Abstract
N-acylhomoserine lactones, known as autoinducers (AIs), are widely conserved signal molecules present in quorum-sensing systems of many Gram-negative bacteria. AIs are involved in the regulation of diverse biological functions, including expression of pathogenic genes in the plant pathogens Pseudomonas solanacearum, several Erwinia species, and the human pathogen Pseudomonas aeruginosa. A bacterial isolate, Bacillus sp. 240B1, is capable of enzymatic inactivation of AIs. The gene (aiiA) for AI inactivation from Bacillus sp. 240B1 has been cloned and shown to encode a protein of 250 amino acids. Sequence alignment indicates that AiiA contains a “HXHXDH” zinc-binding motif that is conserved in several groups of metallohydrolases. Site-directed mutagenesis showed that conserved aspartate and most histidine residues are required for AiiA activity. Expression of aiiA in transformed Erwinia carotovora strain SCG1 significantly reduces the release of AI, decreases extracellular pectolytic enzyme activities, and attenuates pathogenicity on potato, eggplant, Chinese cabbage, carrot, celery, cauliflower, and tobacco. Our results indicate that the AI-inactivation approach represents a promising strategy for prevention of diseases in which virulence is regulated by AIs.
Cell-to-cell communication by means of small signal molecules not only is of vital importance to multicelled organisms such as animals and plants, it also plays important roles in functional coordination among family members of single-celled organisms such as bacteria. Rapid progress over the last few years has established that N-acylhomoserine lactones, known as autoinducers (AIs), are widely conserved signal molecules present in quorum-sensing systems of many Gram-negative bacteria. AIs were originally found in marine bacteria (Vibrio species) in the regulation of bioluminescence (1, 2). In recent years, AIs have been identified in several Gram-negative bacteria. AIs are involved in the regulation of a range of biological functions, including Ti plasmid conjugal transfer in Agrobacterium tumefaciens (3), induction of virulence genes in Erwinia carotovora, Erwinia chrysanthemi, Erwinia stewartii, Pseudomonas aeruginosa, Pseudomonas solanacearum, and Xenorhabdus nematophilus (4–12), regulation of antibiotic production in Pseudomonas aureofaciens and E. carotovora (10, 13), regulation of swarming motility in Serratia liquefaciens (14), and biofilm formation in Pseudomonas fluorescens and P. aeruginosa (15, 16). Many more bacterial species are known to produce AIs, but the relevant biological functions have not yet been established (17–19).
Different bacterial species can produce different AIs. All AI derivatives share identical homoserine lactone moieties but differ in the length and structure of their acyl groups. The biological functions regulated by AIs are of considerable scientific, economic, and medical importance. We are interested in the manipulation of this gene regulatory system for disease control. Although the target genes regulated by AIs are extremely varied, the basic mechanism of AIs biosynthesis and gene regulation seems to be conserved in different bacteria. The general feature of gene regulation by AIs is cell-density dependence, also known as quorum sensing. At low cell densities, the AIs are at low concentrations, and, at high cell densities, the AIs can accumulate to a concentration sufficient for activation of related regulatory genes (20). Because the concentration of AIs is a key factor in determining virulence gene expression in several pathogenic bacteria, it is possible to develop a strategy for disease control by controlling production of AIs or eliminating AIs produced by pathogenic bacteria. To test this possibility, E. carotovora was selected as the target organism. The pathogenicity of these pathogens is correlated with their ability to produce and secrete plant cell wall-degrading enzymes (21, 22). E. carotovora mutants that are defective in the production of AI are avirulent (4). Here, we report that a gene encoding autoinducer inactivation (aiiA) has been cloned from the Gram-positive bacterium Bacillus sp. 240B1. Our results show that the aiiA gene product inhibits virulence of E. carotovora when expressed in the pathogen.
Materials and Methods
Bacterial Strains and Media.
Bacillus sp. 240B1 was isolated from a soil sample. E. carotovora strain SCG1 was isolated from Chinese cabbage leaves showing soft rot symptoms. Escherichia coli strain DH5α was used as a host for DNA cloning and subcloning. A. tumefaciens strain NT1(traR; tra∷lacZ749) was used as an indicator in the bioassay for AI activity (23). Escherichia coli was cultured in LB medium at 37°C, and other strains were cultured in LB at 28°C. A minimal medium for bioassay of AIs has been described (3). The minimal medium supports growth of Agrobacterium and Erwinia but not Escherichia coli. Appropriate antibiotics were added as indicated at the following concentrations: ampicillin, 100 μg/ml; tetracycline, 20 μg/ml; and kanamycin, 50 μg/ml.
Bacteria Screening and Bioassay of AI Activity.
Three previously described AIs, N-β-oxohexanoyl-l-homoserine lactone (OHHL), N-β-oxodecanoyl-l-homoserine lactone (ODHL), and N-β-oxooctanoyl-l-homoserine lactone (OOHL) were used (3), and OOHL was used in routine screening. Soil and plant samples, which were finely chopped when necessary, were suspended in sterilized water with shaking for 1 h before spreading over YEB (yeast extract, 5 g/liter; casein hydrolysate, 10 g/liter; NaCl, 5 g/liter; sucrose, 5 g/liter; MgSO4⋅7H2O, 0.5 g/liter; and agar, 15 g/liter) plates. Visibly distinct colony types from each sample were restreaked to ensure purity of isolates. Bacterial isolates were cultured in 1.5-ml Eppendorf tubes or 96-well assay plates with LB medium plus appropriate antibiotics at 30°C with gentle shaking. After incubation for 16–40 h, the bacterial culture was mixed with an equal volume of fresh medium containing AI (40 μM) at a final volume of 40 μl. The mixtures were incubated at 28°C for 4 h, followed by 60-min sterilization under UV light. Plates containing 20 ml of minimal agar medium supplemented with 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-Gal, 40 μg/ml) were used for the bioassay. The solidified medium in the plates was cut into separated slices (1 cm in width). The sterilized reaction mixture or cell-free suspension (5 μl) was added to one end of an agar slice, and then the cultures of the AI indicator strain were spotted (0.6 μl of OD600 ≈ 0.4) at progressively further distances from the loaded samples. The plates were incubated at 28°C for 24 h. The distance (x) from the last induced blue colony to the origin of the AI sample in each agar slice was measured. The relative amounts of AI were quantified from the distance (x) by using the formula: AI (ng) = 0.673 × 10(1.036x). This relationship was established by adding known amounts of OOHL to the bioassay plates and determining the distance of blue colonies from the origin. The correlation coefficient (r2) of the exponential equation is 0.99. For determination of AI production ability of wild-type and genetically modified Erwinia strains, the same bioassay procedure was used, except that no AI was added to the bacterial culture.
Cloning and Sequencing aiiA Gene.
Genomic DNA from strain 240B1 was digested partially with EcoRI. DNA fragments were ligated to the dephosphorylated EcoRI site of cosmid vector pLAFR3 (24). Ligated DNA was packaged with Gigapack III XL Packaging Extract (Stratagene) and transfected into Escherichia coli DH5α. Cosmid clones with AI inactivation activity were identified by using the bioassay method described above. Subcloning into the sequencing vector pGEM-7Zf(+) was carried out by routine techniques (25). Deletion analysis was carried out by using the DNase I method as described by Lin et al. (26). Sequencing was performed on both strands by using the ABI Prism dRhodamine Terminator Cycle Sequencing Ready Reaction Kit (Perkin–Elmer Applied Biosystems).
Purification of AiiA Protein.
The coding region of the aiiA gene was amplified by PCR using forward primer 5′-ATCGG ATCCA TGACA GTAAA GAAGC TTTAT TTCG-3′, and reverse primer 5′-GTCGA ATTCC TCAAC AAGAT ACTCC TAATG ATGT-3′. The amplified PCR product was digested by BamHI and EcoRI and fused in-frame to the glutathione S-transferase (GST) gene under the control of the isopropyl β-d-thiogalactopyranoside (IPTG)-inducible lac promoter in GST fusion vector pGEX-2T (Amersham Pharmacia). The GST-AiiA fusion protein was purified as described (27).
Site-Directed Mutagenesis.
Site-directed mutagenesis of aiiA was performed by using a QuikChange Site-Directed Mutagenesis kit (Stratagene), following the manufacturer's protocol. Plasmid clone F41, which contains the aiiA gene, was used as a template. See Table 1 for a list of the oligonucleotide primers used for introducing point mutations. Each mutated clone was assayed for its AI-inactivation activity and was sequenced to confirm the mutation.
Table 1.
Oligonucleotide primers for mutagenesis and enzyme activity of mutants
| Mutation* | Primer (5′ to 3′)† | Relative activity, %‡ |
|---|---|---|
| H104 H106 D108 H109 | ||
| Wild type | ATTATTAGTTCT CAC TTG CAT TTT GAT CAT GCAGGAGGAAATGGC | 100.0 |
| H104S/H106S/D108S/H109S | ATTATTAGTTCT TCC TTG TCT TTT TCT TCT GCAGGAGGAAATGGC | 0.0 |
| H104S/H106S/H109S | ATTATTAGTTCT TCC TTG TCT TTT GAT TCT GCAGGAGGAAATGGC | 0.0 |
| D108S/H109S | ATTATTAGTTCT CAC TTG CAT TTT CTT CTT GCAGGAGGAAATGGC | 0.0 |
| H104S/H106S | ATTATTAGTTCT TCC TTG TCT TTT GAT CAT GCAGGAGGAAATGGC | 51.1 |
| H104L/H106L/D108L/H109L | ATTATTAGTTCT CTC TTG CTT TTT CTT CTT GCAGGAGGAAATGGC | 37.9 |
| H104L/H106L/D108L | ATTATTAGTTCT CTC TTG CTT TTT CTT CAT GCAGGAGGAAATGGC | 61.4 |
| H104S | ATTATTAGTTCT TCC TTG CAT TTT GAT CAT GC | 100.0 |
| H106S | ATTATTAGTTCT CAC TTG TCT TTT GAT CAT GC | 61.4 |
| D108S | CT CAC TTG CAT TTT TCT CAT GCAGGAGGAAATGGC | 0.0 |
| D108E | CT CAC TTG CAT TTT GAA CAT GCAGGAGGAAATGGC | 90.8 |
| H109S | CT CAC TTG CAT TTT GAT TCT GCAGGAGGAAATGGC | 0.0 |
| H169 | ||
| Wild type | GCATACACCAGGC CAT ACTCCAGGGCATCAATCG | 100.0 |
| H169S | GCATACACCAGGC TCT ACTCCAGGGCATCAATCG | 61.4 |
The numbers indicate the position of the target amino acids in AiiA peptide; the letters before and after numbers indicate, respectively, the original and substitute amino acids.
† The corresponding conserved amino acids and their positions in AiiA peptide are marked above the primer sequences. Spaces were introduced in the primer sequences for the convenience of inspection. The bases changed were printed boldface and underlined in each primer.
‡ The enzyme bioassay results of each mutant are expressed as activity relative to that of wild-type AiiA; bioassay was repeated three times, and data are means of two replicates.
Genetic Modification of E. carotovora Strain SCG1.
The E7-R3 plasmid, carrying the aiiA gene in the cosmid vector pLAFR3, was transferred into E. carotovora stain SCG1 by triparental mating with the helper strain RK2013. Transconjugants were selected on plates containing minimal medium with tetracycline and were confirmed by PCR with primers specific to aiiA.
Extracellular Pectolytic Enzyme Assay and Virulence Tests.
The activities of extracellular pectate lyase (Pel), pectin lyase (Pnl), and polygalacturonase (Peh) were determined (28). In virulence tests, the actively growing bacteria were centrifuged for 1 min at 3,000 × g and resuspended in LB liquid medium to OD600 = 1.3 (2 × 109 colony-forming units/ml). For inoculation of plants, 4 μl of bacterial suspension was added to a cut surface or wound site. Inoculated plant tissues were incubated in a Petri dish at 28°C for 48 h. For determining pathogenesis of E. carotovora strains on Chinese cabbage, cauliflower, and tobacco plants, the inoculated plants were incubated at 28°C for 3–7 days, and symptoms were recorded daily.
Results
Identification and Cloning of the Gene Responsible for Inactivation of AI.
More than 400 field bacterial isolates and about 100 strains of the laboratory bacterial culture collection have been screened for AI inactivation activity. Twenty-four isolates showed different levels of enzymatic activities for AI inactivation. Bacterial isolate 240B1, which showed a strong ability to eliminate AI activity, was selected for further study. Total protein extracts from isolate 240B1 eliminated AI activity completely during a 1-h incubation, and the capacity of the protein extract to inactivate AI was abolished by treatment with proteinase K for 1 h or boiling for 5 min. These observations indicate enzymatic inactivation of AI by bacterial isolate 240B1. The isolate was taxonomically characterized as Bacillus sp. because of the following characteristics: Gram-positive, flagella peritrichous, catalase positive, facultatively anaerobic, straight rod, endospore formation, and 16S rRNA sequence homology with the members of Bacillus cereus group in the genus Bacillus (X.-Z.L. and L.-H.Z., unpublished observations).
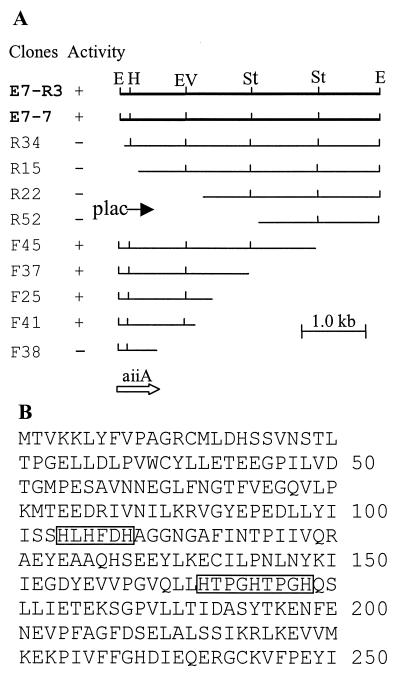
To identify the gene encoding AI inactivation, a cosmid library was constructed in Escherichia coli with the genomic DNA of Bacillus sp. strain 240B1. Twelve hundred clones were screened for AI inactivation activity. Three clones showing AI inactivating function were identified. Restriction analysis showed that the three clones shared one common band of 4.3 kb generated by EcoRI digestion. The bioassay with the subclone E7-7 containing this 4.3-kb EcoRI fragment confirmed that this fragment encodes the AI inactivation function (Fig. 1A). To identify the minimum size and location of the AI inactivation gene (aiiA), a series of deletion clones were generated by deletion from both ends of this 4.3-kb fragment. The results indicated that the aiiA gene is contained in a 1.2-kb fragment in clone F41 (Fig. 1A).
Figure 1.
Deletion analysis of AI inactivation region of Bacillus sp. 240B1 (A) and the predicted amino acid sequence of the aiiA gene product (B). Clone E7-R3 is contained in cosmid vector pLAFR3, whereas the others are in cloning vector pGEM-7Zf(+). The location and direction of Plac promoters in the cosmid and in the pGEM-7Zf(+) clone are indicated by solid arrows. AI inactivation activity of the clones is shown in the second column: +, with AI inactivation activity; −, without AI inactivation activity. Restriction enzymes: E, EcoRI; H, HindIII; Ev, EcoRV; St, StyI. The open arrow indicates the location and transcription direction of the aiiA ORF. In the AiiA protein sequence, two conserved regions are boxed.
aiiA Encodes a Protein Not Found in the Databases.
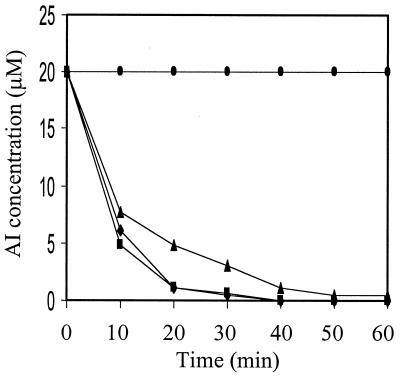
The 1.2-kb DNA insert in clone F41 was completely sequenced from both strands. The predicted amino acid sequence is shown in Fig. 1B. The complete sequence of the DNA insert contains 1,222 bp, and there are four potential ORFs starting from nucleotide positions 1, 42, 156, and 228. Deletion analysis indicated that only the longest ORF encodes AI inactivation function (Fig. 1A). This was confirmed by fusing the longest ORF to the GST gene in the same ORF and testing for AI inactivation activity of the purified fusion protein. The purified AiiA protein effectively inactivated the three AIs tested, i.e., OHHL, ODHL, and OOHL (Fig. 2). This ORF contains a sequence of 750 nt and encodes a protein of 250 aa, with a predicted molecular mass of 28,036 Da and an isoelectric point at 4.7 because of 19 strongly basic and 39 strongly acidic amino acid residues. AiiA is unlikely to be a secretary protein because no hydrophobic signal peptide has been found at its N terminus. This agrees with our observation that no AI-inactivation activity was detected in the supernatants of bacterial cultures of strain 240B1 and Escherichia coli DH5α containing the aiiA gene. The putative initiation codon is preceded at a spacing of 7 bp by a potential ribosome-binding sequence (AAGGTGG), which is complementary to the 3′ end of the Escherichia coli 16S rRNA. The best sequence match (TATTGT) to the consensus −10 promoter element (TATAAT) occurs 35 bp upstream of the initiation codon. A TCTT box following a T-rich region resembling the potential factor-independent termination site is found downstream of the termination codon (29). Database search showed that aiiA has no significant similarity to known sequences in the major databases (GenBank, European Molecular Biology Laboratory, Protein Information Resource, and Swiss-Prot) by fasta and blast analysis at either nucleotide or peptide sequence level. The general homology level to known enzymes is <28%. However, both fasta and blast search data indicated two small conserved regions among the AiiA sequence and several known enzymes, including glyoxalase II, metalloβ-lactamase, and arylsulfatase. The first region is “104HLHFDHAG111” and the second is “165HTPGHTPGH173” (Fig. 1B). Sequence alignment of the first conserved region with 19 peptides in these three families indicates an “HXHXDH” pattern existing among AiiA, glyoxalases II, and arylsulfatases. The same pattern is also found in metallo-β-lactamases, except the histidine residue after aspartic acid residues is not conserved (Fig. 3). The HXHXD sequence is proposed as a zinc-binding motif in glyoxalase II, β-lactamase, and arylsulfatase (30). Sequence alignment of the second region locates one invariable histidine (H169) among AiiA and the three metalloenzymes (Fig. 3).
Figure 2.
Time course of AI inactivation by purified AiiA protein. The purified AiiA protein was diluted to a concentration of 50 ng/μl with 1/15 M phosphate buffer (pH 8.0). Equal volume of diluted AiiA protein and 40 μM OHHL (⧫), ODHL (■), or OOHL (▴) was added in a 1.5-ml Eppendorf centrifuge tube. The reaction was conducted at 28°C. The same concentration of the denatured AiiA (boiling for 5 min) and OHHL in the phosphate buffer was used as control (●). Samples were taken at 10-min intervals for a total of 60 min, and the reaction was stopped by boiling for 3 min. The samples were assayed for AI activity as described (3).
Figure 3.
Multiple alignment of the conserved regions among AiiA and other protein sequences. The numbering is based on the sequence of AiiA. The dashes indicate amino acids identical to AiiA sequence. The consensus amino acids are indicated. In species column, abbreviations are as follows: E, Escherichia; R, Rhodobacter; S, Schistosoma; B, Buchnera; Pse. carr., Pseudoalteromonas carrageenovora; M, Mycobacterium; C, Caenorhabditis; P, Pseudomonas; and Se, Serratia. In the protein column, the abbreviations are: Gly, glyoxalase II; Ary, arylsulfatase; Lac, β-lactamase.
The Conserved Histidine and Aspartate Residues Are Required for AiiA Activity.
Site-directed mutagenesis was used to identify the role of the conserved histidines and aspartate residues in these two regions. We replaced all four invariable residues in the HXHXDH motif with either four non-metal-binding serine residues or four leucine residues. A leucine residue can interact with zinc but with a lower affinity than a histidine residue (31, 32). Table 1 shows that mutating HXHXDH to SXSXSS in the AiiA protein abolished AI inactivation, whereas substitution with LXLXLL led to about 62% loss of enzyme activity, suggesting a critical role played by this motif and that AiiA is likely a metalloenzyme.
Single-residue mutagenesis showed that H106, D108, H109, and H169 are necessary for AiiA activity (Table 1). Single replacement of D108 and H109, respectively, by a serine resulted in complete loss of enzyme activity, whereas mutation of H106 and H169 to serine showed a loss of activity of about 38%. Replacement of the aspartate with glutamine (D108E) led to <10% activity loss, presumably because a glutamine residue can also act as a zinc ligand (33). However, mutant H104S showed the same activity as the wild-type enzyme, indicating that this histidine residue is not involved in AiiA catalytic activity.
Expression of aiiA in E. carotovora Decreases Release of AI and Extracellular Pectolytic Enzymes.
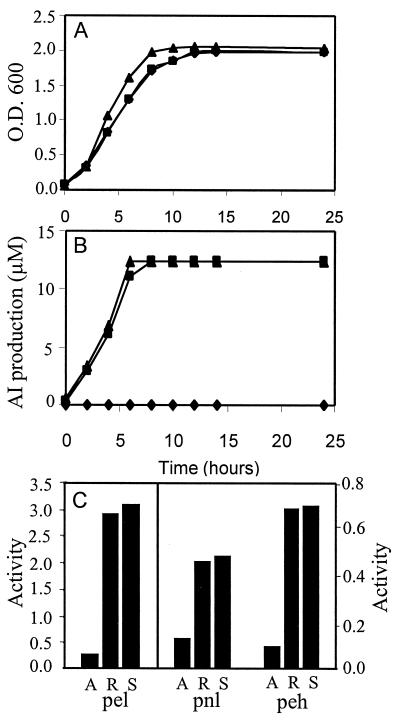
The cosmid clone E7-R3 containing aiiA was transferred into E. carotovora strain SCG1 by using triparental mating. The pLAFR3 vector was stably maintained in E. carotovora without selection pressure. Fig. 4A shows that the bacterial growth of strains SCG1(pLAFR3) and SCG1(E7-R3) was similar but slightly slower than that of wild-type strain SCG1, possibly because of the extra metabolic load for maintaining the cosmid vector. The AI produced by strains SCG1 and SCG1(pLAFR3) reached a maximal level of 12 μM after about 8 h of culture, whereas no AI was detected in the culture supernatant of strain SCG1(E7-R3) (Fig. 4B). After 20 h of culture, the enzyme activities of three extracellular pectolytic enzymes in culture supernatants were determined. Fig. 4C shows that expression of aiiA in E. carotovora SCG1(E7-R3) reduced production of extracellular pectolytic enzymes. The activities of three pectolytic enzymes, Pel, Pnl, and Phe, are 3–10 times lower in E. carotovora SCG1(E7-R3) than in the wild-type strain SCG1 and strain SCG1(pLAFR3).
Figure 4.
Effect of aiiA gene product on cell growth (A), AI production (B), and extracellular pectolytic enzyme activity (C) in E. carotovora strains SCG1(E7-R3) (♦ in A and B, A in C), SCG1(pLAFR3) (■, R), and SCG1 (▴, S). The enzyme activities were expressed as the increase of absorbance at 235 nm (for Pel and Pnl) or 500 nm (for Peh) per minute per milliliter of culture supernatant.
E. carotovora SCG1 Expressing AiiA Loses Virulence in Planta.
E. carotovora SCG1(E7-R3) that expressed AiiA failed to cause or caused only minor soft rot disease symptoms in different detached tissues of potato, eggplant, Chinese cabbage, carrot, and celery, whereas its parental strain caused severe tissue-macerating symptoms. E. carotovora strain SCG1(pLAFR3) that contains only the cosmid vector caused the same level of disease severity as its parental strain E. carotovora strain SCG1 (Table 2). To determine the effect of infection time on symptom development, the bacterial strains were inoculated into intact plants of Chinese cabbage, cauliflower, and tobacco. Soft rot symptoms appeared 1 day after inoculation with wild-type strain SCG1 and strain SCG1(pLAFR3). No significant tissue maceration was detected in plants inoculated with strain SCG1(E7-R3) 1 wk after inoculation. Fig. 5 shows inoculated Chinese cabbages 3 and 6 days after inoculation. Similar results were obtained from inoculated cauliflower and tobacco plants.
Table 2.
Virulence assay of E. carotovora strains on plant tissues
| Plant | Tissue |
Inoculum* | Maceration area, mm2†
|
||
|---|---|---|---|---|---|
| SCG1 (aiiA) | SCGI (pLAFR3) | SCG1 | |||
| Potato | Tuber | 100 | 0 | 136 ± 33 | 151 ± 16 |
| 10−1 | 0 | 98 ± 22 | 156 ± 0 | ||
| Celery | Petiole | 100 | 12 ± 0 | 503 ± 10 | 406 ± 15 |
| 10−1 | 0 | 283 ± 52 | 294 ± 13 | ||
| Carrot | Root | 100 | 0 | 130 ± 28 | 176 ± 50 |
| 10−1 | 0 | 101 ± 21 | 128 ± 21 | ||
| Eggplant | Fruit | 100 | 0 | 380 ± 15 | 480 ± 75 |
| 10−1 | 0 | 349 ± 29 | 370 ± 44 | ||
| Chinese cabbage | Leaf | 100 | 6 ± 1 | 308 ± 29 | 320 ± 39 |
| 10−1 | 0 | 148 ± 23 | 153 ± 5 | ||
The 100 inoculum equals to 2 × 109 colony-forming units/ml, which was diluted 10 times to prepare 10−1 inoculum.
† The maceration areas were measured 48 h after inoculation by taking 1:1 photographs; data are means of three replicates.
Figure 5.
Effect of aiiA gene expression on E. carotovora pathogenicity. From left to right, Chinese cabbage inoculated, respectively, with 10 μl of bacterial inoculum (2 × 109 colony-forming units/ml) of SCG1(E7-R3), SCG1(pLAFR3), and SCG1. The photographs were taken 3 (Upper) and 6 (Lower) days after inoculation.
Discussion
Bacterial isolate 240B1, identified as a Bacillus sp., produces an enzyme that can effectively inactivate the three AIs tested. The gene (aiiA) encoding the AI inactivation enzyme has been cloned and fully sequenced. Expression of aiiA in transformed Escherichia coli and the plant pathogenic bacterium E. carotovora results in AI inactivation and significantly reduces AI release from E. carotovora. To our knowledge, it is the first protein identified capable of enzymatic inactivation of N-acylhomoserine lactones, the AIs for global gene regulation in diverse bacterial species.
AiiA has no significant homology to known proteins in major databases. However, a conserved HXHXDH motif was found in AiiA and members of glyoxalase II and arylsulfatase families, and in β-lactamases as well but less conserved (Fig. 3). Glyoxalase II and β-lactamase are known to be zinc metalloenzymes. Crystal structure of β-lactamase from Bacillus cereus revealed only a single zinc atom. It showed that the two histidine residues (H86 and H88) in the motif 86HXHXD90 were involved in zinc binding, and the aspartic acid residue (D90) was involved in catalysis (34). Structural knowledge of glyoxalase II is lacking. Metal analysis of Arabidopsis glyoxalase II showed it contains two zinc atoms per monomer (35). Based on these observations, Crowder et al. (35) proposed a potential active site model for glyoxalase II. In this model, two histidines (H135 and H137) and one aspartic acid (D139) in the motif 135HXHXDH140 of glyoxalase II were suggested to be involved in binding to zinc ion. However, this has not been confirmed by site-directed mutagenesis. The same motif 104HXHXDH109 was also found in the AiiA sequence. Our mutagenesis analysis, however, showed that mutating the first histidine (H104S) in the motif did not affect AiiA activity, whereas the last histidine (H109) is important for AiiA catalysis. The results suggest that different enzymes may have subtle changes in active center layout to accommodate different substrates.
Commonly, zinc metalloenzymes require three ligands directly coordinating the zinc ion as well as a fourth ligand, usually H2O (33). The first two ligands are usually separated by a short space of one to three amino acid residues. A third ligand is usually distanced from the second by 20–120 amino acids (33). In the AiiA sequence, several invariant histidines with glutamate residue show a pattern of 104HXHXDH109–≈60 aa–H169. This pattern agrees with the metallohydrolase criterion. These data suggest that AiiA is a metal-binding protein and it may use a reaction mechanism to perform its catalytic function similar to that used by other related metallohydrolases. It is known that glyoxalase II hydrolyzes the thioester linkage between d-lactic acid and glutathione, metallo-β-lactamase cleaves the amide bond of the penicillin β-lactam ring, and arylsulfatase hydrolyzes the sulfate-ester bond of sulfatides. We speculate that AiiA either hydrolyzes the amide linkage between homoserine lactone and its acyl side chain or cleaves the ester bond of the homoserine lactone ring. However, more work needs to be done on AiiA substrate specificity, enzyme kinetics, and reaction products for full characterization of AiiA.
E. carotovora as a plant pathogen produces and secretes exoenzymes that act as virulence determinants for soft rot diseases of various plants, including potato, cabbage, tomato, chili, carrot, celery, onion, lettuce, etc. (22). Mutants defective in production of OHHL were also defective in synthesis of the pectinase, cellulase, and protease exoenzymes. These mutants failed to induce soft rot disease in potato tubers (4). It was found that the expI gene, which is homologous to the luxI gene of Vibrio fischeri, encodes AI production in E. carotovora. An expI mutant was avirulent when inoculated into tobacco leaves, but virulence was restored by external autoinducer addition (6). To determine the impact of aiiA gene on virulence, the cosmid clone containing the aiiA gene was introduced into E. carotovora strain SCG1. Expression of the AiiA enzyme in E. carotovora significantly reduced the release of AIs, decreased extracellular pectolytic enzyme activities, and attenuated soft rot disease symptoms on all plants tested, including potato, eggplant, Chinese cabbage, carrot, and celery (Table 2 and Fig. 5). Our results further support the important role of AIs in the regulation of expression of virulence genes in E. carotovora and the potential of the aiiA gene to confer resistance in plants to soft rot disease and other diseases in which the AIs are involved in regulation of pathogenic gene expression.
The aiiA gene could also be a useful tool for investigation of the role of AIs in those bacteria where the biological functions regulated by AIs have not been established. In recent years, many more bacterial species have been shown to produce AIs (11, 17, 19, 36). Some of them are important plant pathogens such as Pseudomonas and Xanthomonas species. The gene knock-out approach, based on sequence homology, could be difficult. The overall levels of sequence similarity of AI synthase and the related regulatory proteins from different genera are rather low, often no higher than 28–35% identity between LuxI-type proteins and 18–25% identity for LuxR-type proteins (20). However, it is feasible and simple to introduce the aiiA gene into these bacteria to probe the biological functions regulated by AIs.
Acknowledgments
We thank C. K. Dumenyo and A. K. Chatterjee (University of Missouri, Columbia) for kindly providing us the methods for extracellular enzyme assay.
Abbreviations
- AI
autoinducer
- OHHL
N-β-oxohexanoyl-l-homoserine lactone
- ODHL
N-β-oxodecanoyl-l-homoserine lactone
- OOHL
N-β-oxooctanoyl-l-homoserine lactone
- Pel
pectate lyase
- Pnl
pectin lyase
- Peh
polygalacturonase
- GST
glutathione S-transferase
- IPTG
isopropyl β-d-thiogalactopyranoside
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF196486).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.060023897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.060023897
References
- 1.Eberhard A, Burlingame A L, Eberhard C, Kenyon G L, Nealson K H, Oppenheimer N J. Biochemistry. 1981;20:2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- 2.Cao J G, Meighen E A. J Biol Chem. 1989;264:21670–21676. [PubMed] [Google Scholar]
- 3.Zhang L-H, Murphy P J, Kerr A, Tate M E. Nature (London) 1993;362:446–447. doi: 10.1038/362446a0. [DOI] [PubMed] [Google Scholar]
- 4.Jones S M, Yu B, Bainton N J, Birdsall M, Bycroft B W, Chhabra S R, Cox A J R, Golby P, Reeves P J, Stephens S, et al. EMBO J. 1993;12:2477–2482. doi: 10.1002/j.1460-2075.1993.tb05902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Passador L, Cook J M, Gambello M J, Rust L, Iglewski B H. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 6.Pirhonen M, Flego D, Heikinheimo R, Palva E. EMBO J. 1993;12:2467–2476. doi: 10.1002/j.1460-2075.1993.tb05901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearson J P, Gray K M, Passador L, Tucker K D, Eberhard A, Iglewski B H, Greenberg E P. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beck von Bodman S, Farrand S K. J Bacteriol. 1995;177:5000–5008. doi: 10.1128/jb.177.17.5000-5008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flavier A B, Schell M A, Denny T P. Mol Microbiol. 1998;28:475–486. doi: 10.1046/j.1365-2958.1998.00804.x. [DOI] [PubMed] [Google Scholar]
- 10.Costa J M, Loper J E. Can J Microbiol. 1997;43:1164–1171. doi: 10.1139/m97-165. [DOI] [PubMed] [Google Scholar]
- 11.Dunphy G, Miyamoto C, Meighen E. J Bacteriol. 1997;179:5288–5291. doi: 10.1128/jb.179.17.5288-5291.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nasser W, Bouillant M L, Salmond G, Reverchon S. Mol Microbiol. 1998;29:1391–1405. doi: 10.1046/j.1365-2958.1998.01022.x. [DOI] [PubMed] [Google Scholar]
- 13.Pierson L S, III, Keppenne V D, Wood D W. J Bacteriol. 1994;176:3966–3974. doi: 10.1128/jb.176.13.3966-3974.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eberl L, Winson M K, Sternberg C, Stewart G S A B, Christiansen G, Chhabra S R, Bycroft B, Williams P, Molin S, Givskov M. Mol Microbiol. 1996;20:127–136. doi: 10.1111/j.1365-2958.1996.tb02495.x. [DOI] [PubMed] [Google Scholar]
- 15.Allison D G, Ruiz B, SanJose C, Jaspe A, Gilbert P. FEMS Microbiol Lett. 1998;167:179–184. doi: 10.1111/j.1574-6968.1998.tb13225.x. [DOI] [PubMed] [Google Scholar]
- 16.Davies D G, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 17.Bassler B L, Greenberg E P, Stevens A M. J Bacteriol. 1997;179:4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dumenyo C K M, Chun A W, Chatterjee A K. Eur J Plant Pathol. 1998;104:569–582. [Google Scholar]
- 19.Cha C, Gao P, Chen Y C, Shaw P D, Farrand S K. Mol Plant–Microbe Interact. 1998;11:1119–1129. doi: 10.1094/MPMI.1998.11.11.1119. [DOI] [PubMed] [Google Scholar]
- 20.Fuqua C, Winans S C. J Bacteriol. 1996;178:435–440. doi: 10.1128/jb.178.2.435-440.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collmer A K, Keen N T. Annu Rev Phytopathol. 1986;24:383–409. [Google Scholar]
- 22.Kotoujansky A. Annu Rev Phytopathol. 1987;25:405–430. [Google Scholar]
- 23.Piper K R, Beck von Bodman S, Farrand S K. Nature (London) 1993;362:448–450. doi: 10.1038/362448a0. [DOI] [PubMed] [Google Scholar]
- 24.Staskawicz B D, Keen N T, Napoli C. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J F, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 26.Lin H C, Lei S P, Wilcox G. Anal Biochem. 1985;147:114–119. doi: 10.1016/0003-2697(85)90016-8. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L-H, Xu J, Birch R G. Microbiology. 1998;144:555–559. doi: 10.1099/00221287-144-2-555. [DOI] [PubMed] [Google Scholar]
- 28.Chatterjee A, Cui Y, Liu Y, Dumenyo C K, Chatterjee A K. Appl Environ Microbiol. 1995;61:1959–1967. doi: 10.1128/aem.61.5.1959-1967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brendel V, Trifonov E N. Nucleic Acids Res. 1984;12:4411–4427. doi: 10.1093/nar/12.10.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melino S. Trends Biochem Sci. 1998;23:381–382. doi: 10.1016/s0968-0004(98)01264-x. [DOI] [PubMed] [Google Scholar]
- 31.Omburo G A, Jacobitz S, Torphy T J, Colman R W. Cell Signalling. 1998;10:491–497. doi: 10.1016/s0898-6568(97)00175-7. [DOI] [PubMed] [Google Scholar]
- 32.Amrallah A H, Abdalla N A, El-Haty E Y. Talanta. 1998;46:491–500. doi: 10.1016/s0039-9140(97)00218-x. [DOI] [PubMed] [Google Scholar]
- 33.Vallee B L, Galdes A. Adv Enzymol Relat Areas Mol Biol. 1984;56:263–430. doi: 10.1002/9780470123027.ch5. [DOI] [PubMed] [Google Scholar]
- 34.Carfi A, Pares S, Duee E, Galleni M, Duez C, Frere J M, Dideberg O. EMBO J. 1995;14:4914–4921. doi: 10.1002/j.1460-2075.1995.tb00174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crowder M W, Maiti M K, Banovic L, Makaroff C A. FEBS Lett. 1997;418:351–354. doi: 10.1016/s0014-5793(97)01416-6. [DOI] [PubMed] [Google Scholar]
- 36.Surette M G, Bassler B L. Proc Natl Acad Sci USA. 1998;95:7046–7050. doi: 10.1073/pnas.95.12.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]