Abstract
Posttranslational modifications of histones play important roles in chromatin structure and genomic stability. Distinct lysine residues in histones are targets for covalent binding of biotin, catalyzed by holocarboxylase synthetase (HCS) and biotinidase (BTD). Histone biotinylation has been implicated in heterochromatin structures, DNA repair, and mitotic chromosome condensation. To test whether HCS and BTD deficiency alters histone biotinylation, and to characterize phenotypes associated with HCS and BTD deficiency, HCS- and BTD-deficient flies were generated by RNA interference (RNAi). Expression of HCS and BTD decreased by 65–90% in RNAi-treated flies, as judged by mRNA abundance, BTD activity, and abundance of HCS protein. Decreased expression of HCS and BTD caused decreased biotinylation of K9 and K18 in histone H3. This was associated with altered expression of 201 genes in HCS-deficient flies. Lifespan of HCS- and BTD-deficient flies decreased by up to 32% compared to wild-type controls. Heat tolerance decreased by up to 55% in HCS-deficient flies compared to controls, as judged by survival times; effects of BTD deficiency were minor. Consistent with this observation, HCS deficiency was associated with altered expression of 285 heat-responsive genes. HCS and BTD deficiency did not affect cold tolerance, suggesting stress-specific effects of chromatin remodeling by histone biotinylation. This is the first study to provide evidence that HCS-dependent histone biotinylation affects gene function and phenotype, suggesting that the complex phenotypes of HCS- and BTD-deficiency disorders may reflect chromatin structure changes.
Keywords: biotinidase, Drosophila melanogaster, holocarboxylase synthetase, lifespan, stress resistance
In eukaryotes, biotin serves as a covalently bound coenzyme for acetyl-CoA carboxylase α, acetyl-CoA carboxylase β, pyruvate carboxylase (PC)3, propionyl-CoA carboxylase (PCC), and 3-methylcrotonyl-CoA carboxylase. Carboxylases play essential roles in the metabolism of fatty acids, glucose, and some amino acids. The attachment of biotin to specific lysine (K) residues in carboxylases is catalyzed by holocarboxylase synthetase (HCS) in an ATP-dependent reaction (1). Recently, our view of potential biological functions of biotin has been expanded by the observation that histones contain covalently bound biotin (2, 3).
The following lysine (K) residues in histones are targets for biotinylation: K9, K13, K125, K127, and K129 in histone H2A; K4, K9 and K18 in histone H3; and K8 and K12 in histone H4 (4–6). Evidence suggests that K8-biotinylated histone H4 and K12-biotinylated histone H4 are enriched in heterochromatin and participate in gene silencing (Camporeale, G., Oommen, A. M., Griffin, J. B., Sarath, G. & Zempleni, J., unpublished data). Moreover, the abundance of K8- and K12 biotinylated histone H4 is cell cycle dependent and reaches peak levels during mitotic chromatin condensation (7). Finally, K12-biotinylated histone H4 might participate in the cellular response to DNA double-strand breaks (8).
Histone biotinylation is mediated by HCS (9) and perhaps biotinidase (BTD) (3). Covalent modifications of histones are typically reversible (10); enzymes mediating debiotinylation of histones are largely unknown. Evidence suggests that BTD may mediate debiotinylation of histones (11) in addition to acting as a histone biotinyl transferase (3). It is uncertain how cells control these two opposing enzymatic activities of BTD. Variables such as the microenvironment in confined regions of chromatin, posttranslational modifications of BTD, and alternate splicing of BTD might determine whether BTD acts as biotinyl histone transferase or histone debiotinylase (12). In addition, BTD-binding proteins might contribute to regulation.
The biotin protein ligases are evolutionarily conserved. A subset of these enzymes (exemplified by E. coli BirA) is known to function as transcriptional repressors (13, 14). The effector of BirA transcriptional repression is biotinyl-5′-adenylate (bio-5′-AMP). The BirA-bio-5′-AMP complex is active in both enzymatic transfer of biotin and site-specific DNA binding. When biotin is abundant, BirA-bio-5′-AMP accumulates and binds to the operator sequence of the biotin biosynthesis operon to repress transcription. Thus, BirA couples the intracellular demand for and synthesis of biotin in E. coli. HCS is a structural and functional homolog of BirA (15, 16).
Inborn mutations of human HCS and BTD decrease the histone biotinyl transferase activity of these enzymes (3, 5, 9). For example, biotinylation of histones is decreased in HCS-deficient human fibroblasts (5, 9) and lymphoma (Jurkat) cells (Camporeale, G. & Zempleni, J., unpublished data). Notwithstanding the potential importance of these observations, effects of hcs and btd gene mutations on chromatin structure and phenotypes are largely unknown.
The Drosophila melanogaster genome encodes orthologs of human HCS (GenBank accession number AAF52022) and BTD (GenBank accession number AAF46130). Consistent with this observation, all five major classes of histones are biotinylated in Drosophila (17). Biotin deficiency is associated with decreased lifespan and fertility in flies (17), but it remains uncertain whether this is caused by altered biotinylation of histones.
In the present study, Drosophila was used as a model to investigate whether knockdown of HCS and BTD expression is associated with altered chromatin structure, as evidenced by decreased biotinylation of histone H3. Histone H3 was chosen as a model because it is biotinylated more abundantly than other histones in flies (17). We used HCS- and BTD-deficient flies to investigate effects of altered histone biotinylation on phenotypes in flies. Here, we focused on lifespan and temperature tolerance as markers for phenotypic changes, given that previous studies had linked biotin status to these variables (17). We identified genes whose expression is altered by loss of HCS expression.
MATERIALS AND METHODS
Immunofluorescent staining of polytene chromosomes
Larval polytene chromosome squashes were prepared essentially as described (18). Anti-HCS serum was used at 1:50 dilution; FITC-labeled anti-rabbit secondary antibody was used at 1:200 dilution.
Generation of transgenic flies
HCS- and BTD-deficient flies were generated by using RNA interference (RNAi) by inserting gene-specific cassettes into the pUAST-sp plasmid (19). For generation of a vector targeting HCS, a 1.4kb DNA fragment of the Drosophila CG14670 gene (coordinates 35696 to 37133, GenBank accession no. AE003602) was PCR amplified using the following PCR primers: EcoRI-5′-HCS (5′- ATATGAATTCGGAAGATTACGGTAAGCTAATTGC-3′) and EcoRI-3’-HCS (5′-ATATGAATT CGCGACTTGGTGGACTCCTCGC-3′) to generate the EcoRI-HCS-EcoRI cassette; and XbaI-5′-HCS (5′-CGCTCTAGATTACGGTAAGCTAATTGC-3′) and Xho-3′-HCS (5′-CGCCTCGAGTGCGACTT GGTGGACTCC-3′) to generate the XbaI-HCS-XhoI cassette. Both the EcoRI-HCS-EcoRI and the XbaI-HCS-XhoI cassettes were inserted as inverted repeats (IR) into the pUAST-sp vector to generate a plasmid denoted pUAST-IRsp-HCS; in the pUAST-IRsp-HCS plasmid the HCS cassettes are separated by a 200 bp spacer to enhance stability of the IR (19). For generation of a vector targeting BTD, a 1.5 kb DNA fragment of the Drosophila CG3599 gene (coordinates 300621 to 302187, GenBank accession no. AE003436) was PCR amplified using the following PCR primers: EcoRI-5′-BTD (5′-CGCGAATTCGCCGCCTCTGGTGATTCAATCC-3′) and BglII-3′-BTD (5′-CGCAGATC TGTCAGTGCTCTTCAGCAGTTCG-3′) to generate the EcoRI-BTD-BglII cassette; and XbaI-5′-BTD (5′-TTGTCTTGTCTAGATTATTCGCCGCC-3′) and XhoI-3′-BTD (5′-CGCCTCGAGTGTCAGTG CTCTTCAGCAGTTCG-3′) to generate the XbaI-BTD-XhoI cassette. Both the EcoRI-BTD-BglII and the XbaI-BTD-XhoI cassettes were inserted into the pUAST-sp vector as described above to generate a plasmid denoted pUAST-IRsp-BTD (19).
Germline transformations with pUAST-IRsp-HCS and pUAST-IRsp-BTD were conducted using P elements as described (20). In transformed flies, transcription of siRNA is driven by Gal4. Hence, the experiments described below were conducted by mating pUAST-IRsp-HCS and pUAST-IRsp-BTD transgenic virgin female flies to male Actin5C-Gal4 driver lines. Flies with straight wings produce siRNA and were selected for further experimentation. As a control, we used F1 crosses of wild-type (y w) virgin females with Actin5C-Gal4 male drivers (denoted as Actin5C-Gal4 control flies). The following additional controls were used in some experiments: Act5C-Gal4 driver flies; y w flies; and pUAST-IRsp-HCS and pUAST-IRsp-BTD flies that were not crossed with the Act5C-Gal4 driver.
Drosophila husbandry
Flies were housed at 25ºC on standard cornmeal diet and were transferred to fresh tubes every 72 h unless noted otherwise. Flies were kept on a 12 h light/dark cycle.
Abundance of mRNA coding for HCS and BTD
Total RNA was isolated from 100 flies with TriZol reagent according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). Genomic DNA was removed enzymatically using TURBO DNA-free™ (Ambion, Austin, TX). Briefly, cDNA was synthesized using the ImProm-II reverse transcriptase system from Promega (Madison, USA). Primers for HCS (forward primer = 5′-AGTATTGGAATTGGAGAATGC-3′; reverse primer = 5′-AACTTACATATTGATGGGAACC-3′) and BTD (forward primer = 5′-CTACTACACCGATATACCCAGCAC-3′; reverse primer = 5′-CGCTGATGAAACAGTTGAGGAC-3′) were designed using the Beacon designer 4.0 software (Premier Biosoft International, Palo Alto, CA). Real-time PCR was performed using the iCycler IQ™ multicolor real-time detection system (Bio-Rad, Palo Alto, CA). Histone H4 (GenBank accession number NM_165383) was used as reference gene (forward primer = 5′-GTGCTGCGTGATAACATC-3′; reverse primer = 5′-TGGCTGTAACTGTCTTCC-3′).
Biotinylated carboxylases
The abundance of biotinylated carboxylases and the activity of PCC were quantified as markers for HCS deficiency in transgenic flies. Briefly, 50 flies were homogenized as described (17) and proteins (~ 100 μg) were resolved using 4–8% Tris-acetate gels (Invitrogen). Transblots were probed with streptavidin peroxidase (21). Band intensities were quantified by gel densitometry (22). Activity of PCC in fly homogenates was quantified as described (17).
Biotinidase activity
Whole fly homogenates were prepared as described above; enzyme activity in homogenates was quantified by monitoring the BTD-mediated hydrolysis of biotinyl-p-aminobenzoic acid in a colorimetric assay (2).
HCS abundance
Abundance of HCS in whole fly homogenates was quantified by Western blot analysis using rabbit anti-human HCS (23); this antibody cross-reacts with HCS from Drosophila. Proteins were resolved using 4–8% Bis-Tris gels (Invitrogen), and transblots were probed with anti-HCS serum (20,000-fold dilution) and goat anti-rabbit IgG peroxidase conjugate (Sigma, St. Louis, MO; 10 ng/mL).
Biotinylated histones
Histone extracts from whole fly homogenates were prepared as described (24) with minor modifications: 5 mol/L HCl was added to fly homogenates to produce a final concentration of 0.25 mol/L. Histones were extracted overnight (4ºC); insoluble proteins were removed by centrifugation and histones in the supernatant were precipitated with trichloroacetic acid (1.2 mol/L final concentration). Histones were washed with acetone and dissolved in 8 M urea. Histones were resolved by gel electrophoresis (6) and probed using either streptavidin-peroxidase (2) or antisera (250-fold dilution) to K9-biotinylated histone H3 (K9BioH3) and K18-biotinylated histone H3 (K18BioH3) (5). Histone loading was equalized based on protein assays, densitometric quantitation of gels stained with Commassie blue and Western blot analysis using antibody to histone H3 C-terminal tail.
DNA microarray
RNA from 50 flies was extracted with 1 mL of TriZol reagent as described above. RNA was purified using the RNeasy mini kit (Qiagen, Valencia, CA) and hybridized to the Drosophila melanogaster Genome 2.0 Array (Affymetrix, Santa Clara, CA) at the Genomics Core Research Facility, University of Nebraska-Lincoln (25). GeneChip operating software 1.4 (Affymetrix) was used for normalization and analysis of microarray data.
Lifespan
The lifespan of flies was determined as described previously with minor modifications (17). Briefly, newly eclosed female and male flies were housed separately in groups of 20 flies/tube (n = 10 tubes per treatment group). Dead flies were counted and removed daily.
Temperature stress
Resistance of flies to cold and heat stress was determined as described previously with minor modifications (17). In cold stress experiments, male and female flies were housed separately at 4°C for up to 16 h; at timed intervals flies were transferred to room temperature and surviving flies were counted after 24 h. At each time point four tubes (25 flies each) were collected (n=4). For heat stress experiments, four tubes (25 flies each) were kept at 34°C up to 9 h; dead flies were counted every 0.5 h.
Statistics
Homogeneity of variances among groups was tested using Bartlett’s test (26). If variances were heterogeneous, data were log-transformed before further statistical analysis. Significance of differences among groups was tested by one-way ANOVA. Fisher’s Protected Least Significant Difference procedure was used for posthoc testing (26). Effects of treatment over time (e.g., resistance to heat stress) were analyzed by repeated measures ANOVA. If effects of treatment over time were statistically significant, treatment effects at individual time points were assessed by one-way ANOVA and Fisher’s Protected Least Significant Difference procedure for individual time points. StatView 5.0.1 (SAS Institute; Cary, NC) was used to perform all calculations. Differences were considered significant if P < 0.05. Data are expressed as means ± SD.
RESULTS
HCS is a chromosomal protein in Drosophila
Previous studies showed that BTD and HCS are found in the nuclear as well as the cytoplasmic fractions (23, 27). To test whether HCS is bound to chromosomes, we immunostained fixed giant polytene chromosomes from Drosophila third instar larvae with antibody to HCS. HCS was broadly distributed across the euchromatic arms of all chromosomes, with particular concentrations at certain bands, some puffs (sites of active transcription) and some telomeres (Fig. 1). Pre-immune serum used at the same concentration produced no significant staining of polytene chromosomes. This result supports a model in which HCS plays a direct role in chromatin remodeling and histone biotinylation at specific chromosomal loci.
FIGURE 1.
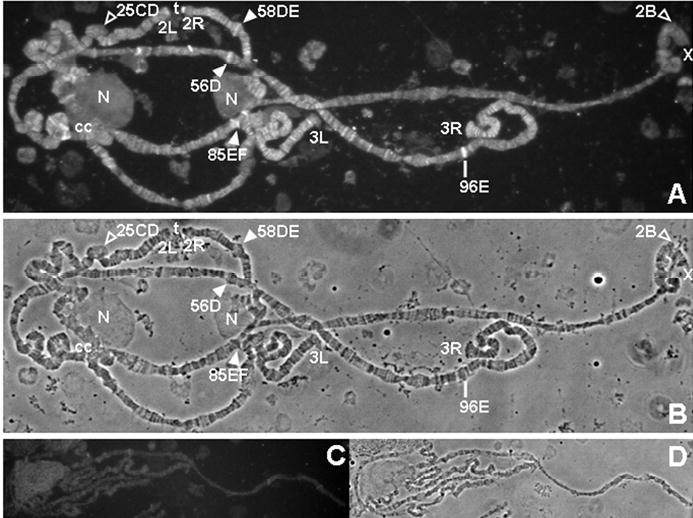
Holocarboxylase synthetase (HCS) binds to distinct chromosomal sites in wild-type Drosophila melanogaster. Panel A: Immunostaining of fixed polytene chromosomes with anti-HCS serum. Panel B: Phase contrast image of chromosomes in A. Panel C: Immunostaining of fixed polytene chromosomes with pre-immune serum. Panel D: Phase contrast image of chromosomes in C. Legend: filled arrowheads = puffs that stain for HCS; open arrowheads = puffs that do not stain for HCS; line = non-puffed site that stains for HCS; t = staining telomere; N = nucleolus; cc = heterochromatic chromocenter; X = X chromosome; 2L and 2R = left and right arms of chromosome 2; 3L and 3R = left and right arms of chromosome 3.
Expression of HCS and BTD in RNAi knockdown flies
Expression of HCS and BTD RNAi in transformed flies caused a substantial decrease in the abundance of HCS and BTD, respectively. The abundance of mRNA encoding HCS and BTD was smaller in knockdown flies compared to Act5C-Gal4 control flies (Fig. 2A). The following additional controls were tested and produced amounts of HCS and BTD mRNA similar to those observed in Act5C-Gal4 control flies: Act5C-Gal4 driver flies, y w flies, and pUAST-IRsp-HCS and pUAST-IRsp-BTD flies not crossed with the Act5C-Gal4 driver (data not shown). HCS protein was barely detectable in HCS-RNAi flies compared with BTD-RNAi and Act5C-Gal4 control flies; β-actin was used as control and was not affected by HCS knockdown (Fig. 2B). BTD activity in homogenates from BTD-RNAi flies decreased compared with Act5C-Gal4 control flies; BTD activity was unaffected in HCS-RNAi flies (Fig. 2C).
FIGURE 2.
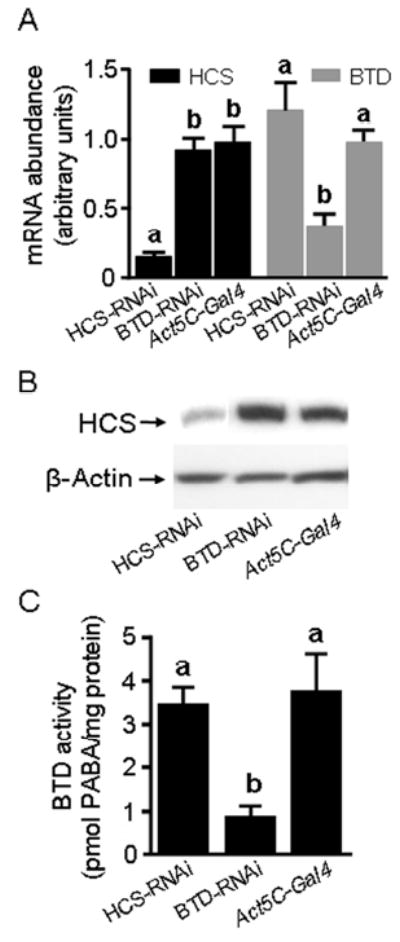
RNAi to HCS and biotinidase (BTD) decreases the expression of HCS and BTD, respectively, in Drosophila melanogaster compared to control flies. Panel A: mRNA levels of HCS and BTD in HCS-RNAi flies, BTD-RNAi flies, and Act5C-Gal4 control flies. Panel B: Quantification of HCS protein by Western blot analysis; β-actin was used as a loading control. Panel C: Quantification of BTD activity in fly homogenates. PABA = para-amino benzoic acid. a,bColumns not sharing the same letter are significantly different (P < 0.05). Values are means ± SD (n = 3).
Biotin-dependent carboxylases
Biotinylation of carboxylases decreased significantly in HCS-RNAi and BTD-RNAi flies compared with Act5C-Gal4 controls, as judged by streptavidin blotting (Fig. 3A) and gel densitometric analysis (Fig. 3B). 3-Methylcrotonyl-CoA carboxylase and PCC co-migrated as one single band and were not considered for analysis by gel densitometry; acetyl-CoA carboxylase was barely detectable in any of the treatment groups and was also not considered for analysis by gel densitometry. Decreased biotinylation of carboxylases in HCS- and BTD-deficient flies was associated with decreased PCC activity compared to Act5C-Gal4 controls (Figure 3C). The abundance of biotinylated carboxylases and PCC activity was similar in homogenates from Act5C-Gal4 control flies and various other controls such as the Act5C-Gal4 driver stock and y w flies (data not shown).
FIGURE 3.
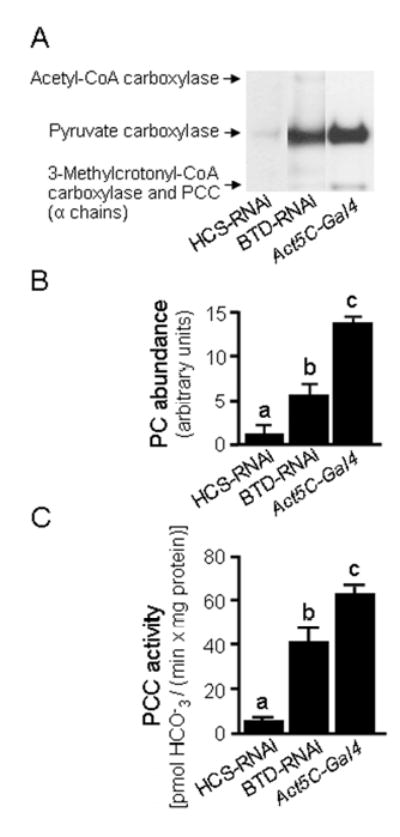
RNAi knockdown of HCS and BTD is associated with decreased biotinylation of carboxylases in Drosophila melanogaster. Panel A: Abundance of biotinylated carboxylases in HCS-deficient (HCS-RNAi) and BTD-deficient (BTD-RNAi) flies compared with Act5C-Gal4 control flies. PCC = Propionyl-CoA carboxylase. Panel B: Gel densitometric quantification of pyruvate carboxylase (PC). Panel C: PCC activity in fly homogenates. a,b,cColumns not sharing the same letter are significantly different from other groups (P < 0.05). Values are means ± SD (n = 3).
Decreased biotinylation of carboxylases in HCS-RNAi and BTD-RNAi flies is likely mediated by two distinct mechanisms. In HCS-RNAi flies, the decreased biotinylation of carboxylases is mediated by decreased biotinyl ligase activity. In BTD-RNAi flies, biotinidase deficiency leads to biotin deficiency known from individuals affected with this inborn error (28). The mechanism is speculated to be increased urinary excretion of biocytin (the biotinyl lysine remnant of proteolytic breakdown of biotinylated proteins) and failure to efficiently digest and absorb biotin covalently bound to dietary proteins.
Biotinylation of histones
Previous studies provided evidence for gender specificity of histone biotinylation in flies (17). Hence, we investigated the effects of HCS and BTD knockdown on histone biotinylation separately for males and females. In males, both HCS and BTD deficiency caused a decrease in histone biotinylation. However, effects were more pronounced for HCS deficiency than for BTD deficiency. When biotinylated histones were probed with streptavidin-peroxidase as a general probe for biotin, the signal intensity was substantially decreased in HCS-RNAi flies and was moderately decreased in BTD-RNAi flies compared with Act5C-Gal4 controls (Fig. 4A). Biotinylated histone H4 was barely detectable in both transgenic flies and controls, consistent with previous studies in our laboratory (17). An unidentified biotinylated protein migrated between histones H1 and H3; this protein is denoted “unknown” in Fig. 4A. Based on size and acid solubility of this protein, we speculate that this band corresponds to an unidentified species of biotinylated histone or histone variant. In contrast to males, HCS and BTD deficiency had only minor effects on biotinylation of histones in female flies (Fig. 4A).
FIGURE 4.
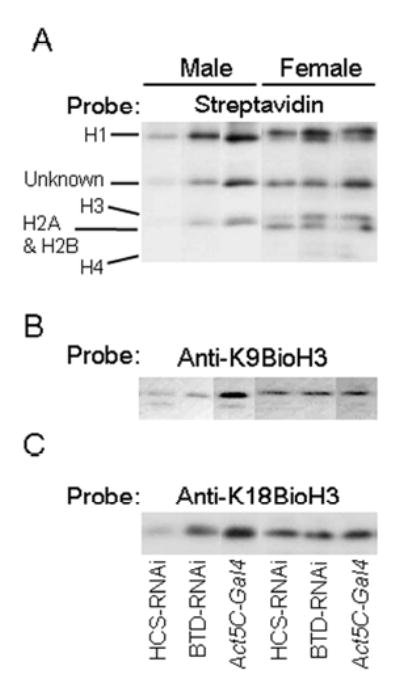
RNAi knockdown of HCS and BTD is associated with decreased biotinylation of histones in Drosophila melanogaster. Panel A: Abundance of biotinylated histones in HCS-RNAi and BTD-RNAi flies compared with Act5C-Gal4 control flies, as judged by streptavidin blotting. Panel B: Transblots probed with anti-K9BioH3. Panel C: Transblots probed with anti-K18BioH3. Note that all lanes shown were from the same blot, although the order presented is changed in order to facilitate comparisons.
Next, we determined whether HCS and BTD deficiency had effects on the biotinylation of specific lysine residues. K9BioH3 and K18BioH3 were substantially decreased in male HCS-RNAi flies, and were moderately decreased in male BTD-RNAi flies compared with Act5C-Gal4 controls (Fig. 4B and 4C). In female flies, the abundance of K9BioH3 and K18BioH3 was decreased moderately in HCS-RNAi flies, whereas no decrease was detectable in BTD-RNAi flies compared with Act5C-Gal4 controls (Fig. 4B and 4C).
Theoretically, the small magnitude of effects of HCS and BTD deficiency in females compared with males could have been caused by inefficient knockdown of enzymes in females. We sought to exclude this possibility by quantifying the abundance of biotinylated carboxylases separately for males and females. The percent decrease of biotinylated carboxylases was similar in males and females. For example, the activity of biotinylated PC decreased by about 90% in male and female HCS-deficient flies compared with Act5C-Gal4 controls. Likewise, the abundance of biotinylated PC decreased by about 50% in male and female BTD-deficient flies compared with controls (data not shown). These data suggest that the quantitatively minor decrease of biotinylated histones in female HCS-RNAi and BTD-RNAi flies compared with males was not caused by insufficient knockdown of HCS and BTD.
Gene expression is affected by HCS deficiency
We used expression profiling of knockdown flies to look for evidence that patterns of gene expression were altered in response to changes in chromatin modification. As a model, we selected the treatment group that exhibits the greatest alteration of histone biotinylation in the above experiments: HCS-deficient male flies. Gene expression profiles in these flies and Act5C-Gal4 controls were obtained by using DNA microarrays. The expression of 77 genes decreased by at least 50% in HCS-RNAi flies compared with Act5C-Gal4 controls, whereas the expression of 124 genes increased by at least 100% compared to controls (online supplemental Table S1). We observed some clusters of HCS-dependent genes, based on biological process and molecular function. For example, HCS deficiency was associated with substantially increased expression of genes that play roles in signal transduction, transport, and cell death, and is associated with substantially decreased expression of genes that play roles in defense response (Table 1).
SUPPLEMENTAL TABLE S1.
Genes with increased or decreased expression in HCS-deficient (HCS-RNAi) male Drosophila compared with Act5C-Gal4 control flies
|
Expression increased by >200% in HCS-RNAi flies compared with controls:1
defective chorion 1 (CG2175); lethal (3) malignant blood neoplasm (CG12755); held out wings (CG10293); vitelline membrane 34Ca (CG9271); pfam01151 (CG8534); vitelline membrane 26Ab (CG9046); RAB (CG32776); ventral veins lacking (CG10037); Rieske iron-sulfur protein (CG7361); cytochrome P450-4e3 (CG4105); vitelline membrane 26Aa (CG9048); chorion protein 36 (CG1478); DUF229 (CG9168); chitin binding Peritrophin-A (CG9307); nanos (CG5637); unknown genes (CG17042; CG5480; CG17298; CG14118; CG14934); unidentified transcripts (Affymetrix probe identifiers 1639811_at; 1635146_at; 1635036_at; 1624084_s_at). |
|
Expression increased by 100 to 200% in HCS-RNAi flies compared with controls:
cutlet (CG33122); Saccharomyces cerevisiae UAS construct a of Yu (CG18405); male fertility factor kl3 (AAG29546); oskar (CG10901); mitochondrial carrier protein (CG18327); follicle cell protein 3C (CG18508); Kelch (CG17754); emp24/gp25L/p24 family (CG33105); CHK (CG5126); widerborst (CG5643); LeuB (CG7755); odorant-binding protein 83cd (CG15582); DUF233 (CG2650); transferrin 1 (CG6186); predicted Na+-dependent transporter (CG9903); white (CAA53795); Nhe2 (CG9256); LRR_RI (CG5096); longest ORF (CG13606); Ccp84Aa (CG2360); Tryp_SPc (CG31267); larval serum protein 1 (CG6821); misexpression suppressor of KSR 4 (CG31274); AMP deaminase (CG32626); zf-C2H2 (CG10309); lysozyme P (CG9116); fat body protein 1 (CG17285); shaking B (CG32508); LRR_RI (CG2471); tropomyosin 1 (CG4898); Tryp_SPc (CG11911); longitudinals lacking (CG12052); yolkless (CG1372); peptidase S10 (CG32483); ACBP (CG8628); RH40749p (CG6131); nuclear factor I (CG2380); bicoid (CG1034); peptidase C48 (CG11023); Rgk1 (CG9811); Ecdysone-induced protein 74EF (CG32180); His4 (CG31611); misexpression suppressor of KSR 4 (CG31274); Myb-interacting protein 130 (CG3480); fat body protein 2 (CG3763); LP19160p (CG13044); PycA (CG1516); Sec14p-like lipid-binding domain (CG1211); glass multimer reporter construct of Nolan (CG8556); pr-set7 (CG3307); RE12806p (CG17086); chorion protein 38 (CG11213); Usf (CG17592); zf-C3HC4 (CG14435); Gid (CG4610); RE05031p (CG17475); IP03589p (CG14957); cAMP-dependent protein kinase 1 (CG4379); twin of eyeless (CG11186); IP05101p (CG32024); pasilla (CG8144); FReD (CG30281); Pleckstrin homology (PH) domain (CG32982); alkPPc (CG3290); female sterile (1) homoerotic (CG2252); peptidase S9 (CG11034); checkpoint suppressor homologue (CG12690); brain tumor (CG10719); retrotransposon gag protein (CG10102); SEC14 (CG33514); trypsin (CG18211); MICAL (CG33208); unknown genes (CG18348; CG13165; CG13043; CG30395; CG3805; CG14406; CG12998; CG33204; CG32694; CG15263; CG13042; CG14752; CG31038); unidentified transcripts (Affymetrix probe identifiers 1634519_at; 1639606_at; 1639054_s_at; 1641367_at; 1624379_s_at; 1626205_s_at; 1635258_s_at; 1640955_s_at). |
|
Expression decreased by >75% in HCS-RNAi flies compared with controls:
PGRP-SB2 (CG9697); dissatisfaction (CG9019); dopamine receptor 2 (CG18741); larval cuticle protein 1 (CG11650); chorion protein 18 (CG6517); chorion protein 16 (CG6533); chromosome segregation ATPase (CG13083); chorion protein 19 (CG6524); IP04471p (CG30096); FReD (CG32496); Sugar_tr (CG6640); unknown genes (CG32644; CG13084; CG18607; CG14632); unidentified transcripts (Affymetrix probe identifiers 1631178_at; 1632150_at; 1628898_at). |
|
Expression decreased by 50 to 75% in HCS-RNAi flies compared with controls:
yolk protein 2 (CG2979); abhydro lipase (CG2772); strawberry notch (CG1903); yolk protein 3 (CG11129); trypsin-like serine protease (CG17404); GH03623p (CG4784); trypsin-like serine protease (CG1304); yolk protein 1 (CG2985); SAK (CG7186); PSF1 (CG9187); cecropin C (CG1373); LP22678p (CG17738); GH01676p (CG17352); serine protease 6 (CG2071); UDPGT (CG17322); Ugt86Dd (CG6633); trypsin-like serine protease (CG18179); IGcam (CG15214); SSM4 (CG15059); galactose-specific C-type lectin (CG9976); Smc (CG17177); IP07781p (CG14315); attacin-A (CG10146); cecropin B (CAA34845); DUF171 (CG12128); trypsin-like serine protease (CG17571); CBM_14 (CG13675); peritrophin-15a (CG17814); turandot C (CG31508); (CG40485); pastrel (CG8588); cytochrome P450-6a8 (CG10248); unknown genes (CG31606; CG7296; CG12439; CG32602; CG7567; CG11671; CG32510; CG13305); unidentified transcripts (Affymetrix probe identifiers 1632647_s_at; 1627513_at; 1628989_at; 1623579_at; 1626392_s_at; 1628534_at; 1632882_at; 1639231_at; 1624069_at; 1637692_at). |
Gene symbols are provided in parentheses; for unidentified transcripts the Affymetrix probe identifier has been substituted for gene symbols.
TABLE 1.
Select clusters of genes with increased or decreased expression in male HCS-deficient (HCS-RNAi) Drosophila compared with Act5C-Gal4 control flies
| Expression (HCS-RNAi vs. Act5C-Gal4)
|
||
|---|---|---|
| Gene Ontology category | Increased | Decreased |
| Biological process | ||
| Signal transduction | 7 | 1 |
| Cell cycle | 2 | 0 |
| Cell organization | 3 | 0 |
| Cell death | 3 | 1 |
| Catabolism | 6 | 2 |
| Defense response | 2 | 10 |
| Transport | 15 | 2 |
| Nucleic acid metabolism | 2 | 0 |
| Regulation of transcription | 14 | 2 |
| Proteolysis and peptidolysis | 18 | 6 |
| Morphogenesis | 5 | 1 |
| Molecular function | ||
| Protein binding | 7 | 0 |
| DNA/nucleotide binding | 18 | 1 |
| Transcription factor binding | 8 | 1 |
| Kinase | 12 | 4 |
| Transferase | 3 | 10 |
Knockdown of HCS and BTD cause reduced lifespan
Low abundance of HCS and BTD was associated with a decreased lifespan compared to Act5C-Gal4 controls (Table 2). The relative decreases in lifespan were similar for 75%, 50%, 25%, and 5% survival times in both genders. Note that HCS deficiency had a more pronounced effect on lifespan than BTD deficiency.
TABLE 2.
Lifespan of HCS-deficient (HCS-RNAi) and BTD-deficient (BTD-RNAi) male and female Drosophila melanogaster compared with Act5c-Gal4 control flies
| Days of survival
|
||||||
|---|---|---|---|---|---|---|
| Male
|
Female
|
|||||
| % Survival | HCS-RNAi | BTD-RNAi | Control | HCS-RNAi | BTD-RNAi | Control |
| 75 | 17 ± 2.7a | 23 ± 3.1b | 25 ± 2.6c | 37 ± 4.2a | 43 ± 2.8b | 50 ± 6.5c |
| 50 | 28 ± 2.0a | 30 ± 2.6a | 33 ± 2.0b | 47 ± 3.2a | 49 ± 3.1a | 55 ± 2.4b |
| 25 | 32 ± 2.4a | 35 ± 2.3b | 41 ± 1.6c | 57 ± 3.0a | 56 ± 2.1a | 63 ± 2.0b |
| 5 | 40 ± 1.9a | 39 ± 2.4a | 47 ± 2.0b | 63 ± 1.8a | 64 ± 1.8a | 68 ± 1.6b |
Values are means ± SD (n = 10 vials containing 20 flies each).
Values not sharing the same superscript are significantly different from each other for the same sex and % survival category; P < 0.05 as determined by one-way ANOVA and Fisher’s Protected Least Significant Difference procedure for posthoc testing. Males were not compared to females.
Knockdowns of HCS and BTD caused increased sensitivity to heat stress
Knockdown of HCS and BTD is associated with decreased survival of male flies exposed to heat stress. Consistent with the sex-specific differences in histone biotinylation, effects of knockdowns on stress resistance were greater in male than in female flies, and were greater in HCS-deficient flies than in BTD-deficient flies. For example, the 50% survival time was 4.6 ± 0.3 h for male Act5C-Gal4 flies, but only 3.1 ± 0.3 h and 4.3 ± 0.4 h in male HCS-RNAi and BTD-RNAi flies, respectively (Fig. 5A). In female flies, HCS deficiency but not BTD deficiency increased the susceptibility to heat stress (Fig. 5B); the 50% survival time was 5.1 ± 0.3 h for female Act5C-Gal4 flies, 4.3 ± 0.1 h for female HCS-RNAi flies, and 5.5 ± 0.3 h for female BTD-RNAi flies (Fig. 5B). HCS and BTD deficiency specifically decreased heat tolerance as opposed to globally decreasing temperature tolerance. Consistent with this notion, the survival of flies exposed to cold temperatures (4ºC for up to 16 hr) did not depend on HCS and BTD (data not shown).
FIGURE 5.
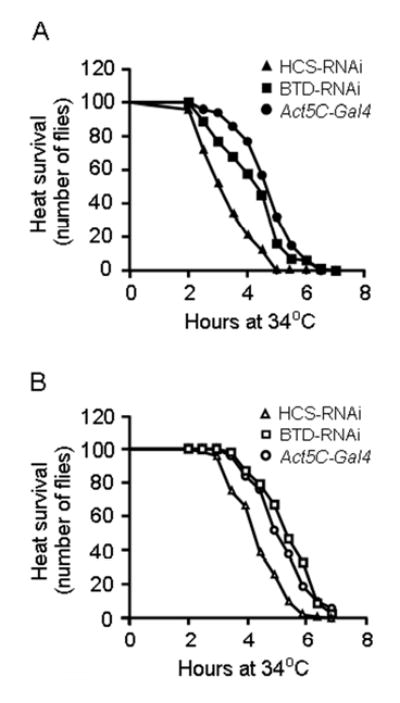
Effects of HCS and BTD RNAi knockdown on heat tolerance in HCS-deficient (HCS-RNAi) and BTD-deficient (BTD-RNAi) flies compared with Act5C-Gal4 control flies. Panel A: HCS-RNAi and BTD-RNAi male flies exhibited decreased heat tolerance compared with control flies. Panel B: HCS deficiency but not BTD deficiency decreased the heat tolerance in female knockdown flies.
Finally, microarray analysis was used to identify heat-responsive genes that were affected by HCS deficiency. Heat stress altered the expression of 398 genes in Act5C-Gal4 flies. In addition we identified 285 heat-responsive genes that were uniquely affected in HCS-deficient flies. Specifically, 167 heat-responsive genes were down-regulated by at least 50% in HCS-deficient flies, and 118 heat-responsive genes were up-regulated by at least 100% in HCS-deficient flies (online supplemental Table S2). Heat-responsive, HCS-dependent genes were clustered based on biological process and molecular function. HCS deficiency was associated with a substantial increase in the expression of genes that play a role in proteolysis, and with a substantial decrease of genes that play a role in chitin metabolism and cuticle structure (Table 3).
SUPPLEMENTAL TABLE S2.
Heat-responsive genes that were upregulated or downregulated in HCS-deficient male flies
|
Expression increased by >200% in HCS-RNAi flies compared with controls:1
Stellate 12D orphon (CG33236); yolk protein 2 (CG2979); yolk protein 3 (CG11129); female-specific independent of transformer (CG17820); LipA (CG10383); LP21536p (CG14963); ANK (CG8679). |
|
Expression increased by 100 to 200% in HCS-RNAi flies compared with controls:
Jonah 66Ci (CG7118); black cells (CG5779); trypsin 29F (CG9564); Drosocin (CG10816); PEX11 (CG33474); emocyanin (CG8193); PAP2 wunen (CG11425); takeout (CG11853); CBM14 (CG10725); SMP2 (CG8709); LP11437p (CG10462); Tryp SPc (CG8329); death associated molecule related to Mch2 (CG18188); FRQ1 (CG32103); astray (CG3705); Rho (CG12102); Thor (CG8846); adenosine 3 (CG31628); DltE (CG13833); Tryp SPc (CG4927); LD43023p (CG8321); DUF727 (CG14505); Cyp4ac2 (CG17970); Jonah 99Fi (CG18030); imaginal disc growth factor 1 (CG4472); GH12638p (CG3672); RE61294p (CG8112); suppressor of cytokine signaling at 36E (CG15154); serine pyruvate aminotransferase (CG3926); RH05653p(CG13116); Sugar_tr (CG31106); metchnikowin (CG8175); BAH (CG32529); adenosine 2 (CG9127); sugarless (CG10072); naked cuticle (CG11614); att-ORFB (CG33488); ia2 (CG31795); Delilah (CG5441); ANK (CG9121); PepO (CG14528); MA3 (CG10990); CLECT (CG8343); sterile (CG1441); SCP (CG6628); lethal (2) essential for life (CG4533); serpin (CG9460); Cyp12a5 (CG11821); PcbC (CG5346); GH08205p (CG3906); gamma-glutamyltranspeptidase (CG17636); Cyp6a22 (CG10240); midline fasciclin (CG3359); Pal (CG5472); GNS1/SUR4 (CG33110); ornithine decarboxylase 1 (CG8721); Ecdysone-induced protein 75B (CG8127); DM8 (CG14456); Tryp_SPc (CG10587); pancreatic lipase like protein (CG6283); Atf-2 (CG30420); hemolysin (CG33173); BTB/POZ (CG31160); Ecdysone-induced protein 75B (CG8127); bunched (CG5461); COG0842 (CG14559); PepO (CG5527); scratch (CG1130); odorant-binding protein 8a (CG12665); raptor (CG4320); protein tyrosine phosphatase 99A (CG2005); haem peroxidase (CG8913); adh_short (CG3842); paired (CG6716); adh_short (CG2064); martik (CG3361); Jun-related antigen (CG2275); deformed epidermal autoregulatory factor-1 (CG8567); X box binding protein-1 (CG9415); Tryp_SPc (CG9672); DSPc (CG10089); Jonah 65Ai (CG10475); ab-hydrolase associated lipase (CG5932); CLECT (CG12111); carbonic anhydrase alpha (CG18673); mitochondrial ribosomal protein L51 (CG15434); predicted GTPase (CG8801); MADF (CG8281); chitin-binding domain type 2 (CG7874); heat shock construct of Kidd (CG17342); unknown genes (CG1648; CG13654; CG32185; CG12065; CG15784; CG4285; CG11686; CG13868; CG5597; CG12726; CG10433; CG13947; CG9776; CG31829; CG14499; CG13946; CG4577; CG11112); unidentified transcripts (Affymetrix probe identifiers 1638496_at; 1625050_s_at). |
|
Expression decreased by >75% in HCS-RNAi flies compared with controls:
four-jointed (CG10917); MdlB (CG10505); fat body protein 1 (CG17285); odorant binding protein (CG2650); GH05104p (CG13607); larval serum protein 1 (CG6821); LIM domain (CG30179); LIM domain (CG30174); yellow-d2 (CG9891); unknown genes (CG13049; CG15080; CG8515; CG13043; CG12998; CG31775). |
|
Expression decreased by 50 to 75% in HCS-RNAi flies compared with controls:
ACBP (CG8628); krotzkopf verkehrt (CG2666); RKIP (CG18594); peptidase M28 (CG30049); CBM14 (CG31973); Stretchin-Mlck (CG18255); RH01578p (CG15006); larval serum protein 1 (CG6821); CBM14 (CG32499); vanin-like (CG32754); Zim2 (CG3777); MH2 (CG13183); sterile (CG17562); sickle (CG13701); His1 (CG31617); MH2 (CG13188); CBM14 (CG13676); predicted trypsin-like serine protease (CG31265); LRRRI (CG2471); glyco18 (CG1869); wrinkled (CG5123); PDZ signaling (CG14168); Tryp_SPc (CG17475); CBM14 (CG2989); cd00190 (CG4053); DUF233 (CG11852); ecdysis triggering hormone (CG18105); BCM14 (CG17052); myosin light chain 2 (CG2184); smart00631 (CG14820); Gasp (CG10287); Cyp4d14 (CG3540); ninaD (CG31783); cp309 (CG33957); Duf227 (CG11893); GILT (CG13822); IP07649p (CG13155); CLECT (CG4115); UDPGT (CG5724); CD36 family (CG3829); peroxidase (CG3477); crystallin (CG16963); exportin 6 (CG3923); Orc1 (CG10667); TRP (CG11396); Kelch (CG17754); DUF1193 (CG3631); Dimp (CG9850); LD12613p (CG1919); HL04910p (CG7759); mRpL52 (CG1577); SD18375p (CG3773); dachs (CG10595); peritrophin-A (CG17058); DltE (CG31810); HL05804p (CG30437); formyl transferase (CG1750); LDLa (CG32209); sugar_tr (CG33281); predicted GTPase (CG18528); arginase (CG18104); Obp83g (CG31558); SMC4 (CG18304); shark (CG18247); CO esterase (CG7529); aret (CG31762); sugar_tr (CG31530); KU (CG3513); Cks85A (CG9790); longest ORF LD45181p (CG6912); msl-3 (CG8631); aminopeptidase N (CG31198); Pex2/Pex12 (CG3639); alpha amylase (CG33138); Ice (CG7788); Tub84D (CG2512); peptidase S28 (CG18493); Jonah 44E (CG8579); Peptidase S28 RE36938p (CG3734); SMC4 LD41224p (CG196); GH12942p (CG8179); trans-isoprenyl diphosphate synthase (CG31005); mRpL55 (CG14283); RAB GTPase (CG4789); Niemann-Pick type C2 (CG31410); alanine dehydrogenase/PNT (CG7144); ribosome recycling factor (CG4447); DUF227 (CG31087); MOSC (CG1665); AraJ (CG18281); pellucida (CG2467); RH50269p (CG6639); RH32892p (CG1674); branchless (CG4608); SD05384p CNH domain (CG8060); Paramyosin (CG5939); GalNAc-T2 (CG6394); leucine-rich repeats (CG8561); RE03018p (CG13003); Glutathione S-transferase (CG4688); eel-Fucolectin Tachylectin-4 (CG9095); HDc (CG11900); actinin (CG4376); Arabinose efflux permease (CG8051); purple (CG16784); flightin (CG7445); HL07915p (CG8501); leucine-rich repeats (CG7896); RE05963p (CG15884); jagunal (CG10978); Serine/Threonine kinase (CG7028); sugar_tr (CG4462); chitin binding Peritrophin-A (CG4778); unknown genes (CG6131; CG12045; CG17562; CG15213; CG15515; CG5494; CG4962; CG8443; CG1136; CG11760; CG3984; CG16884; CG2444; CG14795; CG13067; CG11413; CG16885; CG9299; CG8736; CG13056; CG11380; CG13489; CG13689; CG14959; CG13210; CG10563; CG17124; CG9422; CG15282; CG13484; CG33256; CG13288); unidentified transcripts (Affymetrix probe identifier 1630614_s_at; 1627236_s_at; 1639591_at; 1637055_s_at; 1639778_at; 1640675_at). |
Gene symbols are provided in parentheses; for unidentified transcripts the Affymetrix probe identifier has been substituted for gene symbols.
TABLE 3.
Select clusters of heat-responsive genes that were upregulated or downregulated in HCS-deficient male flies
| Gene Ontology category | Upregulated | Downregulated |
|---|---|---|
| Biological process | ||
| Signal transduction | 6 | 6 |
| Cell cycle | 1 | 2 |
| Cell proliferation | 5 | 3 |
| Proteolysis and peptidolysis | 13 | 12 |
| Chitin metabolism | 2 | 9 |
| Cell motility | 0 | 3 |
| Transport | 12 | 17 |
| Biosynthesis | 7 | 8 |
| Regulation of transcription | 10 | 3 |
| Molecular function | ||
| Protein binding activity | 2 | 8 |
| DNA/nucleotide binding activity | 8 | 4 |
| Transcription factor activity | 9 | 2 |
| Receptor activity | 6 | 9 |
| Cuticle structure | 1 | 10 |
DISCUSSION
Biotinylation of histones by HCS and BTD is hypothesized to participate in heterochromatin structure, gene silencing, and the cellular response to DNA damage. The phenotypes associated with HCS and BTD deficiency in humans have been extensively described (29). While deficiency for either enzyme is viable, there is a bewildering array of seemingly disparate symptoms associated with both conditions. In the present study, we sought new insights into roles of histone biotinylation in gene function and chromatin structure, and to characterize phenotypes associated with HCS and BTD deficiency in Drosophila melanogaster. The demonstration that HCS is a chromosomal protein in Drosophila supports the inference that HCS directly affects histone biotinylation in chromatin.
Expression of HCS and BTD was dramatically knocked down by RNAi, as judged by the abundance of mRNA coding for HCS and BTD, levels of HCS protein and biotinylated carboxylases, and activity of BTD. HCS deficiency was associated with a global decrease in histone biotinylation and also with a decrease of specific biotinylation sites in histone H3. The reduction in histone biotinylation seen in HCS knockdown flies is consistent with our finding that HCS is a chromosomal protein, and suggests that the effect is direct. Effects of HCS deficiency on histone biotinylation were more modest in females than in males. Generally, effects of BTD deficiency on histone biotinylation are smaller than the effects observed for HCS deficiency. However, the fact that both knockdowns have significant effects demonstrates that these proteins have at least some non-redundant activity in vivo.
Both HCS and BTD deficiency decrease histone biotinylation to a greater extent in males than in females. The reason for this gender specificity is unknown. We excluded the possibility of insufficient knockdown of HCS and BTD in females by demonstrating that biotinylated carboxylases were decreased to a similar level in transgenic flies of both genders. Why, then, is histone biotinylation decreased to a greater extent in males than in females? One possibility is that the nuclear translocation of biotin might be more efficient in females than in males, or that the turnover of biotinylated histones might be greater in males than in females. Another possibility is that the Y chromosome, which is male-specific and is entirely heterochromatic in somatic tissue, represents a major target for histone biotinylation in Drosophila; reduction of either HCS or BTD would thus be expected to have a disproportionate effect in males. Finally, we cannot exclude the possibility that a deficiency of either of the two histone biotinyl transferases was compensated for by the other biotinyl transferase in females. We sought to investigate this possibility by creating HCS and BTD double knockdowns. However, these flies were non-viable.
Previous studies provided evidence for roles of histone biotinylation in cell proliferation (2, 7), DNA repair (8, 30), and gene silencing (Camporeale, G., Oommen, A. M., Griffin, J. B., Sarath, G. & Zempleni, J., unpublished data). Consistent with the biological significance of these processes, a previous study suggested that biotin deficiency decreases lifespan and fertility in flies (17). However, in this previous study a clear link of biotin deficiency and lifespan to histone biotinylation was not evident. Here, for the first time we provide evidence that reduced biotinylation of histones in HCS-deficient flies is linked to phenotypes such as decreased lifespan and heat tolerance. Also we provide evidence that alterations of chromatin structure by decreased biotinylation of histones is associated with unique patterns of gene expression. Gene expression analysis indicates that HCS deficiency alters the expression of 201 genes. We also identified 285 heat-responsive genes that depend on HCS for normal expression. Currently, it is unknown which of these genes mediate the increased heat susceptibility in HCS-deficient flies.
A caveat to these findings is that HCS and BTD deficiency decreases biotinylation of both histones and carboxylases. Theoretically, some of the effects described here might be due to decreased biotinylation of carboxylases rather than histones. To address this uncertainty we are currently investigating genotypes and phenotypes of flies in which individual carboxylases were knocked down. These ongoing studies provided evidence that knockdown of pyruvate carboxylase and 3-methylcrotonyl-CoA carboxylase is not associated with increased susceptibility to heat stress (Camporeale, G., Eissenberg, J. C. & Zempleni, J., unpublished data). Based on these ongoing studies we hypothesize that the increased heat susceptibility observed in HCS-deficient flies is a direct result of changes in histone biotinylation and chromatin structure, rather than being a result of secondary changes due to reduced activities of biotin-dependent enzymes.
The hypothesis that many of the effects of HCS and BTD deficiency act at the level of chromatin modification offers a novel explanation for the diverse and apparently unrelated deficits seen in HCS and BTD deficiency in patients. Rather than being connected through metabolic pathways, as in classical vitamin deficiencies, the symptoms of HCS and BTD deficiency may reflect, to a significant degree, the misregulation of genes connected by their use of histone biotinylation as a regulatory mechanism. The development of Drosophila models for HCS and BTD deficiency, combined with the favorable cytology and genetics of Drosophila, will help define the role of histone biotinylation in gene regulation and disease.
Footnotes
This work was supported by NIH grants DK 60447, DK 063945, and 1 P20 RR16469, and by NSF grants EPSCoR EPS-0346476, MCB 0131414, and MCB 6552870. This paper is a contribution of the University of Nebraska Agricultural Research Division, Lincoln, NE 68583 (Journal Series No. 15192).
Supplemental Tables S1 and S2 are available as Online Supporting Material with the online posting of this paper at http://jn.nutrition.org.
The abbreviations used are: BTD, biotinidase; HCS, holocarboxylase synthetase; K9BioH3, lysine-9 biotinylated histone H3; K18BioH3, lysine-18 biotinylated histone H3; IR, inverted repeat; K, lysine; PC, pyruvate carboxylase; PCC, propionyl-CoA carboxylase; RNAi, RNA interference; y w, wild type.
References
- 1.Camporeale G, Zempleni J. Biotin. In: Bowman BA, Russell RM, editors. Present Knowledge in Nutrition. 9th ed. Washington, D.C.: International Life Sciences Institute; 2006 (in press).
- 2.Stanley JS, Griffin JB, Zempleni J. Biotinylation of histones in human cells: effects of cell proliferation. Eur J Biochem. 2001;268:5424–9. doi: 10.1046/j.0014-2956.2001.02481.x. [DOI] [PubMed] [Google Scholar]
- 3.Hymes J, Fleischhauer K, Wolf B. Biotinylation of histones by human serum biotinidase: assessment of biotinyl-transferase activity in sera from normal individuals and children with biotinidase deficiency. Biochem Mol Med. 1995;56:76–83. doi: 10.1006/bmme.1995.1059. [DOI] [PubMed] [Google Scholar]
- 4.Camporeale G, Chew YC, Kueh A, Sarath G, Zempleni J. Use of synthetic peptides for identifying biotinylation sites in human histones. In: McMahon RJ, editor. Avidin-Biotin Technology in the Life Sciences. Totowa, N.J.: Humana Press; 2005.
- 5.Kobza K, Camporeale G, Rueckert B, Kueh A, Griffin JB, Sarath G, Zempleni J. K4, K9, and K18 in human histone H3 are targets for biotinylation by biotinidase. FEBS J. 2005;272:4249–59. doi: 10.1111/j.1742-4658.2005.04839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camporeale G, Shubert EE, Sarath G, Cerny R, Zempleni J. K8 and K12 are biotinylated in human histone H4. Eur J Biochem. 2004;271:2257–63. doi: 10.1111/j.1432-1033.2004.04167.x. [DOI] [PubMed] [Google Scholar]
- 7.Kothapalli N, Zempleni J. Biotinylation of histones depends on the cell cycle in NCI-H69 small cell lung cancer cells. FASEB J. 2005;19:A55. [abstract] [Google Scholar]
- 8.Kothapalli N, Sarath G, Zempleni J. Biotinylation of K12 in histone H4 decreases in response to DNA double strand breaks in human JAr choriocarcinoma cells. J Nutr. 2005;135:2337–42. doi: 10.1093/jn/135.10.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narang MA, Dumas R, Ayer LM, Gravel RA. Reduced histone biotinylation in multiple carboxylase deficiency patients: a nuclear role for holocarboxylase synthetase. Hum Mol Genet. 2004;13:15–23. doi: 10.1093/hmg/ddh006. [DOI] [PubMed] [Google Scholar]
- 10.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 11.Ballard TD, Wolff J, Griffin JB, Stanley JS, Calcar Sv, Zempleni J. Biotinidase catalyzes debiotinylation of histones. Eur J Nutr. 2002;41:78–84. doi: 10.1007/s003940200011. [DOI] [PubMed] [Google Scholar]
- 12.Zempleni J. Uptake, localization, and noncarboxylase roles of biotin. Annu Rev Nutr. 2005;25:175–96. doi: 10.1146/annurev.nutr.25.121304.131724. [DOI] [PubMed] [Google Scholar]
- 13.Eisenberg MA, Prakash O, Hsiung SC. Purification and properties of the biotin repressor. A bifunctional protein. J Biol Chem. 1982;257:15167–73. [PubMed] [Google Scholar]
- 14.Beckett D. Energetic methods to study bifunctional biotin operon repressor. Methods Enzymol. 1998;295:424–50. doi: 10.1016/s0076-6879(98)95052-2. [DOI] [PubMed] [Google Scholar]
- 15.Leon-Del-Rio A, Leclerc D, Akerman B, Wakamatsu N, Gravel RA. Isolation of a cDNA encoding human holocarboxylase synthetase by functional complementation of a biotin auxotroph of Escherichia coli. Proc Natl Acad Sci U S A. 1995;92:4626–30. doi: 10.1073/pnas.92.10.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dupuis L, Campeau E, Leclerc D, Gravel RA. Mechanism of biotin responsiveness in biotin-responsive multiple carboxylase deficiency. Molec Genet Metabol. 1999;66:80–90. doi: 10.1006/mgme.1998.2785. [DOI] [PubMed] [Google Scholar]
- 17.Landenberger A, Kabil H, Harshman LG, Zempleni J. Biotin deficiency decreases life span and fertility but increases stress resistance in Drosophila melanogaster. J Nutr Biochem. 2004;15:591–600. doi: 10.1016/j.jnutbio.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 18.Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- 19.Giordano E, Rendina R, Peluso I, Furia M. RNAi triggered by symmetrically transcribed transgenes in Drosophila melanogaster. Genetics. 2002;160:637–48. doi: 10.1093/genetics/160.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spradling AC. P element-mediated transformation. In: Roberts DB, editor. Drosophila, a practical approach. Oxford: IRL Press; 1986. p. 175–97.
- 21.Manthey KC, Griffin JB, Zempleni J. Biotin supply affects expression of biotin transporters, biotinylation of carboxylases, and metabolism of interleukin-2 in Jurkat cells. J Nutr. 2002;132:887–92. doi: 10.1093/jn/132.5.887. [DOI] [PubMed] [Google Scholar]
- 22.Griffin JB, Rodriguez-Melendez R, Zempleni J. The nuclear abundance of transcription factors Sp1 and Sp3 depends on biotin in Jurkat cells. J Nutr. 2003;133:3409–15. doi: 10.1093/jn/133.11.3409. [DOI] [PubMed] [Google Scholar]
- 23.Chew YC, Camporeale G, Kothapalli N, Sarath G, Zempleni J. Lysine residues in N- and C-terminal regions of human histone H2A are targets for biotinylation by biotinidase. J Nutr Biochem. 2006;17:225–33. doi: 10.1016/j.jnutbio.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang HL, Benzer S, Min KT. Life extension in Drosophila by feeding a drug. Proc Natl Acad Sci U S A. 2002;99:838–43. doi: 10.1073/pnas.022631999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiedmann S, Rodriguez-Melendez R, Ortega-Cuellar D, Zempleni J. Clusters of biotin-responsive genes in human peripheral blood mononuclear cells. J Nutr Biochem. 2004;15:433–9. doi: 10.1016/j.jnutbio.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 26.SAS Institute. StatView Reference. 3rd ed. Cary, NC: SAS Publishing; 1999.
- 27.Pispa J. Animal biotinidase. Ann Med Exp Biol Fenniae. 1965;43:4–39. [PubMed] [Google Scholar]
- 28.Wolf B, Grier RE, Allen RJ, Goodman SI, Kien CL. Biotinidase deficiency: An enzymatic defect in late-onset multiple carboxylase deficiency. Clin Chim Acta. 1983;131:273–81. doi: 10.1016/0009-8981(83)90096-7. [DOI] [PubMed] [Google Scholar]
- 29.Wolf B. Disorders of biotin metabolism: Treatable neurological syndromes. In: Rosenberg RN, Prusiner BS, Mauro SD, Barchi RL, Kunkel LM, editors. The Molecular and Genetic Basis of Neurological Disease. Stoneham, MA: Butterworth; 1992. p. 569–81.
- 30.Peters DM, Griffin JB, Stanley JS, Beck MM, Zempleni J. Exposure to UV light causes increased biotinylation of histones in Jurkat cells. Am J Physiol Cell Physiol. 2002;283:C878–C84. doi: 10.1152/ajpcell.00107.2002. [DOI] [PubMed] [Google Scholar]


