Abstract
A gapped genome sequence of the biomining bacterium Thiobacillus ferrooxidans strain ATCC23270 was assembled from sheared DNA fragments (3.2-times coverage) into 1,912 contigs. A total of 2,712 potential genes (ORFs) were identified in 2.6 Mbp (megabase pairs) of Thiobacillus genomic sequence. Of these genes, 2,159 could be assigned functions by using the WIT-Pro/EMP genome analysis system, most with a high degree of certainty. Nine hundred of the genes have been assigned roles in metabolic pathways, producing an overview of cellular biosynthesis, bioenergetics, and catabolism. Sequence similarities, relative gene positions on the chromosome, and metabolic reconstruction (placement of gene products in metabolic pathways) were all used to aid gene assignments and for development of a functional overview. Amino acid biosynthesis was chosen to demonstrate the analytical capabilities of this approach. Only 10 expected enzymatic activities, of the nearly 150 involved in the biosynthesis of all 20 amino acids, are currently unassigned in the Thiobacillus genome. This result compares favorably with 10 missing genes for amino acid biosynthesis in the complete Escherichia coli genome. Gapped genome analysis can therefore give a decent picture of the central metabolism of a microorganism, equivalent to that of a complete sequence, at significantly lower cost.
Keywords: genome analysis, metabolic reconstruction, gapped genomes
Starting with Haemophilus influenzae in 1995 (1), the total genomic DNA of >20 microbial organisms has been sequenced (http://www.tigr.org/tdb/mdb/mdb.html). Computational tools for genome analysis have been developed along with expanding genome sequence data and merged into shared analytical environments, such as http://WIT.mcs.anl.gov/WIT2/CGI/index.cgi and http://igweb.integratedgenomics.com/IGwit/, GeneQuiz (http://columba.ebi.ac.uk:8765/), Pedant (http://pedant.mips.biochem.mpg.de/), and Entrez Genomes (http://www.ncbi.nlm.nih.gov/Entrez/Genome/org.html). These tools have increased gene recognition rates from <50% (2, 3) to >70% (4, 5). Today, the remaining 30% of “hypothetical” genes separate us from a complete description of the genomic content of an organism. New experimental approaches, including various types of probe arrays (6) and systematic gene knock-outs (7, 8) assist the annotation of these unassigned parts of the genome.
Computational approaches based on various types of clustering of potential genes, whether with regard to positions on the chromosome (such as in operons; ref. 9) or clusters of orthologous genes from different organisms (COGs; ref. 10) in what might be called “phylogenetic space,” permit further gene assignments. The next stage of genome analysis is the integration of gene assignments into an overview of metabolism via metabolic reconstruction, the conceptual assembly of functional multi-component systems such as metabolic pathways, transport units, and signal transduction pathways (11). The identification of functional subsystems goes beyond what is achieved solely by similarity based assignment of functions to ORFs. It allows resolution of inconsistencies between different types of analysis and often results in changes from the initial gene function assignments based on similarity scoring (11). By using the WIT-Pro/EMP genome analysis system, a major part of the central metabolism of a microbe can be reconstructed in silico without input of direct experimental data.
Two strategies have been used for bacterial genome sequencing: whole-genome shot-gun (1) and ordered assembly (3, 12). Both of these approaches were intended to produce a highly accurate complete sequence without gaps or ambiguous areas. The complete sequence usually requires an 8- to 10-fold average sequence coverage with extensive effort needed to bridge the final gaps. A major purpose of the present study is to determine whether a gapped genomic sequence, in which 95–98% of the total genome sequence is represented as a set of unlinked contigs, resulting from a lower redundancy of coverage, can provide a useful way to study microbial genomes. Current sets of bioinformatics tools and data from an increasing number of available genomes appear to have shifted the balance from a need for complete genomes to a situation in which gapped genomes are sufficient for many purposes.
The genomic sequence of T. ferrooxidans strain ATCC23270 was used to demonstrate the utility of gapped genome data. T. ferrooxidans is a major component in the consortia of microorganisms used in biomining (13–16) and a contributor to acid mine runoff, which results in pollution near metal and coal mines and other related environments. Microbial biomining has become an established and major part of copper ore extraction worldwide. Recently, microbes have been successfully exploited for the recovery of gold by bioleaching from arsenopyrite ores and also zinc bioleaching. Microbial gold biomining has grown from nothing 10 years ago to perhaps 30% of the world's gold production today (14, 15). T. ferrooxidans generally functions in a consortium of several microbes (14, 16). As the first biomining microbe used for detailed laboratory molecular biology studies (17), T. ferrooxidans was an ideal target microbe for sequencing and analysis (http://igweb.integratedgenomics.com/IGwit/). T. ferrooxidans is a chemoautotroph that gains energy by oxidative phosphorylation, nitrogen from N2 in the air, and carbon exclusively from CO2 fixation via ribulose-1,5-bisphosphate carboxylase and a classic Calvin cycle (14, 15, 17). It derives energy from oxidation of reduced sulfur to H2SO4 and oxidation of Fe2+ to Fe3+ [which precipitates as insoluble Fe(OH)3]. T. ferrooxidans strains have single chromosomes, which may range in genome size from 2.2 Mbp to 2.9 Mbp (18).
Reconstruction of the biosynthesis of the 20 standard amino acids by T. ferrooxidans was chosen as a test of the ability to predict the properties of an organism based on its gapped-genome sequence analyzed in the WIT environment. Although most genes needed for the enzymes of biosynthesis for all 20 amino acids could be identified, 10 genes are currently missing from the list of 150 genes coding for the metabolic transformations necessary for amino acid biosynthesis from glucose, sulfide, and ammonia. The 150 polypeptides catalyze 114 separate reactions because there are many multi-heterosubunit enzymes. An important question as this work progresses is whether completion of the predicted missing 100 kbp (3.7%) of DNA sequence (sequencing of the same T. ferrooxidans strain has begun at The Institute for Genome Research) will yield these 10 missing genes or whether our inability to recognize them is due to nonorthologous alternative gene products carrying out the same or equivalent reactions (11, 19).
Materials and Methods
Chromosomal DNA Extraction and Cloning.
Chromosomal DNA from a culture of T. ferrooxidans strain ATCC23270 (provided by Robert C. Blake II, Xavier University, New Orleans) was prepared as described (20). DNA (10 mg) were digested by using Sau3A endonuclease under conditions of partial cleavage. Approximately 40-kbp fragments were size-selected by using sucrose gradient centrifugation and cloned into dephosphorylated cosmid vector Lorist 6 (21) using cosmid arm cloning (22). Hybrid cosmids were individually harvested in 96-well plates and stored at −70°C. Plasmid cloning was done similarly except DNA fragmentation for approximately one-half of the clones was done by using the HydroShear device (GeneMachines, San Carlos, CA). Size-selected 4-kbp DNA fragments were separated by agarose gel electrophoresis and inserted into the dephosphorylated plasmid vector pGEM3. Transformant colonies were tested for the lack of α-complementation of defective β-galactosidase in Escherichia coli strain DH5, individually collected, and stored.
Plasmid DNA Extraction and Sequencing.
Plasmid and cosmid DNA were extracted using the REAL method (Qiagen, Valencia, CA) with modifications. A first round of sequences was generated from the ends of these subclones using specifically designed “forward” and “reverse” primers located close to the cloning site. Sequencing reactions used a Perkin–Elmer (Foster City, CA) Big Dye kit for fluorescent sequencing. Products of the reactions were analyzed by using an Applied Biosystems model 377 automated sequencer. Average read lengths used were 534 bases. After this step, 864 primer-walking sequencing reactions were added. Primers were designed with the help of the PrimerSelect module of the lasergene (DNA Star, Madison WI) program and by an automated primer-selecting module for sequencher (GeneCodes, Ann Arbor, MI) developed by Randal Cox (University of Chicago). Primers were synthesized by Genosys (The Woodlands, TX) and Operon Technologies (Alameda, CA).
Results
Genome Cloning, Sequencing, ORF Identification, and Primary Functional Assignment.
Genomic DNA of T. ferrooxidans strain ATCC23270 was digested and cloned as described above. Then 9,600 plasmid and 960 cosmid clones were individually transferred to 96-well plates and their DNA was extracted and subjected to dye-terminator sequencing by using two insert-flanking oligonucleotide primers. The success rate (measured as the number of sequencing reactions included in the assembly process) was 91% and the average reading length used was 534 bases, which provided 3.2-times coverage for the 2.7-Mbp predicted genome size of T. ferrooxidans (18).
Sequence assembly was performed first using the sequencher 3.0 program (GeneCodes, Ann Arbor, MI); this was substituted by the phred/phrapp system (http://genome.washington.edu/) subsequently. Primary contigs (1,912) were generated by this assembly process. Primers (864) complementary to the ends of the primary contigs were used for two rounds of primer walking, which closed 559 gaps, leaving the final 1,353 contigs. The size distribution of these contigs is shown in Table 1. With 1,353 contigs, there are 1,353 gaps remaining in the expected single circular chromosome of T. ferrooxidans. With an expected 100 kbp of sequence remaining to be completed, by comparing the 2,611 kbp total in Table 1 with the expected total of 2.7 Mbp (ref. 18), we conclude that the average remaining gap is ≈75 bp, consistent with our experience with other gapped sequences, in which the coverage of genes is nearly complete and the gaps surprisingly small (analysis not shown).
Table 1.
Size distribution of the assembled contigs
| Size range | No. of contigs | Total DNA, kb | Genome fraction, % |
|---|---|---|---|
| >10 kb | 37 | 484 | 18.6 |
| 5–10 kb | 84 | 578 | 22.1 |
| 3–5 kb | 100 | 365 | 14.0 |
| 1–3 kb | 375 | 721 | 27.6 |
| <1 kb | 757 | 463 | 17.7 |
| Total | 1,353 | 2,611 | 100 |
A gene-searching program called critica (23) was used to translate the DNA sequences of each contig in all six reading frames and to tentatively identify the initial, most reliable subset of potential genes based on their high similarity to proteins in the nonredundant database. These ORFs were used as a learning set to reveal codon biases in coding sequences, which, together with potential ribosome-binding sequences (24), were then used to detect all potential genes in the T. ferrooxidans DNA sequence.
Potential coding regions recognized in the DNA contigs were subjected to a fasta search against the nonredundant database of assigned genes and loaded into the WIT-Pro system together with the computed tables of best hits. WIT-Pro is a derivative of the WIT-Pro/EMP genome analysis system (available on http://igweb.integratedgenomics.com/IGwit/), which provides an organized set of tools for the characterization of gene structures and functions. One can formulate queries based on the metabolic content predicted for the sequenced organism by looking for specific functions missing from the metabolic pathways or by separating alternative gene functions derived from similarities found for a putative gene. Chromosomal clustering of functionally related genes is another component of the system (9).
An Overview of the T. ferrooxidans Genome.
The 1,353 contigs comprising 2,611,378 bp were assembled for the T. ferrooxidans genome sequence (Table 1). A comparison of genome statistics for T. ferrooxidans, E. coli, and Bacillus subtilis is shown in Table 2. Then, 2,712 potential genes were recognized in the T. ferrooxidans sequence, which gives about the same gene density (1 ORF per 963 bp) as in the completely sequenced E. coli and B. subtilis genomes with 4,289 and 4,083 genes, respectively. However, some of the T. ferrooxidans ORFs are in fact only gene fragments separated by stop codons introduced by sequencing errors or by contig ends. Our study of amino acid biosynthesis found ≈10% of such disrupted ORFs.
Table 2.
Comparison of the T. ferrooxidans ATCC 23270 genome with those of E. coli K-12 and B. subtilis 168
| T. ferrooxidans | E. coli | B. subtilis | |
|---|---|---|---|
| Genome size, Mb | 2.7 | 4.7 | 4.2 |
| DNA assembled, % | 96 | 100 | 100 |
| Total ORFs | 2,712 | 4,289 | 4,083 |
| Assigned ORFs | 2,159 | 3,499 | 3,016 |
| Asserted pathways | 305 | 906 | 82 |
| Missing assignments* | 95 | 102 | 178 |
| No sequences† | 49 | 233 | 173 |
These ORFs cannot yet be assigned functions by using all the annotation tools in WIT.
† These represent known physiological functions for which no ORF can be assigned at present.
The 2,159 (80%) of the T. ferrooxidans genes could be given a functional assignment, based on an assigned similar ORF in the data base. This ratio of assigned genes to the total number of potential genes is close to that seen for the complete genomes of E. coli (82%) and B. subtilis (73%). These high values reflect the power of the iterative WIT-Pro analytical process together with the rapidly growing database. Approximately 900 of the assigned genes were connected into functional pathways. The remaining identified genes include those coding for structural proteins, for proteins involved in macromolecular biosynthesis and degradation, and some membrane transporters. The 900 genes coding for metabolic enzymes are mostly for pathways that are well defined. The assigned genes could be connected into 305 functional pathways. A linear pathway is a sequence of biochemical transformations from one metabolic branching point to another. A branch point occurs when a metabolite is produced by or consumed in more than a single biochemical transformation. The 95 steps in the required predicted functions for T. ferrooxidans do not have assigned ORFs, which could encode the corresponding enzyme or structural gene. This number is less than that for E. coli or B. subtilis (Table 2) and about the same relative to genome size. This comparison suggests that most of these missing genes are not likely to result from genome gaps. The largest number of missing genes probably results from nonorthologous gene displacements (19), so that functionally active genes are present but cannot be recognized. Another small group of missing genes marked as “no sequences” in Table 2 encode metabolic functions without associated sequence data in any organism; these represent cases where the biochemical activity has been demonstrated or predicted, but for which no corresponding sequence information is available. These last two classes constitute an unrecoverable hole in the current metabolic reconstruction process.
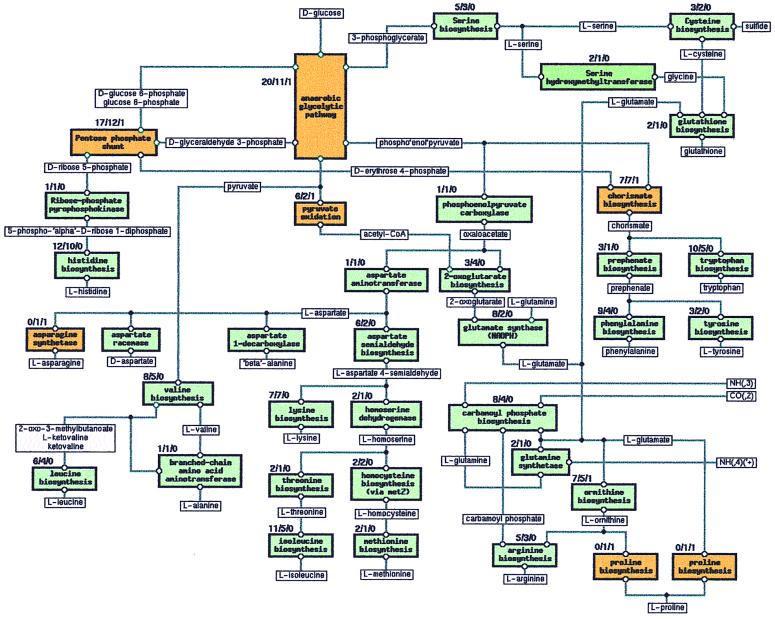
An overview of amino acid metabolism in T. ferrooxidans is shown in Fig. 1. This view can also be found on our web site, together with connections to each pathway of metabolism (by clicking on each box in Fig. 1) and, through these, to each of the 167 individual ORFs involved in amino acid biosynthesis. Each ORF name is listed along with its corresponding EC number and descriptive name, the DNA sequence, and the translated amino acid sequence. Fig. 1 displays the biosynthesis of all 20 amino acids from glucose, ammonia, CO2, and sulfide. Amino acid metabolism is split into 35 pathways (boxes in Fig. 1), according to the definition given above. Biosynthesis of the 20 amino acids requires 114 biochemical reactions (i.e., EC numbers), catalyzed by 150 polypeptides. Each box in Fig. 1 contains three numbers: to the left are the numbers of ORFs currently assigned to each pathway and available on http://www.integratedgenomics.com/FE; in the middle are the number of steps in the pathway (which can also be seen on the web site, by clicking first on the box and then on “diagram picture” in the upper left of the frame). The 167 ORFs determine 150 distinct polypeptides due to the presence of isozymes and split genes. Approximately 20 of the 167 potential genes recognized in T. ferrooxidans to be involved in amino acid metabolism are fragments of genes split by sequencing errors, which generate false stops. Ten genes are missing in our reconstruction of the amino acid metabolism: EC 2.7.1.11 in glycolysis; EC 1.8.1.4 in pyruvate dehydrogenase; EC 2.7.1.71 in chorismate biosynthesis; EC 2.6.1.57 and 1.3.1.43 in tyrosine biosynthesis; EC 2.7.1.39 in threonine biosynthesis; EC 1.2.1.38 in ornithine biosynthesis; EC 4.3.1.12 and EC 1.5.1.2 for proline biosynthesis from ornithine and from glutamate respectively, and EC 6.3.5.4 for asparagine synthetase.
Figure 1.
Block diagram of amino acid metabolism of T. ferrooxidans. Numbers on top of each pathway box are: total number of related ORFs in the T. ferrooxidans genome/number of steps in the pathway/number of currently missing ORFs. Yellow boxes represent metabolic blocks with one or more missing ORFs. Green boxes represent metabolic blocks with all required ORFs present. Each box, displayed at the IG Internet site, can be selected to show detailed data.
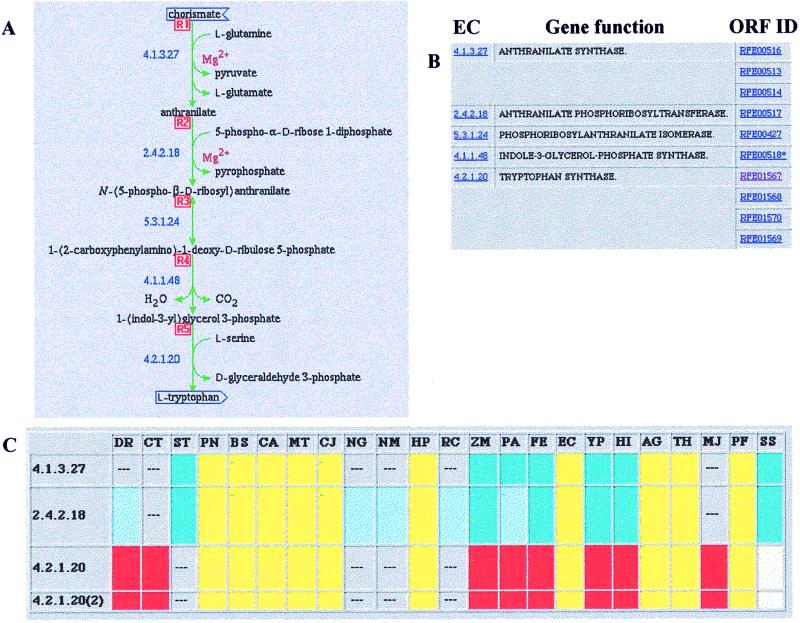
Each box in Fig. 1 can be converted (by selecting the corresponding box displayed on the Internet site) to a more detailed picture such as those shown in Figs. 2 and 3, in which the biosynthesis of tryptophan and histidine are displayed. The metabolic pathway diagrams in Figs. 2A and 3A are also available on the Internet site, as are the EC enzyme assignments, gene functions, and ORF identification numbers. In some cases, the corresponding gene in E. coli (EC) or B. subtilis (BS) is listed in Figs. 2B and 3B. The asterisk on ORF RFE00518* in Fig. 2B has no meaning within the context of this paper. The gene clustering patterns for 22 (Fig. 2C) or 28 additional microbes (Fig. 3C) are not currently linked in these diagrams, but they are available on the WIT/EMP site (http://igweb.integratedgenomics.com/IGwit/). For the example of tryptophan biosynthesis in T. ferrooxidans, the assignments remained the same after a second metabolic reconstruction step. The process of refining assignments is ongoing. However, even some weak initial hits by similarity searches were confirmed both by chromosomal clustering of the genes and by the position of their products in the metabolic pathway. The data in this report, on the http://www.integratedgenomics.com/FE site, and access to the remainder of the T. ferrooxidans data have been made available to T. ferrooxidans researchers. Interested parties are encouraged to contact the corresponding author.
Figure 2.
Tryptophan synthesis in T. ferrooxidans. (A) The substrates and products of different enzymatic reactions together with the EC numbers for the enzymes involved. (B) The EC enzyme numbers, names of the enzymes, and “RFE” ORF assignment number for the gene product in the Thiobacillus database. Any item in blue can be selected on the IG Internet site to reveal deeper information. (C) Functional coupling of genes for tryptophan biosynthesis in different microorganisms. Identical colors indicate genes in corresponding clusters in each organism; the colors are arbitrary. The dashes indicate corresponding assignments not clustered with others in that organism. Note also that the diagram of A and ORF assignments from B can be brought up by way of Fig. 1. DR, Dienococcus radiodurans; CT, cyanobacterium Synechocystis sp.; ST, Streptococcus pyogenes; PN, Streptococcus pneumoniae; BS, Bacillus subtilis; CA, Clostridium acetobutylicum; MT, Mycobacterium tuberculosis; CJ, Campylobacter jejuni; NG, Neisseria gonorrhoeae; NM, Neisseria meningitidis; HP, Helicobacter pylori; RC, Rhodobacter capsulatus; ZM, Zymomonas mobilis; PA, Pseudomonas aeruginosa; FE, Thiobacillus ferrooxidans; EC, Escherichia coli; YP, Yersinia pestis; HI, Haemophilus influenzae; AG, Archaeoglobus fulgidus; TH, Methanobacterium thermoautotrophicum; MJ, Methanococcus jannaschii; PF, Pyrococcus furiosus; and SS, Sulfolobus solfataricus.
Figure 3.
Histidine synthesis in T. ferrooxidans. Legend as in Fig. 2. Additional microorganism data bases used were BA, Buchnera aphidicola; BP, Bordetella pertussis; CL, Cervus elaphus;; KL, Kluyveromyces lactis; LL, Lactococcus lactis; MB, Mycobacterium bovis; MI, Mycobacterium smegmatis; MN, Streptococcus mutans; SA, Staphylococcus aureus; SX, Staphylococcus xylosus; SY, Synechococcus sp.; TY, Salmonella typhimurium; and XA, Xanthomonas campestris.
Discussion
A gapped genome sequence was used to reconstruct some of the metabolic pathways of T. ferrooxidans, with the biosynthesis of the 20 amino acids of proteins shown in detail. The value of gapped genomes, which can be obtained at a fraction of the cost of complete genomes, has been debated. An equivalent discussion is occurring for the human genome sequence and it has recently been recognized that the first 10% of dollars spent can generate ≈90% of the total data (25, 26). The conclusion from these discussions can be different for academic, government, and industrial genome communities because the needs and goals are different. Clearly, there are specific problems, such as the characterization of phylogenetic diversity amongst microbial types, for which a large number of sequenced genomes is critical. However, the cost of complete and error-free genome sequencing is unnecessarily high for some of these applications. The emphasis on complete genome sequences was important at the beginning of these studies 5 years ago because it produced an accurate reference data set, which provides the basis for analysis of gapped genomes today.
The current paper shows that gapped genome sequencing provides an effective approach to microbial genome analysis. It is clear that such sequencing, here with 3.2-times coverage, produces more errors than complete genome sequencing with 8- to 10-fold coverage. The current error rate is estimated to be 1 per 1,000–2,000 base pairs vs. 1 in 10,000 base pairs for complete sequencing. These numbers are based on our calculations, our experience with recently corrected errors, and comparisons of comparable but redundant data for partially sequenced genomes from different groups (analysis not shown). The sequencing errors that produce frameshifts and truncated ORFs are most readily identified by similarity analysis, which includes consideration of predicted product length.
The gapped sequence contains part of almost every ORF, which allows the assignment of functions to almost all ORFs and the accurate reconstruction of the metabolism of the organism. Given the pattern of short intervals between genes in other microbial genomes and in the current T. ferrooxidans sequence, most gaps occur within genes. With only 3.6% of the DNA remaining to be sequenced, one expects <10% of the gaps to be found between genes. Of course, considerations of the distribution of intergenic spacings and of gaps associated with localized cloning and sequencing difficulties make a more precise calculation based on random statistics inappropriate. The inter-gene regions of complete genomes have been subject to quantitatively greater reinterpretation, due to mis-callings of start sites, than have intra-gene sequences (compare refs. 2 and 5).
The functional reconstruction of the metabolism of T. ferrooxidans from its genome sequence provides a foundation for research aimed at the modification of metabolism for more efficient use of T. ferrooxidans in the mining industry. It may be possible to predict experimental approaches to improving cell growth, based on the understanding of central metabolism, which in turn would be based on the reconstruction of metabolism from the genome. Predicted growth rate-limiting metabolic pathways and steps might be altered by transferring mutant genes or increasing gene number, given the possibility of better methods for introducing genes into T. ferrooxidans. Improving cell growth by altering amino acid metabolism, as analyzed in this report, or carbon flow and hence the yield of substances produced via secondary pathways, could alleviate limited growth and bio-leaching associated with wild-type T. ferrooxidans. The data in this report, the Internet site http://www.integratedgenomics.com/FE, and the remainder of this database have already been made available to researchers with interests in T. ferrooxidans.
Abbreviation
- Mbp
megabase pairs
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF246306–AF246444). Additional detailed information about these sequences is available at http://www.integratedgenomics.com/FE.
References
- 1.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 2.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 3.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, et al. DNA Res. 1996;3:185–209. doi: 10.1093/dnares/3.3.185. [DOI] [PubMed] [Google Scholar]
- 4.Vlcek C, Paces V, Maltsev N, Haselkorn R, Fonstein M. Proc Natl Acad Sci USA. 1997;94:9384–9388. doi: 10.1073/pnas.94.17.9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selkov E, Maltsev N, Olsen G J, Overbeek R, Whitman W B. Gene. 1997;197:GC11–GC26. doi: 10.1016/s0378-1119(97)00307-7. [DOI] [PubMed] [Google Scholar]
- 6.Winzeler E A, Richards D R, Conway A R, Goldstein A L, Kalman S, McCullough M J, McCusker J H, Stevens D A, Wodicka L, Lockhart D J, et al. Science. 1998;281:1194–1197. doi: 10.1126/science.281.5380.1194. [DOI] [PubMed] [Google Scholar]
- 7.Kumar V, Fonstein M, Haselkorn R. Nature (London) 1996;381:653–654. doi: 10.1038/381653a0. [DOI] [PubMed] [Google Scholar]
- 8.Oliver S G. Curr Opin Genet Dev. 1997;7:405–409. doi: 10.1016/s0959-437x(97)80156-6. [DOI] [PubMed] [Google Scholar]
- 9.Overbeek R, Fonstein M, D'Souza M, Pusch G D, Maltsev N. Proc Natl Acad Sci USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tatusov R L, Koonin E V, Lipman D J. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 11.Selkov E, Basmanova S, Gaasterland T, Goryanin I, Gretchkin Y, Maltsev N, Nenashev V, Overbeek R, Panyushkina E, Pronevitch L, et al. Nucleic Acids Res. 1996;24:26–28. doi: 10.1093/nar/24.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blattner F R, Plunkett G, III, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 13.Rawlings D E, Silver S. Nat Biotechnol. 1995;13:773–778. [Google Scholar]
- 14.Rawlings D E, editor. Biomining: Theory, Microbes and Industrial Processes. Berlin: Springer; 1997. [Google Scholar]
- 15.Holmes D S. In: Bioconversion of Waste Materials to Industrial Products. 2nd Ed. Martin A M, editor. London: Blackie Academic and Professional; 1998. pp. 517–545. [Google Scholar]
- 16.Rawlings D E, Tributsch H, Hansford G S. Microbiology. 1999;145:5–13. doi: 10.1099/13500872-145-1-5. [DOI] [PubMed] [Google Scholar]
- 17.Rawlings D E, Kusano T. Microbiol Rev. 1994;58:39–55. doi: 10.1128/mr.58.1.39-55.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amils R, Irazabal N, Moreira D, Abad J P, Marvin I. Biochimie. 1998;80:911–921. doi: 10.1016/s0300-9084(00)88888-4. [DOI] [PubMed] [Google Scholar]
- 19.Koonin E V, Mushegian A R, Bork P. Trends Genet. 1996;12:334–336. [PubMed] [Google Scholar]
- 20.Sambrook J, Fritch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 21.Gibson T J, Rosenthal A, Waterston R H. Gene. 1987;53:283–286. doi: 10.1016/0378-1119(87)90017-5. [DOI] [PubMed] [Google Scholar]
- 22.Fonstein M, Zheng S, Haselkorn R. J Bacteriol. 1992;174:4070–4077. doi: 10.1128/jb.174.12.4070-4077.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Badger J H, Olsen G J. Mol Biol Evol. 1999;16:512–524. doi: 10.1093/oxfordjournals.molbev.a026133. [DOI] [PubMed] [Google Scholar]
- 24.Shine J, Dalgarno L. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venter J C, Adams M D, Sutton G G, Kerlavage A R, Smith H O, Hunkapiller M. Science. 1998;280:1540–1542. doi: 10.1126/science.280.5369.1540. [DOI] [PubMed] [Google Scholar]
- 26.Collins F S, Patrinos A, Jordan E, Chakravarti A, Gesteland R, Walters L. Science. 1998;282:682–689. doi: 10.1126/science.282.5389.682. [DOI] [PubMed] [Google Scholar]