Abstract
Among the many proteases associated with human cancer, seprase or fibroblast activation protein alpha (FAP-α)1, a type II transmembrane glycoprotein, has two types of EDTA-resistant protease activities: dipeptidyl peptidase (DP) and a 170-kDa gelatinase activity. To test if activation of gelatinases associated with seprase could be involved in malignant tumors, we used a mammalian expression system to generate a soluble recombinant seprase (r-seprase). In the presence of putative EDTA-sensitive activators, r-seprase was converted into 70- to 50-kDa shortened forms of seprase (s-seprase), which exhibited a 7-fold increase in gelatinase activity while levels of DP activity remained unchanged. In malignant human tumors, seprase is expressed predominantly in tumor cells as shown by in situ hybridization and immunohistochemistry. Proteins purified from experimental xenografts and malignant tumors using antibody- or lectin-affinity columns in the presence of 5 mM EDTA were assayed for seprase activation in vivo. Seprase expression and activation occur most prevalently in ovarian carcinoma, but were also detected in four other malignant tumor types, including adenocarcinoma of the colon and stomach, invasive ductal carcinoma of the breast, and malignant melanoma. Together, these data show that, in malignant tumors, seprase is proteolytically activated to confer its substrate specificity in collagen proteolysis and tumor invasion.
Keywords: seprase, fibroblast activation protein alpha (FAP-α), gelatinase, activation, malignant tumor
Introduction
Seprase was originally identified as a 170-kDa transmembrane glycoprotein with gelatinolytic activity present at the invadopodia of human malignant melanoma LOX cells (1, 2). In fact, cloning of the seprase gene and characterization of its protein showed that seprase was identical to FAP-α, a protein proposed to be expressed only on reactive stromal fibroblasts in common human epithelial cancers (3-7). It has been shown by immunohistochemistry that seprase is present in tumor cells and stromal fibroblasts in invasive breast (8-10), gastric (11), colonic (12, 13), and cervical (14) carcinomas, but is absent or undetectable in all normal tissue cells except in the early stage of wound healing (15).
Seprase has dual functions in tumor progression. The proteolytic activity of seprase was shown to promote cell invasiveness toward the ECM (1, 2, 15, 16), and support tumor growth and proliferation (9, 17, 18). Other studies showed that the proteolytically inactive protein suppressed the transformation of cell lines and inhibit tumor growth supported by stromal cells (19, 20). Although different domains of seprase appear to have opposing effects on tumor progression, the active structure responsible for tumor invasiveness remains to be elucidated.
Similar to what has been found in dipeptidyl peptidase IV (DP4), the closest homolog to seprase, seprase is capable of cleaving amino terminal prolyl dipeptides from polypeptides and exhibits DP activity in the presence of 5 mM EDTA (3-5, 15, 21, 22). Seprase has also been shown to effectively degrade macromolecule substrates such as gelatin. Experiments that affinity-labeled the catalytic site with [3H]diisopropyl fluorophosphates (3) and experiments utilizing site-directed mutagenesis, Ser624 --> Ala624 (5, 17), showed that both DP and gelatinase activities are EDTA resistant and require a non-classical serine protease catalytic center. In addition, the proteolytic activity of seprase depends on the homodimerization of two 97-kDa monomers that form a 170-kDa dimer with a centralized catalytic pocket (3, 23). However, structural and substrate kinetics studies of the 170-kDa dimeric apoenzyme showed that its catalytic site was effective in cleaving dipeptides, but might be not adequate to degrade gelatin (23, 24).
To explore the potential function of seprase in the invasion of tumor cells into the ECM, we used a novel mammalian expression system to generate a soluble recombinant seprase (r-seprase) that lacks seprase cytoplasmic and transmembrane domains. This r-seprase is able to dimerize and exhibits an increase in gelatinase activity compared to full-length native seprase (n-seprase). In addition, the gelatinase activity of r-seprase can be increased further after more extensive truncation by EDTA-sensitive proteases. EDTA is an effective inhibitor to the activation of r-seprase gelatinase activity. We show, for the first time, the expression of seprase in the tumor cells of ovarian carcinoma by in situ hybridization and immunohistochemistry. To look for the presence of shortened seprase (s-seprase) in vivo, we examined seprase proteins derived from xenografts and malignant tumors that were purified by antibody- or lectin-affinity columns in the presence of EDTA. We found that n-seprase was activated to form 70- to 50-kDa forms in both xenografts and malignant tumors, suggesting that proteolytic truncation is a biologically relevant mechanism for seprase activation. In light of the finding that proteolytically activated seprase is truncated from the amino terminus, we discuss a truncation mechanism for seprase activation, which reduces steric hindrance and increases accessibility of macromolecular substrates such as gelatin. Because seprase is present at very low levels in differentiated epithelium and normal tissues, but up-regulated in malignant tumors, it is an attractive therapeutic target for tumor progression.
Materials and Methods
Plasmid constructs and cells producing r-seprase. Utilizing the pA15 plasmid DNA, which contained seprase cDNA (GenBank accession number U76833) as a PCR template (4), the cDNA fragment encoding the seprase extracellular domain (amino acids 27-760) was amplified with a forward primer (5’- AAG GAT CCC GCC CTT CAA GAG TTC ATA ACT -3’) and a reverse primer (5’- AAC TCG AGG TCT GAC AAA GAG AAA CAC TG -3’). The PCR product, excluding the coding sequences of both the short cytoplasmic (amino acids 1-6) and hydrophobic transmembrane domains of seprase (amino acids 7-26), was inserted into a modified pCEP4 vector (Invitrogen). This backbone is an Epstein- Barr Virus (EBV) based vector that employs the cytomegalovirus (CMV) immediate early enhancer/promoter for high level transcription of recombinant genes and carries an EBV replication origin (oriP) to permit its extrachromosomal replication in human cells. Compared to pCEP4, the modified vector additionally contains the coding sequences of a secretion signal from the V-J2-C region of the mouse Ig kappa-chain and a V5-His fusion tag, derived from pSecTag/FRT/V5-His-TOPO vector (Invitrogen). The cDNA sequence of the seprase extracellular domain was inserted, in frame, along with the N-terminal secretion signal and the C-terminal V5-His tag, allowing efficient secretion, easy detection (by Anti-V5 Antibody; Invitrogen) and rapid purification (by His·Bind Resin columns; Novagen) of recombinant seprase. The final construct was verified by sequencing both DNA strands and named pE15.
293-EBNA monkey kidney cells (Invitrogen) that were intended for use with vectors containing an EBV origin of replication (oriP) were maintained in complete medium [DMEM (GBCO-BRL), 10% fetal bovine serum (GBCO-BRL), 2 mM L-glutamine, 100 U/ml penicillin and 100 mg/ml streptomycin] supplemented with 250 μg/ml G418. Plasmid pE15 was transfected into 293-EBNA cells using LipofectAMINE Reagent (GBCO-BRL) according to the manufacturer’s instruction. After transfection, cells were initially cultured in complete medium supplemented with 250 μg/ml G418 and 200 μg/ml Hygromycin B (GBCO-BRL), then cultured in protein-free HyQ PF-293 medium (HyClone) containing 250 μg/ml G418 and 200 μg/ml Hygromycin B. Cell viability was checked with a Trypan Blue (GBCO- BRL) exclusion test. Freshly collected culture medium was filtered with four layers of filter paper (Whatman) at room temperature and loaded into an equilibrated DEAE Sepharose Fast Flow column (Sigma) at 4 °C. R-seprase was eluted with a NaCl gradient from 0 M to 1.0 M in 10 mM phosphate buffer, pH 7.0, and then absorbed by a Wheat Germ Agglutinin (WGA) affinity chromatography column (Amersham Pharmacia Biotech). After being eluted with 0.5 M N-Acetylglucosamine (Sigma) in PBS, r- seprase was further purified by the charged His·Bind Resin column (Novagen). The following elution was performed either with the elution buffer containing 1 M imidazole or with the stripping buffer containing 0.1 M EDTA. Eluted protein was concentrated to 400 μl with an ULTRAFREE-15 Centrifugal Filter Device (Millipore) and fractionated with a Superdex 200 Prep grade gel filtration column (Pharmacia Biotech). R-seprase was tracked throughout the procedure by the soluble DP assay (15).
In vitro invasion assay. For the evaluation of cell invasiveness, glutaraldehyde-cross-linked gelatin films were prepared with a small modification of the method described previously (25). Briefly, 16-well Lab-Tek Chamber Slides (Nalgene Nunc International) were coated with a layer of 5% gelatin (Sigma) and 2.5% sucrose in PBS, air dried and then fixed with 0.5% glutaraldehyde in PBS on ice for 10 min. FITC-labeled human fibronectin (Becton Dickinson Labware; 50 μg/ml) was coupled on the surface of the cross-linked gelatin film. Cells (1000 cells/well) were seeded on the films and cultured for 2 days in the complete medium. After being fixed by 4% formaldehyde and permeablilized by 0.1% Triton X-100, the cells were stained by Phalloidin-CPITC (Sigma; 50 μg/ml in PBS). Cells were photographed with a Plan Fluor ELWD 40X/0.60 objective on an ECLIPSE TE300 microscope (Nikon, Japan) coupled to a SONY Digital Camera DKC-5000 where the FITC and CPITC labels were photographed using epifluorescence microscopy.
MAbs recognizing seprase. Rat mAbs D8, D28 and D43 are directed against human placental n- seprase (3, 4). Rat mAb E97 was developed using heat-denatured human placental seprase as an immunogen and screened by Western immunoblotting. Rat mAb E97 is particularly useful in identifying both monomeric and dimeric forms of denatured seprase on immunoblots, but it does not recognize non-denatured seprase dimers in ELISA or soluble enzymatic assays (see Fig. 1D).
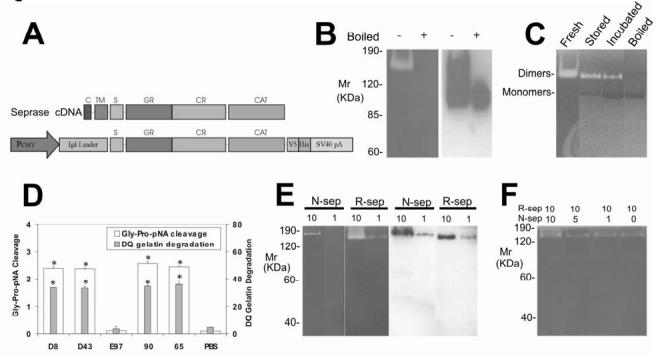
Figure 1Isolation and characterization of r-seprase. A, R-seprase expression vector pE15. The cDNA regions that encode predicted domains of seprase are indicated: C, cytoplasmic domain, amino acids 1-6; TM, transmembrane domain, amino acids 7-26; S, stalk region, amino acids 27-48; GR, glycosylation rich region, amino acids 49-314; CR, cysteine rich domain, amino acids 305-466; CAT, catalytic domain, amino acids 500-760. The cDNA fragment that encodes r-seprase lacks the cytoplasmic and transmembrane domains and was inserted into a CMV promoter driven expression cassette containing an N-terminal mouse Igk secretion signal and a C-terminal V5-His tag. B, A gelatin zymogram (left panel) and a corresponding Western immunoblot (right panel) of r-seprase. The culture medium conditioned by the pE15 transfected 293-EBNA cells was prepared under non-boiled (-) or boiled (+) conditions, and subsequently resolved on a gelatin zymogram and a Western immunoblot using anti-V5 antibody. The purified protein, when boiled for 3 min, completely dissociates into 90-kDa inactive monomers, whereas the proteins remain mostly in dimeric form if not subjected to boiling. The serine gelatinase activity of r-seprase was detected by incubating the zymogram in the presence of 5 mM EDTA to quench metalloprotease activity. Note that both r-seprase dimers and monomers were present in the medium conditioned by transfectants; however, the monomers were gelatinolytically inactive. C, Dissociation of the active 160-kDa r-seprase into the inactive 90-kDa monomer detected by gelatin zymography. Freshly purified r-seprase is active at 160-kDa (Fresh). Moderate activity is detected in the dimerized portion of r-seprase samples stored at -20 °C because a portion has dissociated into 90-kDa inactive monomers (Stored). Samples stored at -20 °C and then subsequently incubated at 37 °C overnight in the presence of 5 mM EDTA also have moderate activity in the portion that remained dimeric; however, a large portion dissociated into 90-kDa inactive monomers (Incubated). Note that r-seprase dimers create white bands against the dark grey background due to their gelatinase activity, whereas the monomers have no gelatinase activity and therefore appear as black bands compared to the gelatin background. D, The proteolytic activities of r-seprase. The seprase specific DP and gelatinase activities were measured using immunocaptured protein in the soluble enzymatic assays under non-denaturing conditions containing 5 mM EDTA as described (15). Seprase was captured from culture medium conditioned with pE15 transfected 293-EBNA cells using rat mAbs D8 and D43 (against n-seprase), mAb E97 (against denatured seprase monomeric subunits and their shortened forms) and mouse mAbs 90 and 65 (against r-seprase dimer). The DP (Gly-Pro-pNA cleavage) and gelatinase (DQ gelatin degradation) activities of the isolated r-seprase were measured in parallel. The symbol * above the bars indicate a significant increase in activity compared with the PBS control (P<0.05). E, Specificity of gelatinase activity of n-seprase and r-seprase revealed by parallel gelatin zymography and Western immunoblotting analyses. 10 and 1 μl of LOX cell Triton X-100 detergent lysate (n-sep) and the concentrated culture medium conditioned by the pE15 transfected 293-EBNA cells (r-sep) were loaded into parallel SDS gels: one was subjected to gelatin zymography in 5 mM EDTA conditions (left panel) for serine-type gelatinase detection and the other by Western immunoblotting using mAb 90 (right panel) for assessment of n- and r-seprase protein. F, Mixing of the r-seprase with different amounts of the detergent extract containing n-seprase to test for the presence of endogenous seprase inhibitors. From 1-10 μl of LOX cell Triton X-100 detergent lysate (n-sep) were mixed with 10 μl of the concentrated culture medium conditioned by the pE15 transfected 293-EBNA cells (r-sep). Mixtures were subjected to gelatin zymography in 5 mM EDTA conditions for assessment of possible inhibition of gelatinase activity specific for r-seprase.
To generate mouse mAbs directed against r-seprase, BALB/c inbred mice (Taconic) were immunized using 10 μg of purified r-seprase. Splenocytes and Sp2/0-Ag14 cells (ATCC) were fused. Hybridomas were screened by ELISA. Briefly, Microtiter® U bottom Polyvinyl Chloride 96 well plates (Dynex Technologies) were coated with r-seprase and blocked with 5% BSA in PBS. Coated r-seprase was subjected to sequential incubations with Hybridoma supernatant and Anti-Mouse IgG Peroxidase-Conjugates (Sigma). The bound secondary antibody was detected with 2,2’-Azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) Diammonium Salt solution (Sigma) and absorbance at 410 nm was detected with a Microplate Spectrophotometer System equipped with SOFTmax Pro version 1.3.1 (Molecular Devices). Hybridomas secreting mAbs against r-seprase were confirmed by soluble enzymatic assays and Western immunoblotting analyses. The isotype of each mAb was determined by ImmunoType Mouse Monoclonal Antibody Isotyping Kit (Sigma). Each of mAb 65, mAb 68, mAb 82 and mAb 90 was created as described above; all were characterized as immunoglobulin isotype IgG1. Hybridoma cells were cultured in Cellgro Protein Free Medium (Mediatech) to prepare mAbs for isolation with a Protein G Sepharose 4 column (Amersham Pharmacia Biotech) and a Superdex 200 Prep Grade Gel Filtration column.
Studies on human tumor tissues. The human subject protection protocol for collection of tumor tissues from patients with malignant diseases has been reviewed and approved by the institutional review board at Stony Brook University. Malignant tumors and adjacent normal tissues were also obtained from National Disease Research Interchange (Philadelphia). Sample preparations and methods involving antibody staining of formalin-fixed, paraffin-embedded tissue samples for immunohistochemistry, as well as the parallel analysis of gelatinolytic activities and proteins specific for seprase by gelatin zymography and Western immunoblotting were performed as described previously (11, 12). Seprase was isolated from tumor lysates using WGA lectin- or immuno-affinity purification procedures as described (15). Soluble enzymatic assays determining DP and gelatinase activities of purified protein were performed as described (15).
In situ Hybridization. In situ hybridization was performed to reveal seprase mRNA expression in tumor tissue sections. Briefly, a DNA template for transcription of an anti-sense RNA probe was amplified with PCR using seprase cDNA as template. The primers (5’- GAT TCT TCC TCC TCA ATT TG -3’ and 5’- TAA TAC GAC TCA CTA TAG GGT CAC CTT GGA AAG CTG TTC -3’) were designed so that the resulting PCR product of 190bp had no significant homology to other genes, and that a T7 bacteriophage promoter was added to one end. Similarly, a DNA template for control sense RNA probe was amplified by using two primers (5’- TAA TAC GAC TCA CTA TAG GGA TTC TTC CTC CTC AAT TTG -3’ and 5’- GTC ACC TTG GAA AGC TGT TC -3’) that added the T7 promoter to the other end of the PCR product. Specific FITC-labeled anti-sense and sense RNA probes were synthesized by in vitro transcription. Template DNA was removed by digestion with RNase-free DNase I. RNA probes were hybridized to sections of formalin-fixed, paraffin-embedded tumor tissues that were obtained as described above for immunohistochemistry. The FITC-labeled probes were revealed with anti-FITC mouse antibodies, anti-mouse antibodies and APAAP (a mixture of alkaline phosphatase and mouse anti- alkaline phosphatase antibodies), followed by colorimetric substrates NBT and BCIP. Finally, the tissue sections were counterstained with Nuclear Fast Red, dehydrated and mounted.
Results and Discussion
Isolation of r-seprase. We used a novel approach to express seprase with an active gelatinase activity, in which the cytoplasmic and transmembrane domains were deleted, a secretion signal was added and a novel mammalian expression system was utilized (Fig. 1A). Specifically, the seprase cDNA sequence that encodes the entire extracellular domain of 734 amino acids was cloned downstream of a CMV promoter and inserted between an N-terminal secretion signal and a C-terminal V5-His tag in a modified pCEP4 vector (Fig. 1A). Seprase cDNA sequences, encoding the predicted cytoplasmic domain and transmembrane domain, were excluded. The newly constructed r-seprase expression vector was named pE15. 293-EBNA cells were transfected with pE15 and cultured in the HyQ PF-293 protein-free medium. R-seprase was affinity-purified and subjected to Western immunoblotting and proteolysis assays.
To identify 170 kDa n-seprase dimers and 97 kDa monomeric subunits, gelatin zymography incubated in conditions containing 5 mM EDTA and on Western immunoblots using antibodies directed against seprase subunits can be used (3, 4). Similarly, r-seprase was found to be a 160-kDa dimer with gelatinase activity that can be dissociated into two proteolytically inactive 90-kDa subunits (Fig. 1B-C). Parallel SDS PAGE gelatin zymography and Western immunoblotting showed that the 160-kDa dimer degraded gelatin, but the 90-kDa monomer could not (Fig. 1B-C). In addition, the 160-kDa dimers captured by mAbs D8, D43, 90 or 65 exhibited DP and gelatinase activities, but the dissociated subunits isolated by mAb E97 did not (Fig. 1D). In contrast to the previously proposed role of the transmembrane domain holding subunits together (3, 4), the present data demonstrate that the cytoplasmic and transmembrane domains of seprase are not required for its subunit dimerization, nor for its enzymatic activities.
Proteolytic truncation of seprase activates its gelatinase activity but not DP activity. We have obtained several lines of evidence to suggest that N-terminal truncation of seprase activates its gelatinase activity, but does not affect its DP activity. Initially, we compared the gelatinase activity of n-seprase with that of r-seprase. Since the dimeric forms of both n-seprase and r-seprase were recognized by mAb 90 (Fig. 1E), mAb90 was used in Western immunoblotting to quantify relative amounts of the r-seprase and n-seprase protein. When similar amounts of soluble r-seprase and membrane bound n-seprase protein were loaded on gelatin zymograms to compare the gelatinase activity, the gelatinase activity of r-seprase dimer is considerably higher than that of the n-seprase dimer (Fig. 1E). Alternatively, the low gelatinase activity in the membrane bound n-seprase could be due to the presence of endogenous inhibitors. To test this, different amounts of the detergent extract containing n-seprase were mixed with a given amount of r-seprase and the mixtures were subjected to gelatin zymography in 5 mM EDTA conditions (Fig. 1F). The gelatinase activity specific for r-seprase remained at similar levels, suggesting the absence of endogenous seprase inhibitors associated with the membrane extract. Together these data demonstrate that the greater gelatinase activity of r-seprase is a result of truncation of the native form.
Previous investigations that utilized detergents to extract the membrane bound n-seprase showed a specific gelatinase activity of n-seprase at 170-kDa (1, 3, 7). However, the use of detergents for n-seprase extraction might confer the detected gelatinase activity by opening the protein to increase the availability of its catalytic site for macromolecular substrates, whereas n-seprase in its cellular environment in vivo may not exhibit any activity until it is proteolytically activated.
Proteolytic truncation of r-seprase resulted in further increase in gelatinase activity (Fig. 2). The gelatinase activation occurs readily in r-seprase purified by DEAE Sepharose and WGA affinity chromatography columns that contain greater than 10% of impurity (Fig. 2A-B), but is not noticeable in samples of r-seprase purified by His·Bind Resin column that are over 95% pure (Fig. 2A-B). Proteolytically activated seprase forms have molecular masses of 100-85 kDa and 70-50 kDa on gelatin zymograms (Fig. 2B) that can be recognized by mAb E97 on a parallel Western immunoblot (Fig. 2C). The smaller, activated gelatinases reduce in fraction with increasing purification: DEAE > WGA > His, suggesting the involvement of other proteases necessary for seprase activation.
Figure 2.
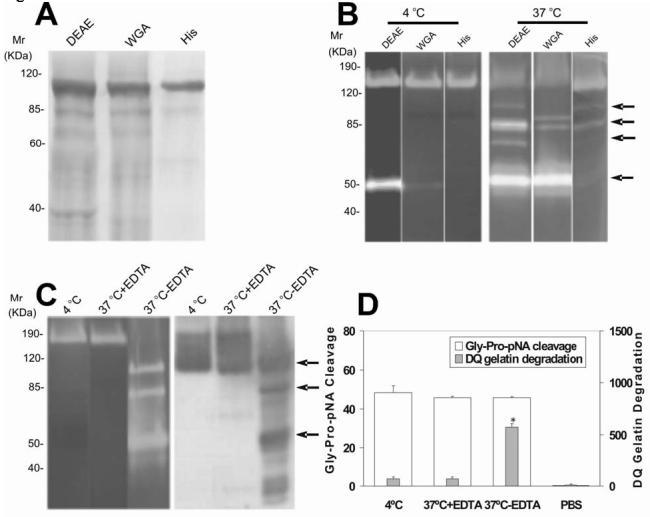
Purification and proteolytic activities of r-seprase. A, SDS PAGE analysis on purified r-seprase. The r-seprase samples indicated were increasingly and sequentially purified by the DEAE Sepharose column (DEAE), the WGA affinity chromatography column (WGA) and the His·Bind Resin column (His) from the culture medium conditioned by pE15 transfected 293-EBNA cells. The samples were then heated to 100 °C for 3 min in SDS sampling buffer and separated by SDS PAGE. The gel was stained with Coomassie Brilliant Blue. Each lane corresponds to the dissociated monomers of r-seprase present in 10-mL of original culture medium. B, Increased gelatinase activity of purified r-seprase incubated at 37 °C. R-seprase samples were purified as above, but not heated, were incubated at 4 °C (left panel) or 37 °C (right panel) overnight, and then subjected to gelatin zymography in 5 mM EDTA conditions. The top two arrows show the 100-85 kDa gelatinases and the lower two arrows indicate the 70-50 kDa gelatinases that were present only in the less purified fractions, which were potentially exposed to other proteases. Each lane corresponds to the r-seprase present in 10-mL of original culture medium. C, Activation of the 160-kDa r-seprase into 100-85 kDa and 70-50 kDa gelatinases (arrows) by EDTA sensitive protease. R-seprase was enriched by WGA-affinity chromatography column and incubated at 4 °C or 37 °C for 1 day in the presence or absence of 5 mM EDTA (indicated by 4 °C, 37°C+EDTA and 37°C-EDTA, respectively), and then subjected to gelatin zymography in 5 mM EDTA conditions and Western immunoblotting using mAb E97 (against seprase subunits and shortened forms that were denatured by Western immunoblotting transfer buffer). The top two arrows show the 100-85 kDa gelatinases and the lowest arrow indicates the 50 kDa gelatinases. D, Activation of r-seprase increases gelatinase activity, but not DP activity. R-seprase was enriched by a WGA-affinity chromatography column and incubated at 4 °C or 37 °C for 1 day in the presence or absence of 5 mM EDTA (indicated by 4 °C, 37°C+EDTA and 37°C-EDTA, respectively), and then subjected to the soluble enzymatic assays as described (15). PBS was used as a negative control in the soluble enzymatic assay. The values shown are the mean ± SD. The symbol * above the bar is used to indicate a significant increase (p<0.001) in gelatinase activity of truncated seprase.
The involved protease for r-seprase activation is an EDTA-sensitive endogenous enzymatic activator (Fig. 2C). When the r-seprase samples, partially purified by DEAE Sepharose and WGA-affinity chromatography columns, were incubated at 4 °C or 37 °C for 1 day in the presence or absence of 5 mM EDTA (Fig. 2C) and then subjected to gelatin zymography developed in the presence of 5 mM EDTA and Western immunoblotting using mAb E97, the activated seprase forms only appeared in the r-seprase sample that was incubated at 37 °C in the absence of EDTA (Fig. 2C). Similarly, when these same r-seprase samples were subjected to soluble enzymatic assays for DP and gelatinase activities specific for seprase, the r-seprase sample incubated at 37 °C in the absence of EDTA exhibited a seven-fold increase in gelatinase activity, but did not increase its DP activity (Fig. 2D). Incubation of r-seprase at 4 °C or 37 °C in the presence of EDTA did not significantly change the DP and gelatinase activities of r-seprase (Fig. 2D). Note that both gelatin zymography and soluble enzymatic assays for DP and gelatinase activities specific for seprase were developed in the presence of 5 mM EDTA to suppress metalloprotease activity.
Overall, the gelatinase activity of the activated r-seprase was elevated as shown by gelatin zymography (Fig. 2B-C) and soluble DP and gelatinase assays (Fig. 2D). However, the DP activity of different forms of r-seprase was not increased by proteolytic truncation (Fig. 2D). These data demonstrate that proteolytic truncation of seprase reduces steric hindrance for the gelatin substrate, but not the DP substrate, and increases the gelatinolytic activity of seprase. Truncation is likely to occur at the N-terminal region because the catalytic center of seprase is localized to the distal end of C-terminus and catalytic activity is retained in these shortened forms.
Interestingly, the anti-V5 antibody and the His.Bind.Resin column were not able to capture the truncated 50-kDa form (data not shown). Since the anti-V5 antibody and His.Bind.Resin column are able to bind to the full-length r-seprase dimer and monomer, it is possible that the 50-kDa form might have lost its C-terminal V5 and His tags during proteolytic cleavage and production of the shorter forms. The cleavage site of the C-terminal truncation is not known. It is likely to be within the 26 amino acid residues after His734 of the catalytic triad, as the truncated 50-kDa form remains active.
Expression and activity of n-seprase. We established LOX melanoma cells with contrasting levels of seprase expression, sephigh and seplow. LOX cells with low seprase expression (seplow) were generated by RNAi knockdown using the vector pGUS-SEP1384, followed by stable cell selection, and cells with high seprase (sephigh) were similarly transfected with a control pGUS vector. The sephigh cells produced high levels of n-seprase (Fig. 3A) and invaded fibronectin-coated gelatin films in vitro (Fig.3D); whereas the seplow cells did not. Seprase is only prominent in the cell lysate derived from sephigh cells, in which the dimers and intact 97-kDa subunits of n-seprase could be demonstrated by Western immunoblotting (Fig. 3A).
Figure 3.
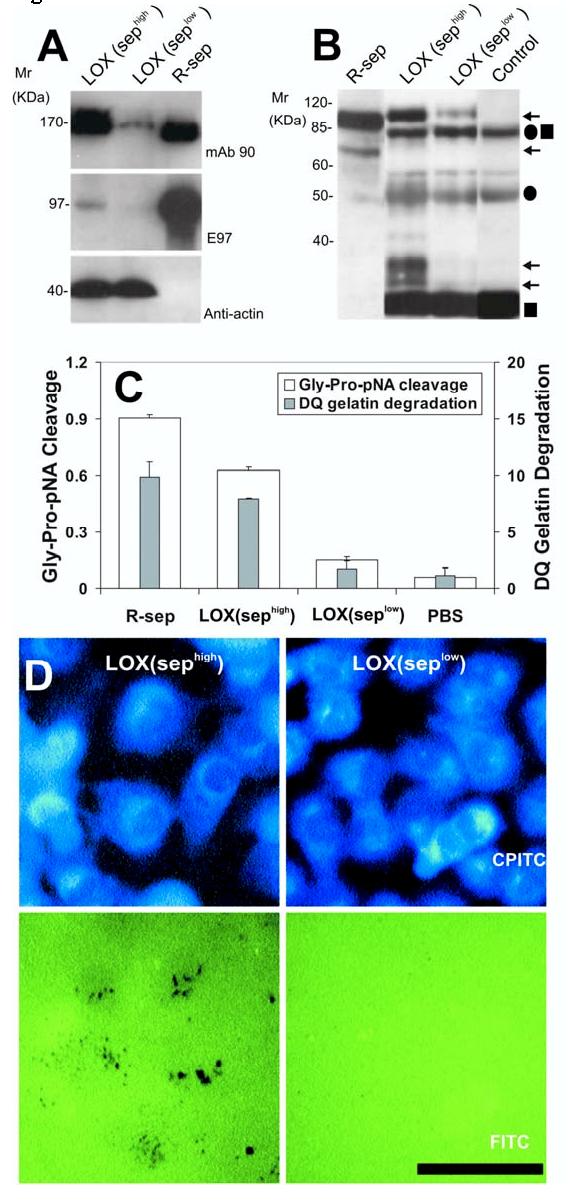
Expression and activity of n-seprase in LOX melanoma cell lines and tumor tissues from a xenograft model. A, N-seprase in LOX cell lines, sephigh and seplow. LOX cells transfected with the control pGUS vector express high levels of seprase, whereas transfection with the pGUS-SEP 1384 vector utilized RNA interference (RNAi) creates a cell line with limited seprase expression. Cell lysates of the two LOX cell lines, sephigh and seplow, were subjected to SDS-PAGE followed by Western immunoblotting under non-boiling (for mAb 90) and boiling conditions (for mAb E97 and mAb anti-actin). The lane labeled r-sep is the positive control r-seprase; the band labeled anti-actin serves as a protein loading control. B, N-seprase in experimental tumors created from the two LOX cell lines, sephigh and seplow. Tumors were lysed with EDTA-containing buffer. N-seprase was isolated from tumor tissue lysates by immunoprecipitation using mAb D8-D28-D43 conjugated beads that specifically target seprase antigen. The beads-antigen complexes were boiled in SDS-PAGE loading buffer to release seprase polypeptides, and separated via SDS-PAGE. Subsequent Western immunoblotting using rat mAb E97 revealed bands corresponding to n-seprase polypeptides of 97-, 65-, 35- and 25-kDa (arrows). Antibody IgG fragments released from the beads, including linked heavy and light chains (•■), heavy chain alone (•) and light chain alone (■), were also detected. R-seprase (r-sep) was used as a positive control. The negative control (control) was a protein sample derived from the antibody beads that were not mixed with tumor lysates. C, Seprase specific DP and gelatinase activity in experimental tumors created from the two LOX cell lines, sephigh and seplow. Tumor tissue lysates were applied to plates coated with mAb D8 to specifically capture active seprase forms. The soluble enzymatic assays were conducted in the presence of 5 mM EDTA to inhibit any metalloprotease activity. The experiment was performed in triplicate and the values are the mean ± SD. A significant increase (p<0.001) in seprase activity compared to the seprase knockdown tumors is indicated by the * above the bar. D, Degradation/invasion of fibronectin-coated cross-linked gelatin substrata comparing two conditions of LOX cells, LOX (sephigh) and LOX (seplow). LOX cells transfected with pGUS and pGUS-sep1384, respectively, were cultured on FITC-fibronectin-coated crosslinked gelatin films. After fixation and Phalloidin-CPITC staining, cells were visualized by epifluorescence microscopy of CPITC to visualize cell location, and the same fields were then photographed by epifluorescence microscopy of FITC to visualize the sites of matrix degradation left behind the migrating cells which are shown by dark spots on the green background. Bar, 50 μm.
We also established experimental human tumors from sephigh and seplow cells and determined the subunit composition of s-seprase in the human tumors developed in immunodeficient mice (Fig. 3B). We found that tumors derived from sephigh cells produced seprase peptides of 97-, 65-, 35- and 25-kDa but those from seplow cells did not (Fig. 3B); consistently the former also exhibited higher DP and gelatinase activities specific for seprase than the latter (Fig. 3C).
Detection of s-seprase in human tumors. We detected seprase mRNA expression in tumor tissues using in situ hybridization. In Figures 4A-B, an example of ovarian adenocarcinomas shows that seprase mRNA is mainly localized to tumor cells (Fig. 4A, open arrows) and is less prominent in stromal cells (Fig. 4A, arrows), suggesting that the majority of seprase is produced by tumor cells. Activated seprase may attach to stromal matrices during its proteolytic degradation of the substrates.
Figure 4.
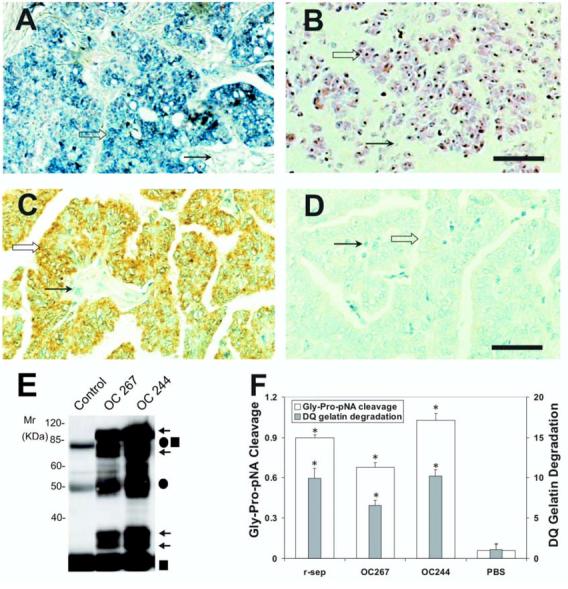
Expression and activity of n-seprase in human ovarian carcinoma. A and B: N-seprase mRNA expression in human ovarian carcinoma tissues. In situ hybridization was performed on formalin-fixed, paraffin-embedded tumor sections using FITC-labeled anti-sense (A) or sense (B) RNA probes for seprase. FITC-labeling was revealed using the alkaline phosphatase colorimetric substrates BCIP/NBT, which show a purple/blue staining. Counterstaining using nuclear fast red appears red. Seprase mRNA is mainly localized in tumor cells (open arrows) and is less prominent in stromal cells (arrows). Bar, 100 μm. C and D: N-seprase protein expression in human ovarian carcinoma tissues. Immunohistochemistry was carried out on formalin-fixed, paraffin-embedded tumor sections using mAb D8 against seprase (C) or a control rat antibody (D). The positive staining is brown, while the counterstaining using hematoxylin appears blue. Seprase protein was mainly detected in tumor cells (open arrows) instead of stromal cells (arrows). Bar, 100 μm. E: Isolation of n-seprase truncated forms from human ovarian carcinoma tissues. Tumor tissues were lysed in the presence of 5mM EDTA. N-seprase in tumor lysates (OC 267 and OC244) was isolated and identified as described in (3B) above. Note that the top two arrows indicate dissociated seprase polypeptides at 97- and 65-kDa; the lower two arrows show dissociated seprase polypeptides at 35- and 25-kDa. F: Seprase specific DP and gelatinase activity in human ovarian carcinoma tissues. Tumor tissue lysates were applied to plates coated with mAb D8 to specifically capture active seprase forms. The soluble enzymatic assays were conducted in the presence of 5 mM EDTA to inhibit any metalloprotease activity. * denotes a significance of P<0.001 compared to the PBS negative control.
Immunohistochemical techniques were used to evaluate the expression of seprase protein in surgically removed tumors using a panel of mAbs: D8, D28 and D43, that recognized seprase as described (11-14). In Figures 4C-D, an example of ovarian carcinoma reveals positive immuno-staining at tumor cells, indicated by their brown membrane/cytoplasm, against cellular nuclei counterstained by hematoxylin in blue. This is compared to the control antibody staining that only shows a blue hematoxylin stain (Fig. 4D).
Affinity-purification of seprase protein from ovarian adenocarcinomas revealed seprase activation in malignant ovarian tumors with shortened seprase forms (s-seprase) composed of 65-, 35- and 25-kDa polypeptides (Fig. 4E). The tumors with seprase activation exhibited high DP and gelatinase activities specific for seprase (Fig. 4F).
Similar findings were made in other types of malignant tumors. In Figures 5A-B, an example of malignant melanoma shows positive immunostaining for seprase, indicated by a pink or red cytoplasmic stain, against cellular nuclei counterstained by hematoxylin in blue, as compared to the control that only shows the blue hematoxylin stain. In order to confirm that the s-seprase forms found in the tumor tissues were actively degrading gelatin, WGA-binding proteins purified from paired tumor and adjacent tissues from the same patients were analyzed in parallel by gelatin zymography and immunoblotting using both mAbs D8 (directed mainly against seprase 170-kDa dimeric form and 97-kDa dissociated monomeric form) and E97 (against all seprase forms including active s-seprase forms, in Western immunoblots, in which seprase forms were partially denatured by protein transfer buffer) as described (11, 12). Figures 5C-F illustrate examples of carcinomas of the colon (Fig. 5C), stomach (Fig. 5D) and breast (Fig. 5E), and malignant melanoma (Fig. 5F). This direct comparison of gelatinolytic activities and proteins specific for seprase showed that: (i) in the assay conditions, in which matrix metalloproteinases (MMPs) were suppressed by EDTA to resolve serine-type gelatinase activities (Fig. 5C-F), gelatinolytic activities and proteins specific for seprase were found to be more prominent in tumors than in adjacent normal tissues; (ii) 100-85 kDa and 70-50 kDa active s-seprase forms recognized by mAb E97 were abundant in the tumors (Fig. 5C-F). The activated s-seprase forms are likely dimeric and composed of 65-, 35- and 25-kDa subunits that were resolved under denaturing conditions (Fig. 4E).
Figure 5.
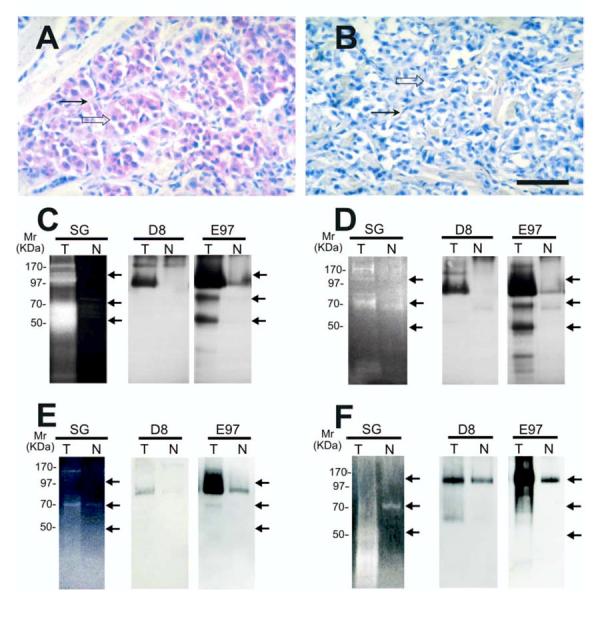
Gelatinase activity and localization of n-seprase in other human tumors. A and B, Immunohistochemical localization of n-seprase in human malignant melanoma. Formalin-fixed, paraffin-embedded human malignant melanoma tissue was sectioned and stained using rat mAb D8 (A) and the control rat mAb (B). The slides were then incubated with biotin-conjugated goat anti-rat antibody (Jackson Immunoresearch, West Grove, PA), stained with strepavidin/alkaline phosphatase conjugate (Jackson Immunoresearch), followed by Fast red chromogen in napthol phosphate reconstitution buffer (Biogenex) for the color development and counterstained with hematoxylin. Note that seprase positive regions of melanoma cells (open arrow) and stromal cells (arrow) are stained red. Bar, 50 μm. C -F, Detection of n-seprase activated forms of different size in invasive carcinomas of the colon (C), stomach (D) and breast (E), and in malignant melanoma (F). Paired tumor (T) and adjacent normal (N) tissues from the same patient were lysed with EDTA-containing lysis buffer. Glycosylated protein captured by the WGA column from the tissue lysates were analyzed in parallel by gelatin zymography (SG) and immunoblotting (D8 and E97). The protein samples were loaded under non-boiling, non-reducing conditions to retain gelatinase activity of seprase active forms. The gelatinase activity of serine-type gelatinases (SG) was detected by incubating the zymogram with 5 mM EDTA to inhibit metalloprotease activity as described (15). Note that two major groups of active s-seprase forms (100-85 kDa and 70-50 kDa indicated by arrows) were found by parallel analysis of gelatin zymography and Western immunoblotting. In immunoblotting, mAb D8 recognizes mainly seprase 170-kDa dimeric form and 97-kDa dissociated monomeric form, while mAb E97 recognizes all seprase forms, including seprase 170-kDa dimeric form, 97-kDa monomeric subunit, activated forms of 100-85 kDa and 70-50 kDa (arrows), and other inactive shortened forms.
We detected the presence of s-seprase forms in all of the malignant tumor tissues examined, including ovarian carcinoma (Fig. 4), adenocarcinoma of the colon and stomach, invasive ductal carcinoma of the breast, and malignant melanoma (Fig. 5). After examining a number of malignant tumor and adjacent normal tissue samples, we found that, overall, seprase expression and activation occur more frequently in malignant tumor tissues (P<0.0001) (Table 1). These data demonstrate the prevalence of activated s-seprase forms in malignant tumors that corresponds to increased gelatinase activity (Table 1). Activated seprase in malignant tumors may be contributing to tumor invasion, in a way similar to that of the urokinase plasminogen activator (26).
Table 1.
Detection of active seprase in malignant tumor and adjacent normal tissues.
| Tumor types | Tumor tissue with active seprase expression/total tumor samples, (%) | Normal tissue with active seprase expression/total normal samples, (%) | P- value |
|---|---|---|---|
| Ovarian carcinoma | 15/15*(100) | 0/6 (0) | <0.0001 |
| Breast carcinoma | 6/8 (75) | 3/8 (37.5) | † |
| Colon carcinoma | 3/4(75) | 1/4 (25) | † |
| Gastric carcinoma | 7/8 (87.5) | 2/8 (25) | 0.0117 |
| Melanoma | 12/13*(92.3) | 4/9 (44.4) | 0.0132 |
| Total | 43/48 (89.6) | 10/35 (28.6) | <0.0001 |
NOTE: In tumor and adjacent normal tissue samples seprase was isolated using WGA column and identified by parallel gelatin zymography and Western immunoblotting, as shown in Fig. 5C-F.
indicates that six of 15 ovarian carcinoma samples and four of 13 melanoma samples were analyzed by isolating active seprase via immunoprecipitation and subjected to soluble enzymatic assay for detection of activity, as represented in Fig. 4E-F. Significance of active seprase expression in tumor versus normal samples, indicated by P-value, was determined by Chi-square test.
indicates that an accurate P value could not be calculated due to small sample size; however these values contribute to the overall significance calculation.
Potential mechanism: Proteolytic truncation reduces steric hindrance for substrates. The construction of seprase is such that the amino-terminal end contains a very short cytoplasmic and transmembrane region that anchors the protease to the surface of a cell while allowing for the bulk of seprase, including its putative catalytic region, to interact with the extracellular milieu. Our data suggests that reduction of steric hindrance via truncation for activation can be a useful mechanism to heighten the activity of seprase. Not only will truncation facilitate access of substrates to the active site, but it is also a potential means to release soluble s-seprase. Our model (Fig. 6) reflects the idea that extensive N-terminal truncation reduces steric hindrance and allows for large substrate molecules to have greater accessibility to the catalytic site.
Figure 6.
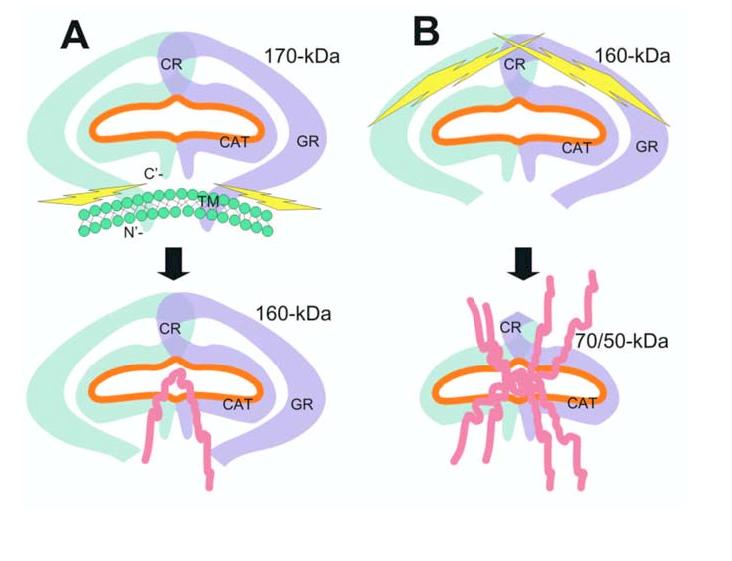
Schematic illustration of a potential seprase activation process. A, Release of membrane-bound n-seprase. The dimeric form of n-seprase (170-kDa) is bound to the cell membrane by the N-terminal stalk region and creates a catalytic pocket (orange line). Likely there is more than one overlapping region that contributes to the dimerization of this protein. The membrane blocks access to the active pocket when the 3-D structure of the dimer is considered. Activating proteases cleave the dimer at the stalk region and release it from the membrane, thereby creating a soluble form like r-seprase (160-kDa). The separation of the protein from the membrane reduces steric hindrance and increases accessibility to the catalytic pocket from below. B, Further N-terminal truncation of seprase and increase of gelatinase activity. Release of N-terminal peptides from the 160-kDa seprase into the 70-/50-kDa form extensively reduces steric hindrance and permits even greater accessibility of substrates to the catalytic site. The amino acid terminus and domains of seprase are indicated: C’, carboxyl terminus; N’, amino terminus; TM, transmembrane domain, amino acids 7-26; GR, glycosylation rich region, amino acids 49-314; CR, cysteine rich domain, amino acids 305-466; CAT, catalytic domain, amino acids 500-760.
Recent structural analysis suggests that it is possible that the 15Å, triple helical collagen is able to feed through the 24Å lateral opening thereby gaining access to the active site (23). Although the structure meets the diameter requirement, there are other considerations. Classical type I collagen is a 300 nm long three-stranded coil with limited flexibility. All previous studies that examine n-seprase activity rely on short peptides to assess DP activity or the denatured form of collagen, gelatin. A single strand of gelatin is a small fraction of the bulk of the triple helix and there is a significant reduction in the rigidity. To interact with the active site of seprase, gelatin would need to navigate a relatively small opening in order to be strung through both monomers for seprase to exhibit its characteristic endopeptidase activity.
It is possible that, like interstitial collagenase (27), a Brownian ratchet-like mechanism feeds the collagen or gelatin through the lateral opening of both monomers in seprase; however, the initial process would require an exogenous source of energy and this energy requirement is not reflected in the activity of seprase in vitro. Also, similar to interstitial collagenase (27), seprase can liberate from its pro-domain (i.e., N-terminal transmembrane domain) to interact directly with the collagen that has been modified by other collagenases (Fig. 6A). In the case of seprase, perhaps there is a modification of this mechanism, which involves increasing the accessibility of the active site by further proteolytic truncation in a manner similar to the zymogen activation of the MMPs (Fig. 6B). In this way seprase activation may facilitate the invasion of malignant tumor cells through a localized degradation of components of the ECM.
Acknowledgments
Grant support: This research was supported by grants from the National Institutes of Health R01CA0039077 and R01EB002065 to WTC, and GCRC Grant MO1RR10710.
References
- 1.Aoyama A, Chen W-T. A 170-kDa membrane-bound protease is associated with the expression of invasiveness by human malignant melanoma cells. Proc.Natl.Acad.Sci.U.S.A. 1990;87:8296–300. doi: 10.1073/pnas.87.21.8296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Monsky WL, Lin C-Y, Aoyama A, Kelly T, Mueller SC, Akiyama SK, Chen W-T. A potential marker protease of invasiveness, seprase, is localized on invadopodia of human malignant melanoma cells. Cancer Res. 1994;54:5702–10. [PubMed] [Google Scholar]
- 3.Pineiro-Sanchez ML, Goldstein LA, Dodt J, Howard L, Yeh Y, Tran H, Argraves WS, Chen W-T. Identification of the 170-kDa melanoma membrane-bound gelatinase (seprase) as a serine integral membrane protease. J.Biol.Chem. 1997;272:7595–601. doi: 10.1074/jbc.272.12.7595. [DOI] [PubMed] [Google Scholar]; J. Biol. Chem. 1998;273:13366. Correction. [PubMed] [Google Scholar]
- 4.Goldstein LA, Ghersi G, Piñeiro-Sánchez ML, Salamone M, Yeh YY, Flessate D, Chen W-T. Molecular cloning of seprase: A serine integral membrane protease from human melanoma. Biochimica et Biophysica Acta. 1997;1361:11–9. doi: 10.1016/s0925-4439(97)00032-x. [DOI] [PubMed] [Google Scholar]
- 5.Park JE, Lenter MC, Zimmermann RN, Garin-Chesa P, Old LJ, Rettig WJ. Fibroblast activation protein, a dual specificity serine protease expressed in reactive human tumor stromal fibroblasts. J Biol.Chem. 1999;274:36505–12. doi: 10.1074/jbc.274.51.36505. [DOI] [PubMed] [Google Scholar]
- 6.Scanlan MJ, Raj BK, Calvo B, Garin-Chesa P, Sanz-Moncasi MP, Healey JH, Old LJ, Rettig WJ. Molecular cloning of fibroblast activation protein alpha, a member of the serine protease family selectively expressed in stromal fibroblasts of epithelial cancers. Proc.Natl.Acad.Sci.U.S.A. 1994;91:5657–61. doi: 10.1073/pnas.91.12.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldstein LA, Chen W-T. Identification of an alternatively spliced seprase mRNA that encodes a novel intracellular isoform. J Biol.Chem. 2000;275:2554–9. doi: 10.1074/jbc.275.4.2554. [DOI] [PubMed] [Google Scholar]
- 8.Ariga N, Sato E, Ohuchi N, Nagura H, Ohtani H. Stromal expression of fibroblast activation protein/seprase, a cell membrane serine proteinase and gelatinase, is associated with longer survival in patients with invasive ductal carcinoma of breast. Int.J Cancer. 2001;95:67–72. doi: 10.1002/1097-0215(20010120)95:1<67::aid-ijc1012>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 9.Huang Y, Wang S, Kelly T. Seprase promotes rapid tumor growth and increased microvessel density in a mouse model of human breast cancer. Cancer Res. 2004;64:2712–6. doi: 10.1158/0008-5472.can-03-3184. [DOI] [PubMed] [Google Scholar]
- 10.Kelly T, Kechelava S, Rozypal TL, West KW, Korourian S. Seprase, a membrane-bound protease, is overexpressed by invasive ductal carcinoma cells of human breast cancers. Mod.Pathol. 1998;11:855–63. [PubMed] [Google Scholar]
- 11.Okada K, Chen W-T, Iwasa S, Jin X, Yamane T, Ooi A, Mitsumata M. Seprase, a membrane-type serine protease, has different expression patterns in intestinal- and diffuse-type gastric cancer. Oncology. 2003;65:363–70. doi: 10.1159/000074650. [DOI] [PubMed] [Google Scholar]
- 12.Mori Y, Kono K, Matsumoto Y, Fujii H, Yamane T, Mitsumata M, Chen W-T. The expression of a type II transmembrane serine protease (Seprase) in human gastric carcinoma. Oncology. 2004;67:411–9. doi: 10.1159/000082926. [DOI] [PubMed] [Google Scholar]
- 13.Iwasa S, Okada K, Chen W-T, Jin X, Yamane T, Ooi A, Mitsumata M. ′Increased expression of seprase, a membrane-type serine protease, is associated with lymph node metastasis in human colorectal cancer′. Cancer Lett. 2005;227:229–36. doi: 10.1016/j.canlet.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 14.Jin X, Iwasa S, Okada K, Mitsumata M, Ooi A. Expression patterns of seprase, a membrane serine protease, in cervical carcinoma and cervical intraepithelial neoplasm. Anticancer Res. 2003;23:3195–8. [PubMed] [Google Scholar]
- 15.Ghersi G, Dong H, Goldstein LA, Yeh Y, Hakkinen L, Larjava HS, Chen W-T. Regulation of fibroblast migration on collagenous matrix by a cell surface peptidase complex. J.Biol.Chem. 2002;277:29231–41. doi: 10.1074/jbc.M202770200. [DOI] [PubMed] [Google Scholar]
- 16.Mueller SC, Ghersi G, Akiyama SK, Sang QX, Howard L, Pineiro-Sanchez M, Nakahara H, Yeh Y, Chen W-T. A novel protease-docking function of integrin at invadopodia. J.Biol.Chem. 1999;274:24947–52. doi: 10.1074/jbc.274.35.24947. [DOI] [PubMed] [Google Scholar]
- 17.Cheng JD, Valianou M, Canutescu AA, Jaffe EK, Lee HO, Wang H, Lai JH, Bachovchin WW, Weiner LM. Abrogation of fibroblast activation protein enzymatic activity attenuates tumor growth. Mol Cancer Ther. 2005;4:351–60. doi: 10.1158/1535-7163.MCT-04-0269. [DOI] [PubMed] [Google Scholar]
- 18.Cheng JD, Dunbrack RL, Jr., Valianou M, Rogatko A, Alpaugh RK, Weiner LM. Promotion of tumor growth by murine fibroblast activation protein, a serine protease, in an animal model. Cancer Res. 2002;62:4767–72. [PubMed] [Google Scholar]
- 19.Ramirez-Montagut T, Blachere NE, Sviderskaya EV, Bennett DC, Rettig WJ, Garin-Chesa P, Houghton AN. FAPalpha, a surface peptidase expressed during wound healing, is a tumor suppressor. Oncogene. 2004;23:5435–46. doi: 10.1038/sj.onc.1207730. [DOI] [PubMed] [Google Scholar]
- 20.Rettig WJ, Su SL, Fortunato SR, Scanlan MJ, Raj BK, Garin-Chesa P, Healey JH, Old LJ. Fibroblast activation protein: purification, epitope mapping and induction by growth factors. Int.J.Cancer. 1994;58:385–92. doi: 10.1002/ijc.2910580314. [DOI] [PubMed] [Google Scholar]
- 21.Levy MT, McCaughan GW, Abbott CA, Park JE, Cunningham AM, Muller E, Rettig WJ, Gorrell MD. Fibroblast activation protein: a cell surface dipeptidyl peptidase and gelatinase expressed by stellate cells at the tissue remodelling interface in human cirrhosis. Hepatology. 1999;29:1768–78. doi: 10.1002/hep.510290631. [DOI] [PubMed] [Google Scholar]
- 22.Niedermeyer J, Enenkel B, Park JE, Lenter M, Rettig WJ, Damm K, Schnapp A. Mouse fibroblast-activation protein--conserved Fap gene organization and biochemical function as a serine protease. Eur.J.Biochem. 1998;254:650–4. doi: 10.1046/j.1432-1327.1998.2540650.x. [DOI] [PubMed] [Google Scholar]
- 23.Aertgeerts K, Levin I, Shi L, Snell GP, Jennings A, Prasad GS, Zhang Y, Kraus ML, Salakian S, Sridhar V, Wijnands R, Tennant MG. Structural and kinetic analysis of the substrate specificity of human fibroblast activation protein alpha. J.Biol.Chem. 2005;280:19441–4. doi: 10.1074/jbc.C500092200. [DOI] [PubMed] [Google Scholar]
- 24.Bjelke JR, Christensen J, Branner S, Wagtmann N, Olsen C, Kanstrup AB, Rasmussen HB. Tyrosine 547 constitutes an essential part of the catalytic mechanism of dipeptidyl peptidase IV. J.Biol.Chem. 2004;279:34691–7. doi: 10.1074/jbc.M405400200. [DOI] [PubMed] [Google Scholar]
- 25.Chen W-T, Yeh Y, Nakahara H. An in vitro cell invasion assay: determination of cell surface proteolytic activity that degrades extracellular matrix. J.Tiss.Cult.Meth. 1994;16:177–81. [Google Scholar]
- 26.Gladson CL, Pijuan-Thompson V, Olman MA, Gillespie GY, Yacoub IZ. Up-regulation of urokinase and urokinase receptor genes in malignant astrocytoma. Am.J.Pathol. 1995;146:1150–60. [PMC free article] [PubMed] [Google Scholar]
- 27.Saffarian S, Collier IE, Marmer BL, Elson EL, Goldberg G. Interstitial collagenase is a Brownian ratchet driven by proteolysis of collagen. Science. 2004;306:108–11. doi: 10.1126/science.1099179. [DOI] [PubMed] [Google Scholar]


