Abstract
We have cloned the genomic breakpoints for a balanced t(14;21)(q11.2;q22) chromosomal translocation associated with T-cell acute lymphoblastic leukemia. Sequence analysis of the genomic breakpoints indicated that the translocation had been mediated by an illegitimate V(D)J recombination event that disrupted the T-cell receptor (TCR) α locus and placed the TCR α locus enhancer on the derivative 21 chromosome. We identified a previously unreported transcript, designated BHLHB1 (for basic domain, helix–loop–helix protein, class B, 1) that had been activated by the translocation. BHLHB1 mapped to the region of chromosome 21 that has been proposed to be responsible, at least in part, for the learning deficits seen in children with Down's syndrome. Although BHLHB1 expression normally is restricted to neural tissues, T-cell lymphoblasts with the t(14;21)(q11.2;q22) also expressed high levels of BHLHB1 mRNA. Expression of BHLHB1 dramatically inhibited E2A-mediated transcription activation in NIH 3T3 fibroblasts and Jurkat T cells. This observation suggests that BHLHB1, similar to SCL/TAL1, may exert a leukemogenic effect through a functional inactivation of E2A or related proteins.
Nonrandom chromosomal abnormalities including deletions, inversions, and translocations are found in the majority of acute leukemias and lymphomas (1). In general, the nonrandom translocations associated with hematopoietic malignancies either generate novel fusion oncogenes, such as the BCR-ABL fusion associated with chronic myelogenous leukemia, or deregulate protooncogenes, such as MYC, which becomes deregulated by its juxtaposition to the IGH enhancer in B cells that have undergone a t(8;14) translocation (1, 2). A number of experimental models, including transgenic mice and murine bone marrow reconstitution experiments, have verified that overexpression of many of these presumptive oncoproteins leads to leukemia and/or lymphoma in mice (3).
In patients with T-cell acute lymphoblastic leukemia (T-ALL), the translocations often involve juxtaposition of T-cell receptor (TCR) α, β, or δ elements to known or putative protooncogenes (1, 2). The mechanism underlying these translocations almost surely involves illegitimate V(D)J recombination, as hallmarks of V(D)J recombinase activity often are found at the translocation breakpoints (4). The most common chromosomal translocations recognized in patients with T-ALL result in the activation of latent protooncogenes such as SCL/TAL1 (5–7), LYL1 (8), TAL2 (9), LMO1 (10, 11), LMO2 (11, 12), TAN (13), and MYC (14), usually by juxtaposition of these genes to TCR regulatory elements. These genes are all known or putative transcription regulators, and it is thought that inappropriate expression of these genes in a thymocyte triggers leukemic transformation. Recent reports suggest that the products of several of these genes may be leukemogenic through inhibition of E-proteins such as E2A (E12/E47) or HEB (15–23).
We recently identified a patient with T-ALL and a novel t(14;21)(q11.2;q22) chromosomal translocation. We have cloned the translocation breakpoint and identified a gene, designated BHLHB1 (for basic domain, helix–loop–helix protein, class B, 1), which is activated by the translocation. BHLHB1 is able to inhibit E2A-mediated transactivation, suggesting the possibility that BHLHB1 may be oncogenic through a functional inactivation of E2A.
Materials and Methods
Isolation and Analysis of Nucleic Acids.
The studies carried out were approved by the Roswell Park Cancer Institute Institutional Review Board. A cryopreserved bone marrow specimen containing leukemic cells (#3280) was obtained from the Pediatric Oncology Group cell bank. Genomic DNA and RNA were isolated as described (24). Southern and Northern blots were performed as described (24).
Library Construction and Screening.
Genomic DNA (5 μg) was digested with EcoRI, extracted with phenol-chloroform, ethanol-precipitated, and resuspended in dH2O. EcoRI-digested genomic DNA (0.4 μg) was ligated to 1 μg of lambda FIX II (Stratagene) phage arms. Recombinant phage DNA was packaged by using Stratagene packaging extracts and protocols. One million plaque-forming units were screened with 32P-labeled Jα45 and Jα70 probes. Hybridizing phage clones were purified, and relevant restriction fragments were subcloned into plasmid vectors. One million plaque-forming units of a human fetal brain cDNA library (Stratagene) were screened with the PCR0102 probe.
Probes and Hybridizations.
The Jα45 and Jα70 probes are genomic DNA fragments located 45 kb and 70 kb, respectively, upstream of TCRA Cα. The 0.4NN, 0.7HR, JJ1, and JJ2 probes are repeat-free genomic fragments obtained from chromosome 21 phage clones. The PCR0102 cDNA fragment was generated by reverse transcriptase–PCR (RT-PCR) amplification of total RNA from the A172 glioblastoma cell line using primers 21PCR01 (5′-CCCTGAGGCTTTTCGGAGCG-3′) and 21PCR02 (5′-GCGGCTGTTGATCTTGAGACGC-3′). The 21PCR03 probe was an oligonucleotide (5′-GTAGACGACGAGGTGCTGGACG-3′). Probes were labeled with 32P by the random priming technique (DNA fragments) or terminal deoxynucleotidyl transferase end-labeling technique (oligonucleotides). Hybridization was performed as described (24), the blots were washed twice with 0.1 × SSC (1 × SSC is 0.15 mol/liter sodium chloride and 0.015 mol/liter sodium citrate) and 0.1% SDS at 52°C for random primed DNA fragments or with 6 × SSC and 0.1% SDS at 10°C below the melting temperature for oligonucleotide probes.
RT-PCR Analysis.
RT-PCR was performed by using Life Technology (Grand Island, NY) reagents and protocols. Briefly, first-strand cDNA was synthesized from 1 μg of total RNA using an oligo(dT) primer. Aliquots of first-strand cDNA were amplified with the 21PCR01 and 21PCR02 primers in a volume of 50 μl. After a “hot start,” 40 PCR cycles of 94°C for 1 min, 56°C for 45 s, and 72°C for 1 min were used, followed by a terminal 10-min extension at 72°C. PCR products were analyzed by agarose gel electrophoresis and hybridization to a 32P-end labeled internal oligonucleotide probe (21PCR03). Parallel cDNA aliquots were amplified by using the same cycling profile and human β actin primers (5′-AGGCCGGCTTCGCGGGCGAC-3′ and 5′-CTCGGGAGCCACACGCAGCTC-3′).
Exon Trapping.
P1-artificial chromosome (PAC) clone DNA was digested to completion with BglII and BamHI, purified by phenol-chloroform extraction, and subcloned into the BamHI site of the exon trapping vector pSPL3. Pools of plasmid DNA from each of four PAC clones were transfected into Cos 7 cells by using calcium phosphate. RNA was isolated 24 h posttransfection by using Trizol (Life Technologies), according to the manufacturer's recommended protocol. RT-PCR was performed by using Life Technologies reagents and protocols to amplify trapped exons. The RT-PCR products were subcloned into the pGEM-T-easy vector (Promega) and sequenced.
Reporter Gene Assays.
The E-box/luciferase reporter constructs were a gift from Tom Kadesch (University of Pennsylvania, Philadelphia) (25). The E(5,2,3)4-luc construct contains four concatamerized μE5, μE2, and μE3 E-boxes controlling luciferase expression; the E(5,2)6-luc construct contains six concatamerized μE5 and μE2 E-boxes controlling luciferase expression. The TATA-luc construct was generated by deleting the E-box sequences from the E(5,2)6-luc vector. The pG5BCAT reporter vector, which has a minimal E1b promoter and five concatamerized Gal4 DNA binding sites linked to the chloramphenicol acetyltransferase (CAT) gene, and the pSG4 Gal4 expression vectors were a gift from Mark Ptashne (Harvard University, Boston) (26, 27). An E2A expression vector (named pCE2–5) was generated by cloning an E2–5 cDNA into the pcDNA3.1 (Invitrogen) expression vector as described (18). A BHLHB1 expression vector (pCBHLHB1) was generated by cloning a BHLHB1 cDNA encoding amino acid residues 81–357 of Fig. 2 into a pRC-CMV vector (Invitrogen) that had been modified by introduction of the human β-globin initiation codon and 5′ untranslated region. The cloning junctions of all constructs were sequenced to confirm the correct sequence and orientation. A vector using simian virus 40 promoter/enhancer sequences (pCAT Control, Promega) driving CAT expression was used to control for transfection efficiency.
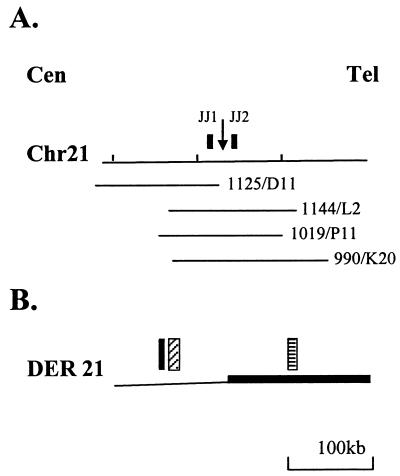
Figure 2.
Transcribed sequences in the region of the breakpoint. (A) Location of PAC clones used for exon trapping experiments. The breakpoint is indicated with a downward arrow. The locations of PAC clones (1125/D11, 1144/L2, 1019/P11, and 990/K20) and probes (JJ1, JJ2) are shown. (B) Transcribed sequences from the derivative 21 chromosome. Solid black box, exon-trapping clone; cross-hatched box, cluster of 12 overlapping EST clones (see text); checked box, TCRA enhancer.
NIH 3T3 fibroblasts were transfected when approximately 50–80% confluent by using Lipofectamine reagent (Life Technologies) and the manufacturer's recommended protocol. The E-box reporter assays used 1 μg of the TATA-luc or E(2,5)-luc reporter vector, 1 μg of the pCE2–5 expression vector or 1–8 μg of the pCBHLHB1 expression vector, and 4 μg of the pCATControl reporter vector, used as a control for transfection efficiency. The Gal4 reporter assays used 2 μg of the pG5BCAT reporter, 2 μg of the pSG4 expression vector, 1 μg of the pCBHLH1 expression vector, and 2 μg of the pRSV-Luc vector as a control for transfection efficiency. Jurkat cells (2–3 × 106) were transfected with Lipofectamine as described above, following the manufacturer's recommended protocol for suspension cells. In transfections where expression and reporter vectors totaled less than 14 μg, the parent pRC-CMV vector was added to total 14 μg. Approximately 42 h after transfection the cells were lysed and assayed for luciferase and CAT activity as described (18).
Results
Case Report.
The patient (#3280) was a 7-year-old Caucasian female who presented with a history of fever, headache, dizziness, and abdominal pain. Complete blood count showed a hemoglobin of 7.9 g/dl, a white blood count of 79,300/μl, and a platelet count of 39,000/μl, with circulating lymphoblasts. Cerebrospinal fluid was negative for malignant cells, and a chest radiogram showed a superior mediastinal mass. A bone marrow aspirate demonstrated L2 ALL, and flow cytometry was positive for T-cell antigens CD2 and CD7. Karyotype was interpreted as 46, XX, del(6)(q21), t(14;21)(q11.2;q22). The patient attained a complete remission with standard chemotherapy but relapsed and died after 4 months of therapy.
Identification of TCRA Gene Rearrangements.
Chromosomal translocations in T-ALL cells that involve chromosome 14q11.2 usually involve either the TCRA or TCRD locus, both of which map to 14q11.2. Because the TCRD locus is contained within the TCRA locus, a productive rearrangement of TCRA deletes TCRD. To identify rearranged restriction fragments that might encompass the translocation breakpoint, we screened genomic DNA isolated from leukemic blasts that displayed a t(14;21) by using a TCRD Jδ1 probe (28); this experiment showed that Jδ1 sequences had been deleted from both alleles (data not shown). This result suggested that one allele had undergone a physiologic TCRA gene rearrangement that had deleted TCRD sequences, whereas the other TCRA allele was involved in the balanced translocation. The same genomic DNA sample was screened with several TCRA probes, and we identified rearrangements of two Jα segments (Jα45 and Jα70, data not shown); presumably, one rearranged fragment represented a physiologic TCRA gene rearrangement, whereas the other rearranged fragment represented the chromosomal translocation.
Cloning the Jα Gene Rearrangements.
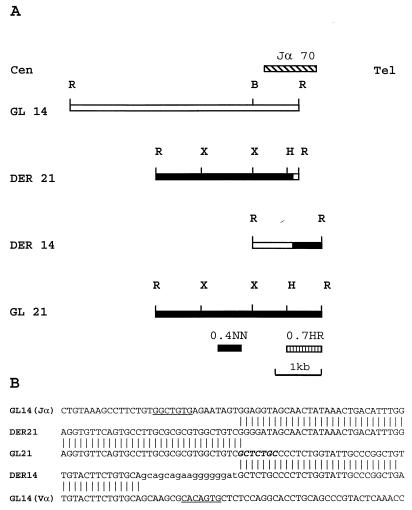
We constructed a lambda phage genomic DNA library from patient lymphoblast DNA and screened the library with the Jα45 and Jα70 probes; positive clones were purified and subcloned into pBluescript II vectors. The rearrangement identified by the Jα45 probe represented a physiologic TCRA Vα-Jα gene rearrangement (data not shown). However, the nucleotide sequence of the rearranged fragment identified by the Jα70 probe diverged from germ-line Jα sequences immediately 5′ of the Jα70 region. The divergent sequence had no similarity to known Vα sequences, suggesting that the fragment identified by the Jα70 probe did not represent a physiologic V(D)J rearrangement, and was produced by the t(14;21) translocation. A fragment isolated from the non-Jα portion of this rearranged fragment mapped to chromosome 21q22 by fluorescence in situ hybridization (data not shown), verifying that the rearrangement identified by the Jα70 probe was the result of a t(14;21) translocation. Germ-line lambda phage clones from this region of chromosome 21q22 were obtained by screening a human placental library with a single-copy probe (0.4NN of Fig. 1A). A single-copy probe (0.7HR) from a germ-line chromosome 21q22 clone was used to clone the reciprocal translocation (Fig. 1A).
Figure 1.
Molecular analysis of the t(14;21) breakpoints. (A) Restriction map of the germ-line (GL) and derivative (DER) alleles. Cen, centromere; tel, telomere; locations of the Jα70, 0.4NN, and 0.7 HR probes are indicated. B, BamHI; H, HindIII; R, EcoRI; X, XbaI. (B) Nucleotide sequence at breakpoint. The germ-line Vα sequence was obtained from GenBank (HUAE000660). The germ-line (GL) and derivative (DER) alleles are presented. Jα and Vα heptamer sequences are underlined, a potential translin binding site is shown in bold. Nontemplated nucleotides from the DER14 are shown in lowercase.
Fig. 1B demonstrates that the translocation involves discontinuous Vα and Jα segments on the derivative 14 and derivative 21 chromosomes, respectively. The presence of nontemplated “N” region nucleotides on the derivative chromosome 14 and consensus heptamer sequences located 9 nt upstream of the Jα breakpoint and 8 nt downstream of the Vα breakpoint strongly suggest that this translocation was mediated by a mistake in normal V(D)J recombination. Additionally, the sequence (5′-GCTCTGC-3′) at the chromosome 21 breakpoint is a 6/7 match for a consensus translin binding site; the translin gene encodes a protein that was identified by virtue of its ability to bind sequences often present at translocation breakpoints in lymphoid malignancies and is thought to play a role in either V(D)J recombination or DNA repair (29, 30).
Identification of a Gene Dysregulated by Translocation.
We used several genomic fragments located near the chromosome 21 breakpoint to screen Northern blots containing RNA from 15 hematopoietic cell lines, 12 nonhematopoietic cell lines, eight normal tissues and several patient samples and to screen human fetal brain, fetal liver, and bone marrow cDNA libraries. No transcripts were detected. We also failed to detect conserved sequences in mouse and chicken DNA with these probes.
We used exon trapping with lambda phage inserts located within 18 kb of the chromosome 21 breakpoint to identify exons near the chromosome 21 breakpoint. Several trapped clones flanked by AG splice acceptor and GT splice donor sequences in the corresponding genomic DNA sequences were recovered; however, these clones had no matches for known genes nor sequences in dbEST, did not identify transcripts on Northern blot, and were not conserved across species (data not shown). These findings suggested that the relevant gene may be located some distance from the translocation breakpoint.
To search for a relevant gene over a larger region, we obtained four human PAC clones that encompassed >200 kb flanking the chromosome 21 breakpoint (Fig. 2A). Each PAC clone was digested with BamHI and BglII, and the fragments were ligated into the pSPL3 exon-trapping vector. Pools of pSPL3 subclones then were used for exon trapping experiments, and a number of clones were obtained and characterized. Several of these clones matched expressed sequence tag (EST) sequences, as well as a completely sequenced region of chromosome 21. A cluster of EST sequences (all derived from neural tissues) and exon-trapping clones mapped to a region 80 kb centromeric to the translocation breakpoint. Because the gene represented by these EST sequences and exon trapping clones was on the derivative 21 chromosome, which contained the TCRA enhancer (Fig. 2B), we evaluated this gene as a candidate for the gene activated by the t(14;21) translocation.
The EST Cluster Encodes a Basic Domain Helix–Loop–Helix (bHLH) Protein Predominantly Expressed in Neural Tissues.
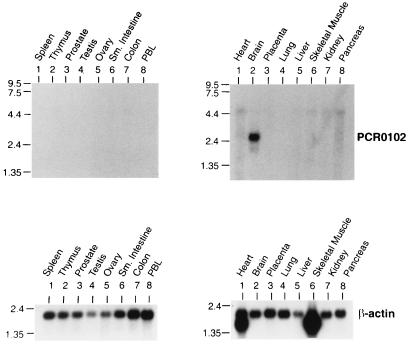
We assembled a composite sequence for this EST cluster and synthesized the 21PCR01 and 21PCR02 primers based on this composite. We used RT-PCR to generate a cDNA probe from this cluster and used this probe (named PCR0102) to screen a fetal brain cDNA library. We recovered a single clone (HB14); nucleotide sequence analysis of the HB14 clone demonstrated that it contained the bHLH motif commonly found in transcription regulators such as MYOD, E2A, and SCL/TAL1 (Fig. 3). This clone had the highest degree of similarity to BETA3 (31), a bHLH protein initially identified in hamster insulin tumor cells. As mentioned above, all of the 12 EST sequences for BHLHB1 had been obtained from neural tissues. Northern blot hybridization with a BHLHB1 cDNA fragment showed that a 2.5-kb transcript was clearly detected only in brain tissue (Fig. 4). A similar result was obtained by analysis of a multiple tissue array dot blot, which contained RNA from 76 different human tissues or cell lines. In this experiment, 22/22 neural tissues showed BHLHB1 expression, whereas none of the 54 nonneural tissues or cell lines showed BHLHB1 expression (data not shown). We conclude that BHLHB1 is expressed primarily in neural tissues.
Figure 3.
BHLHB1 encodes a bHLH protein. (A) Predicted amino acid sequence of BHLHB1. (B) BHLHB1 sequence compared against other bHLH proteins: beta-3 (GenBank accession no. AAB50691), olg-1 (GenBank accession no. AAD34029), NeuroD2 (GenBank accession no. 5453768), atonal (GenBank accession no. P70447), and SCL/TAL1 (GenBank accession no. AAA36598). The consensus (cons) bHLH region is indicated.
Figure 4.
Expression of BHLHB1. Northern blots containing 2 μg of RNA from the indicated tissues were hybridized to a probe (PCR0102) from the 5′ portion of BHLHB1. A clear signal at 2.6 kb is seen only in brain tissue. The blots were rehybridized to a β-actin probe as an internal control.
BHLHB1 Gene Expression Is Activated by Chromosomal Translocation.
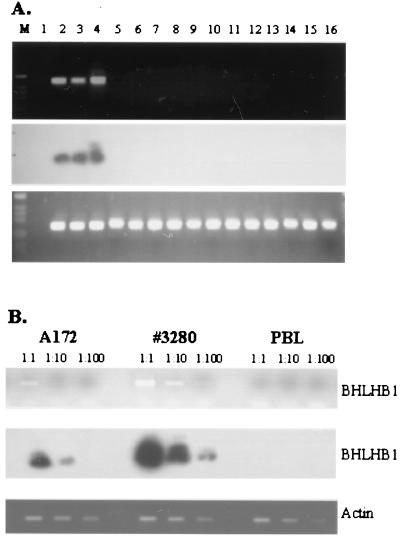
To determine whether BHLHB1 gene expression was activated in cells carrying a t(14;21) translocation, an RT-PCR assay was developed to amplify BHLHB1 mRNA. Fig. 5A demonstrates that whereas BHLHB1 expression can easily be detected by RT-PCR in human glioblastoma cells and the t(14;21) patient sample, only trace amounts at best can be detected in the other samples analyzed. These results indicate that whereas BHLHB1 normally is expressed primarily in neural tissues, it is activated and expressed at levels comparable to those seen in the positive control cell lines in leukemic cells that have the t(14;21) translocation. To determine relative levels of BHLHB1 expression in the t(14;21) leukemic sample compared with the positive control glioblastoma cell lines and peripheral blood lymphocytes, we performed an RT-PCR assay using serial dilutions of first-strand cDNA. Fig. 5B indicates that BHLHB1 mRNA is expressed at levels higher than those detected in the positive control glioblastoma cell line.
Figure 5.
Expression of BHLHB1 in t(14;21) cells. (A) Total RNA was reverse-transcribed and PCR-amplified as described in Materials and Methods. (Top) BHLHB1 primers. (Bottom) Actin primers. (Middle) Hybridization of the gel shown in the Top panel to an internal BHLHB1 oligonucleotide. M, 1-kb ladder size standard; lane 1, dH2O; lanes 2 and 3, glioblastoma cell lines (A172 and U87, respectively); lane 4, t(14;21) leukemic cells; lanes 5–10, T-ALL cell lines (Molt4, HSB-2, CEM, Jurkat, DU528, and SUPT1, respectively); lanes 11 and 16, bone marrow from T-ALL patients without a t(14;21); lanes 12–14, bone marrow from acute myeloid leukemia patients without a t(14;21); lane 15, peripheral blood lymphocytes. A signal from faint bands in lanes 15 and 16 can be detected upon prolonged exposure of the autoradiogram (data not shown), suggesting low-level expression of BHLHB1 in these samples. (B) Serial dilutions of the A172, t(14;21) leukemic cells (#3280), and peripheral blood lymphocyte (PBL) cDNA were amplified to detect BHLHB1 (Upper) and actin (Lower) transcripts. The gel was blotted and hybridized to an internal BHLHB1 oligonucleotide as described above.
Inhibition of E2A Function by BHLHB1.
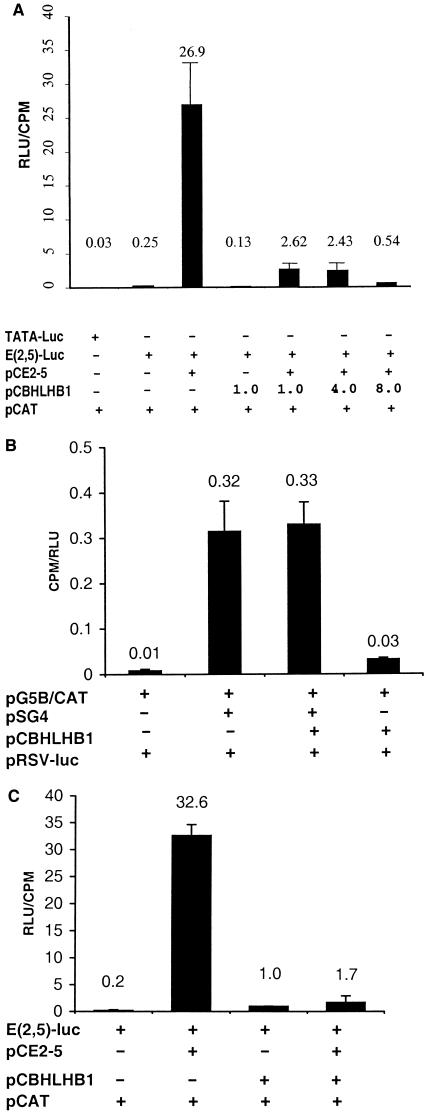
E2A is required for normal T-cell differentiation and seems to serve as a tumor suppressor in thymocytes, because E2a-deficient mice that survive beyond the immediate neonatal period develop aggressive T-cell malignancies (19, 23). Furthermore, it has been shown that the bHLH oncoprotein SCL/TAL1 can bind to and inhibit E2A function in a number of different systems, leading to the hypothesis that SCL/TAL1 exerts its leukemogenic effect through inhibition of E2A (15–18, 21). To determine whether the BHLHB1 gene product can inhibit E2A activity in a similar fashion, we transfected NIH 3T3 fibroblasts with the (E-5,2,3)6-TATA luciferase reporter vector, an E2A expression vector (pCE2–5), and variable amounts of a BHLHB1 expression vector (pCBHLHB1). Fig. 6A shows that the BHLHB1 expression vector dramatically inhibits E2A-dependent luciferase activity, in a dose-dependent fashion.
Figure 6.
BHLHB1 inhibits E2A-induced transactivation. (A) NIH 3T3 cells were transfected in triplicate with the indicated vectors. TATA-luc and E(2,5)-Luc are luciferase reporter vectors, pCAT is the pCATControl vector used to control for transfection efficiency, and pCE2–5 and pCBHLHB1 are expression vectors. The values presented are luciferase activity (in relative luminescence units, RLU) divided by CAT activity (in cpm). Similar results were obtained in four independent transfection experiments. (B) NIH 3T3 cells were transfected in triplicate with the indicated vectors. The pG5B/CAT vector contains a minimal promoter and five concatamerized Gal4 DNA binding sites linked to a CAT reporter cassette. pSG4 is an expression vector that produces a full-length Gal4 protein. All samples were cotransfected with the Rous sarcoma virus-luciferase control vector to control for transfection efficiency; results are expressed as cpm/RLU. Similar results were obtained in two independent experiments. (C) Jurkat T cells were transfected in triplicate as described above for 7A with the indicated vectors. For these experiments, 2 μg of E(2,5)-luc, 2 μg of pCE2–5, and 8 μg of pCBHLHB1 were used. Similar results were obtained in three independent experiments.
To determine whether the ability of BHLHB1 to inhibit E2A-induced transactivation was caused by nonspecific inhibition of transcription by BHLHB1, we tested the possibility that BHLHB1 might inhibit Gal4-mediated transactivation in NIH 3T3 cells. As shown in Fig. 6B, BHLHB1 was unable to inhibit transcription mediated by Gal4. To evaluate whether BHLHB1 was able to inhibit E2A-induced transactivation in human T cells as well as in murine NIH 3T3 fibroblasts, we repeated the experiment outlined in Fig. 6A using the Jurkat T-cell line. Fig. 6C demonstrates that BHLHB1 inhibits E2A-induced transactivation in Jurkat cells.
Discussion
This study reports the molecular characterization of a t(14;21)(q11.2;q22) chromosome translocation present in a patient with T-ALL. A gene, designated BHLHB1, located on 21q22 was cloned, characterized, and shown to be activated in leukemic cells that had undergone a t(14;21)(q11.2;q22) translocation. BHLHB1 was shown to possess a bHLH motif and to inhibit E2A function in in vitro transfection assays.
In all likelihood, the t(14;21) chromosomal translocation occurred as the cell was undergoing a Vα-Jα recombination event. In support of this hypothesis is the finding that one derivative chromosome contains Vα sequences fused to chromosome 21, whereas the other derivative chromosome contains Jα sequences fused to chromosome 21; the region between the Vα and Jα segments, including the TCRD locus, has been deleted. Further support for the involvement of the V(D)J recombinase complex in this translocation is the presence of known Jα and Vα heptamer sequences flanking the breakpoints, and nontemplated (“N” region) nucleotides at the fusion point. With respect to chromosome 21 sequences, the translocation was perfectly reciprocal, with no sequences gained or lost. The relevance of a potential translin binding site at the breakpoint is uncertain.
Chromosomal translocations associated with T-ALL typically activate protooncogenes, most commonly through juxtaposition of TCR regulatory elements (1, 2). Several of the genes (MYC, SCL/TAL1, LYL1, and TAL2) activated by chromosomal translocation in T-ALL patients encode proteins that are members of the bHLH family of transcription regulators; three of these proteins (SCL/TAL1, LYL1, and TAL2) are quite similar throughout their bHLH regions and form a distinct subfamily of bHLH proteins. These three genes are normally either silent or expressed at low levels in thymocytes, but are expressed at high levels in leukemic cells that have undergone translocations involving these genes (8, 9, 32).
Several recent in vivo and in vitro studies support the contention that SCL/TAL1 exerts its leukemogenic effect through a dominant negative inhibition of E2A function (15–18, 21), SCL/TAL1 serves as a powerful inhibitor of E2A-mediated transactivation in cotransfection assays (17, 18, 21), E2a-deficient mice surviving beyond 3 months develop aggressive T-cell lymphomas (19, 23), and transgenic mice that overexpress the E2a inhibitory protein Id1 in the thymus develop T-cell lymphoma (33). In addition, the t(7;9)(q34;q34.3) translocation associated with T-ALL leads to production of an activated form of the Notch protein (13); activated Notch recently has been shown to inhibit E2a activity (20). Therefore, it seems reasonable to speculate that BHLHB1 may be leukemogenic by virtue of its ability to inhibit E2A-mediated transactivation. Formal proof of this hypothesis awaits development of animal models that overexpress BHLHB1 in the thymus.
The t(14;21) translocation led to positioning of the BHLHB1 locus ≈ 130 kb upstream of the TCRA Cα enhancer on the derivative 21 chromosome and the BHLHB1 gene was expressed to relatively high levels in RNA extracted from cells carrying the t(14;21). It is not surprising that TCRA sequences might exert transcription activating effects over this relatively large distance, given the identification of a TCRA locus control region (34). Furthermore, deletion of a 1.1-kb fragment encompassing TCRA enhancer sequences leads to down-regulation of T early alpha (TEA), located >70 kb upstream of the TCRA enhancer (34). Furthermore, the Ig heavy chain enhancer is thought to exert transcription activating effects over distances of >100 kb in patients with either t(14;18) or t(8;14) translocations activating BCL2 or MYC, respectively.
BHLHB1 maps within a 9- to 12-Mb region of chromosome 21q22 that has been proposed to be a critical region for the development of learning defects in Down's syndrome patients. The identification of this critical region was based on genetic analysis of patients with only a partial trisomy 21 (35), and a mouse model in which mice that had an additional copy of the syntenic mouse chromosomal region displayed learning and behavior deficits (36). In this context, it is intriguing that BHLHB1 encodes a bHLH protein whose expression normally is restricted to neural tissues. Because bHLH proteins generally are thought to regulate developmental programs, this mapping data raises the possibility that an extra copy of BHLHB1 might contribute to the learning deficits associated with Down's syndrome.
We have cloned a t(14;21)(q11.2;q22) chromosomal translocation associated with T-ALL. The translocation, which was likely mediated by a mistake in normal V(D)J recombination, activated a gene (BHLHB1) that encodes a bHLH protein. Given that this translocation involves two acrocentric chromosomes, it is possible that cryptic t(14;21) translocations might exist. The observation that ectopic BLHLB1 expression can inhibit E2A activity in vitro suggests that BHLHB1, like several other leukemic oncoproteins, exerts its leukemogenic effects through a functional inhibition of E2A.
Acknowledgments
P.D.A. is a scholar of the Leukemia Society of America. This work was supported in part by a grant from the Association for Research of Childhood Cancer (AROCC) and by grants from the National Institutes of Health (CA16056). The cryopreserved cells were obtained from the Pediatric Oncology Group (POG) ALL cell bank.
Abbreviations
- ALL
acute lymphoblastic leukemia
- T-ALL
T-cell ALL
- TCR
T-cell receptor
- RT-PCR
reverse transcriptase–PCR
- PAC
P1-artificial chromosome
- CAT
chloramphenicol acetyltransferase
- EST
expressed sequence tag
- bHLH
basic domain helix–loop–helix
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF221520).
References
- 1.Rabbitts T H. Nature (London) 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 2.Thandla S, Aplan P D. Semin Oncol. 1997;24:45–56. [PubMed] [Google Scholar]
- 3.Korsmeyer S J. Annu Rev Immunol. 1992;10:785–807. doi: 10.1146/annurev.iy.10.040192.004033. [DOI] [PubMed] [Google Scholar]
- 4.Tycko B, Sklar J. Cancer Cells. 1990;2:1–8. [PubMed] [Google Scholar]
- 5.Chen Q, Cheng J T, Tasi L H, Schneider N, Buchanan G, Carroll A, Crist W, Ozanne B, Siciliano M J, Baer R. EMBO J. 1990;9:415–424. doi: 10.1002/j.1460-2075.1990.tb08126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Begley C G, Aplan P D, Davey M P, Nakahara K, Tchorz K, Kurtzberg J, Hershfield M S, Haynes B F, Cohen D I, Waldmann T A, Kirsch I R. Proc Natl Acad Sci USA. 1989;86:2031–2035. doi: 10.1073/pnas.86.6.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finger L R, Kagan J, Christopher G, Kurtzberg J, Hershfield M S, Nowell P C, Croce C M. Proc Natl Acad Sci USA. 1989;86:5039–5043. doi: 10.1073/pnas.86.13.5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mellentin J D, Smith S D, Cleary M L. Cell. 1989;58:77–83. doi: 10.1016/0092-8674(89)90404-2. [DOI] [PubMed] [Google Scholar]
- 9.Xia Y, Brown L, Yang C Y, Tsan J T, Siciliano M J, Espinosa R, III, Le Beau M M, Baer R J. Proc Natl Acad Sci USA. 1991;88:11416–11420. doi: 10.1073/pnas.88.24.11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGuire E A, Hockett R D, Pollock K M, Bartholdi M F, O'Brien S J, Korsmeyer S J. Mol Cell Biol. 1989;9:2124–2132. doi: 10.1128/mcb.9.5.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boehm T, Foroni L, Kaneko Y, Perutz M F, Rabbitts T H. Proc Natl Acad Sci USA. 1991;88:4367–4371. doi: 10.1073/pnas.88.10.4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Royer-Pokora B, Loos U, Ludwig W D. Oncogene. 1991;6:1887–1893. [PubMed] [Google Scholar]
- 13.Ellisen L W, Bird J, West D C, Soreng A L, Reynolds T C, Smith S D, Sklar J. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 14.Erikson J, Finger L, Sun L, ar-Rushdi A, Nishikura K, Minowada J, Finan J, Emanuel B S, Nowell P C, Croce C M. Science. 1986;232:884–886. doi: 10.1126/science.3486470. [DOI] [PubMed] [Google Scholar]
- 15.Park S T, Sun X H. J Biol Chem. 1998;273:7030–7037. doi: 10.1074/jbc.273.12.7030. [DOI] [PubMed] [Google Scholar]
- 16.Park S T, Nolan G P, Sun X H. J Exp Med. 1999;189:501–508. doi: 10.1084/jem.189.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voronova A F, Lee F. Proc Natl Acad Sci USA. 1994;91:5952–5956. doi: 10.1073/pnas.91.13.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chervinsky D S, Zhao X F, Lam D H, Ellsworth M, Gross K W, Aplan P D. Mol Cell Biol. 1999;19:5025–5035. doi: 10.1128/mcb.19.7.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan W, Young A Z, Soares V C, Kelley R, Benezra R, Zhuang Y. Mol Cell Biol. 1997;17:7317–7327. doi: 10.1128/mcb.17.12.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ordentlich P, Lin A, Shen C P, Blaumueller C, Matsuno K, Artavanis-Tsakonas S, Kadesch T. Mol Cell Biol. 1998;18:2230–2239. doi: 10.1128/mcb.18.4.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu H L, Wadman I, Tsan J T, Baer R. Proc Natl Acad Sci USA. 1994;91:5947–5951. doi: 10.1073/pnas.91.13.5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu H L, Cheng J T, Chen Q, Baer R. Mol Cell Biol. 1991;11:3037–3042. doi: 10.1128/mcb.11.6.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bain G, Engel I, Robanus Maandag E C, te Riele H P, Voland J R, Sharp L, Chun J, Huey B, Pinkel D, Murre C. Mol Cell Biol. 1997;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aplan P D, Chervinsky D S, Stanulla M, Burhans W C. Blood. 1996;87:2649–2658. [PubMed] [Google Scholar]
- 25.Carter R S, Ordentlich P, Kadesch T. Mol Cell Biol. 1997;17:18–23. doi: 10.1128/mcb.17.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sadowski I, Ma J, Triezenberg S, Ptashne M. Nature (London) 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 27.Kakidani H, Ptashne M. Cell. 1988;52:161–167. doi: 10.1016/0092-8674(88)90504-1. [DOI] [PubMed] [Google Scholar]
- 28.Begley C G, Aplan P D, Davey M P, de Villartay J P, Cohen D I, Waldmann T A, Kirsch I R. J Exp Med. 1989;170:339–342. doi: 10.1084/jem.170.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aoki K, Suzuki K, Sugano T, Tasaka T, Nakahara K, Kuge O, Omori A, Kasai M. Nat Genet. 1995;10:167–174. doi: 10.1038/ng0695-167. [DOI] [PubMed] [Google Scholar]
- 30.Kasai M, Matsuzaki T, Katayanagi K, Omori A, Maziarz R T, Strominger J L, Aoki K, Suzuki K. J Biol Chem. 1997;272:11402–11407. doi: 10.1074/jbc.272.17.11402. [DOI] [PubMed] [Google Scholar]
- 31.Peyton M, Stellrecht C M M, Naya F J, Huang H P, Samora P J, Tsai M J. Mol Cell Biol. 1996;16:626–633. doi: 10.1128/mcb.16.2.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Begley C G, Aplan P D, Denning S M, Haynes B F, Waldmann T A, Kirsch I R. Proc Natl Acad Sci USA. 1989;86:10128–10132. doi: 10.1073/pnas.86.24.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim D, Peng X C, Sun X H. Mol Cell Biol. 1999;19:8240–8253. doi: 10.1128/mcb.19.12.8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diaz P, Cado D, Winoto A. Immunity. 1994;1:207–217. doi: 10.1016/1074-7613(94)90099-x. [DOI] [PubMed] [Google Scholar]
- 35.Rahmani Z, Blouin J, Creau-Goldberg N, Watkins P C, Mattei J F, Poissonnier M, Prieur M, Chettouh Z, Nicole A, Aurias A, et al. Proc Natl Acad Sci USA. 1989;86:5958–5962. doi: 10.1073/pnas.86.15.5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reeves R H, Irving N G, Moran T H, Wohn A, Kitt C, Sisodia S S, Schmidt C, Bronson R T, Davisson M T. Nat Genet. 1995;11:177–184. doi: 10.1038/ng1095-177. [DOI] [PubMed] [Google Scholar]