Abstract
The honey bee genome sequence reveals a remarkable expansion of the insect odorant receptor (Or) family relative to the repertoires of the flies Drosophila melanogaster and Anopheles gambiae, which have 62 and 79 Ors respectively. A total of 170 Or genes were annotated in the bee, of which seven are pseudogenes. These constitute five bee-specific subfamilies in an insect Or family tree, one of which has expanded to a total of 157 genes encoding proteins with 15%–99% amino acid identity. Most of the Or genes are in tandem arrays, including one with 60 genes. This bee-specific expansion of the Or repertoire presumably underlies their remarkable olfactory abilities, including perception of several pheromone blends, kin recognition signals, and diverse floral odors. The number of Apis mellifera Ors is approximately equal to the number of glomeruli in the bee antennal lobe (160–170), consistent with a general one-receptor/one-neuron/one-glomerulus relationship. The bee genome encodes just 10 gustatory receptors (Grs) compared with the D. melanogaster and A. gambiae repertoires of 68 and 76 Grs, respectively. A lack of Gr gene family expansion primarily accounts for this difference. A nurturing hive environment and a mutualistic relationship with plants may explain the lack of Gr family expansion. The Or family is the most dramatic example of gene family expansion in the bee genome, and characterizing their caste- and sex-specific gene expression may provide clues to their specific roles in detection of pheromone, kin, and floral odors.
The olfactory abilities of the honey bee have long been admired and studied. Similar to most insects, they employ olfaction in many contexts, most prominently in locating flowers and in social communication. A foraging worker bee may encounter a bewildering number of flowers to choose from, and yet they can discriminate between them using subtle olfactory cues (for review, see Galizia and Menzel 2001), even sensing differences between plant genotypes (see Wright et al. 2002). After only a single visit to a reward, the odor associated with the reward can be correctly identified 90% of the time (Menzel and Erber 1978). Many of the studies of learning and memory in bees therefore employ olfaction as the sensory modality, and much is known about the organization of the antennal lobe and neural processing of olfactory information. In particular, the number of glomeruli in the antennal lobe has been estimated at ∼165 with variation from 160–170 (see Galizia and Menzel 2001). This relatively high number of glomeruli compared with 47 or 48 for Drosophila melanogaster (Laissue et al. 1999; Fishilevich and Vosshall 2005), ∼60 for Anopheles gambiae (R. Ignell, pers. comm.), and 60–66 for moths (Berg et al. 2002; Ai and Kanzaki 2004; Skiri et al. 2005) fits well with the enhanced olfactory range and discriminatory abilities of the honey bee. Drosophila olfactory receptor neurons (ORNs) generally express a single olfactory receptor protein (Or; in addition to the widely-expressed conserved DmOr83b), and each ORN expressing a particular Or converges on a single gomerulus in each antennal lobe, thereby allowing for a map of differential activation of suites of glomeruli to be interpreted in higher brain centers as a particular odor (see Vosshall et al. 2000; Rutzler and Zwiebel 2005; Hallem et al. 2006). There are, however, notable exceptions to the one-to-one relationship of receptors to glomeruli in Drosophila (Fishilevich and Vosshall 2005; Fishilevich et al. 2005; Goldman et al. 2005; Kreher et al. 2005). Nevertheless, the large number of glomeruli in the bee antennal lobe suggests there should be large numbers of odorant receptors encoded in the bee genome.
Insect odorant receptors have long been sought, including in honey bees. Insect Ors were first discovered in the genome sequence of D. melanogaster (Clyne et al. 1999; Gao and Chess 1999; Vosshall et al. 1999; Robertson et al. 2003) as a family of highly divergent proteins with seven-transmembrane (7TM) domains that are a novel lineage of G-protein–coupled receptors (GPCRs) and are expressed at low levels in the antennae and maxillary palps (Rutzler and Zwiebel 2005; Hallem et al. 2006). The extraordinary divergence between these proteins made it clear that Ors in other insects would primarily be discovered from genome sequences. Characterization of the Or family in the mosquito A. gambiae (Hill et al. 2002) revealed that these genes evolve so rapidly that with one exception, there are few simple orthologous relationships between the D. melanogaster and A. gambiae Ors. Given the additional evolutionary distance to honey bee, it seemed apparent that bee Ors would consist almost entirely of lineage-specific subfamilies. The sole exception to this rapid evolution is the unusually widely expressed DmOr83b protein (Vosshall et al. 1999), whose ortholog in A. gambiae (AgOr7) and other insects is highly conserved (Hill et al. 2002; Krieger et al. 2003; Pitts et al. 2004). DmOr83b and its orthologs have a unique function, acting as “chaperones” mediating the function of many of the other Ors in the dendritic membranes of ORNs (Larsson et al. 2004; Jones et al. 2005; Benton et al. 2006). A partial sequence of the honey bee ortholog of DmOr83b and AgOr7 has been described as AmOr2, and the protein sequences share at least 64% amino acid identity (Krieger et al. 2003). The only other insect Ors described are partial sets from the moths Heliothis virescens (Krieger et al. 2002, 2004) and Bombyx mori (Sakurai et al. 2004; Krieger et al. 2005), which, as expected, are moth-specific subfamilies, including the long-sought moth pheromone receptors expressed primarily in males and with ligand specificity for female pheromone components (Nakagawa et al. 2005).
Subsequent to the discovery of the insect Ors, a distantly related and even more highly divergent group of 7TM-domain candidate GPCRs were identified in the D. melanogaster genome. They were termed gustatory receptors (Grs) because most were expressed in gustatory organs such as the mouthparts (Clyne et al. 2000). Subsequent work led to the recognition of 68 Grs encoded by 60 genes through alternative splicing (Dunipace et al. 2001; Scott et al. 2001; Robertson et al. 2003). A similar-sized repertoire of 76 Grs encoded by 52 genes was identified in A. gambiae (Hill et al. 2002). These Ors and Grs are united in an insect chemoreceptor superfamily (Scott et al. 2001; Hill et al. 2002; Robertson et al. 2003) within which the Or family is simply a highly expanded lineage. In fact, some Grs are expressed in olfactory structures and presumably function as olfactory receptors (Scott et al. 2001). Nevertheless, the Grs retain this nominal name and are treated as a family (Robertson et al. 2003). Taste, primarily sucrose responsiveness, has been long studied in bees (see von Frisch 1927); however, little is known about its neurophysiologic bases and nothing is known about the receptor repertoire that mediates gustation in bees.
Here we describe the Or and Gr families in bees that together appear to constitute the entire insect chemoreceptor superfamily repertoire in this genome. Major expansion of the Or repertoire provides the expected diversity of protein sequences that might underlie the expanded olfactory capacities of bees. A very limited Gr repertoire was unexpected and indicates significant shifts in the gustatory mechanisms and capacities of bees.
Results
Or family
A major expansion of the Or family was discovered in the bee genome relative to the Or families in flies and mosquitoes. Altogether, 170 gene models were constructed (Supplemental material), with no evidence for alternative splicing to generate different receptor isoforms as is observed for both the Or and Gr families in D. melanogaster (Clyne et al. 2000; Robertson et al. 2003). Many of these gene models were concatenated in the initial automated annotations summarized in the Official set of 10,157 gene models (The Honey Bee Genome Sequencing Consortium 2006), and thus many lacked GB identification numbers. Our manually annotated versions have been employed for the Assembly 4 version of the bee genome annotation, and the 115 new GB numbers are summarized in Table S1. Seven of the Or gene models appear to be pseudogenes with various defects, including stop codons within an exon, missing start codons, unacceptable intron splice sites, small frame-shifting indels, large indels, and/or terminal truncations. Five gene models that were truncated by the ends of contigs were completed by manual assembly of contig extensions. Another 16 gene models remain truncated by the ends of contigs, but these were all presumed to be intact genes in the actual genome. While this number of apparent pseudogenes is a little higher than seen for D. melanogaster (Robertson et al. 2003) and A. gambiae (Hill et al. 2002), it is still far below the relatively high percentages of odorant receptor pseudogenes in mammalian genomes at 25%–100% (see Young and Trask 2002) and in nematode genomes at 25% (Robertson 1998, 2000; Robertson and Thomas 2005).
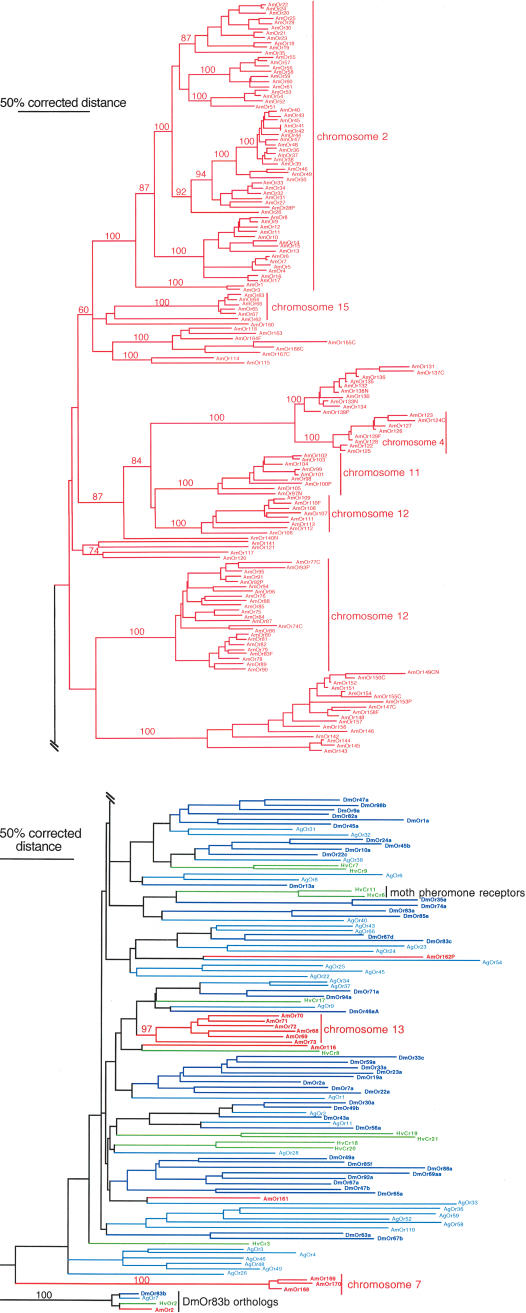
Phylogenetic analysis of the Ors encoded by these 170 genes and pseudogenes (reconstructed and translated with judicious introduction of frame-shifts and ignoring stop codons in alignable exons), along with a reduced set of the Drosophila and Anopheles Ors and the few available H. virescens moth Ors, reveals that they comprise just five lineages or subfamilies (Fig. 1). Four small subfamilies consist of one, one, three, and six proteins, and there is only weak indication of relationships for the single- and six-member subfamilies to Or lineages in the flies. The three-member subfamily forms a basal lineage in the Or tree; however, there is no bootstrap support for this position. This highly divergent cluster of three genes on chromosome 7 was only identified in PSI-BLASTP searches, all others being identified by using TBLASTN searches.
Figure 1.
Phylogenetic relationships of the 170 AmOrs and fly and moth Ors. This corrected distance tree was rooted by declaring the DmOr83b protein and its orthologs in the other species (AgOr7, HvOr2, and AmOr2) as the outgroup, based on the position of DmOr83 at the base of the Or family in phylogenetic analyses of the entire superfamily in Drosophila (Robertson et al. 2003). Relatively closely related fly and moth Ors were removed to facilitate phylogenetic analysis and presentation, hence many of the lineage-specific subfamily expansions in these insects are not obvious (see Hill et al. 2002). Clusters of tandem arrays of AmOrs on particular chromosomes are indicated by vertical lines on the right. Numbers above branches are the percentage of 1000 neighbor-joining bootstrap replicates containing that branch and are shown only for major lineages including bee Ors. Symbols after the AmOr numbers: F indicates the missing genomic sequence was manually assembled to complete the gene model; N, the N terminus of the gene model is missing in a gap or is unidentifiable; C, the C terminus of the gene model is missing in a gap or is unidentifiable; and P, pseudogene with one or more in-frame stop codons, frame-shifting indels, or unacceptable intron splice sites.
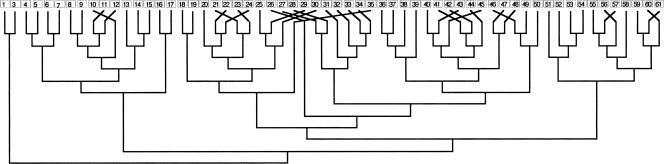
The fifth subfamily is extraordinarily expanded to 157 Ors (Fig. 1), more than twice the entire Or repertoire of D. melanogaster. Most of these AmOrs are encoded by clusters of tandemly arrayed genes, presumably reflecting their origin by unequal crossing-over of neighboring duplicated genes. In one of the largest perfect tandem gene arrays known, 60 AmOrs are encoded on chromosome 2. The evolutionary relationships of these 60 genes, based on their encoded protein sequences, correspond remarkably well to their locations in the tandem array, supporting the notion of origins by unequal crossing-over of neighboring genes (Fig. 2). Some of these 60 AmOrs are rather divergent (down to 20% amino acid identity)—hence this array must have existed for a long time—while others are almost identical, reflecting recent duplication events (or possibly ongoing gene-conversion events).
Figure 2.
Molecular evolution of a 60 AmOr gene array on chromosome 2. The 60 genes are in a perfect uninterrupted tandem array and were numbered from left to right (AmOr2 is the previously named ortholog of the conserved DmOr83b protein and is on chromosome 1). The phylogenetic relationships of these 60 genes are shown in the corrected-distance cladogram of their encoded Ors below the array (AmOr1 and 3 were declared the outgroup based on their location in Fig. 1; branch lengths in this cladogram are meaningless). The few incompatibilities between the tree and the gene location are indicated by diagonal lines.
Gr family
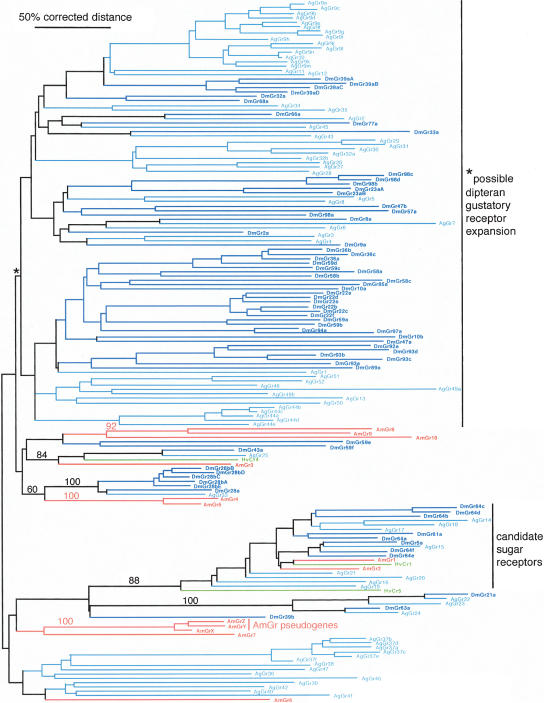
Compared with the two flies, the bee genome encodes very few Grs. Just 10 intact Gr genes, each apparently encoding a single Gr, are present (Supplemental material), compared with 68 Grs encoded by 60 genes in D. melanogaster (Robertson et al. 2003) and the A. gambiae repertoire of 76 Grs encoded by 52 genes (Hill et al. 2002). These 10 bee Grs represent just seven lineages in a phylogenetic analysis of all the fly Grs and the few available from H. virescens (Fig. 3). Two of these receptors, AmGr1 and AmGr2, cluster confidently, along with HvCr1 and HvCr5, with the eight candidate sugar receptors in each of the flies, based on the role of DmGr5a as a trehalose receptor (see Chyb et al. 2003). AmGr3 is orthologous to HvCr4, AgGr25, and DmGr43a, and the fact that each species retains just one member of this lineage suggests that this Gr might serve a particular conserved role. There is weak bootstrap support for the AmGr4 and AmGr5 lineage being orthologous to AgGr33 and the DmGr28a/b complex, which are among the most highly conserved Grs within Drosophila flies (H.M. Robertson, unpubl.), so this Gr lineage might also perform a conserved function. The remaining four bee Gr lineages of one or two Grs each (AmGr6–10) have no apparent orthology to any of the fly lineages.
Figure 3.
Phylogenetic relationships of the 10 AmGrs and three pseudogenes to fly and moth Grs. This corrected distance tree was rooted at the midpoint in the absence of a suitable outgroup. For other details, see Figure 1.
We were unable to identify bee orthologs for the highly conserved lineage of the DmGr21a, DmGr63a, and AgGr22–24 proteins. DmGr21a has been implicated in perception of carbondioxide (Suh et al. 2004), yet bees are able to perceive carbon dioxide (von Lacher 1964; Stange and Diesendorf 1973), so either DmGr21a and its mosquito ortholog AgGr22 are not true carbon dioxide receptors or bees use another receptor to detect this gas.
The overall phylogenetic relationships in Figure 3 suggest that the low number of Grs in the bee genome is more a result of lack of expansion of existing lineages or subfamilies, rather than actual loss of Gr lineages in bee (except the conserved DmGr21a/63a lineage above). Several Gr lineages have instead massively expanded in the fly genomes. For example, the eight candidate sugar receptors in flies appear to result from multiple intradipteran gene duplications from a single ancestor inherited from their common ancestor with the moth lineage (L. Kent and H.M. Robertson, unpubl.). This relationship is not clear in Figure 3 because analysis of this divergent lineage in the context of the entire Gr family does not perfectly resolve these relationships. Other fly-specific expansions include the DmGr28ba–e and AgGr37a–f expansions within alternatively spliced loci. The tree in Figure 3 also suggests that the vast majority of the DmGrs and AgGrs are a dipteran-specific expansion (top two-thirds of the tree indicated by an asterisk). Although there is no bootstrap support for the short branch that leads to this expansion, similar to all the other branches along the backbone, reflecting the extreme divergence of these lineages, it is not an unreasonable possibility given the absence of any AmGr proteins in this entire cluster.
The bee genome also contains ∼50 highly degraded copies of Gr pseudogenes; however, these are rather unusual. We have been able to build only three of these in full-length versions. They are extraordinarily long compared with the intact Gr genes, being ∼10 kb with several long introns each. The remaining copies are all truncated by the ends of contigs; indeed, all of these Gr pseudogenes are in relatively short GroupUn scaffolds (mostly single contigs) that are not currently mapped to any of the 16 bee chromosomes (the single exception involves a 7-kb contig on the end of 73-kb Group6.36; however, this is a misassembly—there are no mate pairs linking it to the rest of Group6.36). Despite being in poorly assembled regions of the genome, the contigs containing these pseudogenes are of high sequence quality, so their pseudogene status is real. For example, the misassembled example in Group6.36 consists of two N-terminal exons encoding ± 190 amino acids; however, this pseudogene fragment has no start codon, a mutated intron splice acceptor site, two in-frame stop codons in these long exons, and a single-base frame-shifting deletion in one exon. The best amino acid reconstructions of the three full-length versions cluster together in the phylogenetic analysis as a highly divergent lineage (AmGrX, Y, and Z in Fig. 3). All the others have DNA sequence similarity to these three and hence reflect an expanded and fairly old set of pseudogenes. These pseudogenes might represent the degraded remnants of a once highly expanded lineage of Grs in the bee genome, but a possible alternative explanation is that at some point a Gr pseudogene became embedded in an unusual portion of the bee genome that is relatively AT-enriched and then became duplicated in the genome. Because the sequencing and assembly of the AT-rich portions of the bee genome were somewhat compromised (The Honey Bee Genome Sequencing Consortium 2006), it remains unclear where these pseudogenes now reside in the genome, and they are probably in the pericentromeric regions.
Expression of AmOrs and AmGrs in olfactory and gustatory organs
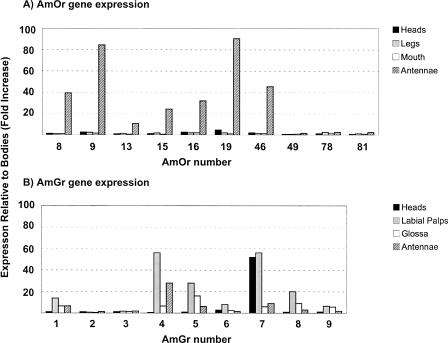
Tissue-specific expression patterns of 10 randomly selected Or genes from the honey bee–specific expansion (Fig. 1), and nine Gr genes, were determined by real-time quantitative PCR. The amplification efficiency of each primer set was validated; standard curves (10 × serial dilutions) yielded regression lines with r2 values >0.95 and slopes ranging from 3.1–3.5 (a slope of 3.3 indicates 100% amplification efficiency). In addition, each primer set produced a single amplicon as judged by the presence of single peaks in the dissociation curve. Expression of seven of 10 Or genes was significantly enriched in worker bee antennae, compared with worker legs, proboscis, head (proboscis and antennae removed), and body (10–100 times greater) (Fig. 4), confirming their likely roles as odorant receptors. Expression levels of all 10 Or genes in tissues other then the antennae were uniformly low. Expression of seven of nine Gr genes was enriched in gustatory organs such as the labial palps and the glossa, each of which has ∼50–70 taste sensilla (10–60 times greater relative to bodies and heads) (Fig. 4; Goodman 2003). Interestingly, AmGr7 was expressed at high levels in the heads (labial palps and glossa removed), so it may be expressed in internal taste organs of the gustatory tract.
Figure 4.
(A) Or and (B) Gr gene expression in honey bee olfactory and gustatory organs measured as fold increases relative to levels in the body as a reference tissue using the 2−ΔΔCT equation (Livak and Schittgen 2001). Gene expression levels were determined by real-time quantitative PCR and normalized to levels of A. mellifera ribosomal protein S8 (GenBank accession NM001011604).
Discussion
Our most remarkable finding is that the bee genome encodes ∼163 intact and presumably functional Ors and just seven pseudogenes. Some of these pseudogenes might be “flatliners,” genes that are intact in other strains (Stewart et al. 2005), something shown already for some D. melanogaster Ors and Grs (Robertson et al. 2003) and A. gambiae Grs (Hill et al. 2002). The similarity of this number of Ors and the number of glomeruli in the bee antennal lobe, which ranges from 160–165 in different individual bees (Galizia and Menzel 2001), is consistent with an approximate one-to-one relationship that may result from an ORN expressing just one Or protein, and its axon projecting to a single glomerulus per antennal lobe.
This expansion of the bee Or family presumably has provided the diversity of odorant receptors that allow bees to recognize diverse floral odors as well as employ complex pheromone blends to coordinate caste-specific tasks within the social colony. The emergence of angiosperm plants and the earliest bees is thought to have coincided ∼100 Myr ago (Michener 1974; Raven and Axelrod 1974). Since that time, the evolution and divergence of bees and angiosperm plants have been closely linked, with the angiosperms evolving new flower characteristics to attract bees (such as odor, color, and food rewards) and with the bees evolving mechanisms to collect and transfer pollen. The expansion of the bee Or family may reflect this coevolution between bees and angiosperms. Determining the expression patterns of these Ors in the various bee castes and life stages should provide indications of their involvement in these various roles and eventually lead to the determination of ligand specificities (e.g., Hallem et al. 2004). For example, the sole task of male drone bees is to mate with virgin queen bees, and their antennae are specifically designed for the detection of queen pheromone. Drone antennae have ∼16,000 placoid sensilla, many of which are sensitive to different components of the queen pheromone, compared with 2700 placoid sensilla on worker antennae (see Brockmann and Brückner 2001). Therefore, we predict that the expression of Ors involved in the detection of the queen pheromone blend should be higher in drone compared with worker antennae.
The lack of expansion of the bee Gr lineages stands in stark contrast to the multiple expansions of different Gr subfamilies in both the Drosophila fly and Anopheles mosquito genomes. It also contrasts with the massive expansion of the bee Ors. We speculate that the coevolution of bees and angiosperm plants, as well as the social nature of bees, may account for this. First, unlike many insects, bees have mutualistic relationships with plants, which provide nectar in return for pollination services. Nectar and pollen alone provide for all of the nutritional requirements of a hive, and 98% of the nectar is composed of sugars and water. Since plants have evolved mechanisms to attract and reward bees, bees have not required the ability to detect and discriminate between the numerous plant secondary chemicals and toxins usually deployed in the chemical ecological arms races between most plants and many insect herbivores. Second, bee larvae are sequestered in cells in the hive and are provisioned by adult nurse bees; hence, they have little need for gustatory receptors to locate and recognize food. Third, bees commonly use their antennae to touch objects, including kin, in a form of contact chemoreception. Therefore, some Ors on the antennae may function in a manner similar to that of contact gustatory receptors and effectively replaced the need for actual Grs.
Our definition of the Or and Gr repertoires of the honey bee has enabled recognition of several lineages of Grs that are unusually conserved and might serve particular functions in diverse insects, as well as massive expansion of the Or family. Determining the expression patterns of these bee Ors and Grs should provide indications of their possible roles and will be the first step toward determining their ligand specificity and defining their roles in bee ecology and social organization.
Methods
Bioinformatics
Assembly 2 of the Apis mellifera genome sequence at the Baylor College of Medicine Human Genome Sequencing Center (R. Gibbs, pers. comm.; http://www.hgsc.bcm.tmc.edu/projects/honeybee/) and BeeBase (http://racerx00.tamu.edu/bee_resources.html) was searched with all available insect Or and Gr proteins as queries using TBLASTN (Altschul et al. 1997). Bee Ors and Grs were in turn employed in searches to find more genes in an iterative process. To find highly divergent, already annotated members of these families, multiple PSI-BLASTP searches were initiated on BeeBase, combining the GenBank non-redundant protein database with the Official and Ab-initio databases of 10,157 and 15,550 proteins, respectively, annotated in the bee genome (The Honey Bee Genome Sequencing Consortium 2006). The genes were reconstructed manually in the PAUP* v4 editor (Swofford 2001) using the expected exon/intron boundaries from fly Ors and Grs (Robertson et al. 2003) as guides, and the Neural Network Splice Predictor program at the Berkeley Drosophila Genome Project was used to locate probable intron splice sites (http://www.fruitfly.org/seq_tools/splice.html). In addition, protein alignments were used to identify irregularities and refine the gene structures. Gene models were checked on the final draft Assembly 4 (see Table S1; The Honey Bee Genome Sequencing Consortium 2006).
Proteins were aligned by using CLUSTAL X (Jeanmougin et al. 1998). Relaxing the pairwise alignment gap and extension penalties for the Gr family by 20%, to eight and 0.08, respectively, yielded the best alignment of the 7TM domains, particularly the relatively conserved TM7 region. Amino acid distances calculated between each pair of proteins were corrected for multiple amino acid changes by using the maximum likelihood model, the BLOSUM62 amino acid exchange matrix, and uniform rates based on the actual sequences in TREE-PUZZLE v5.0 (Schmidt et al. 2002). Phylogenetic trees were constructed by using neighbor-joining followed by a heuristic search for better trees using tree-bisection-reconnection branch-swapping in PAUP* v4.0b10 (Swofford 2001). Bootstrap analysis was performed by using 1000 neighbor-joining replications with uncorrected distances.
Quantitative real-time PCR
Honey bees were collected in the summer months of 2005 from colonies maintained by the University of Illinois Bee Research Facility (Urbana, IL). The antennae, proboscis, legs (mixture of front, middle, and hind legs), heads (without antennae, proboscis, and mandibles), and bodies (without head and legs) were dissected and stored at −80°C. Total RNA was isolated by using an RNeasy Mini Kit (Qiagen), quantified by absorption at a wavelength of 260 nm, and the quality was assessed on a 1% agarose gel. Genomic DNA was digested by using the Ambion DNA-Free kit, and first-strand cDNA (20 μL per reaction) was synthesized from 5 μg of total RNA using an oligo dT primer and the Super-Script III First-Strand Synthesis System for RT-PCR kit (Invitrogen). PCR primers were designed by using ABI Primer Express 2.0 software (Applied Biosystems) set to select for an optimal primer annealing temperature of 59°C (58°C–60°C range), amplicon sizes of 50–150 bp, a 3′GC clamp = 0, and a minimum and maximum GC content of 30% and 80%, respectively. In general, primers were designed by using coding sequence close to the 3′ end of the gene, and where possible, primers spanned an intron. Real-time quantitative PCR was performed by using an ABI Prism 7900HT Sequence Detection System using the SYBR Green PCR Master Mix (Applied Biosystems). The program began with a single cycle for 2 min at 50°C, followed by a single cycle for 10 min at 95°C and 40 cycles of 15 sec at 95°C and 60 sec at 60°C. Afterward, the PCR products were heated to 95°C for 15 sec, cooled to 60°C for 15 sec, and heated to 95°C for 15 sec to measure the dissociation curves. The efficiency of each primer set was first validated by constructing a standard curve, including a no-template control and six 10× serial dilutions of first cDNA (1× dose = 0.33 μL of first cDNA). For each serial dilution, the CT value was plotted against the log(template dilution) and the slope and r2 value of each regression line calculated.
Expression of each Or gene in antennae, proboscis, legs, heads, and bodies was assessed in triplicate by using a template dose equal to ∼0.4 μL of first cDNA per well, along with a no-template control. Dissociation curves were used to assess the purity of the PCR reactions. Or transcript levels were normalized by using the transcript levels of A. mellifera ribosomal protein S8 (GenBank accession NM001011604). Expression levels were calculated relative to the body using the 2−ΔΔCT equation (Livak and Schittgen 2001).
Acknowledgments
We thank Kim Walden for assistance with annotation of the AmOrs, Harland Patch for early work on AmOr2, Ryszard Maleszka for early RT/PCR data on AmGr1 and 2, Chris Elsik and Natalia Milshina at BeeBase for providing new GB numbers for novel and split gene models, the Baylor College of Medicine Human Genome Sequencing Center for public access to the bee genome assemblies before publication, and two anonymous reviewers for their comments on the manuscript. This work was funded by NIH grant AI56081.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5057506.
References
- Ai H., Kanzaki R., Kanzaki R. Modular organization of the silkmoth antennal lobe macroglomerular complex revealed byvoltage-sensitive dye imaging. J. Exp. Biol. 2004;207:633–644. doi: 10.1242/jeb.00784. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang Z., Miller W., Lipman D.J., Miller W., Lipman D.J., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R., Sachse S., Michnick S.W., Vosshall L.B., Sachse S., Michnick S.W., Vosshall L.B., Michnick S.W., Vosshall L.B., Vosshall L.B. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg B.G., Galizia C.G., Brandt R., Mustaparta H., Galizia C.G., Brandt R., Mustaparta H., Brandt R., Mustaparta H., Mustaparta H. Digital atlases of the antennal lobe in two species of tobacco budworm moths, the Oriental Helicoverpa assulta (male) and the American Heliothis virescens (male and female) J. Comp. Neurol. 2002;446:123–134. doi: 10.1002/cne.10180. [DOI] [PubMed] [Google Scholar]
- Brockmann A., Brückner D., Brückner D. Structural differences in the drone olfactory system of two phylogenetically distant Apis species, A. florea and A. mellifera. Naturwissenschaften. 2001;88:78–81. doi: 10.1007/s001140000199. [DOI] [PubMed] [Google Scholar]
- Chyb S., Dahanukar A., Wickens A., Carlson J.R., Dahanukar A., Wickens A., Carlson J.R., Wickens A., Carlson J.R., Carlson J.R. Drosophila Gr5a encodes a taste receptor tuned to trehalose. Proc. Natl. Acad. Sci. 2003;100:14526–14530. doi: 10.1073/pnas.2135339100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne P.J., Warr C.G., Freeman M.R., Lessing D., Kim J., Carlson J.R., Warr C.G., Freeman M.R., Lessing D., Kim J., Carlson J.R., Freeman M.R., Lessing D., Kim J., Carlson J.R., Lessing D., Kim J., Carlson J.R., Kim J., Carlson J.R., Carlson J.R. A novel family of divergent seven-transmembrane proteins: Candidate odorant receptors in Drosophila. Neuron. 1999;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- Clyne P.J., Warr C.G., Carlson J.R., Warr C.G., Carlson J.R., Carlson J.R. Candidate taste receptors in Drosophila. Science. 2000;287:1830–1834. doi: 10.1126/science.287.5459.1830. [DOI] [PubMed] [Google Scholar]
- Dunipace L., Meister S., McNealy C., Amrein H., Meister S., McNealy C., Amrein H., McNealy C., Amrein H., Amrein H. Spatially restricted expression of candidate taste receptors in the Drosophila gustatory system. Curr. Biol. 2001;11:822–835. doi: 10.1016/s0960-9822(01)00258-5. [DOI] [PubMed] [Google Scholar]
- Fishilevich E., Vosshall L.B., Vosshall L.B. Genetic and functional subdivision of the Drosophila antennal lobe. Curr. Biol. 2005;15:1548–1553. doi: 10.1016/j.cub.2005.07.066. [DOI] [PubMed] [Google Scholar]
- Fishilevich E., Domingos A.I., Asahina K., Naef F., Vosshall L.B., Louis M., Domingos A.I., Asahina K., Naef F., Vosshall L.B., Louis M., Asahina K., Naef F., Vosshall L.B., Louis M., Naef F., Vosshall L.B., Louis M., Vosshall L.B., Louis M., Louis M. Chemotaxis behavior mediated by single larval olfactory neurons in Drosophila. Curr. Biol. 2005;15:2086–2096. doi: 10.1016/j.cub.2005.11.016. [DOI] [PubMed] [Google Scholar]
- Galizia C.G., Menzel R., Menzel R. The role of glomeruli in the neural representation of odours: Results from optical recording studies. J. Insect Physiol. 2001;47:115–130. doi: 10.1016/s0022-1910(00)00106-2. [DOI] [PubMed] [Google Scholar]
- Gao Q., Chess A., Chess A. Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics. 1999;60:31–39. doi: 10.1006/geno.1999.5894. [DOI] [PubMed] [Google Scholar]
- Goldman A.L., Van der Goes van Naters W., Lessing D., Warr C.G., Carlson J.R., Van der Goes van Naters W., Lessing D., Warr C.G., Carlson J.R., Lessing D., Warr C.G., Carlson J.R., Warr C.G., Carlson J.R., Carlson J.R. Coexpression of two functional odor receptors in one neuron. Neuron. 2005;45:661–666. doi: 10.1016/j.neuron.2005.01.025. [DOI] [PubMed] [Google Scholar]
- Goodman L. In: Form and function in the honey bee. Cooter R.J., Munn P.A., Munn P.A., editors. International Bee Research Association; Cardiff, UK: 2003. [Google Scholar]
- Hallem E.A., Ho M.G., Carlson J.R., Ho M.G., Carlson J.R., Carlson J.R. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117:965–979. doi: 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Hallem E.A., Dahanukar A., Carlson J.R., Dahanukar A., Carlson J.R., Carlson J.R. Insect odor and taste receptors. Annu. Rev. Entomol. 2006;51:113–135. doi: 10.1146/annurev.ento.51.051705.113646. [DOI] [PubMed] [Google Scholar]
- Hill C.A., Fox A.N., Pitts R.J., Kent L.B., Tan P.L., Chrystal M.A., Cravchik A.F., Collins H., Robertson H.M., Zwiebel L.J., Fox A.N., Pitts R.J., Kent L.B., Tan P.L., Chrystal M.A., Cravchik A.F., Collins H., Robertson H.M., Zwiebel L.J., Pitts R.J., Kent L.B., Tan P.L., Chrystal M.A., Cravchik A.F., Collins H., Robertson H.M., Zwiebel L.J., Kent L.B., Tan P.L., Chrystal M.A., Cravchik A.F., Collins H., Robertson H.M., Zwiebel L.J., Tan P.L., Chrystal M.A., Cravchik A.F., Collins H., Robertson H.M., Zwiebel L.J., Chrystal M.A., Cravchik A.F., Collins H., Robertson H.M., Zwiebel L.J., Cravchik A.F., Collins H., Robertson H.M., Zwiebel L.J., Collins H., Robertson H.M., Zwiebel L.J., Robertson H.M., Zwiebel L.J., Zwiebel L.J. G-protein-coupled receptors in Anopheles gambiae . Science. 2002;298:176–178. doi: 10.1126/science.1076196. [DOI] [PubMed] [Google Scholar]
- The Honey Bee Genome Sequencing Consortium. Insights into social insects from the genome of the honey bee Apis mellifera. Nature. 2006 (in press) [Google Scholar]
- Jeanmougin F., Thompson J.D., Gouy M., Higgins D.G., Gibson T.J., Thompson J.D., Gouy M., Higgins D.G., Gibson T.J., Gouy M., Higgins D.G., Gibson T.J., Higgins D.G., Gibson T.J., Gibson T.J. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- Jones W.D., Nguyen T.A., Kloss B., Lee K.J., Vosshall L.B., Nguyen T.A., Kloss B., Lee K.J., Vosshall L.B., Kloss B., Lee K.J., Vosshall L.B., Lee K.J., Vosshall L.B., Vosshall L.B. Functional conservation of an insect odorant receptor gene across 250 million years of evolution. Curr. Biol. 2005;15:R119–R121. doi: 10.1016/j.cub.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Kreher S.A., Kwon J.Y., Carlson J.R., Kwon J.Y., Carlson J.R., Carlson J.R. The molecular basis of odor coding in the Drosophila larva. Neuron. 2005;46:445–456. doi: 10.1016/j.neuron.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Krieger J., Raming K., Dewer Y.M., Bette S., Conzelmann S., Breer H., Raming K., Dewer Y.M., Bette S., Conzelmann S., Breer H., Dewer Y.M., Bette S., Conzelmann S., Breer H., Bette S., Conzelmann S., Breer H., Conzelmann S., Breer H., Breer H. A divergent gene family encoding candidate olfactory receptors of the moth Heliothis virescens . Eur. J. Neurosci. 2002;16:619–628. doi: 10.1046/j.1460-9568.2002.02109.x. [DOI] [PubMed] [Google Scholar]
- Krieger J., Klink O., Mohl C., Raming K., Breer H., Klink O., Mohl C., Raming K., Breer H., Mohl C., Raming K., Breer H., Raming K., Breer H., Breer H. A candidate olfactory receptor subtype highly conserved across different insect orders. J. Comp. Physiol. A Neuroethol. Sens Neural. Behav Physiol. 2003;189:519–526. doi: 10.1007/s00359-003-0427-x. [DOI] [PubMed] [Google Scholar]
- Krieger J., Grosse-Wilde E., Gohl T., Dewer Y.M., Raming K., Breer H., Grosse-Wilde E., Gohl T., Dewer Y.M., Raming K., Breer H., Gohl T., Dewer Y.M., Raming K., Breer H., Dewer Y.M., Raming K., Breer H., Raming K., Breer H., Breer H. Genes encoding candidate pheromone receptors in a moth (Heliothis virescens) Proc. Natl. Acad. Sci. 2004;101:11845–11850. doi: 10.1073/pnas.0403052101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger J., Grosse-Wilde E., Gohl T., Breer H., Grosse-Wilde E., Gohl T., Breer H., Gohl T., Breer H., Breer H. Candidate pheromone receptors of the silkmoth Bombyx mori . Eur. J. Neurosci. 2005;21:2167–2176. doi: 10.1111/j.1460-9568.2005.04058.x. [DOI] [PubMed] [Google Scholar]
- Laissue P.P., Reiter C., Hiesinger P.R., Halter S., Fischbach K.F., Stocker R.F., Reiter C., Hiesinger P.R., Halter S., Fischbach K.F., Stocker R.F., Hiesinger P.R., Halter S., Fischbach K.F., Stocker R.F., Halter S., Fischbach K.F., Stocker R.F., Fischbach K.F., Stocker R.F., Stocker R.F. Three-dimensional reconstruction of the antennal lobe in Drosophila melanogaster. J. Comp. Neurol. 1999;405:543–552. [PubMed] [Google Scholar]
- Larsson M.C., Domingos A.I., Jones W.D., Chiappe M.E., Amrein H., Vosshall L.B., Domingos A.I., Jones W.D., Chiappe M.E., Amrein H., Vosshall L.B., Jones W.D., Chiappe M.E., Amrein H., Vosshall L.B., Chiappe M.E., Amrein H., Vosshall L.B., Amrein H., Vosshall L.B., Vosshall L.B. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schittgen T.D., Schittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(T) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Menzel R., Erber J., Erber J. Learning and memory in bees. Sci. Am. 1978;239:102–108. [Google Scholar]
- Michener C.D. The social behavior of bees: A comparative study. Harvard University Press; Cambridge, MA: 1974. [Google Scholar]
- Nakagawa T., Sakurai T., Nishioka T., Touhara K., Sakurai T., Nishioka T., Touhara K., Nishioka T., Touhara K., Touhara K. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science. 2005;307:1638–1642. doi: 10.1126/science.1106267. [DOI] [PubMed] [Google Scholar]
- Pitts R.J., Fox A.N., Zwiebel L.J., Fox A.N., Zwiebel L.J., Zwiebel L.J. A highly conserved candidate chemoreceptor expressed in both olfactory and gustatory tissues in the malaria vector Anopheles gambiae . Proc. Natl. Acad. Sci. 2004;101:5058–5063. doi: 10.1073/pnas.0308146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven P.H., Axelrod D.I., Axelrod D.I. Angiosperm biogeography and past continental movements. Ann. Mo. Bot. Gard. 1974;61:539–673. [Google Scholar]
- Robertson H.M. Two large families of chemoreceptor genes in the nematodes Caenorhabditis elegans and Caenorhabditis briggsae reveal extensive gene duplication, diversification, movement, and intron loss. Genome Res. 1998;8:449–463. doi: 10.1101/gr.8.5.449. [DOI] [PubMed] [Google Scholar]
- Robertson H.M. The large srh family of chemoreceptor genes in Caenorhabditis nematodes reveals processes of genome evolution involving large duplications and deletions and intron gains and losses. Genome Res. 2000;10:192–203. doi: 10.1101/gr.10.2.192. [DOI] [PubMed] [Google Scholar]
- Robertson H.M., Thomas J.H., Thomas J.H. WormBook. 2006. The putative chemoreceptor families of C. elegans (Jan. 6, 2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H.M., Warr C.G., Carlson J.R., Warr C.G., Carlson J.R., Carlson J.R. Molecular evolution of the insect chemoreceptor superfamily in Drosophila melanogaster . Proc. Natl. Acad. Sci. 2003;100:14537–14542. doi: 10.1073/pnas.2335847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutzler M., Zwiebel L.J., Zwiebel L.J. Molecular biology of insect olfaction: Recent progress and conceptual models. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 2005;191:777–790. doi: 10.1007/s00359-005-0044-y. [DOI] [PubMed] [Google Scholar]
- Sakurai T., Nakagawa T., Mitsuno H., Mori H., Endo Y., Tanoue S., Yasukochi Y., Touhara K., Nishioka T., Nakagawa T., Mitsuno H., Mori H., Endo Y., Tanoue S., Yasukochi Y., Touhara K., Nishioka T., Mitsuno H., Mori H., Endo Y., Tanoue S., Yasukochi Y., Touhara K., Nishioka T., Mori H., Endo Y., Tanoue S., Yasukochi Y., Touhara K., Nishioka T., Endo Y., Tanoue S., Yasukochi Y., Touhara K., Nishioka T., Tanoue S., Yasukochi Y., Touhara K., Nishioka T., Yasukochi Y., Touhara K., Nishioka T., Touhara K., Nishioka T., Nishioka T. Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori . Proc. Natl. Acad. Sci. 2004;101:16653–16658. doi: 10.1073/pnas.0407596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H.A., Strimmer K., Vingron M., von Haeseler A., Strimmer K., Vingron M., von Haeseler A., Vingron M., von Haeseler A., von Haeseler A. TREE-PUZZLE: Maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Scott K., Brady R., Jr., Cravchik A., Morozov P., Rzhetsky A., Zuker C., Axel R., Brady R., Jr., Cravchik A., Morozov P., Rzhetsky A., Zuker C., Axel R., Cravchik A., Morozov P., Rzhetsky A., Zuker C., Axel R., Morozov P., Rzhetsky A., Zuker C., Axel R., Rzhetsky A., Zuker C., Axel R., Zuker C., Axel R., Axel R. A chemosensory gene family encoding candidate gustatory and olfactory receptors in. Drosophila. Cell. 2001;104:661–673. doi: 10.1016/s0092-8674(01)00263-x. [DOI] [PubMed] [Google Scholar]
- Skiri H.T., Ro H., Berg B.G., Mustaparta H., Ro H., Berg B.G., Mustaparta H., Berg B.G., Mustaparta H., Mustaparta H. Consistent organization of glomeruli in the antennal lobes of related species of heliothine moths. J. Comp. Neurol. 2005;491:367–380. doi: 10.1002/cne.20692. [DOI] [PubMed] [Google Scholar]
- Stange G., Diesendorf M., Diesendorf M. The response of the honeybee antennal CO2-receptors to N2O and Xe. J. Comp. Physiol. 1973;86:139–158. [Google Scholar]
- Stewart M.K., Clark N.L., Merrihew G., Galloway E.M., Thomas J.H., Clark N.L., Merrihew G., Galloway E.M., Thomas J.H., Merrihew G., Galloway E.M., Thomas J.H., Galloway E.M., Thomas J.H., Thomas J.H. High genetic diversity in the chemoreceptor superfamily of Caenorhabditis elegans . Genetics. 2005;169:1985–1996. doi: 10.1534/genetics.104.035329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh G.S., Wong A.M., Hergarden A.C., Wang J.W., Simon A.F., Benzer S., Axel R., Anderson D.J., Wong A.M., Hergarden A.C., Wang J.W., Simon A.F., Benzer S., Axel R., Anderson D.J., Hergarden A.C., Wang J.W., Simon A.F., Benzer S., Axel R., Anderson D.J., Wang J.W., Simon A.F., Benzer S., Axel R., Anderson D.J., Simon A.F., Benzer S., Axel R., Anderson D.J., Benzer S., Axel R., Anderson D.J., Axel R., Anderson D.J., Anderson D.J. A single population of olfactory sensory neurons mediates an innate avoidance behaviour in. Drosophila. Nature. 2004;431:854–859. doi: 10.1038/nature02980. [DOI] [PubMed] [Google Scholar]
- Swofford D.L. PAUP*: Phylogenetic analysis using parsimony and other methods, version4. Sinauer Press; New York: 2001. [Google Scholar]
- von Frisch K. Versuche uber den Geschmacksinn der Bienen. Naturwissenschaften. 1927;14:1–20. [Google Scholar]
- von Lacher V. Elektrophysiologische Unter-suchungen an einzelnen Rezeptoren für Geruch, Kohlendioxid, Luftfeuchtigkeit und Temperatur auf den Antennen der Arbeitsbiene und der Drohne (Apis mellifera L. Z. Vergl. Physiol. 1964;48:587–623. [Google Scholar]
- Vosshall L.B., Amrein H., Morozov P.S., Rzhetsky A., Axel R., Amrein H., Morozov P.S., Rzhetsky A., Axel R., Morozov P.S., Rzhetsky A., Axel R., Rzhetsky A., Axel R., Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- Vosshall L.B., Wong A.M., Axel R., Wong A.M., Axel R., Axel R. An olfactory sensory map in the fly brain. Cell. 2000;102:147–159. doi: 10.1016/s0092-8674(00)00021-0. [DOI] [PubMed] [Google Scholar]
- Wright G.A., Skinner B.D., Smith B.H., Skinner B.D., Smith B.H., Smith B.H. Ability of honeybee, Apis mellifera, to detect and discriminate odors of varieties of canola (Brassica rapa and B. napus) and snapdragon flowers (Antirrhinium majus) J. Chem. Ecol. 2002;28:721–740. doi: 10.1023/a:1015232608858. [DOI] [PubMed] [Google Scholar]
- Young J.M., Trask B.J., Trask B.J. The sense of smell: Genomics of vertebrate odorant receptors. Hum. Mol. Genet. 2002;11:1153–1160. doi: 10.1093/hmg/11.10.1153. [DOI] [PubMed] [Google Scholar]