Toh H. Identification of cryptochrome DASH from vertebrates. Genes Cell. 2004;9:479–495. doi: 10.1111/j.1356-9597.2004.00738.x. [DOI] [PubMed] [Google Scholar]
- Darlington T.K., Wager-Smith K., Ceriani M.F., Staknis D., Gekakis N., Steeves T.D.L., Weitz C., Takahashi J.S., Kay S.A., Wager-Smith K., Ceriani M.F., Staknis D., Gekakis N., Steeves T.D.L., Weitz C., Takahashi J.S., Kay S.A., Ceriani M.F., Staknis D., Gekakis N., Steeves T.D.L., Weitz C., Takahashi J.S., Kay S.A., Staknis D., Gekakis N., Steeves T.D.L., Weitz C., Takahashi J.S., Kay S.A., Gekakis N., Steeves T.D.L., Weitz C., Takahashi J.S., Kay S.A., Steeves T.D.L., Weitz C., Takahashi J.S., Kay S.A., Weitz C., Takahashi J.S., Kay S.A., Takahashi J.S., Kay S.A., Kay S.A. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim . Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- Dissel S., Codd V., Fedic R., Garner K.J., Costa R., Kyriacou C.P., Rosato E., Codd V., Fedic R., Garner K.J., Costa R., Kyriacou C.P., Rosato E., Fedic R., Garner K.J., Costa R., Kyriacou C.P., Rosato E., Garner K.J., Costa R., Kyriacou C.P., Rosato E., Costa R., Kyriacou C.P., Rosato E., Kyriacou C.P., Rosato E., Rosato E. A constitutively active cryptochrome in Drosophila melanogaster . Nat. Neurosci. 2004;7:834–840. doi: 10.1038/nn1285. [DOI] [PubMed] [Google Scholar]
- Drysdale R.A., Crosby M.A., Gelbart W., Campbell K., Emmert D., Matthews B., Russo S., Schroeder A., Smutniak F., Zhang P., Crosby M.A., Gelbart W., Campbell K., Emmert D., Matthews B., Russo S., Schroeder A., Smutniak F., Zhang P., Gelbart W., Campbell K., Emmert D., Matthews B., Russo S., Schroeder A., Smutniak F., Zhang P., Campbell K., Emmert D., Matthews B., Russo S., Schroeder A., Smutniak F., Zhang P., Emmert D., Matthews B., Russo S., Schroeder A., Smutniak F., Zhang P., Matthews B., Russo S., Schroeder A., Smutniak F., Zhang P., Russo S., Schroeder A., Smutniak F., Zhang P., Schroeder A., Smutniak F., Zhang P., Smutniak F., Zhang P., Zhang P., et al. FlyBase: Genes and gene models. Nucleic Acids Res. 2005;33:D390–D395. doi: 10.1093/nar/gki046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap J.C. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Edery I. Circadian rhythms in a nutshell. Physiol. Genomic. 2000;3:59–74. doi: 10.1152/physiolgenomics.2000.3.2.59. [DOI] [PubMed] [Google Scholar]
- Eichinger L., Pachebat J.A., Glockner G., Rajandream M.A., Sucgang R., Berriman M., Song J., Olsen R., Szafranski K., Xu Q., Pachebat J.A., Glockner G., Rajandream M.A., Sucgang R., Berriman M., Song J., Olsen R., Szafranski K., Xu Q., Glockner G., Rajandream M.A., Sucgang R., Berriman M., Song J., Olsen R., Szafranski K., Xu Q., Rajandream M.A., Sucgang R., Berriman M., Song J., Olsen R., Szafranski K., Xu Q., Sucgang R., Berriman M., Song J., Olsen R., Szafranski K., Xu Q., Berriman M., Song J., Olsen R., Szafranski K., Xu Q., Song J., Olsen R., Szafranski K., Xu Q., Olsen R., Szafranski K., Xu Q., Szafranski K., Xu Q., Xu Q., et al. The genome of the social amoeba Dictyostelium discoideum . Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery P., So W.V., Kaneko M., Hall J.C., Rosbash M., So W.V., Kaneko M., Hall J.C., Rosbash M., Kaneko M., Hall J.C., Rosbash M., Hall J.C., Rosbash M., Rosbash M. CRY, a Drosophila clock and light-regulating cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- Field M.D., Maywood E.S., O'Brien J.A., Weaver D.R., Reppert S.M., Hastings M.H., Maywood E.S., O'Brien J.A., Weaver D.R., Reppert S.M., Hastings M.H., O'Brien J.A., Weaver D.R., Reppert S.M., Hastings M.H., Weaver D.R., Reppert S.M., Hastings M.H., Reppert S.M., Hastings M.H., Hastings M.H. Analysis of clock proteins in mouse SCN demonstrates phylogenetic divergence of the circadian clockwork and resetting mechanisms. Neuron. 2000;25:437–447. doi: 10.1016/s0896-6273(00)80906-x. [DOI] [PubMed] [Google Scholar]
- Froy O., Chang D.C., Reppert S.M., Chang D.C., Reppert S.M., Reppert S.M. Redox potential: Differential roles in dCRY and mCRY1 functions. Curr. Biol. 2002;12:147–152. doi: 10.1016/s0960-9822(01)00656-x. [DOI] [PubMed] [Google Scholar]
- Fu Z.W., Inaba M., Noguchi T., Kato H., Inaba M., Noguchi T., Kato H., Noguchi T., Kato H., Kato H. Molecular cloning and circadian regulation of cryptochrome genes in Japanese quail (Coturnix coturnix japonica) J. Biol. Rhythm. 2002;17:14–27. doi: 10.1177/074873002129002302. [DOI] [PubMed] [Google Scholar]
- Gaunt M.W., Miles M.A., Miles M.A. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol. Biol. Evol. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- Gehring W., Rosbash M., Rosbash M. The coevolution of blue-light photoreception and circadian rhythms. J. Mol. Evol. 2003;57:S286–S289. doi: 10.1007/s00239-003-0038-8. [DOI] [PubMed] [Google Scholar]
- Gekakis N., Saez L., Delahayebrown A.M., Myers M.P., Sehgal A., Young M.W., Weitz C.J., Saez L., Delahayebrown A.M., Myers M.P., Sehgal A., Young M.W., Weitz C.J., Delahayebrown A.M., Myers M.P., Sehgal A., Young M.W., Weitz C.J., Myers M.P., Sehgal A., Young M.W., Weitz C.J., Sehgal A., Young M.W., Weitz C.J., Young M.W., Weitz C.J., Weitz C.J. Isolation of timeless by per protein interaction–defective interaction between timeless protein and long-period mutant per(l) Science. 1995;270:811–815. doi: 10.1126/science.270.5237.811. [DOI] [PubMed] [Google Scholar]
- Glossop N.R.J., Houl J.H., Zheng H., Ng F.S., Dudek S.M., Hardin P.E., Houl J.H., Zheng H., Ng F.S., Dudek S.M., Hardin P.E., Zheng H., Ng F.S., Dudek S.M., Hardin P.E., Ng F.S., Dudek S.M., Hardin P.E., Dudek S.M., Hardin P.E., Hardin P.E. VRILLE feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron. 2003;37:249–261. doi: 10.1016/s0896-6273(03)00002-3. [DOI] [PubMed] [Google Scholar]
- Goto S.G., Denlinger D.L., Denlinger D.L. Short-day and long-day expression patterns of genes involved in the flesh fly clock mechanism: period, timeless, cycle and cryptochrome . J. Insect Physiol. 2002;48:803–816. doi: 10.1016/s0022-1910(02)00108-7. [DOI] [PubMed] [Google Scholar]
- Gotter A.L., Manganaro T., Weaver D.R., Kolakowski L.F., Possidente B., Sriram S., MacLaughlin D.T., Reppert S.M., Manganaro T., Weaver D.R., Kolakowski L.F., Possidente B., Sriram S., MacLaughlin D.T., Reppert S.M., Weaver D.R., Kolakowski L.F., Possidente B., Sriram S., MacLaughlin D.T., Reppert S.M., Kolakowski L.F., Possidente B., Sriram S., MacLaughlin D.T., Reppert S.M., Possidente B., Sriram S., MacLaughlin D.T., Reppert S.M., Sriram S., MacLaughlin D.T., Reppert S.M., MacLaughlin D.T., Reppert S.M., Reppert S.M. A time-less function for mouse. Timeless. Nat. Neurosci. 2000;3:755–756. doi: 10.1038/77653. [DOI] [PubMed] [Google Scholar]
- Green C.B. Cryptochromes: Tail-ored for distinct functions. Curr. Biol. 2004;14:R847–R849. doi: 10.1016/j.cub.2004.09.040. [DOI] [PubMed] [Google Scholar]
- Griffin E.A., Jr., Staknis D., Weitz C.J., Staknis D., Weitz C.J., Weitz C.J. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- Hall J.C. Cryptochromes: Sensory reception, transduction, and clock functions subserving circadian systems. Curr. Opin. Neurobiol. 2000;10:456–466. doi: 10.1016/s0959-4388(00)00117-3. [DOI] [PubMed] [Google Scholar]
- Hardin P.E. Transcription regulation within the circadian clock: The E-box and beyond. J. Biol. Rhythm. 2004;19:348–360. doi: 10.1177/0748730404268052. [DOI] [PubMed] [Google Scholar]
- Hardin P.E. The circadian timekeeping system of Drosophila. Curr. Biol. 2005;15:R714–R722. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C., Stengl M., Homberg U., Stengl M., Homberg U., Homberg U. Organization of the circadian system in insects. Chronobiol. Int. 1998;15:567–594. doi: 10.3109/07420529808993195. [DOI] [PubMed] [Google Scholar]
- Higgins D., Thompson J., Gibson T., Thompson J.D., Higgins D.G., Gibson T.J., Thompson J., Gibson T., Thompson J.D., Higgins D.G., Gibson T.J., Gibson T., Thompson J.D., Higgins D.G., Gibson T.J., Thompson J.D., Higgins D.G., Gibson T.J., Higgins D.G., Gibson T.J., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama J., Sassone-Corsi P., Sassone-Corsi P. Structural and functional features of transcription factors controlling the circadian clock. Curr. Opin. Genet. Dev. 2005;15:548–556. doi: 10.1016/j.gde.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Hirayama J., Nakamura H., Ishikawa T., Kobayashi Y., Todo T., Nakamura H., Ishikawa T., Kobayashi Y., Todo T., Ishikawa T., Kobayashi Y., Todo T., Kobayashi Y., Todo T., Todo T. Functional and structural analyses of cryptochrome: Vertebrate CRY regions responsible for interaction with the CLOCK:BMAL1 heterodimer and its nuclear localization. J. Biol. Chem. 2003;37:35620–35628. doi: 10.1074/jbc.M305028200. [DOI] [PubMed] [Google Scholar]
- The Honey Bee Genome Sequencing Consortium. Insights into social insects from the genome of the honey bee Apis mellifera . Nature. 2003 in press. [Google Scholar]
- Hulo N., Bairoch A., Bulliard V., Cerutti L., De Castro E., Langendijk-Genevaux P.S., Pagni M., Sigrist C.J.A., Bairoch A., Bulliard V., Cerutti L., De Castro E., Langendijk-Genevaux P.S., Pagni M., Sigrist C.J.A., Bulliard V., Cerutti L., De Castro E., Langendijk-Genevaux P.S., Pagni M., Sigrist C.J.A., Cerutti L., De Castro E., Langendijk-Genevaux P.S., Pagni M., Sigrist C.J.A., De Castro E., Langendijk-Genevaux P.S., Pagni M., Sigrist C.J.A., Langendijk-Genevaux P.S., Pagni M., Sigrist C.J.A., Pagni M., Sigrist C.J.A., Sigrist C.J.A. The PROSITE database. Nucleic Acids Res. 2006;34:D227–D230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanchenko M., Stanewsky R., Giebultowicz J.M., Stanewsky R., Giebultowicz J.M., Giebultowicz J.M. Circadian photoreception n Drosophila: Functions of cryptochrome in peripheral and central clocks. J. Biol. Rhythms. 2001;16:205–215. doi: 10.1177/074873040101600303. [DOI] [PubMed] [Google Scholar]
- Iwai S., Fukui Y., Fujiwara Y., Takeda M., Fukui Y., Fujiwara Y., Takeda M., Fujiwara Y., Takeda M., Takeda M. Structure and expressions of two circadian clock genes, period and timeless in the commercial silkmoth, Bombyx mori . J. Insect Physiol. 2006;52:625–637. doi: 10.1016/j.jinsphys.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Jeanmougin F., Thompson J.D., Gouy M., Higgins D.G., Gibson T.J., Thompson J.D., Gouy M., Higgins D.G., Gibson T.J., Gouy M., Higgins D.G., Gibson T.J., Higgins D.G., Gibson T.J., Gibson T.J. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- Jones D.T. GenTHREADER: An efficient and reliable protein fold recognition method for genomic sequences. J. Mol. Biol. 1999;287:797–815. doi: 10.1006/jmbi.1999.2583. [DOI] [PubMed] [Google Scholar]
- Kato T., Todo T., Ayaki H., Ishizaki K., Morita T., Mitra S., Ikenaga M., Todo T., Ayaki H., Ishizaki K., Morita T., Mitra S., Ikenaga M., Ayaki H., Ishizaki K., Morita T., Mitra S., Ikenaga M., Ishizaki K., Morita T., Mitra S., Ikenaga M., Morita T., Mitra S., Ikenaga M., Mitra S., Ikenaga M., Ikenaga M. Cloning of a marsupial DNA photolyase gene and the lack of related nucleotide sequences in placental mammals. Nucleic Acids Res. 1994;22:4119–4124. doi: 10.1093/nar/22.20.4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L.A., MacCallum R.M., Sternberg M.J., MacCallum R.M., Sternberg M.J., Sternberg M.J. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- King D.P., Zhao Y., Sangoram A.M., Wilsbacher L., Tanaka M., Antoch M.P., Steeves T.D.L., Vitaterna M., Kornhauser J., Lowrey P., Zhao Y., Sangoram A.M., Wilsbacher L., Tanaka M., Antoch M.P., Steeves T.D.L., Vitaterna M., Kornhauser J., Lowrey P., Sangoram A.M., Wilsbacher L., Tanaka M., Antoch M.P., Steeves T.D.L., Vitaterna M., Kornhauser J., Lowrey P., Wilsbacher L., Tanaka M., Antoch M.P., Steeves T.D.L., Vitaterna M., Kornhauser J., Lowrey P., Tanaka M., Antoch M.P., Steeves T.D.L., Vitaterna M., Kornhauser J., Lowrey P., Antoch M.P., Steeves T.D.L., Vitaterna M., Kornhauser J., Lowrey P., Steeves T.D.L., Vitaterna M., Kornhauser J., Lowrey P., Vitaterna M., Kornhauser J., Lowrey P., Kornhauser J., Lowrey P., Lowrey P., et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Ishikawa T., Hirayama J., Daiyasu H., Kanai S., Toh H., Fukuda I., Tsujimura T., Terada N., Kamei Y., Ishikawa T., Hirayama J., Daiyasu H., Kanai S., Toh H., Fukuda I., Tsujimura T., Terada N., Kamei Y., Hirayama J., Daiyasu H., Kanai S., Toh H., Fukuda I., Tsujimura T., Terada N., Kamei Y., Daiyasu H., Kanai S., Toh H., Fukuda I., Tsujimura T., Terada N., Kamei Y., Kanai S., Toh H., Fukuda I., Tsujimura T., Terada N., Kamei Y., Toh H., Fukuda I., Tsujimura T., Terada N., Kamei Y., Fukuda I., Tsujimura T., Terada N., Kamei Y., Tsujimura T., Terada N., Kamei Y., Terada N., Kamei Y., Kamei Y., et al. Molecular analysis of zebrafish photolyase/cryptochrome family: Two types of cryptochromes present in zebrafish. Genes Cell. 2000;5:725–738. doi: 10.1046/j.1365-2443.2000.00364.x. [DOI] [PubMed] [Google Scholar]
- Krishnan B., Levine J.D., Lynch M.K.S., Dowse H.B., Funes P., Hall J.C., Hardin P.E., Dryer S.E., Levine J.D., Lynch M.K.S., Dowse H.B., Funes P., Hall J.C., Hardin P.E., Dryer S.E., Lynch M.K.S., Dowse H.B., Funes P., Hall J.C., Hardin P.E., Dryer S.E., Dowse H.B., Funes P., Hall J.C., Hardin P.E., Dryer S.E., Funes P., Hall J.C., Hardin P.E., Dryer S.E., Hall J.C., Hardin P.E., Dryer S.E., Hardin P.E., Dryer S.E., Dryer S.E. A new role for cryptochrome in a Drosophila circadian oscillator. Nature. 2001;411:313–317. doi: 10.1038/35077094. [DOI] [PubMed] [Google Scholar]
- Kume K., Zylka M.J., Sriram S., Shearman L.P., Weaver D.R., Jin X., Maywood E.S., Hastings M.H., Reppert M., Zylka M.J., Sriram S., Shearman L.P., Weaver D.R., Jin X., Maywood E.S., Hastings M.H., Reppert M., Sriram S., Shearman L.P., Weaver D.R., Jin X., Maywood E.S., Hastings M.H., Reppert M., Shearman L.P., Weaver D.R., Jin X., Maywood E.S., Hastings M.H., Reppert M., Weaver D.R., Jin X., Maywood E.S., Hastings M.H., Reppert M., Jin X., Maywood E.S., Hastings M.H., Reppert M., Maywood E.S., Hastings M.H., Reppert M., Hastings M.H., Reppert M., Reppert M. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- Lee C., Bae K., Edery I., Bae K., Edery I., Edery I. PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: A basis for circadian transcription. Mol. Cell. Biol. 1999;19:5316–5325. doi: 10.1128/mcb.19.8.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Copley R.R., Schmidt S., Ciccarelli F.D., Doerks T., Schultz J., Ponting C.P., Bork P., Copley R.R., Schmidt S., Ciccarelli F.D., Doerks T., Schultz J., Ponting C.P., Bork P., Schmidt S., Ciccarelli F.D., Doerks T., Schultz J., Ponting C.P., Bork P., Ciccarelli F.D., Doerks T., Schultz J., Ponting C.P., Bork P., Doerks T., Schultz J., Ponting C.P., Bork P., Schultz J., Ponting C.P., Bork P., Ponting C.P., Bork P., Bork P. SMART 4.0: Towards genomic data integration. Nucleic Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine J.D., Funes P., Dowse H.B., Hall J.C., Funes P., Dowse H.B., Hall J.C., Dowse H.B., Hall J.C., Hall J.C. Advanced analysis of a cryptochrome mutation's effects on the robustness and phase of molecular cycles in isolated peripheral tissues of Drosophila. BMC Neurosci. 2002;15:3–5. doi: 10.1186/1471-2202-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.C., Lin M.H., Horvath P., Reddy K.L., Storti R.V., Lin M.H., Horvath P., Reddy K.L., Storti R.V., Horvath P., Reddy K.L., Storti R.V., Reddy K.L., Storti R.V., Storti R.V. PDP1, a novel Drosophila PAR domain bZIP transcription factor expressed in developing mesoderm, endoderm and ectoderm, is a transcriptional regulator of somatic muscle genes. Development. 1997;124:4685–4696. doi: 10.1242/dev.124.22.4685. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Anderson J.B., Cherukuri P.F., DeWeese-Scott C., Geer L.Y., Gwadz M., He S., Hurwitz D.I., Jackson J.D., Ke Z., Anderson J.B., Cherukuri P.F., DeWeese-Scott C., Geer L.Y., Gwadz M., He S., Hurwitz D.I., Jackson J.D., Ke Z., Cherukuri P.F., DeWeese-Scott C., Geer L.Y., Gwadz M., He S., Hurwitz D.I., Jackson J.D., Ke Z., DeWeese-Scott C., Geer L.Y., Gwadz M., He S., Hurwitz D.I., Jackson J.D., Ke Z., Geer L.Y., Gwadz M., He S., Hurwitz D.I., Jackson J.D., Ke Z., Gwadz M., He S., Hurwitz D.I., Jackson J.D., Ke Z., He S., Hurwitz D.I., Jackson J.D., Ke Z., Hurwitz D.I., Jackson J.D., Ke Z., Jackson J.D., Ke Z., Ke Z., et al. CDD: A Conserved Domain Database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin L.J., Jones D.T., Jones D.T. Improvement of the GenTHREADER method for genomic fold recognition. Bioinformatics. 2003;19:874–881. doi: 10.1093/bioinformatics/btg097. [DOI] [PubMed] [Google Scholar]
- Mita K., Kasahara M., Sasaki S., Nagayasu Y., Yamada T., Kanamori H., Namiki N., Kitagawa M., Yamashita H., Yasukochi Y., Kasahara M., Sasaki S., Nagayasu Y., Yamada T., Kanamori H., Namiki N., Kitagawa M., Yamashita H., Yasukochi Y., Sasaki S., Nagayasu Y., Yamada T., Kanamori H., Namiki N., Kitagawa M., Yamashita H., Yasukochi Y., Nagayasu Y., Yamada T., Kanamori H., Namiki N., Kitagawa M., Yamashita H., Yasukochi Y., Yamada T., Kanamori H., Namiki N., Kitagawa M., Yamashita H., Yasukochi Y., Kanamori H., Namiki N., Kitagawa M., Yamashita H., Yasukochi Y., Namiki N., Kitagawa M., Yamashita H., Yasukochi Y., Kitagawa M., Yamashita H., Yasukochi Y., Yamashita H., Yasukochi Y., Yasukochi Y., et al. The genome sequence of silkworm Bombyx mori. DNA Res. 2004;11:27–35. doi: 10.1093/dnares/11.1.27. [DOI] [PubMed] [Google Scholar]
- Moore D. Honey bee circadian clocks: Behavioral control from individual workers to whole-colony rhythms. J. Insect Physiol. 2001;47:843–857. [Google Scholar]
- Moore D., Angel J.E., Cheeseman I.M., Fahrbach S.E., Robinson G.E., Angel J.E., Cheeseman I.M., Fahrbach S.E., Robinson G.E., Cheeseman I.M., Fahrbach S.E., Robinson G.E., Fahrbach S.E., Robinson G.E., Robinson G.E. Timekeeping in the honey bee colony: Integration of circadian rhythms and division of labor. Behav. Ecol. Sociobiol. 1998;43:147–160. [Google Scholar]
- Moore-Ede M.C., Sulzman F.M., Fuller C.A., Sulzman F.M., Fuller C.A., Fuller C.A. The clocks that time us, physiology of the circadian timing system. Harvard University Press; Cambridge, MA: 1983. [Google Scholar]
- Moritz R.F.A., Kryger P., Kryger P. Self-organization of circadian rhythms in groups of honeybees (Apis mellifera L.) Behav. Ecol. Sociobiol. 1994;34:211–215. [Google Scholar]
- Page R.E., Laidlaw H.H., Laidlaw H.H. Full sisters and super sisters: A terminological paradigm. Anim. Behav. 1988;36:944–945. [Google Scholar]
- Panda S., Hogenesch J.B., Kay S.A., Hogenesch J.B., Kay S.A., Kay S.A. Circadian rhythms from flies to human. Nature. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- Partch C.L., Clarkson M.W., Ozgur S., Lee A.L., Sancar A., Clarkson M.W., Ozgur S., Lee A.L., Sancar A., Ozgur S., Lee A.L., Sancar A., Lee A.L., Sancar A., Sancar A. Role of structural plasticity in signal transduction by the Cryptochrome blue-light photoreceptor. Biochemistry. 2005;44:3795–3805. doi: 10.1021/bi047545g. [DOI] [PubMed] [Google Scholar]
- Perry J. Weighing in on a timeless controversy. Proteins. 2005;61:699–703. doi: 10.1002/prot.20706. [DOI] [PubMed] [Google Scholar]
- Reppert S.M., Weaver D.R., Weaver D.R. Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- Rice P., Longden I., Bleasby A., Longden I., Bleasby A., Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Richards S., Liu Y., Bettencourt B.R., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Liu Y., Bettencourt B.R., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Bettencourt B.R., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Hubisz M.J., Chen R., Meisel R.P., Chen R., Meisel R.P., Meisel R.P., et al. Comparative genome sequencing of Drosophila pseudoobscura: Chromosomal, gene, and cis-element evolution. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H.M. Insect genomes. Am Entomol. 2005;51:166–171. [Google Scholar]
- Rosato E., Kyriacou C.P., Kyriacou C.P. Flies, clocks and evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001;356:1769–1778. doi: 10.1098/rstb.2001.0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosato E., Codd V., Mazzotta G., Piccin A., Zordan M., Costa R., Kyriacou C.P., Codd V., Mazzotta G., Piccin A., Zordan M., Costa R., Kyriacou C.P., Mazzotta G., Piccin A., Zordan M., Costa R., Kyriacou C.P., Piccin A., Zordan M., Costa R., Kyriacou C.P., Zordan M., Costa R., Kyriacou C.P., Costa R., Kyriacou C.P., Kyriacou C.P. Light-dependent interaction between Drosophila CRY and the clock protein PER mediated by the carboxy terminus of. CRY. Curr. Biol. 2001;11:909–917. doi: 10.1016/s0960-9822(01)00259-7. [DOI] [PubMed] [Google Scholar]
- Rutila J.E., Suri V., Le M., So W.V., Rosbash M., Hall J.C., Suri V., Le M., So W.V., Rosbash M., Hall J.C., Le M., So W.V., Rosbash M., Hall J.C., So W.V., Rosbash M., Hall J.C., Rosbash M., Hall J.C., Hall J.C. CYCLE is a second bHLH-PAS Clock Protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93:805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- Saez L., Young M.W., Young M.W. Regulation of nuclear entry of the Drosophila clock proteins period and timeless. Neuron. 1996;17:911–920. doi: 10.1016/s0896-6273(00)80222-6. [DOI] [PubMed] [Google Scholar]
- Sakakida Y., Miyamoto Y., Nagoshi E., Akashi M., Nakamura T.J., Mamine T., Kasahara M., Minami Y., Yoneda Y., Takumi T., Miyamoto Y., Nagoshi E., Akashi M., Nakamura T.J., Mamine T., Kasahara M., Minami Y., Yoneda Y., Takumi T., Nagoshi E., Akashi M., Nakamura T.J., Mamine T., Kasahara M., Minami Y., Yoneda Y., Takumi T., Akashi M., Nakamura T.J., Mamine T., Kasahara M., Minami Y., Yoneda Y., Takumi T., Nakamura T.J., Mamine T., Kasahara M., Minami Y., Yoneda Y., Takumi T., Mamine T., Kasahara M., Minami Y., Yoneda Y., Takumi T., Kasahara M., Minami Y., Yoneda Y., Takumi T., Minami Y., Yoneda Y., Takumi T., Yoneda Y., Takumi T., Takumi T. Importin α/β mediates nuclear transport of a mammalian circadian clock component, mCRY2, together with mPER2, through a bipartite nuclear localization signal. J. Biol. Chem. 2005;280:13272–13278. doi: 10.1074/jbc.M413236200. [DOI] [PubMed] [Google Scholar]
- Sanada K., Harada Y., Sakai M., Todo T., Fukada Y., Harada Y., Sakai M., Todo T., Fukada Y., Sakai M., Todo T., Fukada Y., Todo T., Fukada Y., Fukada Y. Serine phosphorylation of mCRY1 and mCRY2 by mitogen-activated protein kinase. Genes Cell. 2004;9:697–708. doi: 10.1111/j.1356-9597.2004.00758.x. [DOI] [PubMed] [Google Scholar]
- Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 2003;103:2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- Sauman I., Reppert S.M., Reppert S.M. Circadian clock neurons in the silkmoh Antheraea pernyi: Novel mechanisms of period protein regulation. Neuron. 1996;17:889–900. doi: 10.1016/s0896-6273(00)80220-2. [DOI] [PubMed] [Google Scholar]
- Sauman I., Briscoe A.D., Zhu H., Shi D., Froy O., Stalleicken J., Yuan Q., Casselman A., Reppert S.M., Briscoe A.D., Zhu H., Shi D., Froy O., Stalleicken J., Yuan Q., Casselman A., Reppert S.M., Zhu H., Shi D., Froy O., Stalleicken J., Yuan Q., Casselman A., Reppert S.M., Shi D., Froy O., Stalleicken J., Yuan Q., Casselman A., Reppert S.M., Froy O., Stalleicken J., Yuan Q., Casselman A., Reppert S.M., Stalleicken J., Yuan Q., Casselman A., Reppert S.M., Yuan Q., Casselman A., Reppert S.M., Casselman A., Reppert S.M., Reppert S.M. Connecting the navigational clock to sun compass input in monarch butterfly brain. Neuron. 2005;46:457–467. doi: 10.1016/j.neuron.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Schmidt H.A., Strimmer K., Vingron M., von Haeseler A., Strimmer K., Vingron M., von Haeseler A., Vingron M., von Haeseler A., von Haeseler A. TREE-PUZZLE: Maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Schultz J., Milpetz F., Bork P., Ponting C.P., Milpetz F., Bork P., Ponting C.P., Bork P., Ponting C.P., Ponting C.P. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehadova H., Sauman I., Sehnal F., Sauman I., Sehnal F., Sehnal F. Immunocytochemical distribution of pigment-dispersing hormone in the cephalic ganglia of polyneopteran insects. Cell Tissue Res. 2003;312:113–125. doi: 10.1007/s00441-003-0705-5. [DOI] [PubMed] [Google Scholar]
- Stanewsky R. Genetic analysis of the circadian system in Drosophila melanogaster and mammals. J. Neurobiol. 2003;54:111–147. doi: 10.1002/neu.10164. [DOI] [PubMed] [Google Scholar]
- Swofford D.L. PAUP*: Phylogenetic analysis using parsimony and other methods, version4. Sinauer Press; New York: 2001. [Google Scholar]
- Takahata S., Ozaki T., Mimura J., Kikuchi Y., Sogawa K., Fujii-Kuriyama Y., Ozaki T., Mimura J., Kikuchi Y., Sogawa K., Fujii-Kuriyama Y., Mimura J., Kikuchi Y., Sogawa K., Fujii-Kuriyama Y., Kikuchi Y., Sogawa K., Fujii-Kuriyama Y., Sogawa K., Fujii-Kuriyama Y., Fujii-Kuriyama Y. Transactivation mechanisms of mouse clock transcription factors, mClock and mArnt3. Genes Cell. 2000;5:739–747. doi: 10.1046/j.1365-2443.2000.00363.x. [DOI] [PubMed] [Google Scholar]
- Takumi T., Nagamine Y., Miyake S., Matsubara C., Taguchi K., Takekida S., Sakakida Y., Nishikawa K., Kishimoto T., Niwa S., Nagamine Y., Miyake S., Matsubara C., Taguchi K., Takekida S., Sakakida Y., Nishikawa K., Kishimoto T., Niwa S., Miyake S., Matsubara C., Taguchi K., Takekida S., Sakakida Y., Nishikawa K., Kishimoto T., Niwa S., Matsubara C., Taguchi K., Takekida S., Sakakida Y., Nishikawa K., Kishimoto T., Niwa S., Taguchi K., Takekida S., Sakakida Y., Nishikawa K., Kishimoto T., Niwa S., Takekida S., Sakakida Y., Nishikawa K., Kishimoto T., Niwa S., Sakakida Y., Nishikawa K., Kishimoto T., Niwa S., Nishikawa K., Kishimoto T., Niwa S., Kishimoto T., Niwa S., Niwa S., et al. A mammalian ortholog of Drosophila timeless, highly expressed in SCN and retina, forms a complex with mPER1. Genes Cell. 1999;4:67–75. doi: 10.1046/j.1365-2443.1999.00238.x. [DOI] [PubMed] [Google Scholar]
- Tauber E., Last K.S., Olive P.J.W., Kyriacou C.P., Last K.S., Olive P.J.W., Kyriacou C.P., Olive P.J.W., Kyriacou C.P., Kyriacou C.P. Clock gene evolution and functional divergence. J. Biol. Rhythm. 2004;19:445–458. doi: 10.1177/0748730404268775. [DOI] [PubMed] [Google Scholar]
- Toma D.P., Bloch G., Moore D., Robinson G.E., Bloch G., Moore D., Robinson G.E., Moore D., Robinson G.E., Robinson G.E. Changes in period mRNA levels in the brain and division of labor in honey bee colonies. Proc. Natl. Acad. Sci. 2000;97:6914–6919. doi: 10.1073/pnas.97.12.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst G.T., Muijtjens M., Kobayashi K., Takano R., Kanno S., Takao M., de Wit J., Verkerk A., Eker A.P., van Leenen D., Muijtjens M., Kobayashi K., Takano R., Kanno S., Takao M., de Wit J., Verkerk A., Eker A.P., van Leenen D., Kobayashi K., Takano R., Kanno S., Takao M., de Wit J., Verkerk A., Eker A.P., van Leenen D., Takano R., Kanno S., Takao M., de Wit J., Verkerk A., Eker A.P., van Leenen D., Kanno S., Takao M., de Wit J., Verkerk A., Eker A.P., van Leenen D., Takao M., de Wit J., Verkerk A., Eker A.P., van Leenen D., de Wit J., Verkerk A., Eker A.P., van Leenen D., Verkerk A., Eker A.P., van Leenen D., Eker A.P., van Leenen D., van Leenen D., et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- Velarde R.A., Sauer C.D., Walden K.K., Fahrbach S.E., Robertson H.M., Sauer C.D., Walden K.K., Fahrbach S.E., Robertson H.M., Walden K.K., Fahrbach S.E., Robertson H.M., Fahrbach S.E., Robertson H.M., Robertson H.M. Pteropsin: A vertebrate-like non-visual opsin expressed in the honey bee brain. Insect Biochem. Mol. Biol. 2005;35:1367–1377. doi: 10.1016/j.ibmb.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Vitaterna M.H., Selby C.P., Todo T., Niwa H., Thompson C., Fruechte E.M., Hitomi K., Thresher R.J., Ishikawa T., Miyazaki J., Selby C.P., Todo T., Niwa H., Thompson C., Fruechte E.M., Hitomi K., Thresher R.J., Ishikawa T., Miyazaki J., Todo T., Niwa H., Thompson C., Fruechte E.M., Hitomi K., Thresher R.J., Ishikawa T., Miyazaki J., Niwa H., Thompson C., Fruechte E.M., Hitomi K., Thresher R.J., Ishikawa T., Miyazaki J., Thompson C., Fruechte E.M., Hitomi K., Thresher R.J., Ishikawa T., Miyazaki J., Fruechte E.M., Hitomi K., Thresher R.J., Ishikawa T., Miyazaki J., Hitomi K., Thresher R.J., Ishikawa T., Miyazaki J., Thresher R.J., Ishikawa T., Miyazaki J., Ishikawa T., Miyazaki J., Miyazaki J., et al. Differential regulation of mammalian Period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc. Natl. Acad. Sci. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodovar N., Clayton J.D., Costa R., Odell M., Kyriacou C.P., Clayton J.D., Costa R., Odell M., Kyriacou C.P., Costa R., Odell M., Kyriacou C.P., Odell M., Kyriacou C.P., Kyriacou C.P. The Drosophila clock protein Timeless is a member of the Arm/HEAT family. Curr. Biol. 2002;12:R610–R611. doi: 10.1016/s0960-9822(02)01130-2. [DOI] [PubMed] [Google Scholar]
- von Frisch K. The dance language and orientation of bes. Harvard University Press; Cambridge, MA: 1967. [Google Scholar]
- Williams J.A., Sehgal A., Sehgal A. Molecular components of the circadian system in Drosophila. Annu. Rev. Physiol. 2001;63:729–755. doi: 10.1146/annurev.physiol.63.1.729. [DOI] [PubMed] [Google Scholar]
- Winer J., Jung C.K.S., Shackel I., Williams P.M., Jung C.K.S., Shackel I., Williams P.M., Shackel I., Williams P.M., Williams P.M. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal. Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- Wise S., Davis N.T., Tyndale E., Noveral J., Folwell M.G., Bedian V., Emery I.F., Siwicki K.K., Davis N.T., Tyndale E., Noveral J., Folwell M.G., Bedian V., Emery I.F., Siwicki K.K., Tyndale E., Noveral J., Folwell M.G., Bedian V., Emery I.F., Siwicki K.K., Noveral J., Folwell M.G., Bedian V., Emery I.F., Siwicki K.K., Folwell M.G., Bedian V., Emery I.F., Siwicki K.K., Bedian V., Emery I.F., Siwicki K.K., Emery I.F., Siwicki K.K., Siwicki K.K. Neuroanatomical studies of period gene expression in the hawkmoth, Manduca sexta. J. Comp. Neurol. 2002;447:366–380. doi: 10.1002/cne.10242. [DOI] [PubMed] [Google Scholar]
- Xia Q., Zhou Z., Lu C., Cheng D., Dai F., Li B., Zhao P., Zha X., Cheng T., Chai C., Zhou Z., Lu C., Cheng D., Dai F., Li B., Zhao P., Zha X., Cheng T., Chai C., Lu C., Cheng D., Dai F., Li B., Zhao P., Zha X., Cheng T., Chai C., Cheng D., Dai F., Li B., Zhao P., Zha X., Cheng T., Chai C., Dai F., Li B., Zhao P., Zha X., Cheng T., Chai C., Li B., Zhao P., Zha X., Cheng T., Chai C., Zhao P., Zha X., Cheng T., Chai C., Zha X., Cheng T., Chai C., Cheng T., Chai C., Chai C., et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori) Science. 2004;306:1937–1940. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- Xu D., Xu Y., Uberbacher E.C., Xu Y., Uberbacher E.C., Uberbacher E.C. Computational tools for protein modeling. Curr. Protein Pept. Sci. 2000;1:1–21. doi: 10.2174/1389203003381469. [DOI] [PubMed] [Google Scholar]
- Yoshimura T., Suzuki Y., Makino E., Suzuki T., Kuroiwa A., Matsuda Y., Namikawa T., Ebihara S., Suzuki Y., Makino E., Suzuki T., Kuroiwa A., Matsuda Y., Namikawa T., Ebihara S., Makino E., Suzuki T., Kuroiwa A., Matsuda Y., Namikawa T., Ebihara S., Suzuki T., Kuroiwa A., Matsuda Y., Namikawa T., Ebihara S., Kuroiwa A., Matsuda Y., Namikawa T., Ebihara S., Matsuda Y., Namikawa T., Ebihara S., Namikawa T., Ebihara S., Ebihara S. Molecular analysis of avian circadian clock genes. Brain Res. Mol. Brain Res. 2000;78:207–215. doi: 10.1016/s0169-328x(00)00091-7. [DOI] [PubMed] [Google Scholar]
- Young M.W., Kay S.A., Kay S.A. Time zones: A comparative genetics of circadian clocks. Nat. Rev. Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- Zavodska R., Sauman I., Sehnal F., Sauman I., Sehnal F., Sehnal F. Distribution of PER protein, pigment-dispersing hormone, prothoracicotropic hormone, and eclosion hormone in the cephalic nervous system of insects. J. Biol. Rhythm. 2003;18:106–122. doi: 10.1177/0748730403251711. [DOI] [PubMed] [Google Scholar]
- Zhu H., Conte F., Green C.B., Conte F., Green C.B., Green C.B. Nuclear localization and transcriptional repression are confined to separable domains in the circadian protein CRYPTOCHROME. Curr. Biol. 2003;13:1653–1658. doi: 10.1016/j.cub.2003.08.033. [DOI] [PubMed] [Google Scholar]
- Zhu H., Yuan Q., Froy O., Casselman A., Reppert S.M., Yuan Q., Froy O., Casselman A., Reppert S.M., Froy O., Casselman A., Reppert S.M., Casselman A., Reppert S.M., Reppert S.M. The two CRYs of the butterfly. Curr. Biol. 2005;15:R953–R954. doi: 10.1016/j.cub.2005.11.030. [DOI] [PubMed] [Google Scholar]
- Zylka M.J., Shearman L.P., Levine J.D., Jin X.W., Weaver D.R., Reppert S.M., Shearman L.P., Levine J.D., Jin X.W., Weaver D.R., Reppert S.M., Levine J.D., Jin X.W., Weaver D.R., Reppert S.M., Jin X.W., Weaver D.R., Reppert S.M., Weaver D.R., Reppert S.M., Reppert S.M. Molecular analysis of mammalian timeless. Neuron. 1998;21:1115–1122. doi: 10.1016/s0896-6273(00)80628-5. [DOI] [PubMed] [Google Scholar]