Abstract
The genomic architecture underlying the evolution of insect social behavior is largely a mystery. Eusociality, defined by overlapping generations, parental brood care, and reproductive division of labor, has most commonly evolved in the Hymenopteran insects, including the honey bee Apis mellifera. In this species, the Major Royal Jelly Protein (MRJP) family is required for all major aspects of eusocial behavior. Here, using data obtained from the A. mellifera genome sequencing project, we demonstrate that the MRJP family is encoded by nine genes arranged in an ∼60-kb tandem array. Furthermore, the MRJP protein family appears to have evolved from a single progenitor gene that encodes a member of the ancient Yellow protein family. Five genes encoding Yellow-family proteins flank the genomic region containing the genes encoding MRJPs. We describe the molecular evolution of these protein families. We then characterize developmental-stage-specific, sex-specific, and caste-specific expression patterns of the mrjp and yellow genes in the honey bee. We review empirical evidence concerning the functions of Yellow proteins in fruit flies and social ants, in order to shed light on the roles of both Yellow and MRJP proteins in A. mellifera. In total, the available evidence suggests that Yellows and MRJPs are multifunctional proteins with diverse, context-dependent physiological and developmental roles. However, many members of the Yellow/MRJP family act as facilitators of reproductive maturation. Finally, it appears that MRJP protein subfamily evolution from the Yellow protein family may have coincided with the evolution of honey bee eusociality.
Explaining the evolution of group social behavior in terms of natural selection on individuals is one of the great triumphs of evolutionary biology (Hamilton 1964). Social behavior, the transition from individual survival by competition to integrated societies with cooperation, has multiple evolutionary origins (Wilson 1975). Eusociality, defined by overlapping generations, parental brood care, and reproductive division of labor, has most commonly evolved in the Hymenopteran insects, including bees, ants, and wasps (Wilson 1975).
The honey bee Apis mellifera is an excellent model for understanding the evolutionary genomics of eusociality. The complex social behavior of this species, which includes hive building, feeding of immature (larval) bees, and locating food resources, influences its important agricultural role as a producer of honey, beeswax, and propolis, and as a pollination facilitator (Williams 2000). The recent publication of the complete A. mellifera genome sequence (The Honey Bee Genome Sequencing Consortium 2006) will accelerate the identification and characterization of genes that modulate social behaviors.
A. mellifera caste determination occurs when young worker bees in the hive (nurse bees) produce, secrete, and feed a substance called Royal Jelly (RJ) to developing larvae. RJ is a natural source of essential amino acids, lipids, vitamins, acetylcholine, and other nutrients (Colhoun and Smith 1960; Schmitzova et al. 1998). In the initial 3 d of development, all larvae are fed RJ; but thereafter only larvae designated by workers to become queens receive RJ. In its place, a mixture of honey, pollen, and water is fed to larvae selected to become workers. Since individuals from the previous generation feed the young of the next generation, determining their developmental fate (fertile queen vs. sterile worker), all major aspects of A. mellifera eusociality are considerably influenced by RJ.
Major Royal Jelly Proteins (MRJPs) constitute ∼90% of total RJ protein (Schmitzova et al. 1998; Sano et al. 2004; Santos et al. 2005; Scarselli et al. 2005). Eight A. mellifera loci encoding MRJPs (mrjp1–mrjp8) have been identified (Klaudiny et al. 1994a, b; Schmitzova et al. 1998; Albert and Klaudiny 2004), but little is known about the function of these genes or their protein products. We previously reported that the mrjp1 gene is expressed in the mushroom bodies of the honey bee brain, implicating this gene in behavior (Kucharski et al. 1998). This result suggests that the MRJPs can be multifunctional, performing a nutritional role as a component of RJ and executing additional roles in various tissues including the brain. However, the expression patterns of the mrjp genes across development and in different sexes and castes have not been well-characterized.
MRJPs share a common evolutionary origin with the Yellow protein family, consisting of representatives from insects and some bacteria (Kucharski et al. 1998; Albert et al. 1999a, b; Maleszka and Kucharski 2000; Drapeau 2001). Two genes encoding Yellow-like proteins were recently discovered in A. mellifera, implying that the honey bee genome also contains a Yellow protein family (Albert and Klaudiny 2004). The characterized functions of Yellow-related proteins in flies suggest possible functions of honey bee Yellow-related and MRJP proteins.
The Yellow protein of Drosophila melanogaster, the patriarch of the Yellow protein family, is multifunctional. Since the early days of genetics, its dual requirement for normal pigmentation and behavior has been studied in considerable detail (e.g., Sturtevant 1913, 1915; Lindsley and Zimm 1992; Wittkopp et al. 2002a, b; Drapeau et al. 2003, 2005), and yellow is now known to be controlled, at least in part, by the fly's sex determination pathway (Radovic et al. 2002; Drapeau et al. 2003). These dual functions of Yellow appear to be conserved within Drosophila (e.g., Tan 1946; Wittkopp et al. 2002b; da Silva et al. 2005; Gompel et al. 2005; W.J. Etges, pers. comm.; J. Jaenike, pers. comm.; B. Prud'homme, pers. comm.). Other Yellow-related proteins appear to have evolved diverse functions in D. melanogaster (Fujii and Amrein 2002; Han et al. 2002; Artero et al. 2003; Giot et al. 2003; Claycomb et al. 2004).
In this study, we describe the genomic organization and putative function of all genes encoding honey bee MRJP and Yellow proteins. We identify and describe two and eight novel mrjp and yellow genes, respectively. Then, we show that all 10 mrjp genes are tandemly arrayed in an ∼60-kb cluster, and are flanked by five yellow genes. An analysis of the intron/exon structures of these genes and the sequences of their protein products suggests that the mrjp gene array evolved via multiple, rapid duplications of a specific yellow precursor, yellow-e3. Transcriptional profiles of mrjp and yellow genes in the honey bee suggest that despite some similarities, the proteins encoded by these genes have evolved diverse physiological and developmental functions since their origin. However, the data presented here, in combination with the available information from flies and ants, suggest a common theme: a requirement of a noteworthy subset of MRJP and Yellow proteins in sex-specific reproductive maturation. Overall, our findings reveal the recent origin of a novel protein subfamily (MRJP) from an ancient protein family (Yellow), and show how this has affected both sex-specific individual behavior and complex group social behavior. The Yellow/MRJP protein family significantly influences the nature of honey bee social organization.
Results
The A. mellifera genome contains 10 mrjp and 10 yellow genes
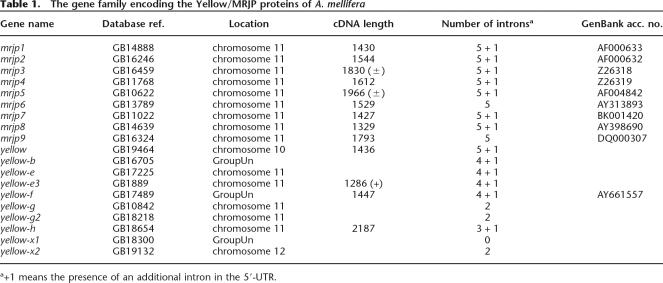
The availability of the A. mellifera genome assembly version 2.0 (The Honey Bee Genome Sequencing Consortium 2006) allowed us to perform BLAST and iterated PSI-BLAST searches to identify genes that encode proteins related to previously known MRJPs. This process recovered two groups of genes. The first group encodes nine highly related proteins with ∼60% identity to each other (BLASTP: E = 10−176 to 10−70). Most of these have been previously characterized at some level, and they were named mrjp1–mrjp9 (Table 1). One additional, novel member of this group is a pseudogene encoding an incomplete polypeptide, which would otherwise encode a tenth member of the MRJP protein group. We name this pseudogene mrjp-Ψ. Although we have not been able to detect a specific mrjp-Ψ transcript, it is conceivable that this pseudogene may be a segregating null allele in the population. The second group of genes with significant similarity to previously known mrjp genes consists of 10 more distantly related genes (BLASTP: E = 10−40 to 10−10) encoding the honey bee orthologs of D. melanogaster Yellow-related proteins (Table 1).
Table 1.
The gene family encoding the Yellow/MRJP proteins of A. mellifera
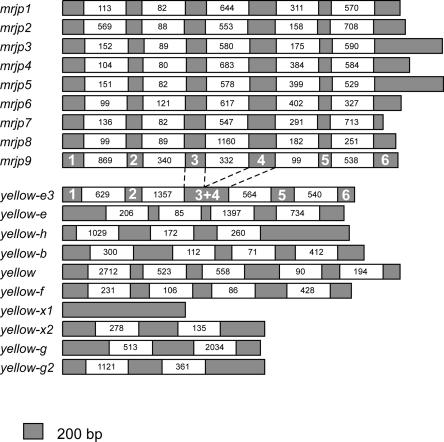
We characterized the intron/exon structure of the 19 protein-encoding mrjp and yellow genes from A. mellifera by sequencing cDNAs from mrjp and yellow genes, and comparing these sequences with the appropriate genomic DNA sequences. Combined with previously collected data on mrjp cDNA sequences (Klaudiny et al. 1994a, b; Kucharski et al. 1998; Albert and Klaudiny 2004), we were able to determine the similarities and differences in intron/exon structure among these 19 evolutionarily related genes (Fig. 1; Supplemental Table S1). There are two noteworthy features. First, the intron/exon structure of the mrjp genes is highly conserved, with each gene having five introns within their coding sequences, located in exactly the same positions. The intron sizes do vary somewhat among mrjp genes, but because they are very similar, and also have the same translational phase, this suggests that this group of genes recently evolved from a common progenitor. Second, the intron/exon structure of the yellow genes is, in general, not conserved within the yellow group or with the mrjp genes. This observation is consistent with the dissimilarity between mrjp and yellow loci in genomic DNA and predicted/actual protein sequences.
Figure 1.
Intron/exon structure of honey bee mrjp and yellow genes. Exons are depicted as black boxes, to scale. To emphasize structural similarities among mrjp genes, introns are depicted with empty boxes of uniform size, displaying actual sizes in base pairs. The intron/exon structure of the yellow-e3 gene is very similar to that of the mrjp genes.
yellow genes are common within arthropods; mrjp genes are rare
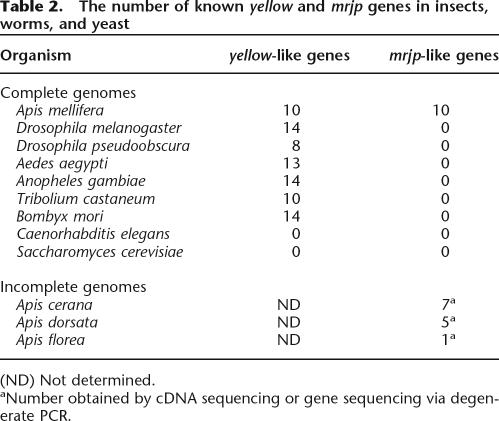
Because the nutritional MRJP component of RJ is essential for eusocial behaviors of the honey bee, we used all publicly available genomic DNA sequences and EST data from other animals to determine whether the distribution of mrjp genes was restricted to a subset of species, in particular, bees. We were able to draw two conclusions from the available data. Every arthropod for which sufficient sequence information exists possesses a “core” group of yellow-like genes (Table 2). We found yellow-like genes in a diverse group of arthropods, including flies, bees, mosquitoes, beetles, and moths, and this group ranges in size from eight to 14 members within a given species. Genes encoding Yellow-like proteins may be largely restricted to arthropods. We found no evidence that these genes exist in the complete genome sequences of the worm Caenorhabditis elegans and the yeast Saccharomyces cerevisiae.
Table 2.
The number of known yellow and mrjp genes in insects, worms, and yeast
Interestingly, however, we discovered that several bacterial species harbor either one or two yellow-like genes (see below). Such bacteria include the species Pseudomonas syringae, Deinococcus radiodurans, Magnetospirillum magnetotacticum, Solibacter usitatus, Burkholderia fungorum, and Geobacter metallireducens. This finding suggests that yellow-like genes are ancient, but have been lost from many lineages (see also Maleszka and Kucharski 2000). Horizontal gene transfer from insects to bacteria could also explain this distribution of yellow-like genes, but we believe such transfer is unlikely in practice.
In great contrast to the wide yellow-like gene distribution among various species, mrjp-like genes are thus far restricted to the honey bee genus Apis (Table 2). In addition to A. mellifera, the genomes of three related honey bee species—Apis cerana, Apis dorsata, and Apis florea—have genes encoding MRJPs (Albertova et al. 2005; Imjongjirak et al. 2005; Su et al. 2005). These data support the following two conclusions: The mrjp duplication events were recent, and the distribution of mrjp genes within animals is very restricted. It remains to be determined, however, precisely how limited this distribution is. A few recently initiated genome sequencing efforts, such as the Nasonia vitripennis project, will be pivotal for understanding the distribution of these genes in Hymenopteran species.
A tandem array of mrjp genes in the honey bee genome
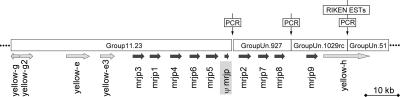
The A. mellifera genome sequence contains spatial information that allowed us to determine the proximity of mrjp and yellow genes to one another. We observed that five mrjp genes and the mrjp-Ψ gene clustered together on one large linkage group (Group 11.23, one of the 16 linkage groups of the honey bee karyotype, representing chromosome 11), and that three others clustered on another (Group Un.927). A third linkage group (Group Un.1029) contained the remaining mrjp gene. Based on the similarities in intron/exon structure and protein sequence among the mrjp genes (see above), it is likely that the mrjp genes evolved via a series of recent duplications. This predicts that they should be in close genomic proximity to each other. This, and additional evidence from cDNA/EST sequences and computer predictions, suggested that the three aforementioned groups and an additional group, Un.51, should comprise one linkage group. This larger linkage group could not have been assembled in the overall genome project because of insufficient trace coverage to date.
We successfully reconstructed an ∼833-kb interscaffold containing the large contig 11.23 and the three previously unoriented smaller contigs by sequencing overlapping DNA fragments obtained by PCR amplification. This “finishing” sequence resulted in a genomic landscape consisting of all 10 mrjp genes and additionally five yellow genes (see Fig. 3, below). All 10 mrjp genes are positioned in a tandem array roughly 60 kb in size. The four most proximal yellow genes—yellow-e, yellow-e3, yellow-g2, and yellow-h—are all oriented in the same direction as the mrjp array. Interestingly, yellow-g2 and the last yellow gene, yellow-g, are close, and in opposite orientation to each other, suggesting that their transcription is controlled via a bidirectional promoter. This finding—that the entire group of 10 mrjp genes is arranged in a relatively small tandem array—supports the theory that this important group of genes evolved from a common ancestral gene. The progenitor of the mrjp genes is most likely a yellow-like gene (see below).
Figure 3.
Genomic landscape of genes encoding MRJPs. Four scaffolds from the A. mellifera genome project were joined by PCR amplification and sequencing, forming an 883-kb interscaffold (see Results). All 10 mrjp genes are aligned in a tandem array ∼60 kb in size. This, in combination with sequence similarity, suggests a recent series of duplication events in the formation of the MRJP protein subfamily. The mrjp array is flanked by five yellow genes, all but one of which read in the same direction as the mrjp loci. The orientation of the yellow-g and yellow-g2 genes suggests the presence of a shared, bidirectional promoter.
In addition to the 15 tightly linked genes on chromosome 11, we localized the remaining five yellow-like genes in the honey bee genome. These genes, in contrast to the five that flank the mrjp array, are distributed in separate scaffolds, and are therefore located throughout the genome.
The mrjp array arose via initial duplication of yellow-e3
The most likely scenario by which the mrjp tandem array evolved is by the duplication of a yellow-like gene located near the extant mrjp cluster. Two specific genes, yellow-e3 and yellow-h, which directly flank the mrjp array, are the best candidates to be the progenitor of all mrjp genes. Because novel genes resulting from recent duplication events will retain the intron/exon structure of their template gene, we used information about the position of introns within the mrjp and yellow genes to determine whether any yellow-like gene could be classified as a strong candidate for the mrjp template. We found that the yellow-e3 intron/exon structure bears a remarkable similarity to that of the mrjp genes: Specifically, they share four out of five introns in the same relative positions, and in the same phase (Fig. 1; Supplemental Table S1). The yellow-h introns are not similar to those in mrjp genes (Fig. 1).
MRJPs are a monophyletic group within the Yellow/MRJP family
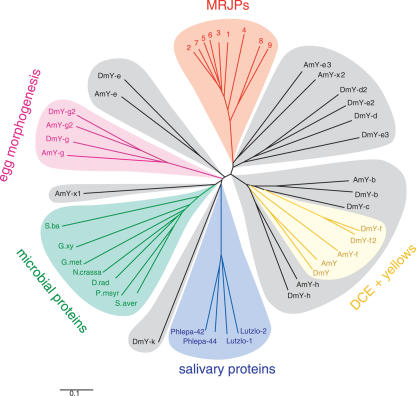
Using the protein sequences of mrjp and yellow genes from insect and microbial species, we constructed an unrooted phylogenetic tree using neighbor-joining (Fig. 2). We observed that MRJPs form a distinct monophyletic branch that appears to have evolved from an insect Yellow-like most recent common ancestor. Our phylogenetic tree suggests that this most recent common ancestor to the MRJPs is Yellow-e3 (Fig. 2). These protein data provide additional support for the hypothesis that the mrjp group of genes evolved recently via rapid and multiple duplication of yellow-e3.
Figure 2.
Phylogeny of the Yellow/MRJP protein family. An unrooted tree was constructed with aligned protein sequences from bees, flies, and bacteria using neighbor-joining (see Methods). The Yellow/MRJP protein family is composed of numerous color-coded subfamilies, each characterized by a group of closely homologous proteins. One of these subfamilies, the MRJPs, has thus far only been found in honey bees. Other subfamilies characterized by similar protein sequences are also known to have similar functions, and are so labeled. (AmY) Apis mellifera yellow; (DmY) Drosophila melanogaster yellow; (Phlepa) Phlebotomus papatasi; (Lutzlo) Lutzomyia longipalpis; (S.ba) Shewanella baltica; (S.aver) Streptomyces avermitilis; (G.met) Geobacter metallireducens; (G.xy) Gluconobacter oxydans; (N.crassa) Neurospora crassa; and (D.rad) Deinococcus radiodurans.
The precise branching pattern of the MRJPs reflects the recent gene duplication in this region of the genome. Specifically, MRJP pairs forming terminal clades in the MRJP branch of the tree, MRJP5 + 6, MRJP2 + 7, and MRJP8 + 9 (Fig. 2), are encoded by neighbor mrjp genes in the tandem array (Fig. 3).
More generally, this phylogenetic analysis demonstrates that for many of D. melanogaster Yellow proteins, close honey bee relatives exist. Because these are likely to represent honey bee orthologs, they were given the same names as their fruit fly counterparts (see Drapeau 2001). Additionally, we discovered two “orphan” Yellow-like proteins that have no obvious D. melanogaster counterparts (Fig. 2). For this reason, we have named the genes that encode them yellow-x1 and yellow-x2. We note in passing that yellow-x1 is the only intronless member of this gene family.
As noted before (Kucharski et al. 1998; Albert et al. 1999a), and as clearly demonstrated by our phylogenetic analysis, the Yellow-like and MRJP protein groups are evolutionarily related. Therefore, we name the total set of proteins the Yellow/MRJP protein family. We also name the proteins belonging only to the MRJP group the MRJP subfamily, reflecting the fact that these proteins are a distinct group within the larger, more ancient Yellow group.
The mrjp and yellow genes have diverse expression patterns
Some of the mrjp genes have a well-characterized nutritive role, and accordingly are highly expressed in worker bee hypopharyngeal glands (HPG) that secrete RJ. Because all nine MRJPs have been found in the RJ proteome (Schmitzova et al. 1998; Sano et al. 2004; Santos et al. 2005; Scarselli et al. 2005), the MRJP subfamily most likely evolved in association with this function. We tested the hypothesis that MRJPs evolved additional, diverse functions following gene duplication.
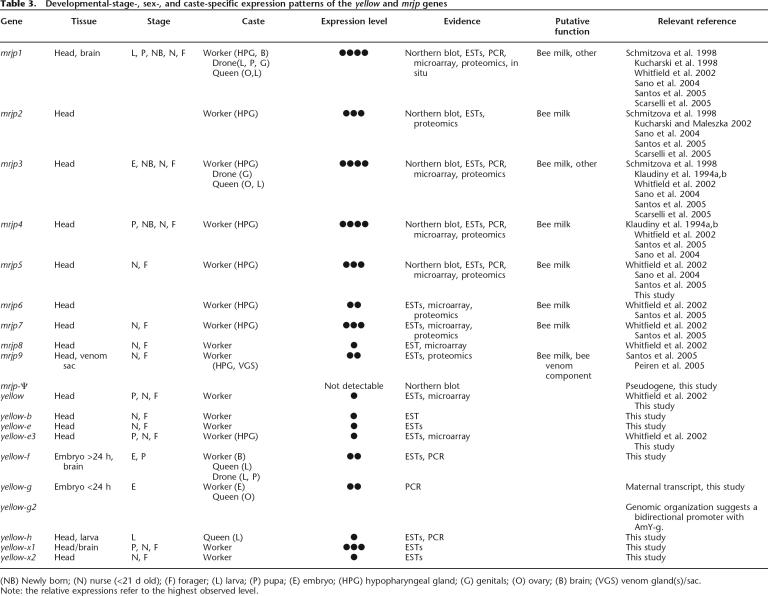
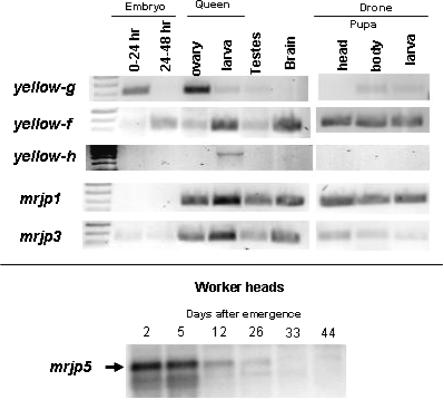
We compiled expression data for the genes encoding the honey bee Yellow/MRJP protein family (Table 3). To determine developmental-stage-, sex-, and caste-specific expression patterns, and to quantify expression levels, we used data from three different EST libraries, PCR amplifications, Northern blots, and microarrays, including expression data from the literature. As expected, we found no evidence for expression of the mrjp-Ψ pseudogene. Expression of a given mrjp gene manifests itself as a single Northern blot band, suggesting that no alternative splicing takes place in these genes. As previously noted, mrjp genes are highly expressed in the HPG, and therefore all show head expression. But, perhaps unexpectedly, expression of mrjp1 and mrjp3 was found not only in workers, but also in queens and drones (males) (Fig. 4; Table 3). The mrjp1 gene is additionally expressed in adult worker brains, suggesting a role in behavior. The mrjp9 gene is expressed in the adult venom gland, where its product performs a currently unknown function.
Table 3.
Developemntal-stage-, sex-, and caste-specific expression patterns of the yellow and mrjp genes
Figure 4.
Developmental-stage-, sex-, and caste-specific expression of a subset of yellow and mrjp genes. Shown are data from yellow-f, yellow-g, and yellow-h, and also mrjp-1, mrjp-3, and mrjp5. RT-PCR products were run on gels for visualization of gene expression. The primers are listed in Supplemental Table S3. The expression of mrjp5 was examined by Northern blotting using a PCR-amplified insert of a cDNA clone as a probe (nucleotides 15–1942 of NM_001011599). See Methods for more details.
We also found expression of mrjp genes throughout honey bee development. Various mrjp genes are expressed in embryos, larvae, and pupae, albeit at lower levels than seen in adults (Fig. 4; Table 3). We also observed several temporal changes in expression, exemplified by mrjp5. This gene has high expression in worker heads until Day 5 after emergence, low expression until Day 26, and undetectable expression thereafter (Fig. 4). Interestingly, the mrjp5 profile is distinct from that of mrjp1 (Kucharski et al. 1998), suggesting that mrjp genes are not coregulated despite being closely arranged in the genome. In total, these data suggest that the nine protein-coding mrjp genes not only have strong expression in the worker HPG for a nutritive function, but have also recently evolved distinct temporal, spatial, sexual, and caste-specific gene expression patterns, and presumably protein functions.
Like their mrjp cousins, the yellow-related honey bee genes also have diverse expression patterns (Table 3). Most are expressed in worker heads at various developmental stages, but more distinct expression patterns are evident. For example, yellow-g shows queen-specific ovary expression, and is also expressed in very young embryos, consistent with a maternal expression pattern (Fig. 4; Table 3). The yellow-h gene is predominantly expressed in developing queens at the larval stage, suggesting a specialized role for this gene's product (Fig. 4; Table 3). In contrast, yellow-f is activated at the late embryonic stage, where its expression continues throughout larval and pupal developmental stages (Fig. 4). The yellow-f gene is also highly expressed in the adult brain. The gland expression of yellow-e3 supports the notion that this gene is a progenitor of the Major Royal Jelly protein family.
A general conclusion that emerges from these analyses is that MRJPs perform context-dependent functions; the nutritional role of MRJP will have different phenotypic implications in the gland as compared to brain functions or developmental processes. The biological significance of a given MRJP product will depend on when and where its message is expressed. Furthermore, we conclude from these data that while a common underlying theme may exist, the honey bee Yellow-related proteins also are largely multifunctional, performing different functions in different tissues at various stages of development. The MRJP subfamily evolved from a Yellow progenitor for the purpose of a nutritive role in RJ. However, the MRJPs have recently evolved diverse roles throughout development and in adults.
The Yellow/MRJP family and sex-specific reproductive maturation
Because most of our knowledge about the functions of proteins in the Yellow/ MRJP family comes from genetic mutant and microarray studies in D. melanogaster, we can use this valuable information as a backdrop for predicting, and understanding, the roles that Yellow-like and MRJP factors perform in honey bees. A review of the literature inevitably leads to the conclusion that Yellow-like proteins have dissimilar expression patterns, mutant effects, protein binding partners, and by implication, functions (Supplemental Table S2; Martin and Ollo 1996; De Gregorio et al. 2001; Diagana et al. 2002; Stanyon et al. 2004). Despite this disparate data, however, a common theme emerges: The Yellow protein family of D. melanogaster mainly appears to be required for sex-specific reproductive maturation and/or development (Supplemental Table S4; Wayne and McIntyre 2002; Gilliland et al. 2005). The classic example of Yellow, required for pigmentation (often sex-specific, particularly in the abdomen and sex-combs) and male sexual behavior characteristics (obviously sexually dimorphic), is representative of this admittedly diverse group of proteins. These data are consistent with what we know about the functions of Yellow/MRJP family members in Apis. Specifically, the MRJPs have a nutritional function in RJ that modulates caste determination, and we have also shown sex-specific expression of numerous mrjp and yellow-like genes, sometimes in reproductive tissues (Fig. 4; Table 3). Thus, we propose a universal role of Yellow-like proteins in sex-specific reproductive maturity.
Discussion
Yellow protein family evolution through metazoans and bacteria
The observation that members of the Yellow/MRJP family are found not only in insects but also scattered through diverse bacterial species suggests that Yellow-like proteins are evolutionarily ancient. Hence, yellow-like genes have been lost from numerous lineages (e.g., enterogenic bacteria), and retained in those where they perform an important function. An alternative, and we believe less probable, hypothesis is that yellow-like genes in bacteria arrived via horizontal transfer from insects (Makarova et al. 2001). The fact that all bacterial Yellow-like proteins, along with the only non-insect eukaryotic N. crassa Yellow, are monophyletic (Fig. 2) militates against horizontal transfer as a viable explanation for the data. The hypothesis proposing multiple, independent transfer events predicts that the transferred bacterial Yellow protein would have a most recent common ancestor among the insects, and across seven proteins this was never the case (see also True et al. 2005 for another example).
Another common aspect of the Yellow/MRJP protein family is that most members have hydrophobic leader peptides at their N termini, suggesting that they are secreted factors. This may make sense in light of the fact that MRJPs have a nutritive function in the RJ that is secreted from the HPG. The D. melanogaster Yellow protein, the best-studied of all the members of the Yellow/MRJP protein family, is a secreted factor that may have hormonal function (Drapeau 2003; see below).
Origin of the MRJP subfamily from an ancient progenitor
Owing to their high level of similarity in intron/exon structure and protein sequence, it has been hypothesized that the mrjp genes arose via multiple nearly simultaneous duplications (Albert et al. 1999a). Gene duplications often give rise to such local clusters of genes (Wagner et al. 2003). An alternative model of evolution is suggested by a local genomic alignment (via the UCSC Genome server; http://genome.ucsc.edu/cgi-bin/hqGateway) between a small region of D. melanogaster chromosome 4, encoding a calcium-binding protein (CG31999), unrelated to Yellows, and MRJP6. In this scenario, some of the mjrp genes may have evolved via mutation accumulation in an ancient pre-existing genomic locus that encodes distinct proteins in modern insects. Indeed, a phylogenetic tree based on DNA alignments confirms that mrjp6 is weakly related to that region of chromosome 4 (Supplemental Fig. S1). However, the tree also shows that all mrjp genes are more closely related to yellow-e3, which is our candidate for the progenitor of the whole family. Furthermore, as shown in Supplemental Figure S2, the genomic alignment is not only interrupted by gaps but is largely confined to stretches of As and Ts with virtually no homologies in the more complex regions. It is therefore difficult to evaluate to what extent it reflects a meaningful evolutionary history.
Our data suggest that the entire mrjp cluster evolved from the yellow-e3 gene. First, yellow-e3 is one of two yellow-related genes directly flanking the mrjp gene cluster (Fig. 3). Second, of all the honey bee yellow-related genes, the intron/exon structure and protein sequence of yellow-e3 are the most similar to those of mrjp genes (Figs. 1, 2). Third, and perhaps most importantly, microarray expression data suggest that the functions of Yellow-e3 and the MRJPs have more in common than Yellow-e3 does to the rest of the Yellow proteins. The yellow-e3 transcript is highly expressed in the head and HPG, and generally follows a developmental pattern typical of mrjp genes. Overall, our findings reveal the recent origin of a novel protein subfamily (MRJP) from an ancient protein family (Yellow). This evolutionary event, combined with the use of MRJP-infused RJ as a nutritional supplement, greatly influenced honey bee behavior and hive social structure.
The function of MRJPs in honey bee Royal Jelly
The hypopharyngeal gland determines honey bee royalty within a structured society. Young worker bees (nurse bees) use the HPG to produce and secrete RJ, critical for the nutritional transformation of an immature female larva into a fertile queen bee. The absence of RJ leads immature females to a sterile worker bee fate. Hence, RJ not only creates colony-level social structure, but also modulates the sex-specific fertility of individuals. At the transition from young to old workers (i.e., nurses to foragers), bees cease the production of RJ as they take on their new role in the hive. Therefore, if MRJPs play an important role in RJ, we would expect that levels of gene expression and protein levels would decrease during and after this transition. This is, indeed, the case (Klaudiny et al. 1994a, b; Ohashi et al. 1997; Kucharski et al. 1998), and the effect has been shown to be reversible (Ohashi et al. 2000).
What is the molecular function of MRJPs in RJ? Recent evidence suggests that repetitive pentapeptide regions concentrated with nitrogen-rich amino acids may function as deposits of biologically accessible nitrogen (Albertova et al. 2005). Because nitrogen is an essential component of biogenic polymers such as nucleic acids and proteins, yet animals must obtain it from exogenous sources, it is often considered a limiting component of eukaryotic diets. Hence, high levels of nitrogen stored in MRJPs may be critical for rapidly growing young larvae and for the development of fertile queens. Other MRJP repeat regions may have additional functions. For example, the methionine-rich tripeptide repeat in the MRJP5 protein is ideal for the storage of sulfur, another limiting nutrient (S. Albert, unpubl.). Regarding other molecular functions of these proteins in the honey bee, a starting hypothesis consists of the assumption that MRJPs retain some of the ancestral roles that are associated with the Yellow proteins.
On the function of D. melanogaster Yellow
The yellow gene and the role(s) of its protein product have been characterized more extensively than any other in the Yellow family. As regards pigmentation (melanization), Yellow is required in cuticle cells for the presence of DOPA-melanin, and ectopically expressed Yellow is sufficient for the formation and deposition of melanin (Wittkopp et al. 2002a, b). Changes in the yellow locus underlie some changes in pigmentation patterns across evolutionary time (Wittkopp et al. 2002a, b; Gompel et al. 2005). Sex-specific abdominal pigmentation in D. melanogaster is Yellow-dependent and dependent on the Bric-a-brac protein, which integrates inputs from the homeotic and sex-determination pathways (Kopp et al. 2000). Yellow accumulation in some cells of the D. melanogaster male central nervous system (CNS) is dependent on sex-limited action of the Fruitless (FRU) protein (Radovic et al. 2002; Drapeau et al. 2003). The yellow gene may also be regulated by the Doublesex (DSX) protein, another transcription factor in the sex-determination cascade (Drapeau et al. 2005). These Yellow+ CNS cells appear to be relevant to male courtship behaviors (Drapeau et al. 2003).
At present, it is not known what the precise biochemical function of the Yellow protein is, but one theory can account for the dual roles of Yellow in pigmentation and behavior, and allows a relationship to be drawn between Yellow and the MRJPs. Yellow protein is secreted from cells because of a signal peptide that commands this action (Hannah 1953; Geyer et al. 1986; Kornezos and Chia 1992; Radovic et al. 2002; Wittkopp et al. 2002a; Drapeau et al. 2003). Yellow may then influence the properties of nearby cells via dopamine-like receptors and a hormone-like mechanism (Drapeau 2003). Activation of different signal transduction pathways in different cell types (cuticle vs. neural) would allow the activation of different downstream genes relevant to particular phenotypes (pigmentation vs. behavior) (Drapeau 2003). Like Yellow, the MRJPs have N-terminal signal peptides directing their secretion from cells, and post-secretion these signals are cleaved (Sano et al. 2004). Additionally, like Yellow (Geyer et al. 1986), MRJPs are glycoproteins with N-linked sugar chains (Kimura et al. 1996), and it is now known that glycosylation can regulate factors involved in extracellular signaling (e.g., Haines and Irvine 2003). Further study of the Yellow/MRJP protein family will lead to a greater understanding of these proteins as extracellular molecules. Because the requirement for Yellow in pigmentation and behavior is conserved within Drosophila (e.g., Drosophila virilis [Wittkopp et al. 2002b]; Drosophila willistoni [da Silva et al. 2005]), and because Yellow-g appears to have a similar function in flies and ants (Claycomb et al. 2004; Tian et al. 2004), we expect that A. mellifera Yellow-like proteins will share many similarities with those of D. melanogaster.
Genetic regulation of complex social behavior
The genes necessary and sufficient for complex social behaviors are largely a mystery. A few noteworthy exceptions include the Gp-9 gene, which encodes an odorant-binding protein in the fire ant Solenopsis invicta (Kreiger and Ross 2002); a locus named caste with unknown function in the red harvester ant Pogonomyrmex barbatus (Volny and Gordon 2002); and the gene encoding the V1a receptor for the neuropeptide vasopressin in voles (Lim et al. 2004).
Although A. mellifera has traditionally been the most intensively studied of all the eusocial insects (e.g., Wilson 1971), little progress has been made toward identifying genes underlying its elaborate social behavior. Honey bee “behavior genes” that encode members of conserved signaling pathways are unlikely to be unique to eusocial insects (see Robinson et al. 2005). In contrast, mrjp genes are not only crucial for A. mellifera colony-level behavior, but are thus far unique to the genus Apis. A comparison of mrjp loci to one of the honey bee behavior genes, the protein kinase foraging, which has been implicated in the transition of workers from hive work to outside-the-hive foraging (Ben-Shahar et al. 2002), reveals a key difference—foraging has a well-characterized homolog in D. melanogaster that controls larval foraging behavior (e.g., Osborne et al. 1997). While this fact makes foraging no less interesting, it follows that the foraging locus predates the speciation of flies and bees, and that the function of foraging in the control of bee social behavior came after the initial evolution of such behavior. Because mrjp genes have thus far only been found in Apis, we speculate that they may have specifically evolved to modulate eusociality via nutrition. Here, nature and nurture converge during the determination of complex behavior.
Methods
Sample collection
Foraging honey bee workers were captured near hive entrances and snap-frozen in liquid nitrogen (LN). To ensure that fully mature workers were harvested, only those that carried pollen or nectar were selected. They are estimated to be 20–35 d old. To obtain newly emerged honey bees, a single brood frame was removed from the hive and incubated at 32°C with 80% humidity. Then, individual insects were collected within 5 min after emergence and snap-frozen in LN. Pupae and larvae were harvested directly from a brood frame and snap-frozen in LN. Eggs and embryos were collected from a standard Jenter device.
Molecular biology
Tissue dissection, RNA extraction, hybridization, and data analyses were carried out as described previously (Kucharski and Maleszka 2002, 2003). For RT-PCR amplification, RNA samples were reverse-transcribed using Superscript II enzyme (Invitrogen) and an anchored d(T)20VN primer following the manufacturer's protocol (Kucharski and Maleszka 2005). Cloning and sequencing of cDNAs were done as in Albert and Klaudiny (2004). Isolated DNA was cloned into PCR.2.1 (Invitrogen) and sequenced by fluorescent (BigDye) sequencing.
Bioinformatics
Sequenced cDNAs were assembled and analyzed by University Wisconsin GCG 10.3 (Accelrys) and EMBOSS program packages. BLAST and iterated PSI-BLAST searches were run either locally or using the NCBI BLAST server (http://www.ncbi.nlm.nih.gov/BLAST). Sequence alignments were done using CLUSTALW with the Gonnet protein weight matrix. Aligned sequences were subjected to phylogenic analyses using the neighbor-joining method; gaps in the sequences were not considered. Branch stability was tested by bootstrap analysis (1000 repetitions). Dendrograms were drawn using the Phylodendron server (http://iubio.bio.indiana.edu/treeapp/treeprint-form.html).
Details of D. melanogaster gene sequences, structures, and functions were obtained through FlyBase (http://www.flybase.org/) and associated external databases (Fly GRID, http://biodata.mshri.on.ca/fly_grid/servlet/SearchPage; Berkeley Drosophila Genome Project, http://www.fruitfly.org/cgi-bin/ex/insitu.pl; Yale Drosophila Developmental Gene Expression Timecourse, http://genome.med.yale.edu/Lifecycle/; Gene Ontology, http://www.geneontology.org/), and additionally from the primary literature. The honey bee genomic resources are available via BeeBase at http://racerx00.tamu.edu/bee_resources.html.
Acknowledgments
We thank Bill Etges, John Jaenike, Benjamin Prud'homme, and Charlie Whitfield for discussing their unpublished data with us; Laurent Keller and John Wang for BLAST-ing the Solenopsis EST database; and Jack Werren for helpful discussions. We thank the coordinators of The Honey Bee Sequencing Project at the Baylor College of Medicine and all of the groups in the consortium who produced data and made them publicly available. This research was supported by U.S. National Institutes of Health Grant F32 GM72197 to M.D.D, Deutsche Forschung Gemeinschaft Grant RA642/11 to S.A., and by the ARC Special Centre for the Molecular Genetics and Development (to R.K. and R.M.).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5012006.
References
- Albert S., Klaudiny J., Klaudiny J. The MRJP/YELLOW protein family of Apis mellifera: Identification of new members in the EST library. J. Insect Physiol. 2004;50:51–59. doi: 10.1016/j.jinsphys.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Albert S., Bhattacharya D., Klaudiny J., Schmitzova J., Simuth J., Bhattacharya D., Klaudiny J., Schmitzova J., Simuth J., Klaudiny J., Schmitzova J., Simuth J., Schmitzova J., Simuth J., Simuth J. The family of major royal jelly proteins and its evolution. J. Mol. Evol. 1999a;49:290–297. doi: 10.1007/pl00006551. [DOI] [PubMed] [Google Scholar]
- Albert S., Klaudiny J., Simuth J., Klaudiny J., Simuth J., Simuth J. Molecular characterization of MRJP3, highly polymorphic protein of honeybee (Apis mellifera) royal jelly. Insect Biochem. Mol. Biol. 1999b;29:427–434. doi: 10.1016/s0965-1748(99)00019-3. [DOI] [PubMed] [Google Scholar]
- Albertova V., Su S., Brockmann A., Gadau J., Albert S., Su S., Brockmann A., Gadau J., Albert S., Brockmann A., Gadau J., Albert S., Gadau J., Albert S., Albert S. Organization and potential function of the mrjp3 locus in four honeybee species. J. Agric. Food Chem. 2005;53:8075–8081. doi: 10.1021/jf051417x. [DOI] [PubMed] [Google Scholar]
- Artero R., Furlong E.E., Beckett K., Scott M.P., Baylies M., Furlong E.E., Beckett K., Scott M.P., Baylies M., Beckett K., Scott M.P., Baylies M., Scott M.P., Baylies M., Baylies M. Notch and Ras signaling pathway effector genes expressed in fusion competent and founder cells during Drosophila myogenesis. Development. 2003;130:6257–6272. doi: 10.1242/dev.00843. [DOI] [PubMed] [Google Scholar]
- Ben-Shahar Y., Robichon A., Sokolowski M.B., Robinson G.E., Robichon A., Sokolowski M.B., Robinson G.E., Sokolowski M.B., Robinson G.E., Robinson G.E. Influence of gene action across different time scales on behavior. Science. 2002;296:741–744. doi: 10.1126/science.1069911. [DOI] [PubMed] [Google Scholar]
- Claycomb J.M., Benasutti M., Bosco G., Fenger D.D., Orr-Weaver T.L., Benasutti M., Bosco G., Fenger D.D., Orr-Weaver T.L., Bosco G., Fenger D.D., Orr-Weaver T.L., Fenger D.D., Orr-Weaver T.L., Orr-Weaver T.L. Gene amplification as a developmental strategy: Isolation of two developmental amplicons in Drosophila. Dev. Cell. 2004;6:145–155. doi: 10.1016/s1534-5807(03)00398-8. [DOI] [PubMed] [Google Scholar]
- Colhoun E.H., Smith M.V., Smith M.V. Neurohormonal properties of royal jelly. Nature. 1960;188:854–855. doi: 10.1038/188854a0. [DOI] [PubMed] [Google Scholar]
- da Silva L.B., Leite D.F., Valente V.L., Rohde C., Leite D.F., Valente V.L., Rohde C., Valente V.L., Rohde C., Rohde C. Mating activity of yellow and sepia Drosophila willistoni mutants. Behav. Proc. 2005;70:149–155. doi: 10.1016/j.beproc.2005.06.004. [DOI] [PubMed] [Google Scholar]
- De Gregorio E., Spellman P.T., Rubin G.M., Lemaitre B., Spellman P.T., Rubin G.M., Lemaitre B., Rubin G.M., Lemaitre B., Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc. Natl. Acad. Sci. 2001;98:12590–12595. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregorio E., Spellman P.T., Tzou P., Rubin G.M., Lemaitre B., Spellman P.T., Tzou P., Rubin G.M., Lemaitre B., Tzou P., Rubin G.M., Lemaitre B., Rubin G.M., Lemaitre B., Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002;21:2568–2579. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diagana T.T., Thomas U., Prokopenko S.N., Xiao B., Worley P.F., Thomas J.B., Thomas U., Prokopenko S.N., Xiao B., Worley P.F., Thomas J.B., Prokopenko S.N., Xiao B., Worley P.F., Thomas J.B., Xiao B., Worley P.F., Thomas J.B., Worley P.F., Thomas J.B., Thomas J.B. Mutation of Drosophila homer disrupts control of locomotor activity and behavioral plasticity. J. Neurosci. 2002;22:428–436. doi: 10.1523/JNEUROSCI.22-02-00428.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapeau M.D. The family of yellow-related Drosophila melanogaster proteins. Biochem. Biophys. Res. Commun. 2001;281:611–613. doi: 10.1006/bbrc.2001.4391. [DOI] [PubMed] [Google Scholar]
- Drapeau M.D. A novel hypothesis on the biochemical role of the Drosophila Yellow protein. Biochem. Biophys. Res. Commun. 2003;311:1–3. doi: 10.1016/j.bbrc.2003.09.106. [DOI] [PubMed] [Google Scholar]
- Drapeau M.D., Radovic A., Wittkopp P.J., Long A.D., Radovic A., Wittkopp P.J., Long A.D., Wittkopp P.J., Long A.D., Long A.D. A gene necessary for normal male courtship, yellow, acts downstream of fruitless in the Drosophila melanogaster larval brain. J. Neurobiol. 2003;55:53–72. doi: 10.1002/neu.10196. [DOI] [PubMed] [Google Scholar]
- Drapeau M.D., Cyran S.A., Viering M.M., Geyer P.K., Long A.D., Cyran S.A., Viering M.M., Geyer P.K., Long A.D., Viering M.M., Geyer P.K., Long A.D., Geyer P.K., Long A.D., Long A.D. A cis-regulatory sequence within the yellow locus of Drosophila melanogaster required for normal male mating success. Genetics. 2005;172:1009–1030. doi: 10.1534/genetics.105.045666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S., Amrein H., Amrein H. Genes expressed in the Drosophila head reveal a role for fat cells in sex-specific physiology. EMBO J. 2002;21:5353–5363. doi: 10.1093/emboj/cdf556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer P.K., Spana C., Corces V., Spana C., Corces V., Corces V. On the molecular mechanism of gypsy-induced mutations at the yellow locus of Drosophila melanogaster . EMBO J. 1986;5:2657–2662. doi: 10.1002/j.1460-2075.1986.tb04548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland W.D., Wayson S.M., Hawley R.S., Wayson S.M., Hawley R.S., Hawley R.S. The meiotic defects of mutants in the Drosophila mps1 gene reveal a critical role of Mps1 in the segregation of achiasmate homologs. Curr. Biol. 2005;15:672–677. doi: 10.1016/j.cub.2005.02.062. [DOI] [PubMed] [Google Scholar]
- Giot L., Bader J.S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Bader J.S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Ooi C.E., Godwin B., Vitols E., Godwin B., Vitols E., Vitols E., et al. A protein interaction map of Drosophila melanogaster . Scienc. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Gompel N., Prud'homme B., Wittkopp P.J., Kassner V.A., Carroll S.B., Prud'homme B., Wittkopp P.J., Kassner V.A., Carroll S.B., Wittkopp P.J., Kassner V.A., Carroll S.B., Kassner V.A., Carroll S.B., Carroll S.B. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- Haines N., Irvine K.D., Irvine K.D. Glycosylation regulates Notch signaling. Nat. Rev. Mol. Cell Biol. 2003;4:786–797. doi: 10.1038/nrm1228. [DOI] [PubMed] [Google Scholar]
- Hamilton W.D. The genetical evolution of social behaviour. I and II. J. Theor. Biol. 1964;7:1–52. doi: 10.1016/0022-5193(64)90038-4. [DOI] [PubMed] [Google Scholar]
- Han Q., Fang J., Ding H., Johnson J.K., Christensen B.M., Li J., Fang J., Ding H., Johnson J.K., Christensen B.M., Li J., Ding H., Johnson J.K., Christensen B.M., Li J., Johnson J.K., Christensen B.M., Li J., Christensen B.M., Li J., Li J. Identification of Drosophila melanogaster yellow-f and yellow-f2 proteins as dopachrome-conversion enzymes. Biochem. J. 2002;368:333–340. doi: 10.1042/BJ20020272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah A. Non-autonomy of yellow in gynandromorphs of Drosophila melanogaster . J. Exp. Zool. 1953;123:523–560. [Google Scholar]
- The Honey Bee Genome Sequencing Consortium. Insights into social insects from the genome of the honey bee Apis mellifera . Nature. 2006 (in press) [Google Scholar]
- Imjongjirak C., Klinbunga S., Sittipraneed S., Klinbunga S., Sittipraneed S., Sittipraneed S. Cloning, expression and genomic organization of genes encoding major royal jelly protein 1 and 2 of the honey bee (Apis cerana) J. Biochem. Mol. Biol. 2005;38:49–57. doi: 10.5483/bmbrep.2005.38.1.049. [DOI] [PubMed] [Google Scholar]
- Kimura Y., Kajiyama S., Kanaeda J., Izukawa T., Yonekura M., Kajiyama S., Kanaeda J., Izukawa T., Yonekura M., Kanaeda J., Izukawa T., Yonekura M., Izukawa T., Yonekura M., Yonekura M. N-linked sugar chain of 55-kDa royal jelly glycoprotein. Biosci. Biotechnol. Biochem. 1996;60:2099–2102. doi: 10.1271/bbb.60.2099. [DOI] [PubMed] [Google Scholar]
- Klaudiny J., Kulifajova J., Crailsheim K., Simuth J., Kulifajova J., Crailsheim K., Simuth J., Crailsheim K., Simuth J., Simuth J. New approach to the study of division of labour in the honeybee colony (Apis mellifera L.) Apidologie (Celle) 1994a;25:596–600. [Google Scholar]
- Klaudiny J., Hanes J., Kulifajova J., Albert S., Simuth J., Hanes J., Kulifajova J., Albert S., Simuth J., Kulifajova J., Albert S., Simuth J., Albert S., Simuth J., Simuth J. Molecular cloning of two cDNAs from the head of the nurse honey bee (Apis mellifera L.) for coding related proteins of royal jelly. J. Apic. Res. 1994b;33:105–111. [Google Scholar]
- Kopp A., Duncan I., Godt D., Carroll S.B., Duncan I., Godt D., Carroll S.B., Godt D., Carroll S.B., Carroll S.B. Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature. 2000;408:553–559. doi: 10.1038/35046017. [DOI] [PubMed] [Google Scholar]
- Kornezos A., Chia W., Chia W. Apical secretion and association of the Drosophila yellow gene product with developing larval cuticular structures during embryogenesis. Mol. Gen. Genet. 1992;235:397–405. doi: 10.1007/BF00279386. [DOI] [PubMed] [Google Scholar]
- Kreiger M.J., Ross K.G., Ross K.G. Identification of a major gene regulating complex social behavior. Science. 2002;295:328–332. doi: 10.1126/science.1065247. [DOI] [PubMed] [Google Scholar]
- Kucharski R., Maleszka R., Maleszka R. Evaluation of differential gene expression during behavioral development in the honeybee using microarrays and Northern blots. Genome Biol. 2002;3:research0007. doi: 10.1186/gb-2002-3-2-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharski R., Maleszka R., Maleszka R. Transcriptional profiling reveals multifunctional roles for transferrin in the honeybee (Apis mellifera) J. Insect Sci. 2003;3:27–36. doi: 10.1093/jis/3.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharski R., Maleszka R., Maleszka R. Microarray and rtPCR analyses of gene expression in the honey bee brain following caffeine treatment. J. Mol. Neurosci. 2005;27:269–276. doi: 10.1385/JMN:27:3:269. [DOI] [PubMed] [Google Scholar]
- Kucharski R., Maleszka R., Hayward D.C., Ball E.E., Maleszka R., Hayward D.C., Ball E.E., Hayward D.C., Ball E.E., Ball E.E. A royal jelly protein is expressed in a subset of Kenyon cells in the mushroom bodies of the honey bee brain. Naturwissenschafter. 1998;85:343–346. doi: 10.1007/s001140050512. [DOI] [PubMed] [Google Scholar]
- Lim M.M., Wang Z., Olazabal D.E., Ren X., Terwilliger E.F., Young L.J., Wang Z., Olazabal D.E., Ren X., Terwilliger E.F., Young L.J., Olazabal D.E., Ren X., Terwilliger E.F., Young L.J., Ren X., Terwilliger E.F., Young L.J., Terwilliger E.F., Young L.J., Young L.J. Enhanced partner preference in a promiscuous species by manipulating the expression of a single gene. Nature. 2004;429:754–757. doi: 10.1038/nature02539. [DOI] [PubMed] [Google Scholar]
- Lindsley D.L., Zimm G.G., Zimm G.G. Academic Press; San Diego, CA: 1992. The genome of Drosophila melanogaster. [Google Scholar]
- Makarova K.S., Aravind L., Wolf Y.I., Tatusov R.L., Minton K.W., Koonin E.V., Daly M.J., Aravind L., Wolf Y.I., Tatusov R.L., Minton K.W., Koonin E.V., Daly M.J., Wolf Y.I., Tatusov R.L., Minton K.W., Koonin E.V., Daly M.J., Tatusov R.L., Minton K.W., Koonin E.V., Daly M.J., Minton K.W., Koonin E.V., Daly M.J., Koonin E.V., Daly M.J., Daly M.J. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 2001;65:44–79. doi: 10.1128/MMBR.65.1.44-79.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleszka R., Kucharski R., Kucharski R. Analysis of Drosophila yellow-B cDNA reveals a new family of proteins related to the Royal Jelly proteins in the honeybee and to an orphan protein in an unusual bacterium Deinococcus radiodurans . Biochem. Biophys. Res. Commun. 2000;270:773–776. doi: 10.1006/bbrc.2000.2506. [DOI] [PubMed] [Google Scholar]
- Martin J.R., Ollo R., Ollo R. A new Drosophila Ca2+/calmodulin-dependent protein kinase (Caki) is localized in the central nervous system and implicated in walking speed. EMBO J. 1996;15:1865–1876. [PMC free article] [PubMed] [Google Scholar]
- Ohashi K., Natori S., Kubo T., Natori S., Kubo T., Kubo T. Change in the mode of gene expression of the hypopharyngeal gland cells with an age-dependent role change of the worker honeybee Apis mellifera L. Eur. J. Biochem. 1997;249:797–802. doi: 10.1111/j.1432-1033.1997.t01-1-00797.x. [DOI] [PubMed] [Google Scholar]
- Ohashi K., Sasaki M., Sasagawa H., Nakamura J., Natori S., Kubo T., Sasaki M., Sasagawa H., Nakamura J., Natori S., Kubo T., Sasagawa H., Nakamura J., Natori S., Kubo T., Nakamura J., Natori S., Kubo T., Natori S., Kubo T., Kubo T. Functional flexibility of the honey bee hypopharyngeal gland in a dequeened colony. Zool. Sci. 2000;17:1089–1094. doi: 10.2108/zsj.17.1089. [DOI] [PubMed] [Google Scholar]
- Osborne K.A., Robichon A., Burgess E., Butland S., Shaw R.A., Coulthard A., Pereira H.S., Greenspan R.J., Sokolowski M.B., Robichon A., Burgess E., Butland S., Shaw R.A., Coulthard A., Pereira H.S., Greenspan R.J., Sokolowski M.B., Burgess E., Butland S., Shaw R.A., Coulthard A., Pereira H.S., Greenspan R.J., Sokolowski M.B., Butland S., Shaw R.A., Coulthard A., Pereira H.S., Greenspan R.J., Sokolowski M.B., Shaw R.A., Coulthard A., Pereira H.S., Greenspan R.J., Sokolowski M.B., Coulthard A., Pereira H.S., Greenspan R.J., Sokolowski M.B., Pereira H.S., Greenspan R.J., Sokolowski M.B., Greenspan R.J., Sokolowski M.B., Sokolowski M.B. Natural behavior polymorphism due to a cGMP-dependent protein kinase of Drosophila. Science. 1997;277:834–836. doi: 10.1126/science.277.5327.834. [DOI] [PubMed] [Google Scholar]
- Peiren N., Vanrobaeys F., De Graaf D.C., Devreese B., Van Beeumen J., Jacobs F.J., Vanrobaeys F., De Graaf D.C., Devreese B., Van Beeumen J., Jacobs F.J., De Graaf D.C., Devreese B., Van Beeumen J., Jacobs F.J., Devreese B., Van Beeumen J., Jacobs F.J., Van Beeumen J., Jacobs F.J., Jacobs F.J. The protein composition of honeybee venom reconsidered by a proteomic approach. Biochim. Biophys. Act. 2005;1752:1–5. doi: 10.1016/j.bbapap.2005.07.017. [DOI] [PubMed] [Google Scholar]
- Radovic A., Wittkopp P.J., Long A.D., Drapeau M.D., Wittkopp P.J., Long A.D., Drapeau M.D., Long A.D., Drapeau M.D., Drapeau M.D. Immunohistochemical colocalization of Yellow and male-specific Fruitless in Drosophila melanogaster neuroblasts. Biochem. Biophys. Res. Commun. 2002;293:1262–1264. doi: 10.1016/S0006-291X(02)00366-2. [DOI] [PubMed] [Google Scholar]
- Robinson G.E., Grozinger C.M., Whitfield C.W., Grozinger C.M., Whitfield C.W., Whitfield C.W. Sociogenomics: Social life in molecular terms. Nat. Rev. Genet. 2005;6:257–260. doi: 10.1038/nrg1575. [DOI] [PubMed] [Google Scholar]
- Sano O., Kunikata T., Kohno K., Iwaki K., Ikeda M., Kurimoto M., Kunikata T., Kohno K., Iwaki K., Ikeda M., Kurimoto M., Kohno K., Iwaki K., Ikeda M., Kurimoto M., Iwaki K., Ikeda M., Kurimoto M., Ikeda M., Kurimoto M., Kurimoto M. Characterization of Royal Jelly Proteins in both Africanized and European Honeybees (Apis mellifera) by two-dimensional gel electrophoresis. J. Agric. Food Chem. 2004;52:15–20. doi: 10.1021/jf030340e. [DOI] [PubMed] [Google Scholar]
- Santos K.S., Dos Santos L.D., Mendes M.A., De Souza B.M., Malaspina O., Palma M.S., Dos Santos L.D., Mendes M.A., De Souza B.M., Malaspina O., Palma M.S., Mendes M.A., De Souza B.M., Malaspina O., Palma M.S., De Souza B.M., Malaspina O., Palma M.S., Malaspina O., Palma M.S., Palma M.S. Profiling the proteome complement of the secretion from hypopharyngeal gland of Africanized nurse-honeybees (Apis mellifera L.) Insect Biochem. Mol. Biol. 2005;35:85–91. doi: 10.1016/j.ibmb.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Scarselli R., Donadio E., Giuffrida M.G., Fortunato D., Conti A., Balestreri E., Felicioli R., Pinzauti M., Sabatini A.G., Felicioli A., Donadio E., Giuffrida M.G., Fortunato D., Conti A., Balestreri E., Felicioli R., Pinzauti M., Sabatini A.G., Felicioli A., Giuffrida M.G., Fortunato D., Conti A., Balestreri E., Felicioli R., Pinzauti M., Sabatini A.G., Felicioli A., Fortunato D., Conti A., Balestreri E., Felicioli R., Pinzauti M., Sabatini A.G., Felicioli A., Conti A., Balestreri E., Felicioli R., Pinzauti M., Sabatini A.G., Felicioli A., Balestreri E., Felicioli R., Pinzauti M., Sabatini A.G., Felicioli A., Felicioli R., Pinzauti M., Sabatini A.G., Felicioli A., Pinzauti M., Sabatini A.G., Felicioli A., Sabatini A.G., Felicioli A., Felicioli A. Towards royal jelly proteome. Proteomics. 2005;5:769–776. doi: 10.1002/pmic.200401149. [DOI] [PubMed] [Google Scholar]
- Schmitzova J., Klaudiny J., Albert S., Schroeder W., Schreckengost W., Hanes J., Judova J., Simuth J., Klaudiny J., Albert S., Schroeder W., Schreckengost W., Hanes J., Judova J., Simuth J., Albert S., Schroeder W., Schreckengost W., Hanes J., Judova J., Simuth J., Schroeder W., Schreckengost W., Hanes J., Judova J., Simuth J., Schreckengost W., Hanes J., Judova J., Simuth J., Hanes J., Judova J., Simuth J., Judova J., Simuth J., Simuth J. A family of major royal jelly proteins of the honeybee Apis mellifera L. Cell. Mol. Life Sci. 1998;54:1020–1030. doi: 10.1007/s000180050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanyon C.A., Liu G., Mangiola B.A., Patel N., Giot L., Kuang B., Zhang H., Zhong J., Finley R.L., Jr., Liu G., Mangiola B.A., Patel N., Giot L., Kuang B., Zhang H., Zhong J., Finley R.L., Jr., Mangiola B.A., Patel N., Giot L., Kuang B., Zhang H., Zhong J., Finley R.L., Jr., Patel N., Giot L., Kuang B., Zhang H., Zhong J., Finley R.L., Jr., Giot L., Kuang B., Zhang H., Zhong J., Finley R.L., Jr., Kuang B., Zhang H., Zhong J., Finley R.L., Jr., Zhang H., Zhong J., Finley R.L., Jr., Zhong J., Finley R.L., Jr., Finley R.L., Jr. A Drosophila protein-interaction map centered on cell-cycle regulators. Genome Biol. 2004;5:R96. doi: 10.1186/gb-2004-5-12-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A.H. The linear arrangement of six sex-linked factors in Drosophila, as shown by their mode of association. J. Exp. Zool. 1913;14:43–59. [Google Scholar]
- Sturtevant A.H. Experiments on sex recognition and the problem of sexual selection in Drosophila. J. Anim. Behav. 1915;5:351–366. [Google Scholar]
- Su S.K., Albert S., Chen S., Zhong B., Albert S., Chen S., Zhong B., Chen S., Zhong B., Zhong B. Molecular cloning and analysis of four cDNAs from the heads of Apis cerana cerana nurse honeybees coding for Major Royal Jelly Proteins. Apidologie (Celle) 2005;36:389–401. [Google Scholar]
- Tan C.C. Genetics of sexual isolation between Drosophila pseudoobscura and Drosophila persimilis . Genetics. 1946;31:558–573. doi: 10.1093/genetics/31.6.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H.S., Vinson S.B., Coates C.J., Vinson S.B., Coates C.J., Coates C.J. Differential gene expression between alate and dealate queens in the red imported fire ant, Solenopsis invicta Buren (Hymenoptera: Formicidae) Insect Biochem. Mol. Biol. 2004;34:937–949. doi: 10.1016/j.ibmb.2004.06.004. [DOI] [PubMed] [Google Scholar]
- True J.R., Yeh S.-D., Hovemann B.T., Kemme T., Meinertzhagen I.A., Edwards T.N., Liou S.-R., Han Q., Li J., Yeh S.-D., Hovemann B.T., Kemme T., Meinertzhagen I.A., Edwards T.N., Liou S.-R., Han Q., Li J., Hovemann B.T., Kemme T., Meinertzhagen I.A., Edwards T.N., Liou S.-R., Han Q., Li J., Kemme T., Meinertzhagen I.A., Edwards T.N., Liou S.-R., Han Q., Li J., Meinertzhagen I.A., Edwards T.N., Liou S.-R., Han Q., Li J., Edwards T.N., Liou S.-R., Han Q., Li J., Liou S.-R., Han Q., Li J., Han Q., Li J., Li J. Drosophila tan encodes a novel hydrolase required in pigmentation and vision. PLoS Genet. 2005;1:e63. doi: 10.1371/journal.pgen.0010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volny V.P., Gordon D.M., Gordon D.M. Genetic basis for queen–worker dimorphism in a social insect. Proc. Natl. Acad. Sci. 2002;99:6108–6111. doi: 10.1073/pnas.092066699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G.P., Amemiya C., Ruddle F., Amemiya C., Ruddle F., Ruddle F. Hox cluster duplications and the opportunity for evolutionary novelties. Proc. Natl. Acad. Sci. 2003;100:14603–14606. doi: 10.1073/pnas.2536656100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne M.L., McIntyre L.M., McIntyre L.M. Combining mapping and arraying: An approach to candidate gene identification. Proc. Natl. Acad. Sci. 2002;99:14903–14906. doi: 10.1073/pnas.222549199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C.W., Band M.R., Bonaldo M.F., Kumar C.G., Liu L., Pardinas J.R., Robertson H.M., Soares M.B., Robinson G.E., Band M.R., Bonaldo M.F., Kumar C.G., Liu L., Pardinas J.R., Robertson H.M., Soares M.B., Robinson G.E., Bonaldo M.F., Kumar C.G., Liu L., Pardinas J.R., Robertson H.M., Soares M.B., Robinson G.E., Kumar C.G., Liu L., Pardinas J.R., Robertson H.M., Soares M.B., Robinson G.E., Liu L., Pardinas J.R., Robertson H.M., Soares M.B., Robinson G.E., Pardinas J.R., Robertson H.M., Soares M.B., Robinson G.E., Robertson H.M., Soares M.B., Robinson G.E., Soares M.B., Robinson G.E., Robinson G.E. Annotated expressed sequence tags and cDNA microarrays for studies of brain and behavior in the honey bee. Genome Res. 2002;12:555–566. doi: 10.1101/gr.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D.L. A veterinary approach to the European honey bee (Apis mellifera) Vet. J. 2000;160:61–73. doi: 10.1053/tvjl.2000.0474. [DOI] [PubMed] [Google Scholar]
- Wilson E.O. The insect societies. Harvard University Press; Cambridge, MA: 1971. [Google Scholar]
- Wilson E.O. Sociobiology: The new synthesis. Harvard University Press; Cambridge, MA: 1975. [Google Scholar]
- Wittkopp P.J., True J.R., Carroll S.B., True J.R., Carroll S.B., Carroll S.B. Reciprocal functions of the Drosophila Yellow and Ebony proteins in the development and evolution of pigment patterns. Development. 2002a;129:1849–1858. doi: 10.1242/dev.129.8.1849. [DOI] [PubMed] [Google Scholar]
- Wittkopp P.J., Vaccaro K., Carroll S.B., Vaccaro K., Carroll S.B., Carroll S.B. Evolution of yellow gene regulation and pigmentation in Drosophila. Curr. Biol. 2002b;12:1547–1556. doi: 10.1016/s0960-9822(02)01113-2. [DOI] [PubMed] [Google Scholar]