Abstract
The remarkable olfactory power of insect species is thought to be generated by a combinatorial action of two large protein families, G protein-coupled olfactory receptors (ORs) and odorant binding proteins (OBPs). In olfactory sensilla, OBPs deliver hydrophobic airborne molecules to ORs, but their expression in nonolfactory tissues suggests that they also may function as general carriers in other developmental and physiological processes. Here we used bioinformatic and experimental approaches to characterize the OBP-like gene family in a highly social insect, the Western honey bee. Comparison with other insects shows that the honey bee has the smallest set of these genes, consisting of only 21 OBPs. This number stands in stark contrast to the more than 70 OBPs in Anopheles gambiae and 51 in Drosophila melanogaster. In the honey bee as in the two dipterans, these genes are organized in clusters. We show that the evolution of their structure involved frequent intron losses. We describe a monophyletic subfamily of OBPs where the diversification of some amino acids appears to have been accelerated by positive selection. Expression profiling under a wide range of conditions shows that in the honey bee only nine OBPs are antenna-specific. The remaining genes are expressed either ubiquitously or are tightly regulated in specialized tissues or during development. These findings support the view that OBPs are not restricted to olfaction and are likely to be involved in broader physiological functions.
Olfaction plays a role in almost every aspect of insect life. In a highly social species, like the honey bee, olfaction is not only used to recognize a huge variety of airborne molecules, but also to provide the 50,000 members of a colony with a sensory network that maintains the internal cohesion of the hive. In this context, the ability to perceive several pheromone blends and to receive kin recognition signals are particularly important.
The recognition and discrimination of thousands of odorous compounds is mediated by olfactory sensory neurons. In many terrestrial animals, like mammals and insects, the chemo-sensory neurons are surrounded by an aqueous milieu acting as a barrier for volatile, primarily lipophilic molecules. Consequently, many airborne molecules, such as hydrophobic odorants and pheromones, must first be recognized by a specialized class of proteins that facilitate their delivery to the olfactory receptors (OR). It is now widely accepted that in both insects and vertebrates this function is provided by odorant binding proteins (OBPs) (Pelosi 1996; Krieger and Breer 1999; Deyu and Leal 2002). In spite of bearing the same names and performing similar functions, insect OBPs and vertebrate OBPs appear to be phylogenetically unrelated (Vogt et al. 1990; Hildebrand and Shepherd 1997). Insect OBPs are small, water soluble molecules expressed in both olfactory and gustatory sensilla, as well as in other specialized tissues (Pelosi et al. 2005). Several studies have demonstrated selective binding of odorants and/or pheromones to different OBPs (Danty et al. 1999; Plettner et al. 2000; Pophof 2002, 2004; Zhou et al. 2004b). It has been proposed that, in addition to playing a role in the activation of odorant-responsive chemo-sensory neurons, OBPs might work as selective filters in odor recognition (Kim et al. 1998) or even participate in signal termination by inactivating odorant molecules (Pelosi and Maida 1995). This notion is supported by a recent study on Drosophila melanogaster OBP76a that implicates this protein directly in pheromone signal transduction (Xu et al. 2005).
Recent genomic projects have offered new insights into the molecular mechanisms of olfaction by revealing the full repertoire of OBPs and ORs in a number of animal species (e.g., Hekmat-Scafe et al. 2002; Robertson et al. 2003). Two main strategies are seen in the animal genomes that have been sequenced to date. Nematodes and mammals possess a large number (∼1000) of G protein-coupled ORs (Prasad and Reed 1999), but very few OBPs (around five in mammals and none in nematodes). In these animals, odorant discrimination seems to be based entirely on a combinatorial utilization of ORs while OBPs, if present, act only as generic carriers (Löbel et al. 2002). By contrast, insects have a much smaller number of ORs (around 70 in D. melanogaster and Anopheles gambiae) and more OBPs (more than 50 in each of these two dipterans). To reconcile these genomic differences between different groups of animals, it has been proposed that odorant detection in insects might be mediated by a combinatorial usage of both ORs and OBPs (Hekmat-Scafe et al. 2002). According to this model, a subset of ORs (Goldman et al. 2005) and a subset of OBPs (Shanbhag et al. 2005) expressed in each sensillum would increase the discriminatory potential of the insect olfactory machinery. However, the extent to which OBPs are critical for olfactory discrimination remains unclear, largely because OBPs have also been found in nonolfactory tissues, suggesting that their roles may be restricted to general carrier capabilities with broad specificity for lipophilic compounds. Some of the OBPs implicated in nonolfactory functions include the B proteins of Tenebrio molitor accessory glands (Paesen and Happ 1995), the male specific serum proteins of Ceratitis capitata (Thymianou et al. 1998), and the heme-binding protein of Rhodnius prolixus (Paiva-Silva et al. 2002).
With an aim to accelerate our understanding of the molecular basis of chemosensory pathways in insects, we have annotated the honey bee gene family encoding OBP-like proteins. Comparison with D. melanogaster and A. gambiae shows that the honey bee possesses the smallest OBP repertoire. Several honey bee OBPs are found in olfactory tissues, but only a minority is olfactory-specific. Our study also casts some light on the evolution of this gene family by suggesting that it has a relatively recent origin, and showing that such emerging lineage-specific expansions can diversify under positive selection pressure.
Results
Annotation of the honey bee genes encoding OBP-like proteins
In total, we have identified 21 genes encoding putative OBPs in the honey bee genomse assembly v.2.0, including five that have already been known from previous studies (obp1, obp2, obp4, obp5, and obp6). We believe that this set represents the real number of OBP-like genes in this species. The genome assembly was tested against available honey bee sequence data sets (ESTs, cDNAs, and STS markers) for extent of completeness. About 97% of the STS markers and 98% of the EST sequences and 96% of the cDNAs are represented in the assembly (ftp://ftp.hgsc.bcm.tmc.edu/pub/data/Tcastaneum; Tribolium genomic sequences). Thus, judging from the completeness of the assembly, the likelihood of finding more OBP-like genes in the honey bee is very low.
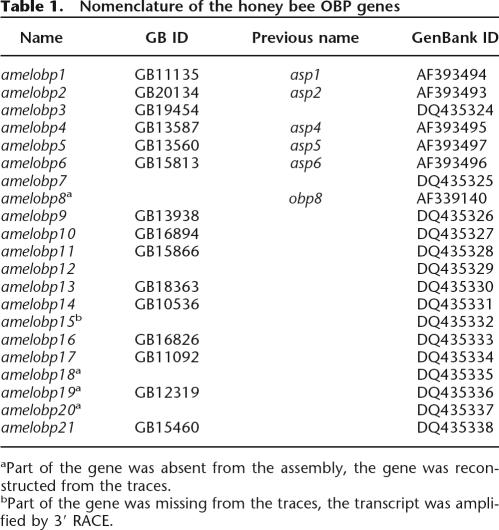
All these genes are listed in Table 1. Because many of the genes encoding OBPs reside in relatively AT-rich regions with poorer trace coverage, we found several of them to be incomplete or fragmented in the genome assembly. We therefore used a combination of experimental and in silico approaches to reconstruct the full-length sequences of all the members of this family in the honey bee.
Table 1.
Nomenclature of the honey bee OBP genes Name GB ID
The cDNA deposited in GenBank as obp8 (acc. no. AF339140) has a coding sequence very similar to that of obp6, suggesting a very recent segmental duplication. From position 28 in OBP6, their coding sequence differs only by one residue and nine synonymous substitutions in the 118 amino acids segment. In contrast, their 3′ UTRs do not show any noticeable similarities. In the assembly v.2.0, the gene encoding OBP8 was incomplete, but its 3′ UTR was found to lie at the end of a contig adjacent to another one encoding OBP6. We reconstructed the gap between these two contigs and found that it encodes the missing part of OBP8. By Southern blot hybridization (data not shown) we confirmed that these two very closely related genes are bona fide paralogs. Interestingly, EST data suggest that obp6 is alternatively spliced (acc. no. BE844326 and AF393496). Using RT-PCR amplification and sequencing, we confirmed that both variants are expressed in the antennae (see Supplemental Table 1). So far this is the only documented case of alternative splicing in the honey bee OBP family.
Another gene that had to be manually assembled encodes obp18. In this case, only the first and last of its five exons are present in the genome assembly. Fortunately, the large number of ESTs available for this gene allowed us to reconstruct the entire genomic landscape of this transcription unit.
The penultimate exon and part of the last exon of obp15 are missing from the assembly and from the honey bee genomic traces. We used 3′ RACE to sequence the missing part of this gene's transcript.
Two genes, obp19 and obp20, encode highly similar OBPs. They are tandemly arranged 5 kb apart on each end of the same contig. Because the first exon of obp19 and the last exon of obp20 are missing from the genome assembly, we obtained the entire sequences of both genes by manually extending this contig with a number of traces that have not been used for automatic assembly.
All honey bee OBP genes were found to have consensus GT/AG splice sites with the exception of obp6 and obp8, which have a GC/AG splice site in the fourth intron.
To test the robustness of our annotations, and to rule out the possibility that some of the genes belonging to this family might have been missed by our search algorithm, we applied our method to the well characterized genomes of the two dipteran species, A. gambiae and D. melanogaster. Interestingly, we were able not only to find all the previously reported OBPs, but also four new OBP genes in the mosquito (Supplemental Table 5). In addition, we annotated the OBP gene family in the beetle, Tribolium castaneum, using the v.2.0 assembly of this genome available from the Tribolium genome project Web page. We found 46 genes encoding OBPs in this coleopteran's genome. These findings support the notion that OBP gene families in insects are relatively large and vary from ∼50 (fly, beetle) up to 70 members (mosquito). In this context, Apis mellifera possesses an unusually small set of OBP genes, in contrast to its vastly expanded olfac-tory receptor family (Robertson et al. 2006).
OBPs are clustered in the honey bee genome
Like in D. melanogaster and A. gambiae most of the OBP-like genes in the honey bee are organized in clusters in the genome, reflecting the relatively recent expansions of genes belonging to this family. Only three genes, obp1, obp9, and obp12, are represented by single loci that have no other obps in close proximity. Two genes, obp10 and obp11, are arranged in a head to tail tandem, ∼1kb apart, whereas the remaining obps are organized in two clusters.
The first cluster contains nine obps, tandemly arranged in the same orientation within a 40-kb region on chromosome 15. Most contigs that make up this cluster are small, “unoriented,” and have a low sequencing coverage in the traces database. This may be due to the AT content of this region being as high as 81%. In fact, most traces that cover this region come from a genomic library with an enriched AT content. The AT richness of the underlying genomic regions explains the difficulties in the annotation of obp15, obp18, obp19, and obp20. We note in passing that our reconstruction of this region is in agreement with an independent super-scaffolding effort (Robertson et al. 2006). The seven remaining OBPs are organized in a single cluster on chromosome 9. This cluster encompasses all the OBPs described in previous studies. As in the first cluster, OBPs in this group are arranged in the same orientation.
C-minus OBPs in the honey bee form a monophyletic group
Figure 1 shows the alignment of the honey bee OBP proteins. They are all very homogenous in size, ranging from 15 to 18 kDa, and have a predicted signal peptide at the 5′ termini. T. castaneum OBPs display a similar homogeneity of size (Supplemental Fig. 1). This contrasts with OBPs in D. melanogaster and A. gambiae, where the proteins in some subfamilies show substantial increases in size. For example, the C-plus subfamilies found in both dipterans (Zhou et al. 2004a) and the atypical OBPs in the mosquito (Xu et al. 2003) are about twice as long as other OBPs, mostly because of the extended C termini.
Figure 1.
The alignment of the predicted polypeptides encoding OBPs in Apis mellifera. Conserved residues are highlighted and the signal peptides are in boxes. The rectangular shapes above the alignment represent the α-helices in AmelOBP1 secondary structure. The splice sites are labeled with separators: Vertical ones indicate splice sites between codons; backward slanted separators point out splice sites within codons after the first base.
The three-dimensional structures of honey bee OBPs are likely to be very similar as suggested by the alignment of the sixα-helices characteristic of this class of proteins (Pelosi 1998). The six-cysteine signature found in the OBP family is conserved in 13 members with all the honey bee proteins having four conserved cysteines. Following the naming system proposed by Hekmat-Scafe et al. (2002), we refer to OBPs lacking the second and the fifth cysteine as C-minus OBPs.
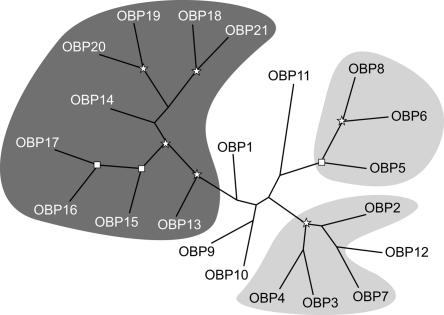
The alignment of honey bee OBPs was used to build a neighbor-joining tree (Fig. 2). In this tree, the C-minus OBPs are grouped together as a monophyletic group with strong bootstrap support. One interesting feature of the tree is the position of the six-cysteine-containing obp13 at the root of the C-minus group. This suggests that obp13 and the C-minus OBPs evolved from a common ancestor containing six cysteines, two of which have been retained only in the obp13 lineage. Consequently, we include obp13 together with the C-minus subfamily in phylogenetic evaluations of the honey bee OBP gene family. The members of the C-minus group make up the cluster on chromosome 15 described in the previous section. They display a rather high level of sequence similarity, with a pairwise median identity of 48%. In contrast, the classic OBPs not belonging to the C-minus group have less similar sequences (16% median identity). This diversity is reflected in the phylogenetic tree, where their origins are more difficult to follow than the C-minus subfamily. Only two subgroups, both belonging to the cluster on chromosome 9, have a good bootstrap support. obp12 belongs to one of these two clades but is on chromosome 12.
Figure 2.
Phylogeny of the OBP protein family in Apis mellifera. An unrooted tree was constructed with aligned protein sequences from the honey bee using neighbor-joining. The OBP protein family is composed of two color-coded subgroups, the C-minus subfamily (dark gray) on chromosome 15 and OBPs clustered on chromosome 9 (light gray). The stars and squares indicate nodes with 95% and 80%–95% bootstrap supports respectively.
A phylogenetic tree based on the alignment of all unique OBP sequences is shown in Supplemental Figure 2. The three honey bee clades mentioned above also appear in this tree, suggesting that they are lineage specific expansions. (Hekmat-Scafe et al. 2002) and (Vogt 2003) already reported that the phylogeny of OBPs consists mostly of lineage specific expansions with few clear orthologies, except in closely related species. Despite the fact that the honey bee C-minus OBPs are grouped together with some of their relatives in other insects, there is no bootstrap support for this clade. Thus, the loss of cysteines 2 and 5 appears to have occurred more than once, as suggested by the presence of obp13 at the root of the honey bee C-minus group.
Selection on the C-minus OBP subfamily
Recombination may result in higher rates of false positives in maximum likelihood tests for positive selection (Anisimova et al. 2003). Therefore, we first checked whether intergenic recombination occurred in the honey bee C-minus OBP subfamily, using the TOPALi software. As we did not find any evidence for recombination, we used the maximum likelihood method of the PAML package to test for positive selection in this paralogous group.
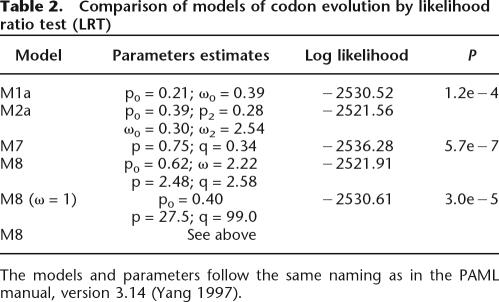
Table 2 shows the comparison by likelihood ratio test (LRT) of different site models of codon evolution. Pairwise comparisons of models accounting for positive selection with alternative neutral selection models show that the positive selection models are significantly more likely. This implies that the honey bee C-minus OBPs lineage is subjected to positive selection. Bayes empirical Bayes (BEB) inference identified eight sites under positive selection with at least 95% confidence. The random effect likelihood (REL) method identifies the same amino acids with a high level of significance (Bayes Factor > 200), as well as other 21 sites (Bayes Factor > 50). We will discuss only these eight sites identified by both methods.
Table 2.
Comparison of models of codon evolution by likelihood ratio test (LRT)
In order to assess whether these sites may be part of the OBP binding pocket, we aligned the C-minus OBPs with four other OBPs, for which the structure of the binding pocket has been determined, namely D. melanogaster LUSH (Kruse et al. 2003), Bombyx mori PBP (Lee et al. 2002), AmelOBP1 (Lartigue et al. 2004), and Leucophaea maderae PBP (Lartigue et al. 2003). Figure 3 highlights the alignment between the sites under positive selection and the amino acid motif of the binding pocket and/or its lip. In most cases, residues under positive selection pressure localize to the binding pocket of at least one insect.
Figure 3.
Positive selection on the C-minus OBP family. The top panel shows the number of non-synonymous substitutions divided by the number of synonymous substitutions when this ratio is >1. Amino acids detected to be significantly under positive selection by both BEB and REL (see Methods) are in black. The bottom panel shows the residues (dots) forming either the binding pocket or its lip in four OBPs for which the secondary structures are known. Those residues that correspond to positively selected amino acids of the C-minus OBPs in the top panel are enclosed.
Conserved splice sites
One feature that is apparent from the alignment in Figure 1 is the high conservation of the splice site locations. Six conserved splice sites can be seen in this alignment. First, an intron is always present close to the predicted signal peptide cleavage site. This intron is situated between 10 and 24 codons upstream of the first cysteine and always occurs between codons. Second, all honey bee OBPs have a splice site 25 bp (eight codons and one base pair) downstream from the first cysteine. The third splice site is only present in obp10 exactly before the second cysteine. The fourth splice site, 21 bp after the third cysteine, is present in all OBP genes, except obp9 and obp10. All honey bee OBP genes have a fifth splice site after the fourth cysteine. In most cases, it lies precisely 10 bp after this cysteine, but in obp7 and obp12 it occurs after 7 bp and 22 bp, respectively. Thus, the number of codons is variable, but the phase is conserved. These discrepancies are most probably caused by amino acid deletions and insertions rather than intron loss and gain. Finally, the sixth splice site can be found 21 bp downstream from the sixth cysteine in obp1, obp6, obp8, obp9, obp10, and obp11.
In a previous study on D. melanogaster OBP genes, Hekmat-Scafe et al. (2002) took a different approach and proposed that six different splice site locations occur near the signal peptide. Since signal peptides show very little sequence conservation, and usually cannot be aligned with confidence, we consider that, in the vicinity of the signal peptide, only a change of phase in a splice site denotes an intron gain event.
A comparison of the splice sites found in the honey bee OBPs with those found in D. melanogaster, A. gambiae, and T. castaneum reveals that the great majority belongs to one of the six classes that we identified in the honey bee (see Supplemental Figs. 1, 3, 4). We did not observe any nonconserved intron positions in T. castaneum. In D. melanogaster, EST and full-length cDNA data support the structure of two OBP genes (dmelobp19b and dmelobp19d) that contain an intron not found in the other insects. Some A. gambiae OBP genes (agamobp29, agamobp56, agamobp59, agamobp63, agamobp66) appear to have introns with a nonconserved position or phase, but only the structure of agamobp56 is supported by experimental data.
The conservation of the majority of splice sites between insects implies that the evolution of gene structure in this family involves predominantly intron losses. Owing to the difficulty of reconstructing the phylogeny of the OBP family, it was impossible to precisely retrace the history of intron losses. Consequently, in order to estimate intron loss rates, we took the admittedly naive approach of comparing the mean number of introns per gene in the four species, D. melanogaster, A. gambiae, T. castaneum, and A. mellifera. While no pairwise comparisons are significantly different between the first three insects, all comparisons with the bee were highly significant (P < 10−9, two tailed t-test with Bonferroni correction). This suggests that intron loss events have been less frequent in the honey bee.
Patterns of expression
Figure 4 shows the expression patterns of the members of the honey bee OBP-like family. We first used semi-quantitative RT-PCR to determine if all OBP genes identified in the bee genome are transcribed and to assess their level of expression. Next, to obtain a more quantitative estimate of their expression levels we used a dot-blot assay. All genes that were detected by RT-PCR produced a signal that was at least twice above that of the background in the dot-blot assay. We were unable to separate the expression profiles of two pairs of almost identical OBPs (obp6/ obp8 and obp19/obp20). obp7 and obp12 were not included in the dot-blot experiments, but their restricted expression in the antenna (obp7) and in the queen ovaries (obp12) has been detected by RTPCR (data not shown). This approach revealed three main categories of expression patterns within this gene family.
Figure 4.
Global expression patterns of Apis mellifera OBPs. The levels of expression are illustrated by three grades of grayscale relative to the ribosomal protein S8: Light gray indicates low level of expression defined as more than 2× background and less than half of the S8 level; dark gray indicates medium level of expression (between half and twice the level of S8), and black stands for high level of expression defined as more than 2× the level of S8. The expression of S8 was always more than 2× the background value. OBPs are shown on the y-axis and the examined tissues on the x-axis: an_fo, forager antennae; an_qu, queen antennae; an_dr, drone antennae; br_d1, newly emerged bee brain; br_fo, forager brain; cu_d1, newly emerged bee head's cuticle; cu_fo, forager head's cuticle; lg_d1, newly emerged bee legs; tx_d1, newly emerged bee thorax; tg_d6, 6-d-old bee tergites; st_d6, 6-d-old bee sternites; fb_d6, 6-d-old bee fat bodies; ov_qu, queen ovaries; eggs, eggs; lary, young larvae, stage 1 and 2; laro, old larvae, stage 5; pupy, young pupae, white-eyed; pupo, old pupae, dark-eyed.
First, as expected, some OBPs are expressed exclusively in the antennae of adult bees (obp1, obp2, obp4, obp5, obp6, obp8, obp11, obp15, and obp12). These genes often gave a weak signal in the head and in the legs, probably due to some chemosensory sensilla present on these body parts, in particular on the proboscis and the pharynx and on the leg tarsi (wings were not included in this study). Within this category of antennal OBPs the most striking gender-related difference is the absence of obp11 from the drones. This result was confirmed by Northern blot (data not shown).
The second category includes OBPs ubiquitously expressed in all adult body parts (obp3, obp16, obp17, obp18, obp19/obp20, and obp21). One particularly interesting case within this group is obp3, which was found in all body parts with the exception of the antennae. More precise dissections of heads into brains and cuticles, and abdomens into sclerites, fat bodies, and ovaries revealed a trend toward higher expression of the ubiquitous class of OBPs in cuticular parts. This suggests that these genes are expressed by epidermal cells. Most of these OBPs are expressed before the imaginal molt, in old pupae.
The last category contains OBPs expressed during relatively narrow developmental stages. The transcript of obp9 is detectable in the queen ovaries and in early embryos—consistent with a maternal expression pattern. Similarly, we found obp7 to be expressed exclusively in queen ovaries. obp14 and obp15 are found in larvae and disappear after pupation. obp13 is highly expressed in the old larvae and throughout the pupal stages. Finally, obp10 appears in pupae and reaches the highest level in the brain of newly emerged bees before declining in older bees.
The majority of OBPs that we found to be restricted to ol-factory tissues belong to the cluster on chromosome 9. However, it is unlikely that these OBPs are under the control of a common regulatory element because the centrally located member of this cluster, obp3, is expressed in all body parts but not in the antennae. Likewise, obp10 and obp11 are linked together, but have markedly different expression patterns.
In the C-minus group, obp13 that is located at the 5′ end of the cluster is expressed in larvae and in pupae. Two other members of this cluster, obp14 and obp15, which reside downstream from obp13, are both expressed in the larvae and in adult bees. The remaining six genes of this cluster are mostly expressed in adults (and some in late pupae). All these examples suggest that, in spite of maintaining tightly clustered arrangements, duplicated OBPs rapidly evolved novel functions. Further studies are needed to gain a better understanding of their functional significance.
Discussion
OBPs and olfaction in the honey bee
We have identified 21 genes encoding OBP-like proteins in the honey bee. This is by far the smallest number for this family observed in any insect to date. D. melanogaster, A. gambiae, and T. castaneum all have a repertoire of more than twice as many OBP genes. The small number of OBPs in the honey bee is even more striking in light of their expression profiles in this species. Only nine OBPs are restricted to olfactory organs. It is conceivable, however, that OBPs that are ubiquitously expressed may have specific olfactory functions when expressed in olfactory organs. If this is the case, the number of OBPs playing a role in chemo-sensation would be at least 16. Drosophila OBP19d was detected in both the inner lumen of taste peg sensilla and in the subcuticular space (Shanbhag et al. 2001a, b) and may have such a dual role. Regardless of this, the number of OBPs potentially involved in olfaction in the honey bee remains low in comparison to the fly or to the mosquito where the majority of OBPs were found expressed in chemosensory tissues (Galindo and Smith 2001; Graham and Davies 2002; Hekmat-Scafe et al. 2002; Biessmann et al. 2005). This suggests differences in the modalities of olfactory discrimination. One possibility is that the honey bee has lower discrimination capabilities than other insects with more OBPs. In the case of the mosquito this may be explained by distinct olfac-tory preferences of females that need to sense animal odors versus males that feed on nectar. Such differential preferences would require additional specialization and perhaps novel carrier proteins. To our knowledge, there is, however, no experimental support for this hypothesis. Another possibility is that in the honey bee other molecular components of the olfactory system provide the “missing” functions performed by the expanded OBP families in other insects. The following three, non-mutually exclusive hypotheses can be considered.
First, some odorant carriers may be encoded by other genes’ families, such as the Chemosensory Protein (CSP) family. It has been suggested (Ishida et al. 2002; Calvello et al. 2005) that in hymenopterans CSPs are more olfactory-specific than OBPs. Inconsistent with this notion is the similar number of CSPs in the honey bee compared with D. melanogaster and A. gambiae, as well as our expression studies (S. Forêt and R. Maleszka, unpubl.) showing that the majority of CSPs in the honey bee are not olfactory-specific.
Second, the unusually high number of olfactory receptors in the bee (∼160–170, Robertson et al. 2006) compared with the fly (62, Robertson et al. 2003) and with the mosquito (79, Hill et al. 2002) could compensate for the restricted discrimination by ol-factory carriers.
Finally, integration of the chemical signal in the antennal lobes may compensate for the deficit of discrimination at more peripheral levels. Interestingly, the honey bees have highly advanced antennal lobes with an estimated 160–170 glomeruli (Galizia et al. 1999) that correspond well with the number of 170 ORs (including 10 pseudo-genes) identified in the bee genome.
OBPs that are expressed in both the antennae and in other body parts seem to be more highly expressed in the epidermis. Here they may participate in the transport of some hydrophobic cuticular compounds, including molecules involved in inter-individual recognition. Such an association of OBPs with the cuticle has already been observed (Shanbhag et al. 2001a). This finding supports the idea that, like vertebrate lipocalins, insect OBPs are involved in the emission as well as in the detection of semiochemicals (Pelosi et al. 2005).
Evolution of the OBP family
Our results suggest that the insect OBP-like family is relatively recent in contrast to the olfactory receptor family that is thought to have an ancient origin (Robertson et al. 2003). Several lines of evidence support this notion. So far, OBPs have only been observed in neopteran insects, and unlike olfactory receptors, which are scattered throughout the D. melanogaster genome (Robertson et al. 2003, but see Robertson et al. 2006), odorant binding proteins are usually found in clusters (this work; Hekmat-Scafe et al. 2002; Newcomb et al. 2002; Xu et al. 2003). In addition, our study reveals highly conserved splice sites in the OBP family with an ancestral structure made of seven exons. The vast majority of the variations from this pattern are caused by intron losses while intron gains appear to have been extremely rare. As argued by some authors intron gains have been markedly infrequent in the last 100–200 Myr (Babenko et al. 2004). The neopteran radiation dates back around 360 Mya (Gaunt and Miles 2002), which is well after the peak of intron gain around 500 Mya (Babenko et al. 2004). It is nevertheless possible that intron loss is under selective pressure in OBPs. If most of these proteins are general transporters with broad specificity for lipophilic compounds and low ligand binding affinity, they are expected to be expressed at high levels. Highly expressed genes are more likely to be selected for intron loss (Jeffares et al. 2005). Our results and those by Biessmann et al. (2005) provide strong support for this view.
The recent origin of OBPs and the high disparity of their sequences imply a rapid rate of evolution of this gene family. One explanation for this accelerated rate of evolution may be that these proteins undergo periods of positive selection. A recent study on the GP-9 OBP (Krieger and Ross 2005) in various Solenopsis species has shown that positive selection drove the evolution of this protein. Similarly, (Willett 2000) reported that directional selection is acting on pheromone binding proteins in some Choristoneura species. Here we report that positive selection has fuelled the diversification of the honey bee C-minus OBP subfamily. Our data suggest two chief mechanisms of diversification in this subfamily. First, they show a significant diversity of expression profiles, including the developmental stage at which they are expressed. Second, it appears that some amino acids of the binding pocket have been the targets of this positive selection, probably resulting in a diversification in the range of ligands bound by these proteins.
Conclusions
Our results lend more support to the combinatorial model of insect olfaction. The relatively small size of the honey bee OBP-like gene family appears to be compensated by the expansion of its ORs repertoire. Consequently, at least at the genomic level the combinatorial power of honey bees’ olfactory systems appears to be similar to that of the other insect species. Unfortunately, we do not know enough about olfactory coding or discrimination to evaluate these numbers in the context of honey bee biology.
Our analyses also shed more light on the evolution of these proteins. OBPs have the structural hallmarks of most lineage specific gene expansion as identified by (Lespinet et al. (2002), in particular an α-helical structure and a conserved cysteine pattern. They seem to be an insect invention and may be restricted to neopterans. Within the insect OBP family, these genes seem to evolve mainly through the development of species, or family-specific expansions, some of which may have undergone positive selection pressure. This implies that OBPs play critical roles in the adaptation of insects to a wide variety of environments and lifestyles. One of these roles is olfaction, a central aspect of insect life. An important task now is to understand the other nonolfac-tory functions of these proteins.
Methods
Identification of insects’ OBPs
A list of 88 unique OBP sequences was obtained from the Pfam database (Bateman et al. 2004) version 17.0 under the category PBP_GOBP (acc. no. PF01395). These sequences were used to find other insect OBPs in the GenBank nonredundant protein database (Benson et al. 2005) by performing three iterations of PSI-BLAST (Altschul et al. 1997). The sequences of D. melanogaster OBPs as described by (Hekmat-Scafe et al. 2002) were downloaded from FlyBase (Drysdale and Crosby 2005). A. gambiae OBPs as described in (Xu et al. 2003) and (Zhou et al. 2004a) were obtained from GenBank.
Annotation of OBP genes
Honey bee genomic regions containing OBP genes were identified with PSI-TBLASTN searches. To identify intron/exon boundaries, these genomic sequences were loaded into the Apollo genome annotation tool (Lewis et al. 2002) together with various gene evidences including BLASTX matches against the GenBank nonredundant protein database, honey bee ESTs and cDNAs, and ab initio gene predictions from Fgenesh (Salamov and Solovyev 2000) and GenScan (Burge and Karlin 1998). In-house scripts were developed to automatically retrieve these data and load them in Apollo. Each predicted OBP protein was checked for the hallmarks of this family: the size (∼16 kDa), a conserved cysteine pattern, and a signal peptide (as predicted by SignalP) (Bendtsen et al. 2004). All annotations were submitted to BeeBase (http://racerx00.tamu.edu/bee_resources.html). A similar procedure was used to annotate T. castaneum OBP genes.
Nomenclature
We are proposing a nomenclature for the honey bee OBPs that is similar to that introduced for A. gambiae by the Anopheles OBP nomenclature committee (Xu et al. 2003). Each OBP gene is given a name starting with the amelobp prefix to denote that it is a honey bee gene belonging to the Odorant Binding Protein-like family, even though it may not be expressed in tissues implicated in olfaction. When it is unambiguous, we use the shorter obp prefix. The previously described honey bee OBPs retain their numerical suffix, so for example ASP1 becomes amelobp1 and OBP8 becomes amelobp8. OBPs residing in a cluster were given consecutive numbers (see Table 1). The same nomenclature was applied to T. castaneum OBPs (Supplemental Fig. 1).
Phylogenetic analysis
The secondary structures of six OBPs have been resolved and are available from the PDB database (Berman et al. 2000) (PDB IDs: 1ls8, 1p28, 1qwv, 1oof, 1r5r, and 1c3z). Since the length and location of the α-helices vary slightly from one OBP to another, we created a custom gap penalty mask by aligning their primary and secondary structures and averaging the probability of α-helices along all sequences. Sequences were aligned to this profile using CLUSTALW (Jeanmougin et al. 1998) in a profile alignment mode. Most similar sequences were aligned first and then combined together as profiles. Alignments were manually refined using the JalView alignment editor (Clamp et al. 2004). Neighbor-joining trees were produced using the Phylip package (Felsenstein 2005). We show trees based on the consensus of 1000 bootstrap replicates.
Tests for positive selection
CLUSTALW alignments of all C-minus OBPs were used as inputs in the following analyses. We checked whether intergenic recombination occurred within the C-minus subfamily using the DSS and PDM methods implemented in TOPALi (Milne et al. 2004) (window size 100, step 2).
We tested for positive selection using the codeml program in the PAML package (Yang 1997). We followed the procedure described in the PAML manual and discussed elsewhere (Swanson et al. 2003; Wong et al. 2004; Yang et al. 2005) to compare “site models” of codon substitution. These models allow us to assess whether a set of sequences, as a whole, is subjected to positive selection and to study the ratio of nonsynonymous to synonymous substitution, ω, at individual sites. Briefly, we used likelihood ratio tests (LRT) to compare model M1a (two categories of codons: ω = 1, neutral, and ω < 1, purifying selection) with model M2a (like M1a but also a positively selected category, ω > 1), model M7 (10 classes of ω, between 0 and 1) with model M8 (like M7, but with an additional class without constraints), and M8 with M8a (like M8 but the additional class is fixed to 1). Sites under positive selection were identified using Bayes empirical Bayes (BEB) inference (Yang et al. 2005) implemented for model M8. An unrooted neighbor-joining tree (Phylip) based on the alignment of all honey bee C-minus OBPs was used. Codon equilibrium frequencies were estimated from the average nucleotide frequencies at each codon position, but other estimates gave similar results.
We also examined the C-minus family for positive selection by the random effect likelihood (REL) method of (Pond and Frost 2005a) available in the HyPhy package (Pond et al. 2005) on the datamonkey server (Pond and Frost 2005b). For this analysis an optimal model of nucleic acid selection was selected by the method available on the same server. Similar results were obtained with other models (HKY85, TN93, and REV).
Other computational methods
Interscaffolding was done using traces and their mate-pair information available at GenBank. Interscaffold sequences were assembled using the CAP3 software (Huang and Madan 1999) kindly provided to us by X. Huang (Michigan Technological University, Houghton, MI). Other statistical tests were performed with the R environment.
Sample collection
Brood frames were taken from the hive and incubated at 32°C, 80% humidity. Eggs, larvae, and pupae were collected from these frames and immediately frozen on dry ice. Larval stages were determined according to Jung-Offmann (1968). Eye and cuticle coloration were used to segregate young and old pupae. Newly emerged bees were collected from the brood frames at 1-h intervals and either dissected immediately or caged in groups of ∼40 individuals for incubator storage for later use. Foragers were captured as they came back to the hive carrying pollen loads. The main body parts (antennae, heads, legs, thoraces, and abdomen) were taken from bees snap-frozen in liquid nitrogen and dissected on dry ice. Finer dissections (brain, head cuticle, abdomen sternites and tergites, fat body) were carried out in bee Ringer solution (Bicker 1995).
Molecular biology
Reverse Northern dot blot hybridization
Gene-specific primers were used to amplify fragments of the coding sequence of each OBP transcript by the RT-PCR approach (the primers and annealing temperatures are described in Supplemental Table 1). The PCR reactions were carried out using the following cycling regime: 94°C for 5 min followed by 35 cycles of 30 sec annealing, 30 sec at 72°C, and 30 sec at 94°C. In the RT-PCR assays aimed at characterizing the expression profiles of OBPs only 20 cycles were used. Intron/exon information was used to design primers amplifying RT-PCR products distinctly different from PCR fragments resulting from genomic contamination. Rapid amplification of 3′ cDNA end (3′ RACE) was conducted under similar conditions except that the 3′ primer (oligo (dT)18VN) was used as the reverse primer. PCR products were sequenced at the Biomolecular Resource Facility of the Australian National University.
For reverse Northern dot blot experiments 1.2% agarose gels were loaded with 16 samples of ∼100 ng per well of reamplified RT-PCR products. Every second well was left empty to avoid cross-well contamination. Electrophoresis was performed in TBE buffer at 20 V/cm and the products were allowed to migrate for 1 cm. The DNA was first denatured by bathing the gels in 0.5 M NaOH, 0.5 M NaCl, then neutralized in 1 M amonium acetate,0.02 M NaOH and blotted onto Hybond N+ nylon membranes (Amersham) by capillary transfer, and cross-linked by UV irradiation. The RNA samples were labeled in a single step by reverse transcription with the addition of P32-cytosine. A mixture of OBP-specific primers was used to produce labeled cDNA. Hybridization and image acquisition were done in a similar fashion as for Northern blots (Kucharski and Maleszka 2003). Briefly, blots were washed three to four times in 2 × SSC, 0.1% SDS at 50°C and exposed to a phosphorstorage screen (Molecular Dynamics) without drying. Computer generated images (MD Phosphor-Imager 400S) of individual gels were analyzed using ImageQuant software. In addition to OBPs, “housekeeping” control honey bee genes (Supplemental Table 2) and negative control vertebrates genes were also added to each membrane (Supplemental Table 3). Since we expected most of the OBPs to be differentially expressed between our experimental conditions, customary microarray analysis methods aimed at gene discovery were not relevant here. Instead, we applied a method similar to Biessmann et al. (2005) and used the intensity of the ribosomal protein S8 as a scaling factor to compare between different membranes. We define background as the signal produced by the negative controls. A gene is considered expressed if its signal is at least twice above the background. The low, medium, and high level of expression is defined in Figure 4. Two OBP genes, obp7 and obp12, were not included on the membranes.
Acknowledgments
We thank Paul Helliwell for his skillful assistance, Joanna Maleszka for help with tissue dissections, and the Baylor College of Medicine Human Genome Sequencing Center for making the Apis mellifera and the Tribolium castaneum genome sequences publicly available prior to publication.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5075706.
References
- Altschul S., Madden T., Schaffer A., Zhang J., Zhang Z., Miller W., Lipman D., Madden T., Schaffer A., Zhang J., Zhang Z., Miller W., Lipman D., Schaffer A., Zhang J., Zhang Z., Miller W., Lipman D., Zhang J., Zhang Z., Miller W., Lipman D., Zhang Z., Miller W., Lipman D., Miller W., Lipman D., Lipman D. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M., Nielsen R., Yang Z., Nielsen R., Yang Z., Yang Z. Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics. 2003;164:1229–1236. doi: 10.1093/genetics/164.3.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babenko V., Rogozin I., Mekhedov S., Koonin E., Rogozin I., Mekhedov S., Koonin E., Mekhedov S., Koonin E., Koonin E. Prevalence of intron gain over intron loss in the evolution of paralogous gene families. Nucleic Acids Res. 2004;32:3724–3733. doi: 10.1093/nar/gkh686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A., Coin L., Durbin R., Finn R., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E., Coin L., Durbin R., Finn R., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E., Durbin R., Finn R., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E., Finn R., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E., Khanna A., Marshall M., Moxon S., Sonnhammer E., Marshall M., Moxon S., Sonnhammer E., Moxon S., Sonnhammer E., Sonnhammer E., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen J., Nielsen H., von Heijne G., Brunak S., Nielsen H., von Heijne G., Brunak S., von Heijne G., Brunak S., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Benson D., Karsch-Mizrachi I., Lipman D., Ostell J., Wheeler D., Karsch-Mizrachi I., Lipman D., Ostell J., Wheeler D., Lipman D., Ostell J., Wheeler D., Ostell J., Wheeler D., Wheeler D. GenBank. Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H., Westbrook J., Feng Z., Gilliland G., Bhat T., Weissig H., Shindyalov I., Bourne P., Westbrook J., Feng Z., Gilliland G., Bhat T., Weissig H., Shindyalov I., Bourne P., Feng Z., Gilliland G., Bhat T., Weissig H., Shindyalov I., Bourne P., Gilliland G., Bhat T., Weissig H., Shindyalov I., Bourne P., Bhat T., Weissig H., Shindyalov I., Bourne P., Weissig H., Shindyalov I., Bourne P., Shindyalov I., Bourne P., Bourne P. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicker G. Transmitter-induced calcium signalling in cultured neurons of the insect brain. J. Neurosci. Method. 1995;69:33–41. doi: 10.1016/S0165-0270(96)00018-0. [DOI] [PubMed] [Google Scholar]
- Biessmann H., Nguyen Q.K., Le D., Walter M.F., Nguyen Q.K., Le D., Walter M.F., Le D., Walter M.F., Walter M.F. Microarray-based survey of a subset of putative olfactory genes in the mosquito Anopheles gambiae . Insect Mol. Biol. 2005;14:575–589. doi: 10.1111/j.1365-2583.2005.00590.x. [DOI] [PubMed] [Google Scholar]
- Burge C., Karlin S., Karlin S. Finding the genes in genomic DNA. Curr. Opin. Struct. Biol. 1998;8:346–354. doi: 10.1016/s0959-440x(98)80069-9. [DOI] [PubMed] [Google Scholar]
- Calvello M., Brandazza A., Navarrini A., Dani F., Turillazzi S., Felicioli A., Pelosi P., Brandazza A., Navarrini A., Dani F., Turillazzi S., Felicioli A., Pelosi P., Navarrini A., Dani F., Turillazzi S., Felicioli A., Pelosi P., Dani F., Turillazzi S., Felicioli A., Pelosi P., Turillazzi S., Felicioli A., Pelosi P., Felicioli A., Pelosi P., Pelosi P. Expression of odorant-binding proteins and chemosensory proteins in some Hymenoptera. Insect Biochem. Mol. Biol. 2005;35:297–307. doi: 10.1016/j.ibmb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Clamp M., Cuff J., Searle S., Barton G., Cuff J., Searle S., Barton G., Searle S., Barton G., Barton G. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Danty E., Briand L., Michard-Vanhee C., Perez V., Arnold G., Gaudemer O., Huet D., Huet J., Ouali C., Masson C., Briand L., Michard-Vanhee C., Perez V., Arnold G., Gaudemer O., Huet D., Huet J., Ouali C., Masson C., Michard-Vanhee C., Perez V., Arnold G., Gaudemer O., Huet D., Huet J., Ouali C., Masson C., Perez V., Arnold G., Gaudemer O., Huet D., Huet J., Ouali C., Masson C., Arnold G., Gaudemer O., Huet D., Huet J., Ouali C., Masson C., Gaudemer O., Huet D., Huet J., Ouali C., Masson C., Huet D., Huet J., Ouali C., Masson C., Huet J., Ouali C., Masson C., Ouali C., Masson C., Masson C., et al. Cloning and expression of a queen pheromone-binding protein in the honeybee: An olfactory-specific, developmentally regulated protein. J. Neurosci. 1999;19:7468–7475. doi: 10.1523/JNEUROSCI.19-17-07468.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyu Z., Leal W., Leal W. Conformational isomers of insect odorant-binding proteins. Arch. Biochem. Biophys. 2002;397:99–105. doi: 10.1006/abbi.2001.2660. [DOI] [PubMed] [Google Scholar]
- Drysdale R., Crosby M., Crosby M. FlyBase: Genes and gene models. Nucleic Acids Res. 2005;33:D390–D395. doi: 10.1093/nar/gki046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.5. Department of Genome Sciences, University of Washington; Seattle, WA: 2005. [Google Scholar]
- Galindo K., Smith D., Smith D. A large family of divergent Drosophila odorant-binding proteins expressed in gustatory and olfactory sensilla. Genetics. 2001;159:1059–1072. doi: 10.1093/genetics/159.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galizia C., McIlwrath S., Menzel R., McIlwrath S., Menzel R., Menzel R. A digital three-dimensional atlas of the honeybee antennal lobe based on optical sections acquired by confocal microscopy. Cell Tissue Res. 1999;295:383–394. doi: 10.1007/s004410051245. [DOI] [PubMed] [Google Scholar]
- Gaunt M., Miles M., Miles M. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol. Biol. Evol. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- Goldman A.L., Van der Goes van Naters W., Lessing D., Warr C.G., Carlson J.R., Van der Goes van Naters W., Lessing D., Warr C.G., Carlson J.R., Lessing D., Warr C.G., Carlson J.R., Warr C.G., Carlson J.R., Carlson J.R. Coexpression of two functional odor receptors in one neuron. Neuron. 2005;45:661–666. doi: 10.1016/j.neuron.2005.01.025. [DOI] [PubMed] [Google Scholar]
- Graham L., Davies P., Davies P. The odorant-binding proteins of Drosophila melanogaster: Annotation and characterization of a divergent gene family. Gene. 2002;292:43–55. doi: 10.1016/s0378-1119(02)00672-8. [DOI] [PubMed] [Google Scholar]
- Hekmat-Scafe D., Scafe C., McKinney A., Tanouye M., Scafe C., McKinney A., Tanouye M., McKinney A., Tanouye M., Tanouye M. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster . Genome Res. 2002;12:1357–1369. doi: 10.1101/gr.239402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand J., Shepherd G., Shepherd G. Mechanisms of olfactory discrimination: Converging evidence for common principles across phyla. Annu. Rev. Neurosci. 1997;20:595–631. doi: 10.1146/annurev.neuro.20.1.595. [DOI] [PubMed] [Google Scholar]
- Hill C., Fox A., Pitts R., Kent L., Tan P., Chrystal M., Cravchik A., Collins F., Robertson H., Zwiebel L., Fox A., Pitts R., Kent L., Tan P., Chrystal M., Cravchik A., Collins F., Robertson H., Zwiebel L., Pitts R., Kent L., Tan P., Chrystal M., Cravchik A., Collins F., Robertson H., Zwiebel L., Kent L., Tan P., Chrystal M., Cravchik A., Collins F., Robertson H., Zwiebel L., Tan P., Chrystal M., Cravchik A., Collins F., Robertson H., Zwiebel L., Chrystal M., Cravchik A., Collins F., Robertson H., Zwiebel L., Cravchik A., Collins F., Robertson H., Zwiebel L., Collins F., Robertson H., Zwiebel L., Robertson H., Zwiebel L., Zwiebel L., et al. G protein-coupled receptors in Anopheles gambiae . Science. 2002;298:176–178. doi: 10.1126/science.1076196. [DOI] [PubMed] [Google Scholar]
- Huang X., Madan A., Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida Y., Chiang V., Leal W., Chiang V., Leal W., Leal W. Protein that makes sense in the Argentine ant. Naturwissenschaften. 2002;89:505–507. doi: 10.1007/s00114-002-0368-1. [DOI] [PubMed] [Google Scholar]
- Jeanmougin F., Thompson J., Gouy M., Higgins D., Gibson T., Thompson J., Gouy M., Higgins D., Gibson T., Gouy M., Higgins D., Gibson T., Higgins D., Gibson T., Gibson T. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- Jeffares D., Mourier T., Penny D., Mourier T., Penny D., Penny D. The biology of intron gain and loss. Trends Genet. 2005;22:16–22. doi: 10.1016/j.tig.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Jung-Offmann I. Développement et croissance. In: Chauvin R., editor. Traité de biologie de l'abeille. Masson; Paris: 1968. pp. 69–144. [Google Scholar]
- Kim M., Repp A., Smith D., Repp A., Smith D., Smith D. LUSH odorant-binding protein mediates chemosensory responses to alcohols in Drosophila melanogaster . Genetics. 1998;150:711–721. doi: 10.1093/genetics/150.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger J., Breer H., Breer H. Olfactory reception in invertebrates. Science. 1999;286:720–723. doi: 10.1126/science.286.5440.720. [DOI] [PubMed] [Google Scholar]
- Krieger M., Ross K., Ross K. Molecular evolutionary analyses of the odorant-binding protein gene Gp-9 in fire ants and other Solenopsis species. Mol. Biol. Evol. 2005;22:2090–2103. doi: 10.1093/molbev/msi203. [DOI] [PubMed] [Google Scholar]
- Kruse S., Zhao R., Smith D., Jones D., Zhao R., Smith D., Jones D., Smith D., Jones D., Jones D. Structure of a specific alcohol-binding site defined by the odorant binding protein LUSH from Drosophila melanogaster . Nat. Struct. Biol. 2003;10:694–700. doi: 10.1038/nsb960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharski R., Maleszka R., Maleszka R. Transcriptional profiling reveals multifunctional roles for transferrin in the honeybee, Apis mellifera . J. Insect Sci. 2003;3:27. doi: 10.1093/jis/3.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartigue A., Gruez A., Spinelli S., Riviere S., Brossut R., Tegoni M., Cambillau C., Gruez A., Spinelli S., Riviere S., Brossut R., Tegoni M., Cambillau C., Spinelli S., Riviere S., Brossut R., Tegoni M., Cambillau C., Riviere S., Brossut R., Tegoni M., Cambillau C., Brossut R., Tegoni M., Cambillau C., Tegoni M., Cambillau C., Cambillau C. The crystal structure of a cockroach pheromone-binding protein suggests a new ligand binding and release mechanism. J. Biol. Chem. 2003;278:30213–30218. doi: 10.1074/jbc.M304688200. [DOI] [PubMed] [Google Scholar]
- Lartigue A., Gruez A., Briand L., Blon F., Bezirard V., Walsh M., Pernollet J., Tegoni M., Cambillau C., Gruez A., Briand L., Blon F., Bezirard V., Walsh M., Pernollet J., Tegoni M., Cambillau C., Briand L., Blon F., Bezirard V., Walsh M., Pernollet J., Tegoni M., Cambillau C., Blon F., Bezirard V., Walsh M., Pernollet J., Tegoni M., Cambillau C., Bezirard V., Walsh M., Pernollet J., Tegoni M., Cambillau C., Walsh M., Pernollet J., Tegoni M., Cambillau C., Pernollet J., Tegoni M., Cambillau C., Tegoni M., Cambillau C., Cambillau C. Sulfur single-wavelength anomalous diffraction crystal structure of a pheromone-binding protein from the honeybee Apis mellifera L. J. Biol. Chem. 2004;279:4459–4464. doi: 10.1074/jbc.M311212200. [DOI] [PubMed] [Google Scholar]
- Lee D., Damberger F., Peng G., Horst R., Guntert P., Nikonova L., Leal W., Wuthrich K., Damberger F., Peng G., Horst R., Guntert P., Nikonova L., Leal W., Wuthrich K., Peng G., Horst R., Guntert P., Nikonova L., Leal W., Wuthrich K., Horst R., Guntert P., Nikonova L., Leal W., Wuthrich K., Guntert P., Nikonova L., Leal W., Wuthrich K., Nikonova L., Leal W., Wuthrich K., Leal W., Wuthrich K., Wuthrich K. NMR structure of the unliganded Bombyx mori pheromone-binding protein at physiological pH. FEBS Lett. 2002;531:314–318. doi: 10.1016/s0014-5793(02)03548-2. [DOI] [PubMed] [Google Scholar]
- Lespinet O., Wolf Y., Koonin E., Aravind L., Wolf Y., Koonin E., Aravind L., Koonin E., Aravind L., Aravind L. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res. 2002;12:1048–1059. doi: 10.1101/gr.174302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, S., Searle, S., Harris, N., Gibson, M., Lyer, V., Richter, J., Wiel, C., Bayraktaroglir, L., Birney, E., Crosby, M., 2002. Apollo: A sequence annotation editor. Genome Biol. 3: research0082. [DOI] [PMC free article] [PubMed]
- Löbel D., Jacob M., Volkner M., Breer H., Jacob M., Volkner M., Breer H., Volkner M., Breer H., Breer H. Odorants of different chemical classes interact with distinct odorant binding protein subtypes. Chem. Senses. 2002;27:39–44. doi: 10.1093/chemse/27.1.39. [DOI] [PubMed] [Google Scholar]
- Milne I., Wright F., Rowe G., Marshall D., Husmeier D., McGuire G., Wright F., Rowe G., Marshall D., Husmeier D., McGuire G., Rowe G., Marshall D., Husmeier D., McGuire G., Marshall D., Husmeier D., McGuire G., Husmeier D., McGuire G., McGuire G. TOPALi: Software for automatic identification of recombinant sequences within DNA multiple alignments. Bioinformatics. 2004;20:1806–1807. doi: 10.1093/bioinformatics/bth155. [DOI] [PubMed] [Google Scholar]
- Newcomb R., Sirey T., Rassam M., Greenwood D., Sirey T., Rassam M., Greenwood D., Rassam M., Greenwood D., Greenwood D. Pheromone binding proteins of Epiphyas postvittana (Lepidoptera: Tortricidae) are encoded at a single locus. Insect Biochem. Mol. Biol. 2002;32:1543–1554. doi: 10.1016/s0965-1748(02)00075-9. [DOI] [PubMed] [Google Scholar]
- Paesen G., Happ G., Happ G. The B proteins secreted by the tubular accessory sex glands of the male mealworm beetle, Tenebrio molitor, have sequence similarity to moth pheromone-binding proteins. Insect Biochem. Mol. Biol. 1995;25:401–408. doi: 10.1016/0965-1748(94)00085-v. [DOI] [PubMed] [Google Scholar]
- Paiva-Silva G., Sorgine M., Benedetti C., Meneghini R., Almeida I., Machado E., Dansa-Petretski M., Yepiz-Plascencia G., Law J., Oliveira P., Sorgine M., Benedetti C., Meneghini R., Almeida I., Machado E., Dansa-Petretski M., Yepiz-Plascencia G., Law J., Oliveira P., Benedetti C., Meneghini R., Almeida I., Machado E., Dansa-Petretski M., Yepiz-Plascencia G., Law J., Oliveira P., Meneghini R., Almeida I., Machado E., Dansa-Petretski M., Yepiz-Plascencia G., Law J., Oliveira P., Almeida I., Machado E., Dansa-Petretski M., Yepiz-Plascencia G., Law J., Oliveira P., Machado E., Dansa-Petretski M., Yepiz-Plascencia G., Law J., Oliveira P., Dansa-Petretski M., Yepiz-Plascencia G., Law J., Oliveira P., Yepiz-Plascencia G., Law J., Oliveira P., Law J., Oliveira P., Oliveira P., et al. On the biosynthesis of Rhodnius prolixus heme-binding protein. Insect Biochem. Mol. Biol. 2002;32:1533–1541. doi: 10.1016/s0965-1748(02)00074-7. [DOI] [PubMed] [Google Scholar]
- Pelosi P. Perireceptor events in olfaction. J. Neurobiol. 1996;30:3–19. doi: 10.1002/(SICI)1097-4695(199605)30:1<3::AID-NEU2>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Pelosi P. Odorant-binding proteins: Structural aspects. Ann. N.Y. Acad. Sci. 1998;855:281–293. doi: 10.1111/j.1749-6632.1998.tb10584.x. [DOI] [PubMed] [Google Scholar]
- Pelosi P., Maida R., Maida R. Odorant-binding proteins in insects. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1995;111:503–514. doi: 10.1016/0305-0491(95)00019-5. [DOI] [PubMed] [Google Scholar]
- Pelosi P., Calvello M., Ban L., Calvello M., Ban L., Ban L. Diversity of odorant-binding proteins and chemosensory proteins in insects. Chem. Senses. 2005;30(Suppl. 1.):i291–i292. doi: 10.1093/chemse/bjh229. [DOI] [PubMed] [Google Scholar]
- Plettner E., Lazar J., Prestwich E., Prestwich G., Lazar J., Prestwich E., Prestwich G., Prestwich E., Prestwich G., Prestwich G. Discrimination of pheromone enantiomers by two pheromone binding proteins from the gypsy moth Lymantria dispar . Biochemistry. 2000;39:8953–8962. doi: 10.1021/bi000461x. [DOI] [PubMed] [Google Scholar]
- Pond S., Frost S., Frost S. A simple hierarchical approach to modeling distributions of substitution rates. Mol. Biol. Evol. 2005a;22:223–234. doi: 10.1093/molbev/msi009. [DOI] [PubMed] [Google Scholar]
- Pond S., Frost S., Frost S. Datamonkey: Rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005b;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- Pond S., Frost S., Muse S., Frost S., Muse S., Muse S. HyPhy: Hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Pophof B. Moth pheromone binding proteins contribute to the excitation of olfactory receptor cells. Naturwissenschaften. 2002;89:515–518. doi: 10.1007/s00114-002-0364-5. [DOI] [PubMed] [Google Scholar]
- Pophof B. Pheromone-binding proteins contribute to the activation of olfactory receptor neurons in the silkmoths Antheraea polyphemus and Bombyx mori . Chem. Senses. 2004;29:117–125. doi: 10.1093/chemse/bjh012. [DOI] [PubMed] [Google Scholar]
- Prasad B., Reed R., Reed R. Chemosensation: Molecular mechanisms in worms and mammals. Trends Genet. 1999;15:150–153. doi: 10.1016/s0168-9525(99)01695-9. [DOI] [PubMed] [Google Scholar]
- Robertson H.M., Wanner K.W., Wanner K.W. Genome Res. 2006. The chemoreceptor superfamily in the honey bee Apis mellifera: Expansion of the odorant, but not gustatory, receptor family. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H., Warr C., Carlson J., Warr C., Carlson J., Carlson J. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster . Proc. Natl. Acad. Sci. 2003;100(Suppl. 2):14537–14542. doi: 10.1073/pnas.2335847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamov A., Solovyev V., Solovyev V. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag S., Hekmat-Scafe D., Kim M., Park S., Carlson J., Pikielny C., Smith D., Steinbrecht R., Hekmat-Scafe D., Kim M., Park S., Carlson J., Pikielny C., Smith D., Steinbrecht R., Kim M., Park S., Carlson J., Pikielny C., Smith D., Steinbrecht R., Park S., Carlson J., Pikielny C., Smith D., Steinbrecht R., Carlson J., Pikielny C., Smith D., Steinbrecht R., Pikielny C., Smith D., Steinbrecht R., Smith D., Steinbrecht R., Steinbrecht R. Expression mosaic of odorant-binding proteins in Drosophila olfactory organs. Microsc. Res. Tech. 2001a;55:297–306. doi: 10.1002/jemt.1179. [DOI] [PubMed] [Google Scholar]
- Shanbhag S., Park S., Pikielny C., Steinbrecht R., Park S., Pikielny C., Steinbrecht R., Pikielny C., Steinbrecht R., Steinbrecht R. Gustatory organs of Drosophila melanogaster: Fine structure and expression of the putative odorant-binding protein PBPRP2. Cell Tissue Res. 2001b;304:423–437. doi: 10.1007/s004410100388. [DOI] [PubMed] [Google Scholar]
- Shanbhag S., Smith D., Steinbrecht R., Smith D., Steinbrecht R., Steinbrecht R. Three odorant-binding proteins are co-expressed in sensilla trichodea of Drosophila melanogaster . Arthropod Structure & Development. 2005;34:153–165. [Google Scholar]
- Swanson W., Nielsen R., Yang Q., Nielsen R., Yang Q., Yang Q. Pervasive adaptive evolution in mammalian fertilization proteins. Mol. Biol. Evol. 2003;20:18–20. doi: 10.1093/oxfordjournals.molbev.a004233. [DOI] [PubMed] [Google Scholar]
- Thymianou S., Mavroidis M., Kokolakis G., Komitopoulou K., Zacharopoulou A., Mintzas A., Mavroidis M., Kokolakis G., Komitopoulou K., Zacharopoulou A., Mintzas A., Kokolakis G., Komitopoulou K., Zacharopoulou A., Mintzas A., Komitopoulou K., Zacharopoulou A., Mintzas A., Zacharopoulou A., Mintzas A., Mintzas A. Cloning and characterization of a cDNA encoding a male-specific serum protein of the Mediterranean fruit fly, Ceratitis capitata, with sequence similarity to odourant-binding proteins. Insect Mol. Biol. 1998;7:345–353. doi: 10.1046/j.1365-2583.1998.740345.x. [DOI] [PubMed] [Google Scholar]
- Vogt R.G. Biochemical diversity of odor detection. In: Blomquist G., Vogt R., Vogt R., editors. Insect pheromone biochemistry and molecular biology. Elsevier; London: 2003. pp. 391–445. [Google Scholar]
- Vogt R.G., Rybczynshi R., Lerner M.R., Rybczynshi R., Lerner M.R., Lerner M.R. The biochemistry of odorant reception and transduction. In: Schild D., editor. Chemosensory information processing. Springer-Verlag; Berlin: 1990. pp. 33–76. [Google Scholar]
- Willett C. Evidence for directional selection acting on pheromone-binding proteins in the genus Choristoneura . Mol. Biol. Evol. 2000;17:553–562. doi: 10.1093/oxfordjournals.molbev.a026335. [DOI] [PubMed] [Google Scholar]
- Wong W., Yang Z., Goldman N., Nielsen R., Yang Z., Goldman N., Nielsen R., Goldman N., Nielsen R., Nielsen R. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics. 2004;168:1041–1051. doi: 10.1534/genetics.104.031153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Zwiebel L., Smith D., Zwiebel L., Smith D., Smith D. Identification of a distinct family of genes encoding atypical odorant-binding proteins in the malaria vector mosquito, Anopheles gambiae . Insect Mol. Biol. 2003;12:549–560. doi: 10.1046/j.1365-2583.2003.00440.x. [DOI] [PubMed] [Google Scholar]
- Xu P., Atkinson R., Jones D., Smith D., Atkinson R., Jones D., Smith D., Jones D., Smith D., Smith D. Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron. 2005;45:193–200. doi: 10.1016/j.neuron.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z., Wong W., Nielsen R., Wong W., Nielsen R., Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Zhou J., Huang W., Zhang G., Pickett J., Field L., Huang W., Zhang G., Pickett J., Field L., Zhang G., Pickett J., Field L., Pickett J., Field L., Field L. Plus-C” odorant-binding protein genes in two Drosophila species and the malaria mosquito Anopheles gambiae . Gene. 2004a;327:117–129. doi: 10.1016/j.gene.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Zhou J., Zhang G., Huang W., Birkett M., Field L., Pickett J., Pelosi P., Zhang G., Huang W., Birkett M., Field L., Pickett J., Pelosi P., Huang W., Birkett M., Field L., Pickett J., Pelosi P., Birkett M., Field L., Pickett J., Pelosi P., Field L., Pickett J., Pelosi P., Pickett J., Pelosi P., Pelosi P. Revisiting the odorant-binding protein LUSH of Drosophila melanogaster: Evidence for odour recognition and discrimination. FEBS Lett. 2004b;558:23–26. doi: 10.1016/S0014-5793(03)01521-7. [DOI] [PubMed] [Google Scholar]