Abstract
Individuals infected with HIV-1 have varying rates of progression to AIDS. Cellular immune responses, comprised of cytolytic and noncytolytic CD8+ T cell effector functions, are considered important for controlling viremia and maintaining the clinically asymptomatic state. Although there is general agreement regarding CD8+ T lymphocyte cytotoxic functions, considerable controversy exists over the nature of the noncytolytic antiviral activity of CD8+ cells. The discovery that RANTES (regulated on activation, normal T cell expressed and secreted), MIP-1α, and MIP-1β (macrophage inflammatory protein 1 α and β) could inhibit HIV-1 replication by blocking viral entry processes led to the notion that these molecules are responsible for the CD8+ cell suppressive activity. However, T tropic HIV isolates requiring the CXCR4 coreceptor for entry are insensitive to the antiviral effects of these β-chemokines. Using a CXCR4-dependent virus, we determined that the mechanism of CD8+ T cell-mediated activity did act after viral entry into the host cell. We also define the kinetics of the HIV life cycle in primary activated human CD4+-enriched T cells by using an HIV-1 reporter virus system pseudotyped with the CXCR4-dependent HIV-1 envelope gene of NL4-3. Analysis of these kinetic data indicates that CD8+ T cell-mediated suppressive activity acts at a stage in the viral life cycle after entry and independently of the HIV envelope. Additionally, we show that the antiviral activity targets stages of the virus life cycle correlating with transcription and early proviral gene expression. These findings not only provide a range of possible targets for the CD8+ T cell-mediated activity but also support the notion that this antiviral activity is multifactorial in nature.
HIV-1 infection of susceptible cells is initiated via interactions between the virus envelope glycoprotein and the cell surface receptor CD4 (1). Fusion of the viral and cell membranes proceeds through subsequent interaction of this complex with a specific chemokine coreceptor, primarily CCR5 or CXCR4 (2–4). HIV-1 isolates that can infect T cell lines and induce syncytia use the CXCR4 receptor and are termed X4 HIV-1 (5). Such isolates are often recovered late in the course of disease, differing from the non-syncytium-inducing strains that are associated with transmission and predominate in the early stages. Non-syncytium-inducing strains gain entry to target cells through use of the CCR5 receptor and are called R5 HIV-1 (5). The course of HIV-1 infection in humans varies between individuals: some show rapid progression, and others lack obvious signs of progressive disease despite many years of infection. HIV-1 strains (6, 7) attenuated by genotypic properties, particularly nef deletions, of the infecting viral quasispecies have been attributed to some cohorts of long-term nonprogressors. In other cohorts, strong immune responses and low virus loads indicate that effective antiviral immune responses confer protection (8). Characterization of the efficient immunity present in these asymptomatic individuals is important for understanding the immunological response to HIV and identification of the immune correlates of protection. One element of the protective response to HIV-1 infection is the cellular immune response, including both CD8+ T cell-mediated cytolytic (9) and noncytolytic (10) suppression of HIV. Many asymptomatic HIV+ individuals possess strong noncytolytic CD8+ T cell-mediated antiviral activities (11, 12). In agreement, we observed a potent inhibitory effect by CD8+ T lymphocytes from asymptomatic HIV+ individuals on both X4- and R5-dependent HIV-1 replication in vitro (13, 14). The natural ligands for these coreceptors—the β-chemokines RANTES (regulated on activation, normal T cell expressed and secreted), MIP-1α, and MIP-1β (macrophage inflammatory protein 1 α and β) for CCR5 and the α-chemokine stromal derived factor-1 for CXCR4—specifically block R5 and X4 HIV infection, respectively (15–17). Although studies have shown that CD8+ cells secrete RANTES, MIP-1α, and MIP-1β as antiviral factors (18), several groups including our own demonstrated that release of β-chemokines by CD8+ cells cannot adequately explain the virus-suppressive activity of CD8+ cells (19–22). Additionally, we previously demonstrated that stromal derived factor-1, a ligand of CXCR4 that blocks HIV entry, does not mediate CD8+ T cell suppression of X4 viruses (23). Thus, the current studies examined not only the effects of CD8+ T cell-mediated suppression on HIV entry but also the subsequent steps in the virus life cycle. In this report, we describe the stages of the virus life cycle that are the key targets for the HIV-1 inhibitory activity of CD8+ cells.
Materials and Methods
Source, Preparation, and Transformation of CD8+ Cells.
Venous blood was obtained from asymptomatic HIV+ individuals through the Duke Infectious Diseases Clinic under the auspices of John A. Bartlett. Peripheral blood mononuclear cells were prepared by standard Ficoll/Hypaque density separation (Sigma). Cells were activated for 3 days with anti-CD3 (OKT3) and anti-CD28 antibodies in AIMV medium (GIBCO/BRL) supplemented with 10% (vol/vol) FBS, recombinant IL-2 (20 units/ml), streptomycin (50 μg/ml), and gentamicin (10 μg/ml) at 37°C in a humidified CO2 incubator. CD4+ cells were removed by positive selection with anti-CD4+ antibody-coated beads (Dynal, Lake Success, NY). CD8+ cells were purified further by positive selection with anti-CD8+ antibody-coated beads (Dynal). Cells were separated from the beads with DetachaBead (Dynal). CD8+ cells from two asymptomatic subjects with potent suppressive activity for X4 viruses were transformed with herpesvirus saimiri (HVS) subgroup C strain 488-77 (a gift from R. Desrosiers, New England Regional Primate Research Center, Southborough, MA) to create continuous cell lines for further characterization. HVS transformation of CD8+ cells was performed as described (21).
Quantitative CD8+ Suppression Assay.
Suppression assays were performed as described (21) with minor modifications as indicated. Peripheral blood mononuclear cells were prepared from freshly drawn blood from a pool of HIV− donors and activated as described above. CD4+- and CD8+-enriched populations were obtained by negative selection with anti-CD8+ or anti-CD4+ antibody-coated beads, respectively. CD4+ cells were centrifuged and resuspended in fresh medium at 2.0 × 106 cells per ml. Duplicate 100-μl aliquots of CD4+ cells were cultured in wells of a 96-well microtiter plate containing 20 μl of serially diluted virus [either QZ4734, a non-syncytia-inducing HIV-1 primary isolate (R5), or IIIB, a syncytia-inducing HIV-1 laboratory strain (X4)]. Fresh medium or CD8+ cells at various effector-to-target ratios (E:T) were added in 80 μl for a total culture volume of 200 μl. Cultures were incubated at 37°C in a humidified CO2 incubator. At 72-h intervals, 90 μl of supernatant was removed and adjusted to 1% with Triton X-100, and cultures were fed with an equal volume of fresh medium. The Triton X-100-treated supernatants were assessed for virus replication by measuring reverse transcriptase activity as described (24). In this assay, the criteria for CD8+ T cell-mediated antiviral activity is a reduction in virus titer by a log or greater compared with that of control (CD4+ T cells alone).
Pseudotyped Virus Production, Infection, and Luciferase Assays.
Pseudotyped viruses were produced by transfection of DNA into the human embryonic kidney cell line 293T by using a modified calcium phosphate method (25). The reporter virus DNA pNL4-3 LUCR−E−, obtained from the National Institutes of Health AIDS Research and Reference Reagent Program (25), was cotransfected with an expression vector for either an HIV-1 envelope, pNL4–3 env (a gift from T. Dragic, Aaron Diamond Aids Research Center, New York), or the amphotropic murine leukemia virus (A-MLV) envelope, S-A-MLV-env (obtained from the National Institutes of Health AIDS Research and Reference Reagent Program). The virus-containing supernatants were harvested 2 days later. CD4+ primary T lymphocytes prepared and activated as described above were infected for 2 h at 37°C with 100 μl of undiluted pseudotyped virus containing supernatant in the presence of 20 μg/ml DEAE. The initial cell concentration was 1.8 × 106 per ml. After the 2-h incubation, fresh medium was added to the cells to create a final concentration of 0.9 × 106 cells per ml. In some experiments, DP178 (20 μg/ml; a gift from T. Matthews, Trimeris, Durham, NC), anti-CD4+ mAb #19 (20 μg/ml; a gift from J. Hoxie, University of Pennsylvania, Philadelphia), nevirapine (0.2 μM), nelfinavir (0.2 μM; both nevirapine and nelfinavir were gifts from D. Lambert, Trimeris), or a tat inhibitor (50 μM; a gift from S. Turk, National Institute of Allergy and Infectious Diseases) was added either simultaneously with the virus or at timed intervals after infection. The tat inhibitor is similar to the compound RO5-3335 [7-chloro-5-(2-pyrryl)-3H-1,4-benzodiazepin-2(H)-one; ref. 26]. For infection with A-MLV, virus was removed after a 2-h incubation, and then CD8+ T cells were added. After 72 h, cells were washed with PBS and luciferase lysis buffer (Promega), followed by one freeze–thaw cycle. The sample was assayed with the Promega Luciferase Assay System and a luminometer (Lumat LB 9501, EG & G Berthold, Gaithersburg, MD).
Cytotoxic T Lymphocyte (CTL) Assay.
CD8+ T cells were tested for the presence of HIV-specific CTL by using autologous Epstein–Barr virus-transformed B lymphocyte cell lines infected with vaccinia constructs expressing the HIV-1 env, gag, pol, or nef genes. Additionally, the CD8+ T cells were cocultured with the CD4+ targets used in this study, and cytolysis was measured as described (14).
Results
Characterization of CD8+ T Cell-Mediated Suppressive Activity.
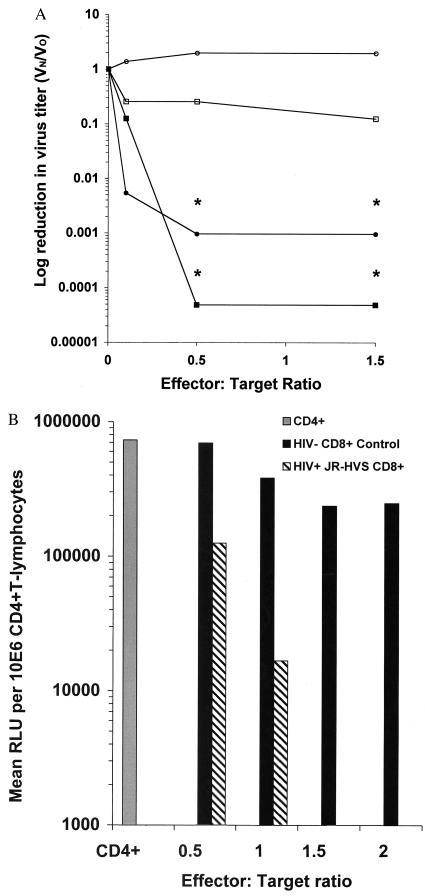
To study β-chemokine-independent mechanisms of suppression, we identified asymptomatic HIV+ patients with robust CD8+ cell inhibitory activity for X4 viruses and transformed their CD8+ T lymphocytes with HVS to develop cell lines with this effector phenotype (21). In a manner analogous to the nontransformed cells from which they were derived (data not shown), the HVS-transformed CD8+ cells from an asymptomatic HIV+ individual potently inhibited infection by both X4- and R5-dependent viruses (Fig. 1A). The CD8+ effector cells at a 0.5:1 E:T completely blocked both R5 HIV and X4 HIV infection at all virus input levels effecting a 3–4 log reduction in viral titer. Nontransformed CD8+-enriched cells from a pool of HIV− donors did not affect the infectious titer of either virus significantly. The suppressive activity was caused by both soluble and contact-associated mediators. Using a transwell system to separate effector and target cell populations, we found that soluble mediators reduced viral reverse transcriptase production by 2- to 3-fold, whereas contact between the effector and target cells elicited a 22-fold reduction in reverse transcriptase (data not shown). It is possible that contact elicited the soluble activities to more potent levels or that close proximity was necessary for the soluble portion to be the most effective. However, because of the insensitivity of the X4 virus to inhibition by RANTES, MIP-1α, and MIP-1β, we infer that CD8+ T cells inhibit virus replication through noncytolytic mechanisms distinct from the release of these β-chemokines.
Figure 1.
(A) Transformed CD8+ T cells from an asymptomatic HIV+ individual suppress CCR5-dependent HIV-1 (QZ4734) and CXCR4-dependent HIV-1 (IIIB) in a quantitative CD8+ suppression assay. Closed symbols, transformed CD8+ T cells from an asymptomatic HIV+ individual; open symbols, CD8+ T cells from a pool of HIV− donors; circles, R5 virus; squares, X4 virus. The vertical axis indicates log reduction in virus titer, defined as virus infectivity in the presence (Vn) divided by the virus infectivity in the absence (Vo) of CD8+ lymphocytes. Asterisks (*) indicate actual tissue culture 50% infective dose per ml below detection limits at <25. These results (day 13) are representative of three experiments performed in duplicate. (B) CD8+ T cells from an HIV+ asymptomatic individual suppress X4 HIV-1 in a single-cycle infection assay. CD4+-enriched T lymphocytes were infected with a reporter virus pseudotyped with the CXCR4-dependent envelope NL4-3. Either transformed CD8+ T cells from an asymptomatic HIV+ individual (hatched bars) or CD8+ T cells from a pool of HIV− donors (solid bars) were added at increasing E:T. The results are representative of three experiments performed in duplicate. RLU, relative light units; JR-HVS, a patient-derived herpesvirus saimiri-transformed cell line.
CD8+ T Cell-Mediated Suppression in a Single-Cycle Infection Assay.
To concentrate on these previously uncharacterized suppressive molecules, we examined the stage or stages of the X4 HIV life cycle that are susceptible to inhibition by CD8+ cells. Reasoning that assays relying on multiple rounds of replication confounded such studies, we developed a CD8+ suppression assay that measures a single cycle of infection. We used an envelope-defective luciferase reporter virus (25) complemented in trans with a X4 envelope, NL4–3, to assess CD8+-mediated suppressive activity. In this system, the luciferase reporter serves as a surrogate for the expression of early proviral genes, requiring only completion of virus entry, reverse transcription, nuclear translocation, and proviral integration. CD8+ T cell effectors suppressed infection of the pseudotyped virus in a dose-dependent manner (Fig. 1B); therefore, they target stages of virus replication occurring before or at the expression of early proviral genes.
Infection Kinetics of HIV in Primary Human CD4+ Enriched T Cells.
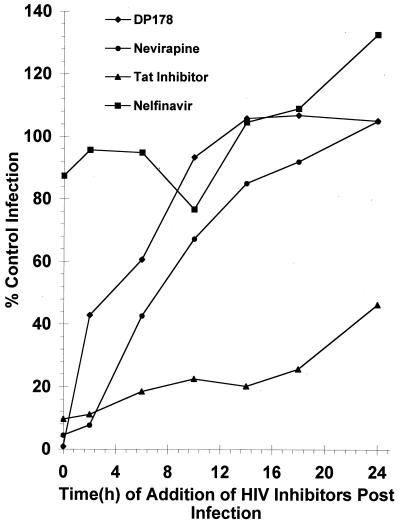
Various inhibitors to the HIV life cycle were used in a single-cycle infection assay to characterize the timing of particular events (Fig. 2). DP178, a gp41 (glycoprotein of Mr 41,000) peptide mimetic, was used as the inhibitor of cell entry, because it blocks fusion (27). By 6 h, entry is largely complete (60% of control infection) for HIV, and this inhibitor has a difficult time blocking replication if added at later time points. Addition of nevirapine, a nonnucleoside reverse transcription inhibitor, as late as 10 h after infection results in 67% infection compared with control, and this percentage is increased to 85% by 14 h. These data indicate that the process of reverse transcription is complete by 10–14 h. To evaluate the timing of transcription and early proviral gene expression, we used an inhibitor of tat (26) that acts at the level of transcriptional transactivation. The addition of the tat inhibitor anywhere from 0 to 24 h after infection significantly inhibited viral replication. At 24 h, the level of infection was only 46% of that of the control. The protease inhibitor nelfinavir was used as a negative control. As expected, nelfinavir was ineffectual at all time points, because a functional protease is not required for the reporter gene (luciferase) to be active.
Figure 2.
Kinetics of X4 HIV-1 infection in primary CD4+ T lymphocytes. Inhibitors of the different stages of the HIV life cycle were added at particular time intervals after HIV-1 infection. Infection in the presence of these inhibitors was compared with the control infection (absence of inhibitors) to determine the relative time of action for each inhibitor. The results shown are an average of two to six experiments performed in duplicate.
CD8+ T Cell-Mediated Suppression Does Not Act on Virus Entry.
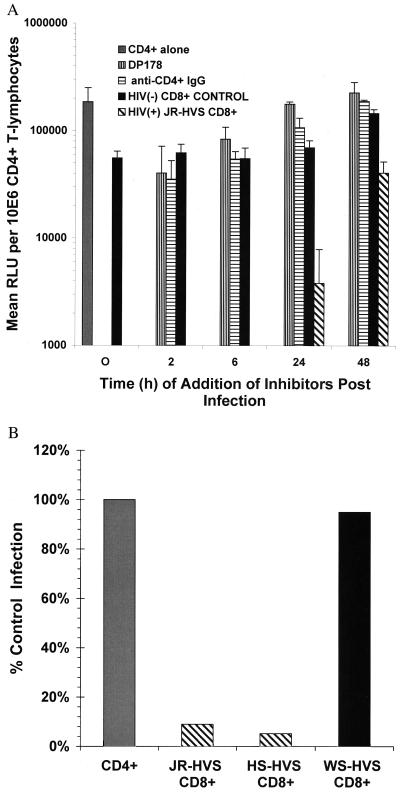
To determine the stage of the virus life cycle at which CD8+ cells act, we added CD8+ effector cells (E:T of 2:1) at various intervals after exposure of CD4+-enriched T cells to the X4 pseudotyped virus (Fig. 3A). The addition of pooled HIV− CD8+ control cells (E:T of 2:1) had little effect on virus replication (Figs. 1B and 3A). Addition of either anti-CD4+ mAb #19, which blocks binding of gp120 to CD4 (28), or a gp41 peptide mimetic (DP178; ref. 27) with the virus at time 0 completely inhibits viral replication. Delaying addition of the entry inhibitors until 2 h after infection allowed virus replication to reach 23–43% of control levels, and a 6-h delay permitted replication levels to reach 30–61% of the control values (Figs. 2 and 3A). Despite the facts that the infection process in this system is not synchronous and that virus is not removed, these data support the conclusion that entry is largely complete within 6 h. Interestingly, the CD8+ effectors completely abrogated viral replication when added as late as 6 h after infection. At 24 h after infection, the effectors still conferred a log reduction in virus replication. Inhibition of virus replication was also observed at these later time points in experiments employing primary nontransformed CD8+ T lymphocytes, indicating that the antiviral activity did not result from the transformation process (data not shown). The capacity of the CD8+ cells to block infection at these later time points suggested that suppression could not be explained by inhibition of viral entry processes. Further evidence that CD8+ suppression does not target HIV-1 envelope-mediated entry processes is presented in Fig. 3B, in which it is shown that CD8+ cells added 2 h after infection completely inhibit replication by a reporter virus pseudotyped with the A-MLV envelope. A-MLV uses the pit-2 receptor, a sodium-dependent phosphate symporter, for cell entry (29, 30). HVS-transformed CD8+ T cells from a single donor lacking X4 HIV suppressive activity were used as a negative control (Fig. 3B, WS-HVS), showing that the transformation process did not induce virus-suppressive activity. Comparison of the results in Figs. 2 and 3A suggests that CD8+ T cell-mediated antiviral activity can act late in the viral life cycle, in much the way that the tat inhibitor can.
Figure 3.
(A) Kinetics of CD8+ T cell-mediated suppression and X4 HIV-1 entry in primary CD4+ T lymphocytes. CD4+-enriched T lymphocytes were infected with X4 HIV pseudotyped virus (NL4–3). Two agents that inhibit viral entry were added at different times after infection. DP178 is an HR2 peptide mimetic that blocks gp41-mediated fusion. Anti-CD4+ mAb #19 blocks binding of gp120 to CD4+. Transformed CD8+ T cells from an asymptomatic HIV+ individual (hatched bars) or CD8+ T cells from a pool of HIV− donors (solid bars) were added at a 2:1 E:T at various times after infection. At 24 h, the CD8+ effectors had a 94.3% inhibition of control infection that was limited to 77% at 48 h. The results shown are the average of two or three experiments performed in duplicate. RLU, relative light units. (B) CD8+ suppression is independent of HIV-1 envelope-mediated entry. CD4+-enriched T lymphocytes were infected with a reporter virus pseudotyped with the A-MLV envelope for 2 h, and then the virus was removed. HVS-transformed CD8+ T cells with X4 suppressive activity (JR-HVS and HS-HVS, hatched bars) or HVS-transformed CD8+ T cells without X4 suppressive activity (WS-HVS, solid bars), were added at a 2:1 E:T after removal of the virus. Percentage of infection is calculated based on infection of the CD4+ targets without CD8+ T cells (100%). Results are an average of three experiments for JR-HVS (5.2%) and HS-HVS CD8+ effectors (9%) and two experiments for the WS-HVS CD8+ cells (95%).
Inhibition of tat-Mediated Transcription and CD8+ T Cell-Mediated Suppressive Activity.
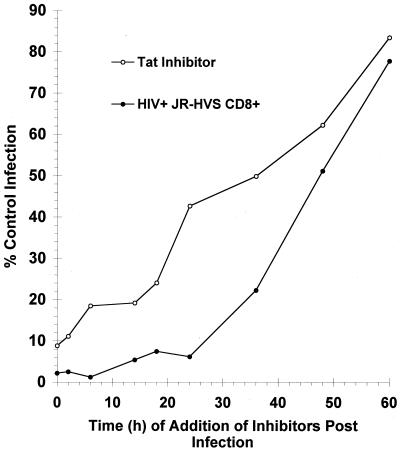
Fig. 4 shows a direct comparison of what happens to viral replication if either the tat inhibitor or the CD8+ cells are added at very late time points in the life cycle. Because the tat inhibitor targets a late stage in the virus life, it potently suppresses viral replication if added to the culture up to 48 h after infection. At 48 h, 62% of the control infection occurs. In comparison, the CD8+ effector cells also inhibit viral replication potently at late time points. It is only between 48 and 60 h after infection that the addition of the CD8+ cells begins to lose its effect significantly, with 51–78% of the control infection occurring. Although the kinetics of inhibition for the tat inhibitor and the CD8+ cells cannot be superimposed, the results do suggest that the mechanisms of action for CD8+ cells act at expression of early proviral gene products and/or tat-mediated transcription. Prior work by our group and others (24, 31, 32) indicated the possibility that CD8+ T cell-mediated suppression could act on the HIV long terminal repeat through the use of transient transfections or integrated reporter assays. Herein, we provide kinetic analysis of a single cycle of HIV infection to support the hypothesis that CD8+ T cell-mediated suppression is acting at tat-mediated transcription and/or expression of early proviral gene products. We demonstrate that CD8+ T cells can act at these time points in an ongoing cycle of HIV replication.
Figure 4.
Late time course of CD8+ T cell-mediated suppression and tat-mediated transactivation. CD4+-enriched T lymphocytes were infected with X4 HIV pseudotyped virus (NL4-3). Either a tat inhibitor or JR-HVS CD8+ effectors were added up to 60 h after infection. The results shown are an average of three experiments performed in duplicate.
Noncytolytic CD8+ T Cell-Mediated Suppression.
Controversy exists over whether CD8+ suppression is cytolytic and HLA class I restricted. One assertion is that class I recognition between the CD8+ cells and the virally infected target cells is a prerequisite for suppressive activity (33). Subsequent to specific MHC recognition, the CD8+ antiviral activity involves not only cytolysis but also a noncytolytic activity mediated through proteoglycan-bound β-chemokine release (34). In contrast, another report showed that HLA histocompatibility is not required but did enhance suppressor function (35). By using CD8+ cell clones, it was shown that HLA class I-restricted CTL activity and virus-suppressive reactivity were separable, although not necessarily mutually exclusive (14). The CD8+ effector cell lines used in this report were devoid of HIV-1-specific CTL activity assessed at high E:T ratios (≥50:1) with targets expressing env, gag, pol, or nef (data not shown), although some alloreactivity was observed with the pool of HIV- CD4+ targets (Table 1, Experiment III). Consequently, we tested the CD8+ effectors for both cytolytic and noncytolytic suppressive activities with several targets. We found that CD8+ effectors could suppress HIV-1 potently in the absence of cytotoxicity (Table 1, Experiment I). Conversely, cytolytic activity (24%) was observed in the absence of significant CD8+ suppression (Table 1, Experiment II). Although a small amount of CD8+-mediated suppression was detected (34.5%) in this case, it was well below our 90% reduction criteria for the noncytolytic suppression assay. To confirm further that the findings in Fig. 3A were not caused by cell lysis after entry, the mix of effector and target cells lacking cytolysis (Table 1, Experiment I) was used in a “time of addition” study (Table 2). In these experiments, complete inhibition by the CD8+ effector cells occurred at all time points beginning from the initiation of infection (T0) to 6 h after infection, with a rise in virus replication beginning as late as 24 h after infection. The concordance of the results for the experiments depicted in Table 2 and Fig. 3A indicates that suppression of HIV-1 replication by CD8+ cells after virus entry is not caused by cytolysis.
Table 1.
CD8+-mediated suppression is separable from cell lysis
| Experiment | CD8+ T cell lines | CD4+ T cell | CD8+ | Cytolysis |
|---|---|---|---|---|
| I | JR-HVS | Single donor (EW) | +* | − |
| II | NS-HVS | Single donor (DS) | − | + |
| III | JR-HVS | Donor pool | + | + |
The plus signs indicate either >90% suppression or >10% cytolysis, both of which are the positive cutoffs for these assays. The negative signs denote <90% suppression or <10% cytolysis. Experiments: I, 98% suppression, 3.4% cytolysis; II, 34.5% suppression, 24% cytolysis; III, >99% suppression, 17.9% cytolysis. Percentage of suppression was calculated based on a control infection in CD4+ targets from a pseudotyped virus CD8+ suppression assay, and percentage of cytolysis was calculated from a standard chromium release assay (38). NS-HVS cells are nonsuppressing CD8+ T cells that were derived from the original JR-HVS cells after long term culture.
Table 2.
Time of addition demonstrates that CD8+-mediated suppression after virus entry is not a result of cell lysis
| Time of addition | Percentage of infection |
|---|---|
| T0 h | 2.2 |
| T2 h | 1.8 |
| T6 h | 1.4 |
| T24 h | 20.5 |
| T48 h | 73.8 |
Discussion
In this report, we provide a kinetic analysis of CD8+ T cell-mediated suppression of HIV-1 infection in primary human CD4+ T lymphocytes. In agreement with studies performed with CD4+ T cell lines (15, 36), we found that viral entry processes in primary CD4+ T lymphocytes are largely complete within 2–6 h of infection (Fig. 3A). Expression of HIV-1 genes occurs by 17–25 h after infection (36). Because CD8+ T cell-mediated suppression was diminished 48 h after infection (Fig. 4), the latest stages in the reporter virus life cycle, such as basal and/or tat-mediated enhancement of transcription, are possible targets of suppression. We demonstrate that potent suppressive activity is independent of the HIV-1 envelope, occurs after viral entry and reverse transcription, and closely mirrors the effects of a tat inhibitor (Figs. 1B, 3 A and B, and 4). As a result, noncytolytic CD8+ T cell-mediated activity inhibits expression of early proviral genes. This work builds on work performed by both our group and others and shows that CD8+ suppressive activity targets gene expression during virus replication in primary T lymphocytes. Our prior work indicated that primary CD8+ cells and to some extent their soluble factors were capable of limiting gene expression, because they inhibited basal as well as tat-enhanced transcriptional activity of the HIV-1 long terminal repeat promoter (24). Potential effects on transcription were also suggested by other groups (19, 31, 32, 37). Therefore, in accordance with preceding work and the discoveries described in this report, we have modeled the mechanism or mechanisms of action of CD8+ T cells to reflect the kinetics of HIV-1 infection in primary CD4+ T lymphocytes (Fig. 5). This model contends that the antiviral activity of noncytolytic CD8+ T cells after virus entry acts late in the virus life cycle during the time of transcription and early proviral gene expression. Considerable attention has focused on the β-chemokines RANTES, MIP-1α and MIP-1β as the elusive CD8+ cell-derived HIV suppressive factors because of their ability to block HIV entry. Although others have also demonstrated that CD8+ suppression of HIV replication is multifaceted (20), our results distinguish between those antiviral effects that inhibit viral entry processes and those described within this report that act at virus gene expression.
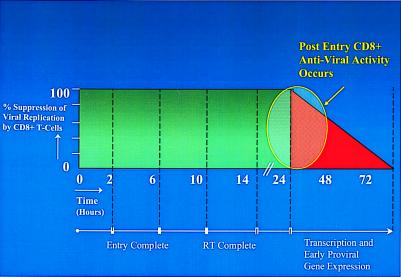
Figure 5.
Kinetic model of the targets of CD8+ T cell-mediated anti-HIV activity. In primary CD4+-enriched T lymphocytes, entry occurs within 2–6 h, and reverse transcription (RT) occurs up to about 14 h. At 24–72 h, transcription from the integrated provirus and expression of gene products take place. CD8+ T cell-mediated suppression of HIV-1 targets those stages of the virus life cycle involving transcription and early gene expression.
Acknowledgments
We thank Derrick Goodman and Anizsa Jones for technical assistance and the patients and staff at the Duke Infectious Disease Clinic. We also thank Dani Bolognesi for many insightful discussions and enduring support. This work was supported by National Institute of Allergy and Infectious Diseases, National Institute of Health, Grants RO1-AI40017 (to M.L.G.), NOI-AI65305, and PO1-AI40237.
Abbreviations
- X4
CXCR4
- R5
CCR5
- MIP-1
macrophage inflammatory protein 1
- RANTES
regulated on activation, normal T cell expressed and secreted
- HVS
herpesvirus saimiri
- CTL
cytotoxic lymphocyte
- A-MLV
amphotropic murine leukemia virus
- E:T
effector-to-target cell ratio
- JR-HVS
HS-HVS, and WS-HVS are each patient-derived herpesvirus saimiri-transformed cell lines
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.070521097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.070521097
References
- 1.Maddon P J, Dalgleish A G, McDougal J S, Clapham P R, Weiss R A, Axel R. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 2.Moore J P, Trkola A, Dragic T. Curr Opin Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 3.Berger E A. AIDS. 1997;11:S3–S16. [PubMed] [Google Scholar]
- 4.Wyatt R, Sodroski J. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 5.Berger E A, Doms R W, Fenyo E M, Korber B T, Littman D R, Moore J P, Sattentau Q J, Schuitemaker H, Sodroski J, Weiss R A. Nature (London) 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 6.Ashton L J, Learmont J, Luo K, Wylie B, Stewart G, Kaldor J M. Lancet. 1994;344:718–720. doi: 10.1016/s0140-6736(94)92210-1. [DOI] [PubMed] [Google Scholar]
- 7.Kirchhoff F, Greenough T C, Brettler D B, Sullivan J L, Desrosiers R C. N Engl J Med. 1995;332:228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- 8.Pantaleo G, Menzo S, Vaccarezza M, Graziosi C, Cohen O J, Demarest J F, Montefiori D, Orenstein J M, Fox C, Schrager L K, et al. N Engl J Med. 1995;332:209–216. doi: 10.1056/NEJM199501263320402. [DOI] [PubMed] [Google Scholar]
- 9.Walker B D, Chakrabarti S, Moss B, Paradis T J, Flynn T, Durno A G, Blumberg R S, Kaplan J C, Hirsch M S, Schooley R T. Nature (London) 1987;328:345–348. doi: 10.1038/328345a0. [DOI] [PubMed] [Google Scholar]
- 10.Walker C M, Moody D J, Stites D P, Levy J A. Science. 1986;234:1563–1566. doi: 10.1126/science.2431484. [DOI] [PubMed] [Google Scholar]
- 11.Mackewicz C E, Ortega H W, Levy J A. J Clin Invest. 1991;87:1462–1466. doi: 10.1172/JCI115153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao Y, Qin L, Zhang L, Safrit J, Ho D D. N Engl J Med. 1995;332:201–208. doi: 10.1056/NEJM199501263320401. [DOI] [PubMed] [Google Scholar]
- 13.Greenberg M L, Lacey S F, Chen C H, Bolognesi D P, Weinhold K J. Springer Semin Immunopathol. 1997;18:355–369. doi: 10.1007/BF00813503. [DOI] [PubMed] [Google Scholar]
- 14.Toso J F, Chen C H, Mohr J R, Piglia L, Oei C, Ferrari G, Greenberg M L, Weinhold K J. J Infect Dis. 1995;172:964–973. doi: 10.1093/infdis/172.4.964. [DOI] [PubMed] [Google Scholar]
- 15.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, et al. Nature (London) 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 16.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. Nature (London) 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 17.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J L, Arenzana-Seisdedos F, Schwartz O, Heard J M, Clark-Lewis I, Legler D F, et al. Nature (London) 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 18.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 19.Moriuchi H, Moriuchi M, Combadiere C, Murphy P M, Fauci A S. Proc Natl Acad Sci USA. 1996;93:15341–15345. doi: 10.1073/pnas.93.26.15341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rubbert A, Weissman D, Combadiere C, Pettrone K A, Daucher J A, Murphy P M, Fauci A S. AIDS Res Hum Retroviruses. 1997;13:63–69. doi: 10.1089/aid.1997.13.63. [DOI] [PubMed] [Google Scholar]
- 21.Lacey S F, Weinhold K J, Chen C H, McDanal C, Oei C, Greenberg M L. AIDS Res Hum Retroviruses. 1998;14:521–531. doi: 10.1089/aid.1998.14.521. [DOI] [PubMed] [Google Scholar]
- 22.Barker E, Bossart K N, Levy J A. Proc Natl Acad Sci USA. 1998;95:1725–1729. doi: 10.1073/pnas.95.4.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lacey S F, McDanal C B, Horuk R, Greenberg M L. Proc Natl Acad Sci USA. 1997;94:9842–9847. doi: 10.1073/pnas.94.18.9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C H, Weinhold K J, Bartlett J A, Bolognesi D P, Greenberg M L. AIDS Res Hum Retroviruses. 1993;9:1079–1086. doi: 10.1089/aid.1993.9.1079. [DOI] [PubMed] [Google Scholar]
- 25.Connor R I, Chen B K, Choe S, Landau N R. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 26.Hsu M C, Schutt A D, Holly M, Slice L W, Sherman M I, Richman D D, Potash M J, Volsky D J. Science. 1991;254:1799–1802. doi: 10.1126/science.1763331. [DOI] [PubMed] [Google Scholar]
- 27.Wild C, Greenwell T, Matthews T. AIDS Res Hum Retroviruses. 1993;9:1051–1053. doi: 10.1089/aid.1993.9.1051. [DOI] [PubMed] [Google Scholar]
- 28.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, et al. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 29.van Zeijl M, Johann S V, Closs E, Cunningham J, Eddy R, Shows T B, O'Hara B. Proc Natl Acad Sci USA. 1994;91:1168–1172. doi: 10.1073/pnas.91.3.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kavanaugh M P, Miller D G, Zhang W, Law W, Kozak S L, Kabat D, Miller A D. Proc Natl Acad Sci USA. 1994;91:7071–7075. doi: 10.1073/pnas.91.15.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Powell J D, Bednarik D P, Folks T M, Jehuda-Cohen T, Villinger F, Sell K W, Ansari A A. Clin Exp Immunol. 1993;91:473–481. doi: 10.1111/j.1365-2249.1993.tb05927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackewicz C E, Blackbourn D J, Levy J A. Proc Natl Acad Sci USA. 1995;92:2308–2312. doi: 10.1073/pnas.92.6.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang O O, Kalams S A, Trocha A, Cao H, Luster A, Johnson R P, Walker B D. J Virol. 1997;71:3120–3128. doi: 10.1128/jvi.71.4.3120-3128.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wagner L, Yang O O, Garcia-Zepeda E A, Ge Y, Kalams S A, Walker B D, Pasternack M S, Luster A D. Nature (London) 1998;391:908–911. doi: 10.1038/36129. [DOI] [PubMed] [Google Scholar]
- 35.Mackewicz C E, Garovoy M R, Levy J A. J Virol. 1998;72:10165–10170. doi: 10.1128/jvi.72.12.10165-10170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Srivastava K K, Fernandez-Larsson R, Zinkus D M, Robinson H L. J Virol. 1991;65:3900–3902. doi: 10.1128/jvi.65.7.3900-3902.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Copeland K F, McKay P J, Rosenthal K L. AIDS Res Hum Retroviruses. 1995;11:1321–1326. doi: 10.1089/aid.1995.11.1321. [DOI] [PubMed] [Google Scholar]
- 38.Ferrari G, King K, Rathbun K, Place C A, Packard M V, Bartlett J A, Bolognesi D P, Weinhold K J. Clin Exp Immunol. 1995;101:239–248. doi: 10.1111/j.1365-2249.1995.tb08345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]