Abstract
Theoretical considerations predict that amplification of expressed gene transcripts by reverse transcription–PCR using arbitrarily chosen primers will result in the preferential amplification of the central portion of the transcript. Systematic, high-throughput sequencing of such products would result in an expressed sequence tag (EST) database consisting of central, generally coding regions of expressed genes. Such a database would add significant value to existing public EST databases, which consist mostly of sequences derived from the extremities of cDNAs, and facilitate the construction of contigs of transcript sequences. We tested our predictions, creating a database of 10,000 sequences from human breast tumors. The data confirmed the central distribution of the sequences, the significant normalization of the sequence population, the frequent extension of contigs composed of existing human ESTs, and the identification of a series of potentially important homologues of known genes. This approach should make a significant contribution to the early identification of important human genes, the deciphering of the draft human genome sequence currently being compiled, and the shotgun sequencing of the human transcriptome.
The identification and sequencing of human expressed sequences (cDNAs) plays a synergistic role to complete genome determination and represents a direct link to functional genomics (1–5). In particular, cDNAs greatly aid exon identification and are essential for determination of tissue and pathology-specific exon usage in the form of alternatively spliced variants (6–8). Furthermore, repeated partial sequencing of expressed sequences, so-called expressed sequence tags (ESTs), have proved a powerful means of identification of genetic polymorphisms (9–11) and for determination of differential gene expression (12–18). To date, more than 1,500,000 human ESTs have been generated and deposited in GenBank, derived principally from the Merck Gene Index Project and the Cancer Genome Anatomy project (refs. 19 and 20; http://www.ncbi.nlm.nih.gov/dbEST). Clustering of these sequences shows that at least some have been derived from an estimated 86,000 different human genes but only approximately 11% of these have a full-length sequence (UniGene build 98, November 3, 1999, http://www.ncbi.nlm.nih.gov/UniGene/index.html). Moreover, approximately 65% of ESTs represent the 3′ extremity of cDNAs and 26% represent the 5′ extremity of cDNAs, resulting in a very biased representation of expressed gene sequences (see Fig. 2). In consequence, a current limitation in analyzing human genes is the relative lack of sequences derived from the central portions of transcripts. We have found, however, that such sequences can be generated systematically and efficiently in a high-throughput format, potentially permitting the rapid, shotgun sequencing of the human transcriptome.
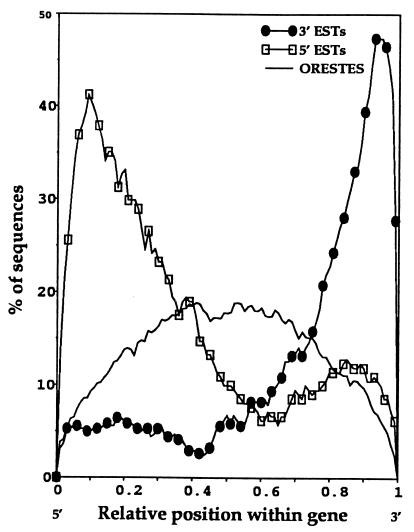
Figure 2.
A comparison of the actual percentage of ORESTES and 5′ and 3′ ESTs that pass through the relative position of full-length cDNA sequences. The figure was constructed by using all human full-length cDNAs of more than 1 kb currently in GenBank, the ORESTES corresponding to these cDNAs, as well as the 3′ and 5′ ESTs available in GenBank corresponding to these genes. With cDNAs of less than 1 kb, the 3′, 5′, and ORESTES reads are highly superimposed, making their relative contributions difficult to distinguish. Small cDNAs thus were not included in the figure.
The basis of the approach we have taken is to generate short cDNA templates of less than twice the length of an average sequencing read by reverse transcription–PCR using arbitrarily selected, nondegenerate primers (either singly or in pairs) under low-stringency conditions as we have described previously (21). It is not possible to predict (in the absence of complete transcript sequence information) with which transcripts an arbitrary primer will bind or the position of primer binding within any given transcript. The position of amplified fragments within transcripts is, in contrast, highly ordered and predictable, with a high percentage of fragments encompassing the midportions of genes. To demonstrate this, we have generated more than 10,000 sequences (which we refer to as ORF ESTs or ORESTES) from PCR fragments derived from the central, coding regions of human breast tumor transcripts by using the protocol described.
Materials and Methods
Template Preparation and DNA Sequencing.
Tissue samples obtained from excised breast tumors, after explicit informed consent of patients, from the Hospital do Câncer A.C. Camargo, São Paulo, were frozen in liquid nitrogen immediately after resection. They then were allowed to partially thaw to −20°C and microdissected to enrich for tumor cells in the sample. Total RNA was extracted with Trizol, and RNA degradation was evaluated by means of a Northern Blot by using a GAPDH cDNA probe. Those samples with intact mRNA were treated with DNaseI (10 units/50 μg of total RNA), and the absence of contaminating genomic DNA was confirmed by PCR using primers for the mitochondrial D loop and for the p53 gene. The amplified product was blotted onto nylon membranes and hybridized with [α-32P]dCTP-labeled probes for the corresponding amplified sequences. Qualified samples, with no detectable DNA, then were processed for isolation of poly(A)+ RNA (MiniMacs; Miltenyi Biotec, Auburn, CA). To produce cDNA templates, samples of 10–100 ng of the purified mRNA were heated at 65°C for 5 min and then subjected to reverse transcription at 37°C for 60 min in the presence of 200 units of mouse murine leukemia virus reverse transcriptase and 15 pmol of a randomly selected primer in a final volume of 20 μl. The criteria for primer selection were GC content of more than 50% and length of 18–25 nt. No specific sequence constraints were imposed. Indeed, almost exclusively, the primers used originally had been designed for specific PCR amplification of DNA sequences in nonhuman genomes and were exploited here if they obeyed the simple criteria listed above. After cDNA synthesis, one microliter of a 1:5 dilution of the single-stranded cDNA then was amplified by PCR by using the same or a single, alternative primer. Amplification profiles were generated by using the following cycling parameters: an initial cycle of 95°C for 5 min, 37°C for 2 min, and 72°C for 2 min followed by 35 cycles of 95°C for 45 sec, 45°C for 1 min, and 72°C for 90 sec. Three microliters of each pool was checked for complexity on 8% silver-stained polyacrylamide gels. Product pools with a single, predominant product (≈1%) reflecting the amplification of a highly abundant gene were not processed further. The remaining amplification pools with multiple bands then were cloned into pUC18 by using the Sureclone kit (Amersham Pharmacia). Minipreps for sequencing the inserts were prepared by alkali lysis or boiling preparations and sequenced by using the Perkin–Elmer Big-Dye reagent kit with ABI377 sequencers. In general, 50–200 sequences were determined from each amplification profile.
Computational Analysis.
Simulation of the amplification process was undertaken by searching for matches (60% identity including the 3′ nucleotide) between five totally random 20 mer sequences and the sequence of all full-length cDNAs currently in GenBank. In all cases in which a match was found, a reverse, complementary match within the same cDNA was sought. In all cases in which two complementary matches were found (taken as indicating a successful PCR amplification), the relative position of the amplicon within the cDNA was noted and used in the compilation of curves showing the percentage of amplicons passing through each percentage point of the genes analyzed. When the effect of amplicon size was analyzed, the whole set of amplicons was searched first for those of the size range desired that then were used to construct the curves against a representative transcript of unit length.
An automated protocol for the analysis of the experimentally generated data was used to: (i) assess sequence quality, (ii) trim vector and primer sequence, (iii) remove undesirable sequences such as bacterial, mitochondrial, and rRNA sequences, (iv) mask repetitive elements, and (v) undertake serial blast searches against existing databases. Sequence quality was determined by counting the number of “N” nucleotides generated by the ABI base caller. We excluded sequences whose level of “N” nucleotides was higher than 20%. We also trimmed sequences by analyzing a window of 30 nt. Windows with more than six “Ns” were deleted. Mitochondrial and rRNA sequences were identified by fasta searches against the GenBank entry corresponding to the human mitochondria complete genome sequence and against a locally developed human rRNA database, respectively. Masking of repetitive elements was performed by using repeatmasker under default parameters. Searches against existing databases were performed with blast (22) by using default parameters. Significant hits were determined by using an E value of 10−5 for searches against protein databases and an E value of 10−15 for searches against DNA databases.
Results and Discussion
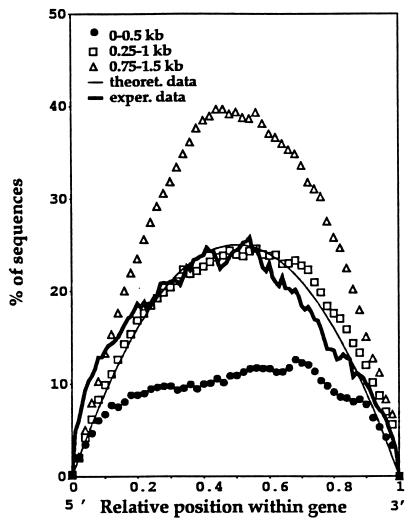
The amplification of any given point within a transcript, by reverse transcription–PCR, requires primer binding on both sides of that point. The chance of each of these events occurring will be proportional to the lengths of the sequences on both sides of the point. Thus, if the size of a transcript is taken as 1 and the distance from the 3′ end of the cDNA to the point in question is taken as S, then the probability of each appropriate primer template interaction will be S and 1 − S, respectively. In consequence, the probability of amplification will be S(1 − S). The chance of any fragment containing the midpoint of the gene will be 0.5 × 0.5 = 0.25, whereas that of any fragment containing a point 1/10th of the way along the gene will be 0.1 × 0.9 = 0.09, etc. Plotting the values of this expression for multiple points along a transcript of unit length results in a symmetrical curve around the midpoint of the transcript (Fig. 1). This simple concept does not take into account the actual proportion of primer template interactions that yield products or the influence of amplicon length to the coverage of the transcript. We therefore explored the concept by undertaking a series of simulations of PCR under low-stringency conditions using the known sequence of all available, full-length human cDNAs in GenBank. The plots generated confirmed the predicted symmetry around the midpoint of the transcripts and revealed, as would be expected, a higher percentage of amplicons passing through the midpoint of the transcript as amplicon size is increased (Fig. 1).
Figure 1.
The predicted, simulated, and experimentally determined position of ORESTES. The smooth, solid curve shows the predicted percentage of ORESTES that should contain the point, with the relative position shown within a hypothetical transcript. The curves described by the symbols indicated the coverage of known, full-length genes by ORESTES of different length generated by computational simulation. The irregular, solid line in bold shows the actual percentage of ORESTES that passed through the relative position of full-length cDNA sequences.
On the basis of the theoretical considerations and simulated amplifications, we generated a set of 10,122 ORESTES from human breast tumor mRNA, of which 1,207 were derived from either mitochondrial transcripts or rRNA, 135 were derived from bacterial contaminants, whereas 855 consisted entirely or almost entirely of repetitive elements precluding their further useful analysis (Table 1). Of the remaining 8,058 sequences, 6,501 were unique. These were divided among sequences that exhibited similarity with known, full-length cDNA sequences, those with similarity to human ESTs, and those without significant similarity to previously identified human transcripts.
Table 1.
Categories of ORESTES
| ORESTES category | Total sequences | Nonredundant sequences |
|---|---|---|
| Mitochondrial transcripts | 803 | ND |
| rRNA | 269 | ND |
| Bacterial sequences | 135 | ND |
| Repetitive sequences* | 855 | ND |
| Similarity with full-length human cDNAs | 3,598 | 2,651 |
| Similarity with human ESTs | 1,649 | 1,393 |
| Similarity with HTG and GSS | 409 | 310 |
| Similarity with nonhuman genes | 18 | 18 |
| Similarity with nonhuman ESTs | 29 | 27 |
| No similarity with sequences in databases | 2,357 | 2,102 |
HTG, high-throughput genome database; GSS, genome sequence survey database. ND, not determined. The total number of reads analyzed was 10,120.
Sequences that are composed entirely or almost entirely of repetitive elements.
We used the nonredundant compilation of those sequences that matched reportedly full-length cDNA sequences to investigate the distribution of ORESTES within transcripts. The percentage of sequences passing through different points along the transcripts followed very closely the predicted distribution with an almost symmetrical curve around the midpoint of the transcripts (Fig. 1). In accord with their centralized distribution, 71% of ORESTES that matched full-length cDNAs were wholly or partially composed of known ORFs as judged by blast against the nonredundant protein database. To investigate whether the ORESTES strategy was likely to add a significant percentage of new sequences to the public databases, we also compared their distribution with those of 5′ and 3′ ESTs against the same full-length genes for which ORESTES sequences were generated (Fig. 2). The data show a clear complementarity of the ORESTES data with that already deposited in GenBank that should permit the rapid construction of contigs covering full-length cDNAs. In the generation of this figure, only complete cDNAs of more than 1 kb were used (excluding some 30% of complete gene sequences) because in very short sequences all the EST data essentially are superimposed, making their relative contributions hard to distinguish. Thus, the ORESTES curve is slightly lower in Fig. 2, as would be predicted from the effect of the relative sizes of the amplicon and transcript.
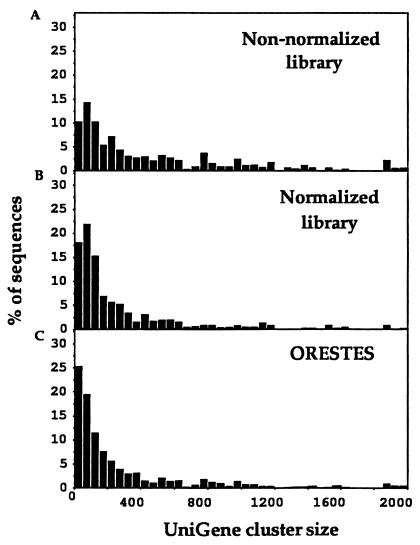
To examine whether the strategy adopted permits sequence analysis of less abundant gene transcripts, as we would expect based on our previous work (21), we listed the UniGene cluster size of the nonredundant compilation of ORESTES that matched fully sequenced human genes. By way of comparison, similar data were generated from nonnormalized and normalized human breast cDNA libraries (Fig. 3). We used only contigs containing full-length cDNAs for the analysis to avoid the strong bias that otherwise would have occurred toward the relative lack of ORESTES matches against small UniGene clusters that contain almost exclusively 3′ reads. The mean cluster size containing ESTs derived from the nonnormalized breast library was 649, the mean cluster size containing ESTs derived from the normalized breast library was 351, and the mean size of clusters against which ORESTES exhibited significant sequence similarity was 318. The median values for the cluster sizes were 317, 138, and 125, respectively. A rigorous comparative analysis of the ORESTES and standard ESTs cannot be pursued because different tissue samples were used. Nevertheless, ORESTES appears to exhibit a performance similar to normalized libraries in terms of accessing genes with lower levels of expression. This is indicated further by the percentage of ORESTES (25.26%) and standard ESTs (10.23% and 18.06% for the nonnormalized and normalized breast libraries, respectively) that exhibited sequence similarity to UniGene clusters of 50 or less entries. The basis of this partial equalization of the frequency of sequences is that the chance of the amplification of a particular transcript is dependent on its sequence and not abundance. Because the percentage of very highly abundant transcripts is very small (23), most amplifications do not result in products derived from very highly expressed genes. This immediately increases the relative abundance of the products derived from the less abundant transcripts in the cell. (Note that in those amplification profiles in which a highly abundant transcript has been amplified this is immediately apparent upon electrophoretic analysis, and the profile is eliminated from further processing.) In addition to this selection, there is also a significant tendency to equalize the relative abundance of amplicons in individual product pools because of the Cot effect of PCR (24).
Figure 3.
Comparison of abundance of ORESTES and ESTs. (A) Nonnormalized breast tumor cDNA library NCI CGAP Br1.1. (B) Normalized breast tumor cDNA library NCI CGAP Br2. (C) ORESTES. The bars show the percentage of nonredundant sequences with similarity to full-length human cDNAs that matched UniGene clusters containing the number of ESTs shown.
The evaluation of the ORESTES strategy provided a considerable amount of new sequence information. Thirty-eight percent of the nonredundant compilation of ORESTES did not exhibit significant similarity with expressed human sequences (Table 1). A small fraction of these exhibits significant similarity against full or partial cDNA sequences from other organisms (Tables 1 and 2) or low levels of similarity against known human transcripts, indicating that they probably derive from orthologous or paralogous genes, respectively (Table 2). A total of 32% of the nonredundant compilation, however, showed no similarity to known expressed genes from any organism. We would expect these sequences to include those of marginal quality, undetected bacterial sequences, sequences derived from immature mRNA, or, possibly, sequences derived from undetected trace amounts of contaminating DNA. Nevertheless, 40% of those ORESTES that exhibited no database matches were predicted to contain ORFs by using estscan (25) or grail (26), and a number of these exhibited a significant match against a variety of domain profiles (Table 3). By way of comparison, 68% of ORESTES that exhibited similarity to known genes were identified as containing coding regions by estscan. Thus, we can predict that the majority of ORESTES with no matches contain high-quality data derived from expressed human genes.
Table 2.
Putative paralogs–orthologs
| Similarity | E value | Identity (%) | GenBank accession no. |
|---|---|---|---|
| AJ006064—R. novergicus coronin-like protein | 6e-16 | 44 /48 (86%) | AI902261 |
| AF045771—Drosophila melanogaster miranda | 4e-5 | 64 /128 (50%) | AI903188 |
| X95466—R. novergicus CPG2 protein | 7e-7 | 42 /71 (59%) | AI904067 |
| A49128—R. novergicus Notch2 protein | 7e-45 | 77 /84 (92%) | AW054433 |
| BAA25687—R. novergicus semaphorin Z | 3e-16 | 39 /39 (100%) | AW054434 |
| U76754—Mus musculus IFN-induced gene | 7e-11 | 32 /61 (52%) | AI904072 |
| U67956—Caenorhabditis elegans F19F9.4 lipolytic enzymes | 4e-8 | 28 /59 (47%) | AI904070 |
| AF067624—C. elegans M01B12.5 | 1e-5 | 23 /44 (52%) | AI904448 |
| Q17598—C. elegans CO3A3.2 | 5e-10 | 23 /44 (52%) | AI905330 |
| X90849—Gallus gallus polybromo 1 (methyltransferase) | 3e-6 | 37 /57 (63%) | AI905381 |
| O61603—D. melanogaster eyelid protein | 4e-23 | 62 /154 (40%) | AI905413 |
| U51032—Saccharomyces cerevisiae SNF2P | 3e-7 | 21 /30 (70%) | AI905587 |
| AF060116—R. novergicus cortactin-binding protein | 1e-11 | 77 /151 (50%) | AI906338 |
| P45481—M. musculus CREB-binding protein | 2e-5 | 29 /57 (50%) | AI902504 |
| O35161—M. musculus cadherin-like protein | 3e-24 | 54 /82 (65%) | AI902505 |
| Z99162—Schizosaccharomyces pombe cleavage and polyadenylation factor | 3e-7 | 36 /137 (26%) | AI902507 |
| U40935—C. elegans putative protein kinase | 2e-5 | 19 /34 (55%) | AI902213 |
| L35604—D. melanogaster ethanolamine kinase | 6e-8 | 28 /71 (39%) | AI905895 |
| U67322—Hepatitis B virus-associated factor (4-aa deletion) | e-133 | 214 /218 (98%) | AI909655 |
| Q23370—C. elegans protein | 1e-17 | 47 /120 (39%) | AI909682 |
| AL049495—S. pombe conserved hypothetical protein | 7e-20 | 47 /120 (39%) | AI910335 |
| D88315—Mouse tetracyclin transporter | 2e-7 | 31 /67 (46%) | AI907566 |
| AF007791—Human HAG-2/C cement gland protein | 3e-34 | 59 /76 (78%) | AI906906 |
| P28370—Human putative global transcription activator SNF2L1 | 3e-10 | 23 /26 (88%) | AI909988 |
| AF041260—AIBC1 (50-aa insert) | 1e-25 | 66 /111 (59%) | AI904532 |
| AF052685—Similar to protocadherin 43 | 4e-12 | 29 /56 (52%) | AI910218 |
| P28160—Zinc finger protein HTF6 | 4e-23 | 51 /76 (67%) | AI908594 |
| AF040990—Similar to human roundabout protein | 1e-10 | 35 /73 (47%) | AI902552 |
| AB002376—Similar to KIAA0378 | 2e-23 | 54 /93 (58%) | AI905704 |
| D64159—Similar to zinc finger protein | 6e-38 | 32 /36 (88%) | AI906359 |
| Q00839—Similar to ribonuclear protein | 1e-40 | 77 /142 (54%) | AI906557 |
| D14480—Calpain-like protein | 7e-35 | 70 /84 (83%) | AI907781 |
| AB018341—Similar to KIAA0798 | 3e-29 | 60 /114 (52%) | AI904640 |
| Y09305—Similar to a human protein kinase | 7e-19 | 38 /56 (68%) | AI909430 |
| AB002376—Similar to KIAA0378 | 3e-37 | 82 /131 (63%) | AI906975 |
| U52965—Human ENX-1 | 5e-6 | 25 /77 (32%) | AI909958 |
| P15586—Human N-acetylglucosamine-6-sulfatase precursor | 4e-22 | 57 /146 (39%) | AI909676 |
Table 3.
ORESTES with matches to protein domains
| Profile ID | Domain description | Score | GenBank accession no. |
|---|---|---|---|
| d111_domain | D111 domain | 14.2158 | AI905887 |
| Brct_domain | BRCT domain | 8.8606 | AI902587 |
| atp_gtp_a2 | P loop nucleotide-binding motif | 8.6225 | AI902167 |
| spec_repeat | Spectrin repeat | 8.5467 | AI902512 |
| sbp_glur | Solute-binding protein/glutamate receptor domain | 8.1420 | AI903015 |
| palp_1 | Pyridoxal-phosphate-dependent enzyme Acyl-carrier phosphopantetheine | 7.2761 | AI908798 |
| acp_domain | BCL-2-like apoptosys inhibitor | 6.9307 | AI903039 |
| bcl2_family | SUR-2-type hidroxylase/desature | 6.8022 | AI903058 |
| sur2_domain | SEA module | 6.7457 | AI903214 |
| sea_domain | Phosphatidyl-inositol phospholipase | 6.4994 | AI905383 |
| pip1c_x_domain | X box | 6.4033 | AI908839 |
| sea_domain | SEA module | 6.3293 | AI910016 |
| tyr_phosphatase_2 | Tyrosine phosphatase domain | 6.2816 | AI909545 |
| pld_domain | Phospholipase D | 6.2544 | AI903067 |
| c_type_lectin_2 | C type lectin | 6.2340 | AI904349 |
| ibn_nt | Importing-β | 6.1850 | AI907745 |
| pas_repeat | PAS-associated domain | 6.1762 | AI903674 |
| acp_domain | Acyl-carrier phosphopantetheine | 6.1551 | AI903034 |
| btb | BTB-TTK domain | 6.1456 | AI909573 |
| cys_prot_calpain | Calpain-type cystein protease | 6.1452 | AI904349 |
| tyr_phosphatase_dual | Tyrosine phosphatase | 6.0742 | AI904256 |
| spec_repeat | Spectrin repeat | 6.0322 | AI906055 |
The list of sequences predicted to contain protein domains was generated by searching estscan-translated ORESTES against the profile library of the prosite database, including the prerelease profiles from ftp://ftp.isrec.isb-sib.ch/sib-isrec/profiles/. Note that the scores represent −Log10 per residue E values, meaning that a match with a score of 8, for example, is expected to occur about once in 108 residues.
The potential of ORESTES to act as the basis of a shotgun approach to the sequencing of human transcripts was demonstrated by the 21% of sequences that partially or wholly matched preexisting human ESTs, from which we were able to construct 783 contigs, each one corresponding to a different UniGene cluster. Of the total number of bases contained in these contigs, 19% represented new sequence contributed by ORESTES. The contigs assembled to date mostly comprise extensions of the sequences contained in 3′ or 5′ ESTs but also a few instances in which the reads from the ends of cDNAs are joined by an ORESTES sequence or, indeed, different clusters are joined.
Knowledge of the complete sequence of the noncoding regions of the human genome will provide the basis of the definition of many facets of gene structure and expression. Nevertheless, it is the coding regions contained within the genome that represent the information of most crucial and immediate importance. These regions probably constitute only about 3% of the human genome. Although 5′ ESTs often fall within coding regions and have been pursued vigorously with this characteristic in mind, ORESTES offer the possibility of significantly extending the coverage of coding regions with ESTs. Using the different complementary EST approaches now available, it eventually may even be possible to contemplate the generation of a shotgun sequence of the human transcriptome. We currently are embarking on the production of approximately 500,000 human ORESTES that will be deposited in the public databases as they are generated. Given the simplicity of the methodology, the small amounts of starting material required, and the speed of data generation, the simultaneous adoption of this approach in diverse laboratories could lead rapidly to the determination of the majority of the human transcriptome.
Acknowledgments
We thank Rui C. Serafim, Ricardo P. Moura, Elisangela Monteiro, Anna Christina de Matos Salim, and Daniel F. Simão for dedicated and expert technical assistance and Juçara Parra for acting as the administrative coordinator of this project. E.D.N., R.G.C., W.S.J., and S.B. were supported by doctoral or postdoctoral fellowships from the Fundaçao de Amparo à Pesquisa do Estado de São Paulo. The work also was supported in part by the Programa de Apoio a Nucleos de Excelencia/Financiadora de Estudos e Projetos and the Fundaçao de Amparo à Pesquisa do Estado de São Paulo.
Abbreviations
- EST
expressed sequence tag
- ORESTES
ORF ESTs
Footnotes
References
- 1.Adams M D, Dubnick M, Kerlavage A R, Moreno R, Kelley J M, Utterback T R, Nagle J W, Fields C, Venter J C. Nature (London) 1992;355:632–634. doi: 10.1038/355632a0. [DOI] [PubMed] [Google Scholar]
- 2.Adams M D, Kerlavage A R, Fleischmann R D, Fuldner R A, Bult C J, Lee N H, Kirkness E F, Weinstock K G, Gocayne J D, White O. Nature (London) 1995;377:3–174. [PubMed] [Google Scholar]
- 3.Hillier L D, Lennon G, Becker M, Bonaldo M F, Chiapelli B, Chissoe S, Dietrich N, DuBuque T, Favello A, Gish W, et al. Genome Res. 1996;6:807–828. doi: 10.1101/gr.6.9.807. [DOI] [PubMed] [Google Scholar]
- 4.Nomura N, Miyajima N, Sazuka T, Tanaka A, Kawarabayasi Y, Sato S, Nagase T, Seki N, Ishikawa K, Tabata S. DNA Res. 1994;1:27–35. doi: 10.1093/dnares/1.1.27. [DOI] [PubMed] [Google Scholar]
- 5.Pandey A, Lewitter F. Trends Biochem Sci. 1999;24:276–280. doi: 10.1016/s0968-0004(99)01400-0. [DOI] [PubMed] [Google Scholar]
- 6.Bailey L C J, Searls D B, Overton G C. Genome Res. 1998;8:362–376. doi: 10.1101/gr.8.4.362. [DOI] [PubMed] [Google Scholar]
- 7.Burke J, Wang H, Hide W, Davison D B. Genome Res. 1998;8:276–290. doi: 10.1101/gr.8.3.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang J, Jacob H J. Genome Res. 1998;8:268–275. doi: 10.1101/gr.8.3.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buetow K H, Edmonson M N, Cassidy A B. Nat Genet. 1999;21:323–325. doi: 10.1038/6851. [DOI] [PubMed] [Google Scholar]
- 10.Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Lane C R, Lim E P, Kalayanaraman N, Nemesh J, et al. Nat Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- 11.Picoult-Newberg L, Ideker T E, Pohl M G, Taylor S L, Donaldson M A, Nickerson D A, Boyce-Jacino M. Genome Res. 1999;9:167–174. [PMC free article] [PubMed] [Google Scholar]
- 12.Gress T M, Muller-Pillasch F, Geng M, Zimmerhackl F, Zehetner G, Friess H, Buchler M, Adler G, Lehrach H. Oncogene. 1996;13:1819–1830. [PubMed] [Google Scholar]
- 13.Huang G M, Ng W, Farkas J, He L, Liang H A, Gordon D, Yu J, Hood L. Genomics. 1999;59:178–186. doi: 10.1006/geno.1999.5822. [DOI] [PubMed] [Google Scholar]
- 14.Jay P, Diriong S, Taviaux S, Roeckel N, Mattei M G, Audit M, Berge-Lefranc J L, Fontes M, Berta P. Genomics. 1997;39:104–108. doi: 10.1006/geno.1996.4470. [DOI] [PubMed] [Google Scholar]
- 15.Malone K, Sohocki M M, Sullivan L S, Daiger S P. Mol Vis. 1999;5:5. [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson P S, Ng W L, Schummer M, True L D, Liu A Y, Bumgarner R E, Ferguson C, Dimak A, Hood L. Genomics. 1998;47:12–25. doi: 10.1006/geno.1997.5035. [DOI] [PubMed] [Google Scholar]
- 17.Neubauer G, King A, Rappsilber J, Calvio C, Watson M, Ajuh P, Sleeman J, Lamond A, Mann M. Nat Genet. 1998;20:46–50. doi: 10.1038/1700. [DOI] [PubMed] [Google Scholar]
- 18.Vasmatzis G, Essand M, Brinkmann U, Lee B, Pastan I. Proc Natl Acad Sci USA. 1998;95:300–304. doi: 10.1073/pnas.95.1.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aaronson J S, Eckman B, Blevins R A, Borkowski J A, Myerson J, Imran S, Elliston K O. Genome Res. 1996;6:829–845. doi: 10.1101/gr.6.9.829. [DOI] [PubMed] [Google Scholar]
- 20.Strausberg R L, Dahl C A, Klausner R D. Nat Genet. 1997;15:415–416. doi: 10.1038/ng0497supp-415. [DOI] [PubMed] [Google Scholar]
- 21.Neto E D, Harrop R, Correa-Oliveira R, Wilson R A, Pena S D, Simpson A J. Gene. 1997;186:135–142. doi: 10.1016/s0378-1119(96)00699-3. [DOI] [PubMed] [Google Scholar]
- 22.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L, Zhou W, Velculescu V E, Kern S E, Hruban R H, Hamilton S R, Vogelstein B, Kinzler K W. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 24.Mathieu-Daude F, Welsh J, Vogt T, McClelland M. Nucleic Acids Res. 1996;24:2080–2086. doi: 10.1093/nar/24.11.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iseli, C., Jongeneel, C. V. & Bucher, P. (2000) Ismb, in press.
- 26.Uberbacher E C, Mural R J. Proc Natl Acad Sci USA. 1991;88:11261–11265. doi: 10.1073/pnas.88.24.11261. [DOI] [PMC free article] [PubMed] [Google Scholar]