Abstract
Neuregulins regulate the expression of ligand- and voltage-gated channels in neurons and skeletal muscle by the activation of their cognate tyrosine kinase receptors, ErbB 1–4. The subcellular distribution and mechanisms that regulate the localization of ErbB receptors are unknown. We have found that ErbB receptors are present in brain subcellular fractions enriched for postsynaptic densities (PSD). The ErbB-4 receptor is unique among the ErbB proteins because its C-terminal tail (T-V-V) conforms to a sequence that binds to a protein motif known as the PDZ domain. Using the yeast two-hybrid system, we found that the C-terminal region of ErbB-4 interacts with the three related membrane-associated guanylate kinases (MAGUKs) PSD-95/SAP90, PSD-93/chapsyn-110, and SAP 102, which harbor three PDZ domains, as well as with β2-syntrophin, which has a single PDZ domain. As with N-methyl-d-aspartate (NMDA) receptors, ErbB4 interacts with the first two PDZ domains of PSD-95. Using coimmunoprecipitation assays, we confirmed the direct interactions between ErbB-4 and PSD-95 in transfected heterologous cells, as well as in vivo, where both proteins are coimmunoprecipitated from brain lysates. Moreover, evidence for colocalization of these proteins was also observed by immunofluorescence in cultured hippocampal neurons. ErbB-4 colocalizes with PSD-95 and NMDA receptors at a subset of excitatory synapses apposed to synaptophysin-positive presynaptic terminals. The capacity of ErbB receptors to interact with PDZ-domain proteins at cell junctions is conserved from invertebrates to mammals. As discussed, the interactions found between receptor tyrosine kinases and MAGUKs at neuronal synapses may have important implications for activity-dependent plasticity.
The neuregulins (Nrgs) are a family of genes encoding growth/differentiation factors, composed of four members (Nrg 1–4), related to the epidermal growth factor (1, 2). Differential splicing of Nrg-1 transcripts generates a series of factors that exert a variety of functions during development of the nervous system (1, 3–5). In neurons, Nrg is synthesized in the soma either as membrane-spanning precursors or proteins lacking transmembrane domains, anterogradely transported down axons, and cleaved or released from the presynaptic terminals in an activity-dependent fashion (6, 7). Several genes that encode neurotransmitter receptors and voltage-gated channels in the postsynaptic cell are regulated by Nrg-1 during muscle and neural development. In muscle, Nrg-1 enhances transcription of nicotinic acetylcholine receptor (AChR) δ and ɛ subunits (8–10) and voltage-gated sodium channels (11), and its reduced levels in heterozygote Nrg-mutant mice result in decreased AChR levels at the junction (12). In neurons, expression of the N-methyl-d-aspartate (NMDA) receptor NR2C subunit in cerebellar granule cells (13) and the neuronal AChR α7 subunit in the superior cervical ganglia (14) are up-regulated by Nrg-1. In these cases, addition of recombinant Nrg mimics the effect of the presynaptic terminals.
Nrgs exert their actions by activating a family of receptor tyrosine kinases, known as the ErbB receptors (ErbB 2–4), which are structurally related to the epidermal growth factor receptor (presently called ErbB-1). The cellular distribution, binding affinities for distinct Nrg isoforms and tyrosine kinase activities vary between the ErbB receptors (3, 15). The ErbB-2 receptor has a high kinase activity but fails to directly bind Nrg-1–4, and ErbB-3 binds Nrg-1 and -2 but has low kinase activity, thus requiring heterodimerization for function. ErbB-4 differs from ErbB-2 and -3 in that it binds Nrg either as a homo- or a heterodimer. Furthermore, the ErbB-4 receptor is preferentially activated by Nrg-3 (16), and possibly Nrg-2, which are the Nrg genes that are predominantly expressed in the nervous system (16–19). The downstream targets of Nrg action are best understood in skeletal muscle. During development, Nrg-1 accumulates with ErbB 2–4 receptors at the neuromuscular junction (NMJ) (20–24). Nrg binding results in receptor dimerization and autophosphorylation at tyrosine residues, signaling by means of the Ras/Raf/MEK/MAPK pathway (3, 9, 25), and the phosphorylation of Ets-related factors that activate transcription of AChR genes (26–28).
In the central nervous system, we have reported that Nrg-1β, which accumulates at the presynaptic mossy fiber terminals in the cerebellum (29), is required for the expression of the NMDA receptor NR2C subunit in the postsynaptic granule cells (13). Interestingly, we found that coactivation of both ErbB and NMDA receptors is required for the Nrg-dependent expression of the NR2C subunit gene in cerebellar slice cultures. Because NMDA receptors are confined to the postsynaptic density (PSD) of excitatory synapses in neurons, this raises the possibility that both receptors cross-talk at synapses. There is mounting evidence that the PSD serves as an important structure that scaffolds ion channels and signaling molecules at sites of synaptic transmission (30, 31), where it may function to couple neural activity to synaptic plasticity. A series of cytoplasmic proteins that interact with and may scaffold NMDA receptors at the PSD have recently been identified; these include: PSD-95/SAP-90 (32), CHAPSYN-110/PSD-93 (33), and SAP-102 (34). Members of this family of membrane-associated guanylate kinases (MAGUKs) harbor three PDZ protein–protein interaction domains that interact with a conserved sequence (S/T-X-V) located at the C-terminal tail of NMDA receptor subunits. Among the ErbB receptors, ErbB-4 is unique in that is has a C-terminal sequence (T-V-V) that conforms to the consensus site necessary for the interaction with PDZ domains. Based on these observations, we have used biochemical and immunohistochemical analyses to determine whether ErbB receptors display a synaptic localization and with which proteins they may interact. Herewith, we demonstrate the interaction of ErbB receptors with MAGUKs and their colocalization in PSDs and at synaptic puncta of cultured hippocampal neurons with NMDA receptors. These results indicate that there is a close association of ErbB and NMDA receptors at synaptic sites by their interaction with PDZ domain-containing proteins, consistent with the idea that both receptors may cross-talk at synapses. This interaction of the ErbB-4 receptor tyrosine kinase with MAGUKs at neuronal synapses increases the repertoire of signaling proteins present at the PSD.
Materials and Methods
Antibodies.
The primary antibodies used were: anti-PSD-95 mouse mAb 7E3-1B8 (Affinity BioReagents, Neshanic Station, NJ) and guinea pig antisera (kindly provided by M. Sheng, Harvard Medical School, Boston), rabbit antisera raised against the C termini of human ErbB-2, -3, and -4 (Santa Cruz Biotechnology) and antisera capable of recognizing the truncated ErbB-4 (kindly provided by C. Lai, Scripps Research Institute, La Jolla, CA), anti-NR1 mouse mAb (PharMingen), anti-synaptophysin mouse mAb (Sigma), glial glutamate transporter-1 antisera (Calbiochem), anti-E2F1 mouse mAb, and normal rabbit serum (Santa Cruz Biotechnology). The secondary antibodies raised against mouse, rabbit, and guinea pig IgG were conjugated either to horseradish peroxidase for Western blots (Amersham Pharmacia), or to rhodamine and FITC for immunofluorescence (Jackson ImmunoResearch).
Yeast Two-Hybrid Screening.
The screening and assays were performed with the Matchmaker cloning system and recommendations from the manufacturer (CLONTECH). The cDNA encoding the C-terminal 238 aa of the human ErbB-4 receptor used for screening was obtained with PCR amplification of an ErbB-4 cDNA (kindly provided by J. Pierce and L. M. Wang, National Institutes of Health, Bethesda, MD) with sense (5′-CGAGACCCGGGTTTTGCTGCTGAACAAGGAGTG-3′) and antisense (5′-CCACAGTCGACCTTACACCACAGTATTCCG-3′) primers containing XbaI and SalI restriction sites, respectively. The XmaI/SalI fragment was subcloned in the pAS2–1 vector (GAL4 DNA binding domain) and verified by sequencing. This fusion construct was used to screen, on histidine-free media, 3 × 105 clones of a pretransformed adult human brain cDNA library in pACT2 (GAL4 activation domain). Positive colonies (>1 mm) were reselected on adenine-free media, and the strength of the interactions was reassessed by using a β-galactosidase assay. The positives clones were rescued in KC8 bacteria, transformed into DH5-α bacteria and, after screening for false positives, analyzed by automated DNA sequencing. To evaluate the interaction of the ErbB-4 C terminus with specific PDZ domains of PSD-95, the individual PDZ domains in the vector pGAD10 (kindly provided by M. Sheng) were transfected into yeast harboring the C-terminal ErbB4 construct (see above), and selected in media lacking histidine and adenine.
Subcellular Fractionation, Immunoblotting, and Immunoprecipitation.
Synaptic membranes and PSDs were obtained from adult Sprague–Dawley rats, as described by Rogers et al. (35). The subcellular fraction obtained after hypotonic lysis and centrifugation of membranes on sucrose gradients was designated the synaptic membrane (SM) fraction. The PSD fraction was obtained by extracting SMs with 1.5% Triton X-100 for 30 min, layering on a 28.5% sucrose Tris acetate solution, and centrifuging at 105,000 × g for 1 h. For Western blotting, the SM and PSD fractions were solubilized with 2% SDS, diluted with sample buffer, separated on 6% Tris-glycine gels (NOVEX, San Diego), and electroblotted onto poly(vinylidene difluoride) membranes (Amersham Pharmacia). Membranes were blocked with 5% dry milk in wash buffer (TBS with 0.1% Tween 20) and then incubated sequentially for 1 h at room temperature (overnight for anti-PSD-95) with the primary and secondary antibodies. Immunoreactivity on the blots was visualized by enhanced chemiluminescence (Amersham Pharmacia). The coimmunoprecipitation from rat forebrain lysates was performed as described by Wenthold et al. (36), only that the deoxycholate homogenates were diluted 1/10 with Triton buffer [1.5% Triton X-100 in 50 mM Tris⋅HCl buffer (pH 7.5)] and recentrifuged before immunoprecipitation. For the coimmunoprecipitation from QT6 cells, the cells were lysed in Triton buffer, homogenized, and centrifuged. The supernatants obtained were incubated overnight with antisera, and the complexes were precipitated with protein-A agarose (Santa Cruz Biotechnology). The immunoprecipitation efficiency was approximately 80%.
Analysis of ErbB-4 and PSD-95 Interactions in Transfected Cells.
Complementary DNAs encoding either the full-length or truncated (missing the C-terminal 48 aa) human ErbB-4 receptor were subcloned in the cytomegalovirus-driven mammalian expression vector pcDNA-AMP (Invitrogen); the PSD-95 expression construct was kindly provided by D. Bredt, University of California, San Diego (37). Quail fibroblast QT-6 cells were grown and transfected by the calcium-phosphate/DNA coprecipitation method as described by Chen and Okayama (38). Cells at 40–60% confluency were transfected with expression vectors for ErbB-4 and PSD-95 by using a total of 4 μg of DNA per 60-mm2 dish, and lysed after 24 h for Western blot or immunoprecipitation assays.
Immunocytochemistry.
Hippocampal neuronal cultures from E18 rats were prepared on glial feeder layers as described (39). After 3 wk, cultures were fixed either with 4% paraformaldehyde/4% sucrose in PBS for 20 min at room temperature or methanol at −20°C. The cells were permeabilized with 0.25% Triton X-100 before incubation overnight at 4°C with primary antibodies diluted in 5% normal goat serum in PBS (NGS/PBS). After extensive washing in NGS/PBS, slides were treated with rhodamine- and FITC-conjugated secondary antibodies (Jackson ImmunoResearch) for 1 h at 35°C. Immunofluorescence was visualized, and images were captured on a Leica microscope equipped with a digital photometric Sensys charge-coupled device camera. Photographs for publication were prepared on adobe photoshop.
Results
ErbB Receptors Are Present in the PSD Fraction.
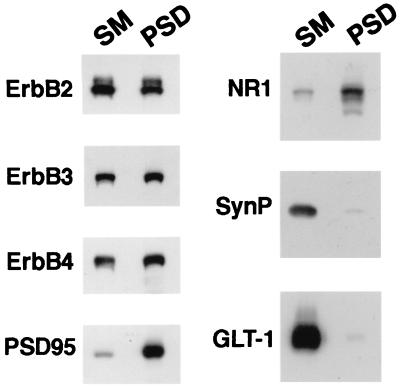
The distribution of ErbB receptors in subcellular fractions made from rat forebrains was analyzed by Western blots. After detergent extraction of SM with 1.5% Triton X-100, ErbB 2–4 receptors remain associated with the PSD fraction, which also contains the NMDA receptor subunits NR1 and PSD-95. The PSD fraction is known to be enriched for the PDZ domain-containing MAGUKs, as well as for NMDA receptors that interact with these proteins (40–42). As shown in Fig. 1, the association of ErbB receptors with the PSD fraction is specific because detergent extraction selectively removes the presynaptic protein synaptophysin and the glutamate transporter GLT-1, an astroglial-specific maker (43), from SM; this fraction has been reported to have both synaptophysin and glial membranes (44). As expected, the relative enrichment observed for NR1 and PSD-95, proteins that are exclusively expressed at postsynaptic sites of neurons, was not seen for ErbB receptors, which have a somatodendritic distribution (see below) and are found in glia (13, 45).
Figure 1.
ErbB receptors are present in the post synaptic density fraction. The subcellular distribution of ErbB receptors in rat forebrain fractions enriched in synaptic membranes (SM) and postsynaptic densities (PSD) was analyzed by Western blot. The blots were probed with specific antisera raised against ErbB-2, ErbB-3, and ErbB-4, and the postsynaptic proteins PSD-95 and NR1. Antisera against synaptophysin (SynP) and glutamate transporter-1 (GLT-1) were used to monitor the presence of presynaptic and glial markers, respectively, in the SM and PSD fractions. All lanes contain 37 μg of protein.
A Yeast Two-Hybrid Screen Reveals Interactions of ErbB-4 with Proteins Containing PDZ Domains.
To investigate the proteins that may interact with ErbB receptors at PSDs, we chose the ErbB-4 receptor. This subunit is highly expressed in neurons (13) and harbors a consensus motif at its C-terminal end (S/T-X-V) necessary for interactions with the PDZ domains in MAGUKs and other PDZ domain-containing proteins (32, 41, 42, 46). A Gal-4 fusion protein containing the ErbB-4 C-terminal 238 aa was used in a yeast two-hybrid system to screen a human brain library; double selection was used (see Materials and Methods). As shown in Table 1, the cDNAs obtained in the yeast two-hybrid screen fell into four groups that encode for PSD-95/SAP90, CHAPSYN110/PSD93, SAP102, and β2-syntrophin. The first three are MAGUKs that harbor three PDZ-interaction domains (32–34), whereas β2-syntrophin harbors a single PDZ domain (47, 48). Additional yeast two-hybrid assays showed that the ErbB-4 C terminus interacts with the first and second, but not the third, PDZ domains of PSD-95 (data not shown). These are the same PDZ domains that interact with the conserved C-terminal motif in NMDA receptor and potassium channel subunits (32, 33, 49), and that is also found in ErbB-4 receptors (T-V-V).
Table 1.
Proteins interacting with ErbB-4 receptor in the yeast two-hybrid system
| Insert encodes | No. of clones recovered | PDZ domains present in
|
|
|---|---|---|---|
| cDNA | Native protein | ||
| PSD-95 | 5 | 1, 2, and 3 (aa 43–767) | 1, 2, and 3 |
| CHAPSYN-110 | 2 | Partial 1, 2, and 3 (aa 149–870) | 1, 2, and 3 |
| SAP-102 | 1 | 1, 2, and 3 (aa 1–689) | 1, 2, and 3 |
| β2-syntrophin | 2 | 1 (aa 1–540) | 1 |
Interactions of ErbB-4 and PSD-95 Are Observed in Vivo.
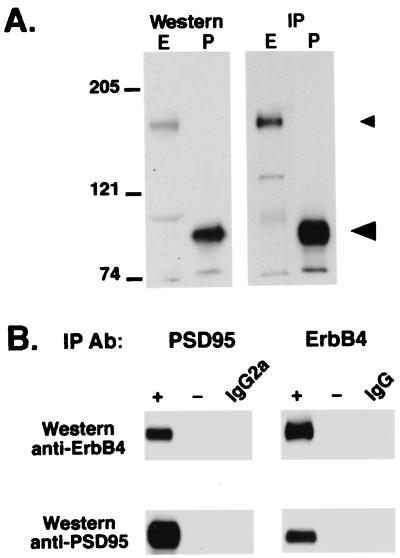
The results of the yeast two-hybrid screen suggested an interaction between ErbB4 and MAGUKS. To test whether ErbB-4 and PSD-95 closely associate in vivo, coimmunoprecipitation experiments were performed from forebrain lysates. Using previously characterized antibodies raised against ErbB-4 (13) and PSD-95 (32), we found that they immunoprecipitate and recognize proteins of approximately 180 and 95 kDa on Western blots from forebrain lysates, respectively (Fig. 2A). We also observed the 180-kDa protein by using antisera generated against other ErbB-4 antigens (24). As shown in Fig. 2B, immunoprecipitation with the PSD-95 monoclonal antibody from a deoxycholate lysate coprecipitates ErbB-4. The inverse, by using the ErbB-4 antiserum for immunoprecipitation, also results in the coprecipitation of the PSD-95/ErbB-4 complex. The ErbB-4/PSD-95 complexes pelleted from the soluble fractions are specific, because immunoprecipitations performed either in the absence of antibody or with nonspecific antisera failed to precipitate either protein.
Figure 2.
Coimmunoprecipitation of ErbB-4 receptor and PSD-95 from forebrain lysates. (A) Total proteins from rat forebrain deoxycholate homogenates (Western, Left), or proteins immunoprecipitated from these lysates (IP, Right), were resolved by SDS/PAGE and probed on Western blots with ErbB-4 (E) and PSD-95 (P) antisera. The antibodies specifically recognize proteins of approximately 180 kDa (small arrowhead) and 95 kDa (large arrowhead), respectively. Migration of the prestained molecular weight markers (myosin, β-galactosidase, and BSA) are indicated. (B) Coimmunoprecipitations were performed from forebrain deoxycholate lysates using PSD-95 (Left) or ErbB-4 (Right) antibodies, and probed on Western blots using anti-ErbB4 or anti-PSD-95 antisera. As negative controls, the immunoprecipitations were performed either in the absence of antibodies (−), normal rabbit immunoglobulins (IgG), or monoclonal antibodies raised against the unrelated transcription factor E2F1 (an IgG2a that is the same isotype as the PSD-95 monoclonal).
Direct Interactions of ErbB-4 and PSD-95 in Heterologous Cells.
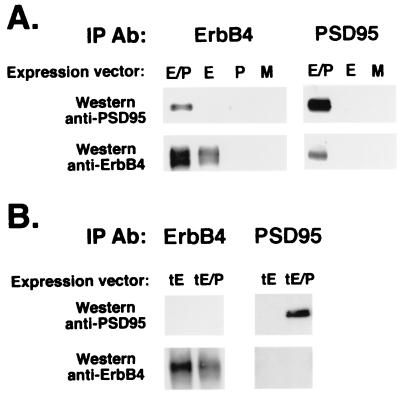
To examine whether ErbB-4 directly interacts with PSD-95 in mammalian cells, coimmunoprecipitation studies were performed by using solubilized fractions from cultured QT-6 quail fibroblasts cotransfected with expression vectors for ErbB-4 and PSD-95. As shown in Fig. 3A, immunoprecipitation of ErbB-4 coprecipitates PSD-95, which was detected on Western blots by using the mouse PSD-95 monoclonal antibody. The converse experiment, using the PSD-95 antibody for the immunoprecipitation, also resulted in the coprecipitation of the ErbB-4 receptor. The interactions between these proteins are mediated through the C-terminal end of ErbB-4, because when a truncated receptor missing 48 aa from its carboxy terminus was coexpressed with PSD-95 in the transfected fibroblasts, we failed to detect coprecipitated complexes even after the autoradiograms were exposed 60 times longer (Fig. 3B). These experiments provide strong evidence for a direct interaction between the C-terminal end of ErbB-4 with at least one member of the MAGUKs.
Figure 3.
ErbB-4 directly associates with PSD-95 in heterologous cells by interactions requiring its C terminus. QT-6 fibroblasts were transiently transfected with ErbB-4 and PSD-95 expression vectors to analyze for interactions between these proteins. (A) Coimmunoprecipitation assays were performed from lysates of cells transfected with constructs encoding PSD-95 (P), ErbB-4 (E), and both proteins (E/P) using ErbB-4 or PSD-95 antisera for immunoprecipitation (IP Ab), and probing the Western blots with anti-PSD-95 and anti-ErbB-4 antisera. A mock transfection (M) using an empty expression vector was used as negative control. (B) A truncated form of ErbB-4, missing the C-terminal 48 amino acids, fails to interact with PSD-95. Coimmunoprecipitation experiments and Western blots were performed as above, using lysates from cells transfected with constructs encoding the truncated ErbB-4 receptor alone (tE) or in combination with PSD-95 (tE/P).
ErbB-4 and PSD-95 Are Colocalized at Synaptic Sites in Cultured Hippocampal Neurons.
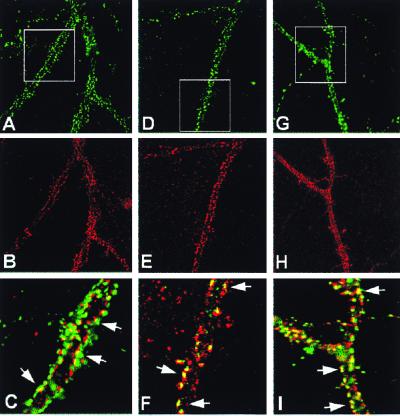
Recently, several groups have found in cultured neurons that different types of PDZ domain proteins cluster at excitatory, but not inhibitory, synapses with NMDA receptors (50, 51). We have used the dissociated hippocampal neuronal cultures to examine whether ErbB-4 receptors cluster at synaptic sites, and whether they colocalize with PSD-95. Cultures exposed to the ErbB-4 receptor antisera showed specific, strong immunofluorescent puncta distributed throughout the length of neurites in a subset of neurons (Fig. 4B); a lighter intradendritic and somitic staining was also observed. These results were obtained with two independent polyclonal antisera and a monoclonal ErbB-4 antibody; no immunofluorescence was observed in the absence of primary antiserum (data not shown). Based on our in situ hybridization studies (I. Karavanova and A.B., unpublished observations), the strong ErbB-4 signals in our hippocampal cultures are likely to be on interneurons. By superimposing the images obtained on double-label immunofluorescence experiments with antibodies raised against the presynaptic marker synaptophysin (Fig. 4A; green) and to the ErbB-4 receptor (Fig. 4B; red), we observe that the ErbB-4 puncta are apposed to synaptophysin-positive terminals (Fig. 4C), indicating that the receptor clusters at synaptic sites. It has been previously reported that PSD-95 is found at puncta that also appose a subset of synaptophysin-positive terminals that contain α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) and NMDA receptors (50). Therefore, we performed double-immunofluorescence experiments to analyze the relative distributions of PSD-95 (Fig. 4D; green), ErbB-4 (Fig. 4 E and H; red), and NMDA (Fig. 4G; green) receptors on the cultured hippocampal neurons. By superimposing the immunofluorescent images, we found that ErbB-4 receptors are colocalized with PSD-95 (Fig. 4F; yellow) and NMDA receptors (Fig. 4I; yellow) at a subset of puncta. Taken together, our results indicate that ErbB tyrosine kinase receptors are present at interneuronal synapses, where they are found in close association with PSD-95 and NMDA receptors at PSDs. These findings could have important implications for synaptic plasticity (see below).
Figure 4.
Immunocytochemical localization of ErbB-4 at synaptic sites of cultured hippocampal cells. Double immunofluorescence was performed on dissociated hippocampal neurons cultured for 3 wk. (A–C) ErbB-4 receptor puncta (red) are located at synaptic sites apposed to a subset of the synaptophysin-positive presynaptic terminals (green). The yellow color in the enlarged view (C) indicates the extent of immunofluorescence overlap. (D–F) ErbB-4 receptors (red) and PSD-95 (green) colocalize in a subset of synaptic puncta (yellow). (G–I) ErbB-4 receptors (red) and the NR1 subunit of the NMDA receptor (green) also colocalize at puncta (yellow). Areas that are boxed are enlarged (Bottom), and the arrows indicate examples of colocalization.
Discussion
Neuregulins have been implicated in the activity-dependent maturation of the neuromuscular junction and interneuronal synapses by regulating the expression of adult isoforms of neurotransmitter receptor subunits and voltage-gated channels (9–14). During NMJ development, ErbB receptors initially are diffusely expressed on the plasma membrane and later aggregate with nicotinic AChRs at the postsynaptic membrane by an agrin-dependent mechanism (52). In contrast to the NMJ, nothing is known about the subcellular distribution and mechanisms regulating the localization of ErbB receptors in neurons. In this report, we have shown that ErbB receptors are present in the PSD fraction of forebrain neurons and that they colocalize at postsynaptic sites of cultured hippocampal neurons with PSD-95 and NMDA receptors, proteins that are present at PSDs. Our results suggest that ErbB-4 receptors may be sequestered or anchored at glutamatergic synapses by their physical interaction with MAGUKs. As discussed below, these results are interesting because of the function of the PSD, and the proposed role of MAGUKs in coupling neurotransmitter receptors and channels to intracellular signaling pathways that regulate activity-dependent synaptic plasticity (31, 40, 42). Thus, Nrgs may influence the properties of central synapses by the activation of ErbB receptors and signaling by means of tyrosine kinase cascades.
Possible Mechanisms Regulating ErbB Receptor Localization at Regions of Signal Transduction.
There is accumulating evidence that the PSD functions as an organizing structure for signal transduction that integrates activity-dependent stimuli to molecules that regulate synaptic plasticity (30, 31). This suggests that components of the PSD may play a central role in regulating processes such as long-term potentiation and long-term depression. The recent discovery that PDZ-domain proteins scaffold receptor and channels at PSDs to kinases and phosphatases that harbor the PDZ interaction domains (42), as well as adhesion molecules like neuroligins and neurexins that can span the synaptic cleft (53), enable the coupling of signals between the pre- and postsynaptic terminals at cellular microdomains (54). In this context, it is important that we have found that the cytoplasmic tail of the ErbB-4 receptor interacts with proteins harboring PDZ domains, which may scaffold ErbB receptors to synaptic sites. We did not investigate whether ErbB-2 and ErbB-3 receptors, which are also present in the PSD fraction (Fig. 1) but lack the consensus C terminus S/T-X-V motif, can interact with MAGUKs. Because these receptors can associate with ErbB-4 to form heterodimers, this could be one possible mechanism by which a portion of the cell's ErbB-2 and -3 receptors localize to the PSD fraction. Another possibility for association of these ErbB receptors with the PSD fraction is that both receptors harbor phosphotyrosine recognition motifs that can interact with proteins containing SH-2 and PTB domains that have been localized at the PSD (see ref. 31). The association of ErbB receptors with MAGUKs, and their colocalization with NMDA receptors at a subset of excitatory synapses, could allow these two signaling pathways to interact at interneuronal synapses (see below).
In contrast to NMDA receptors and PSD-95, ErbB receptors are expressed by cells other than neurons. In our yeast two-hybrid screen, we isolated β2-syntrophin, suggesting that this single PDZ domain-containing protein may interact with ErbB receptors in neural and nonneural tissues. Syntrophins accumulate at cellular junctions in a variety of tissues and have been implicated in coupling transmembrane proteins to signal transduction cascades (47). In muscle, β2-syntrophin is enriched at the NMJ, where it interacts with voltage-gated sodium channels and may participate in their localization by interaction of its PDZ domain with the C terminus of the channel (47, 48). Syntrophins are also found in myocardiocytes, which are cells that also express ErbB receptors. A recent report using subcellular fractionation suggests that ErbB-4 receptors are found in caveoli of cardiac myocytes (55), structures that facilitate ligand-mediated activation of signaling cascades, including those for tyrosine kinase receptors (56, 57). It will be important to determine whether syntrophins participate in the localization of ErbB-4 receptors to the NMJ, caveoli, and interneuronal synapses (i.e., regions of receptor cross-talk).
The mechanisms that regulate ErbB receptor turnover and localization may be more dynamic than those used to restrict NMDA receptors to synapses. For example, the cleavage of the ErbB-4 receptor ectodomain by metaloproteases is regulated by protein kinase C and results in a large pool of cleaved intracellular receptor (58). If this mechanism functions in cultured neurons, it could account for the intradendritic staining that we observed with antisera directed to the C terminus of the receptor but not with an antibody raised against the N terminus. In addition, the location of ErbB-4 receptors is regulated by ligand binding. In cardiomyocytes (55), Nrg causes a rapid translocation of ErbB-4 (but not ErbB-2) receptor out of caveoli, suggesting that the mechanisms that tether ErbB-4 receptors at synapses could also be very dynamic. In this regard, it is interesting that a fraction of PSD-95 localizes to neuronal caveolar-like domains (59). It will be important to determine whether PSD-95 and syntrophins participate in the scaffolding or signaling processes of receptors present in caveolar-like domains in the brain, and whether ErbB receptors preferentially accumulate at these structures.
Evolutionary Conservation of the Interaction of ErbB Receptors with PDZ-Domain Proteins.
There is genetic and molecular evidence that the interaction of ErbB receptors with PDZ-containing proteins has been evolutionarily conserved from invertebrates to mammals. In Caenorhabditis elegans, LET-23 is the single gene encoding for the receptor tyrosine kinase binding the epidermal growth factor-related ligands and is required for vulva differentiation. The products of three genes necessary to target and/or localize LET-23 to the basolateral membrane, LIN-2, -7, -10 and their mammalian homologues, were recently identified. Interestingly, LIN-2/CASK (60), LIN-7/Velis/MAL (61), and LIN-10/Mint/aX11 (62) are proteins containing multiple PDZ domains. Furthermore, the PDZ domain in LIN-7 interacts with the LET-23 tyrosine kinase receptor, which has a T-C-L motif at its C terminus, in a similar fashion that the ErbB-4 receptor interacts with the MAGUKs. At present, it is not known whether CASK, MALS/Velis, and Mint are involved in the localization of ErbB receptors to the PSDs or cellular junctions in mammalian cells. MALS are found at PSDs, and they interact with NMDA receptor subunits; however, an interaction with ErbB-4 receptors could not be detected by protein “pull-down” assays (61). Further experiments will be needed to determine whether LIN homologues or other PDZ domain-containing proteins, such as the MAGUKS and/or β-synthrophin, may participate in the targeting and signaling of ErbB receptors at mammalian synapses. The observation that CASK and LIN-10/Mint1 antibodies stain the trans-Golgi, cytoplasm, and intradendritic region, as we have observed with antisera directed to the cytoplasmic domain of ErbB-4, suggests that these PDZ-domain proteins could be involved in the transport, targeting, and turnover of ErbB receptors.
What May Be the Functional Importance for the Colocalization of ErbB and NMDA Receptors?
Studies on the regulation of AChR gene expression at the NMJ have focused on how ErbB receptor activation signals to the nucleus to modify the heteromeric composition of the nicotinic AChR, but have not addressed the issue of whether the ErbB kinase activity also exerts effects at the membrane. In the central nervous system, the NR2A and NR2B are heavily phosphorylated at tyrosine residues in vivo and in cultured neurons (63, 64), and this posttranslational modification enhances receptor activity by altering the kinetic properties of the channel (65–67). Tyrosine phosphorylation of NMDA receptors is modified by stimuli that induce long-term potentiation (LTP), and its inhibition by phosphatases, inhibits LTP induction (54). The src-related nonreceptor tyrosine kinases, which directly associate with the NMDA receptor, have been implicated in the phosphorylation of the receptor (66). Fyn kinase knockout mice have impaired LTP and hippocampal development (68) and reduced levels of tyrosine-phosphorylated NR2A (69). However, the mechanisms that initiate the signaling cascade that leads to the recruitment of src-related kinases to the NMDA receptor are not known. The recruitment of nonreceptor tyrosine kinases are often initiated by the autophosphorylation of receptor tyrosine kinases (70). In this regard, it is interesting that Nrg activation results in the autophosphorylation of ErbB receptors that are colocalized with NMDA receptors at PSDs. Moreover, Tezuka et al. (69) recently demonstrated that PSD-95 recruits fyn kinase to a complex with NMDA receptors to promote the tyrosine phosphorylation of the NR2A subunit. These observations raise the interesting possibility that the activity-dependent activation of ErbB receptors by Nrg may regulate synaptic plasticity by recruiting tyrosine kinases that regulate NMDA receptor function.
We previously reported that the activation of both ErbB and NMDA receptors is necessary for the expression of the NMDA receptor NR2C subunit during the maturation of cerebellar granule cells (13). The present studies provide strong support for the idea that Nrg and NMDA receptor cross-talk may occur directly at the postsynaptic sites of excitatory synapses. Given that many other neurotrophic factors signal through tyrosine kinase receptors, and there is evidence that cosignaling may expand the specificity of signaling cascades by its combinatorial nature, our studies have important implications to the regulation of other receptor systems during neuronal maturation.
Acknowledgments
We thank E. Ralston and C. Winters for help with hippocampal cultures, Drs. M. Sheng, C. Sala, and C. Lai for antibodies, Betsy Apel for the QT-6 cells, and D. Vullhorst, D. Turetsky, I. Karavanov, and M. Morasso for their invaluable help. R.A.G.G. was partly supported by Consejo Nacional de Investigaciones Científicas y Technologicas, Vollmer Foundation, and Instituto Internacional de Estudios Avanzados.
Abbreviations
- Nrg
neuregulin
- AChR
acetylcholine receptor
- NMDA
N-methyl-d-aspartate
- PSD
postsynaptic density
- MAGUK
membrane-associated guanylate kinase
- SM
synaptic membrane
- NMJ
neuromuscular junction
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.070042497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.070042497
References
- 1.Fischbach G D, Rosen K M. Annu Rev Neurosci. 1997;20:429–458. doi: 10.1146/annurev.neuro.20.1.429. [DOI] [PubMed] [Google Scholar]
- 2.Zhang D, Frantz G, Godowski P J. Mol Psychiatry. 1998;2:112–115. doi: 10.1038/sj.mp.4000369. [DOI] [PubMed] [Google Scholar]
- 3.Lemke G. Mol Cell Neurosci. 1996;7:247–262. doi: 10.1006/mcne.1996.0019. [DOI] [PubMed] [Google Scholar]
- 4.Burden S, Yarden Y. Neuron. 1997;18:847–855. doi: 10.1016/s0896-6273(00)80324-4. [DOI] [PubMed] [Google Scholar]
- 5.Morris J K, Lin W, Hauser C, Marchuk Y, Getman D, Lee K F. Neuron. 1999;23:273–283. doi: 10.1016/s0896-6273(00)80779-5. [DOI] [PubMed] [Google Scholar]
- 6.Loeb J A, Khurana T S, Robbins J T, Yee A G, Fischbach G D. Development (Cambridge, UK) 1999;126:781–791. doi: 10.1242/dev.126.4.781. [DOI] [PubMed] [Google Scholar]
- 7.Han B, Fischbach G D. Philos Trans R Soc London B. 1999;354:411–416. doi: 10.1098/rstb.1999.0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duclert A, Savatier N, Schaeffer L, Changeux J P. J Biol Chem. 1996;271:17433–17438. doi: 10.1074/jbc.271.29.17433. [DOI] [PubMed] [Google Scholar]
- 9.Burden S J. Genes Dev. 1998;12:133–148. doi: 10.1101/gad.12.2.133. [DOI] [PubMed] [Google Scholar]
- 10.Martinou J C, Falls D L, Fischbach G D, Melie J P. Proc Natl Acad Sci USA. 1991;17:7669–7673. doi: 10.1073/pnas.88.17.7669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corfas G, Fischbach G D. J Neurosci. 1993;13:2118–2125. doi: 10.1523/JNEUROSCI.13-05-02118.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandrock A W, Jr, Dryer S E, Rosen K M, Gozani S N, Kramer R, Theill L E, Fischbach G D. Science. 1997;276:599–603. doi: 10.1126/science.276.5312.599. [DOI] [PubMed] [Google Scholar]
- 13.Ozaki M, Sasner M, Yano R, Lu H S, Buonanno A. Nature (London) 1997;390:691–694. doi: 10.1038/37795. [DOI] [PubMed] [Google Scholar]
- 14.Yang X, Kuo Y, Devay P, Yu C, Role L. Neuron. 1998;20:255–270. doi: 10.1016/s0896-6273(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 15.Peles E, Yarden Y. BioEssays. 1993;15:815–824. doi: 10.1002/bies.950151207. [DOI] [PubMed] [Google Scholar]
- 16.Zhang D, Sliwkowski M X, Mark M, Frantz G, Akita R, Sun Y, Hillan K, Crowley C, Brush J, Godowski P J. Proc Natl Acad Sci USA. 1997;94:9562–9567. doi: 10.1073/pnas.94.18.9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Busfield S J, Michnick D A, Chickering T W, Revett T L, Ma J, Woolf E A, Comrack C A, Dussault B J, Woolf J, Goodearl A D, Gearing D P. Moll Cell Biol. 1997;17:4007–4014. doi: 10.1128/mcb.17.7.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carraway K L, III, Weber J L, Unger M J, Ledesma J, Yu N, Gassmann M, Lai C. Nature (London) 1997;387:512–516. doi: 10.1038/387512a0. [DOI] [PubMed] [Google Scholar]
- 19.Chang H, Riese D J, II, Gilbert W, Stern D F, McMahan U J. Nature (London) 1997;387:509–512. doi: 10.1038/387509a0. [DOI] [PubMed] [Google Scholar]
- 20.Jo S A, Zhu X, Marchionni M A, Burden S J. Nature (London) 1995;373:158–161. doi: 10.1038/373158a0. [DOI] [PubMed] [Google Scholar]
- 21.Altiok N, Bessereau J L, Changeux J P. EMBO J. 1995;14:4258–4266. doi: 10.1002/j.1460-2075.1995.tb00100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moscoso L M, Chu G C, Gautam M, Noakes P G, Merlie J P, Sanes J R. Dev Biol. 1995;172:158–169. doi: 10.1006/dbio.1995.0012. [DOI] [PubMed] [Google Scholar]
- 23.Zhu X, Lai C, Thomas S, Burden S J. EMBO J. 1995;14:5842–5848. doi: 10.1002/j.1460-2075.1995.tb00272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodearl A D, Yee A G, Sandrock A W, Corfas G, Fischbach G D. J Cell Biol. 1995;130:1423–1434. doi: 10.1083/jcb.130.6.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tansey M G, Chu G C, Merlie J P. J Cell Biol. 1996;2:465–476. doi: 10.1083/jcb.134.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sapru M K, Florance S K, Kirk C, Goldman D. Proc Natl Acad Sci USA. 1998;95:1289–1294. doi: 10.1073/pnas.95.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaeffer L, Duclert N, Huchet-Dymanus M, Changeux J P. EMBO J. 1998;17:3078–3090. doi: 10.1093/emboj/17.11.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fromm L, Burden S J. Genes Dev. 1998;12:3074–3083. doi: 10.1101/gad.12.19.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandrock A W, Jr, Goodearl A D, Yin Q W, Chang D, Fischbach G D. J Neurosci. 1995;9:6124–6136. doi: 10.1523/JNEUROSCI.15-09-06124.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kennedy M B. Trends Neurosci. 1997;20:264–268. doi: 10.1016/s0166-2236(96)01033-8. [DOI] [PubMed] [Google Scholar]
- 31.Ziff E B. Neuron. 1997;19:1163–1174. doi: 10.1016/s0896-6273(00)80409-2. [DOI] [PubMed] [Google Scholar]
- 32.Kornau H-C, Schenker L T, Kennedy M B, Seeburg P H. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 33.Kim E, Cho K-O, Rothchild A, Sheng M. Neuron. 1996;17:103–113. doi: 10.1016/s0896-6273(00)80284-6. [DOI] [PubMed] [Google Scholar]
- 34.Lau L-F, Mammen A, Ehlers M D, Kindler S, Chung W J, Garner C C, Huganir R L. J Biol Chem. 1996;271:21622–21628. doi: 10.1074/jbc.271.35.21622. [DOI] [PubMed] [Google Scholar]
- 35.Rogers S W, Hughes T E, Hollmann M, Gasic G P, Deneris E S, Heinemann S. J Neurosci. 1991;11:2713–2724. doi: 10.1523/JNEUROSCI.11-09-02713.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wenthold R J, Blahos J, II, Huh K H, Petralia R S. Methods Mol Biol. 1999;128:113–119. doi: 10.1385/1-59259-683-5:113. [DOI] [PubMed] [Google Scholar]
- 37.Topinka J R, Bredt D S. Neuron. 1998;20:125–134. doi: 10.1016/s0896-6273(00)80440-7. [DOI] [PubMed] [Google Scholar]
- 38.Chen C, Okayama H. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayer M L, Vylickly L, Westbrook G L. J Physiol. 1989;415:329–350. doi: 10.1113/jphysiol.1989.sp017724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kornau H-C, Seeburg P H, Kennedy M B. Curr Opin Neurobiol. 1997;7:368–373. doi: 10.1016/s0959-4388(97)80064-5. [DOI] [PubMed] [Google Scholar]
- 41.Sheng M, Wyszynski M. BioEssays. 1997;19:847–853. doi: 10.1002/bies.950191004. [DOI] [PubMed] [Google Scholar]
- 42.Craven S E, Bredt D S. Cell. 1998;93:495–498. doi: 10.1016/s0092-8674(00)81179-4. [DOI] [PubMed] [Google Scholar]
- 43.Chaudhry F A, Lehre K P, van Lookeren Campagne M, Ottersen O P, Danbolt N C, Storm-Mathisen J. Neuron. 1995;15:711–720. doi: 10.1016/0896-6273(95)90158-2. [DOI] [PubMed] [Google Scholar]
- 44.Henn F A, Anderson D J, Rustad D G. Brain Res. 1976;101:341–344. doi: 10.1016/0006-8993(76)90274-2. [DOI] [PubMed] [Google Scholar]
- 45.Carroll S L, Miller M L, Frohnert P W, Kim S S, Corbett J A. J Neurosci. 1997;17:1642–1659. doi: 10.1523/JNEUROSCI.17-05-01642.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Brien R J, Lau L F, Huganir R L. Curr Opin Neurobiol. 1998;8:364–369. doi: 10.1016/s0959-4388(98)80062-7. [DOI] [PubMed] [Google Scholar]
- 47.Gee S H, Madhavan R, Levinson S R, Caldwell J H, Sealock R, Froehner S C. J Neurosci. 1998;18:128–137. doi: 10.1523/JNEUROSCI.18-01-00128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schultz J, Hoffmuller U, Krause G, Ashurst J, Macias M J, Schmieder P, Schneider-Mergener J, Oschnat H. Nat Struct Biol. 1998;5:19–24. doi: 10.1038/nsb0198-19. [DOI] [PubMed] [Google Scholar]
- 49.Kim E, Niethammer M, Rothschild A, Jan Y N, Sheng M. Nature (London) 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 50.Rao A, Craig A M. Neuron. 1997;19:801–812. doi: 10.1016/s0896-6273(00)80962-9. [DOI] [PubMed] [Google Scholar]
- 51.Liao D, Zhang X, O'Brien R, Ehlers D, Huganir R L. Nat Neurosci. 1999;2:37–43. doi: 10.1038/4540. [DOI] [PubMed] [Google Scholar]
- 52.Sanes J R, Lichtman J W. Annu Rev Neurosci. 1999;22:389–442. doi: 10.1146/annurev.neuro.22.1.389. [DOI] [PubMed] [Google Scholar]
- 53.Irie M, Hata Y, Takeuchi M, Ichtchenko K, Toyoda A, Hirao K, Takai Y, Rosahl T W, Sudhof T C. Science. 1997;277:1511–1515. doi: 10.1126/science.277.5331.1511. [DOI] [PubMed] [Google Scholar]
- 54.Buonanno A, Fields R D. Curr Opin Neurobiol. 1999;9:110–120. doi: 10.1016/s0959-4388(99)80014-2. [DOI] [PubMed] [Google Scholar]
- 55.Zhao Y-Y, Feron O, Dessy C, Han X, Marchionni M A, Kelly R A. Circ Res. 1999;84:1380–1387. doi: 10.1161/01.res.84.12.1380. [DOI] [PubMed] [Google Scholar]
- 56.Lisanti M P, Tang Z, Scherer P E, Kubler E, Koleske A J, Sargiacomo M. Mol Membr Biol. 1995;12:121–124. doi: 10.3109/09687689509038506. [DOI] [PubMed] [Google Scholar]
- 57.Wu C, Butz S, Ying Y, Anderson R G. J Biol Chem. 1997;272:3554–3559. doi: 10.1074/jbc.272.6.3554. [DOI] [PubMed] [Google Scholar]
- 58.Vecchi M, Baulida J, Carpenter G. J Biol Chem. 1996;271:18989–18995. doi: 10.1074/jbc.271.31.18989. [DOI] [PubMed] [Google Scholar]
- 59.Perez A-S, Bredt D S. Neurosci Lett. 1998;258:121–123. doi: 10.1016/s0304-3940(98)00846-5. [DOI] [PubMed] [Google Scholar]
- 60.Hata Y, Butz S, Sudhof T C. J Neurosci. 1996;16:2488–2494. doi: 10.1523/JNEUROSCI.16-08-02488.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jo K, Derin R, Li M, Bredt D S. J Neurosci. 1999;19:4189–4199. doi: 10.1523/JNEUROSCI.19-11-04189.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Whitfield C W, Benard C, Barnes T, Hekimi S, Kim S K. Mol Biol Cell. 1999;10:2087–2100. doi: 10.1091/mbc.10.6.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Omkumar R V, Kiely M J, Rosenstein A J, Min K T, Kennedy M B. J Biol Chem. 1996;271:31670–31678. doi: 10.1074/jbc.271.49.31670. [DOI] [PubMed] [Google Scholar]
- 64.Lau L F, Huganir R L. J Biol Chem. 1995;270:20036–20041. doi: 10.1074/jbc.270.34.20036. [DOI] [PubMed] [Google Scholar]
- 65.Wang Y T, Salter M W. Nature (London) 1994;369:233–235. doi: 10.1038/369233a0. [DOI] [PubMed] [Google Scholar]
- 66.Kohr G, Seeburg P H. J Physiol (London) 1996;492:445–452. doi: 10.1113/jphysiol.1996.sp021320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng F, Gingrich M B, Traynelis S F, Conn P J. Nat Neurosci. 1998;1:185–191. doi: 10.1038/634. [DOI] [PubMed] [Google Scholar]
- 68.Grant S G, O'Dell T J, Karl K A, Stein P L, Soriano P, Kandel E R. Science. 1992;258:1903–1910. doi: 10.1126/science.1361685. [DOI] [PubMed] [Google Scholar]
- 69.Tezuka T, Umemori H, Akiyama T, Nakanishi S, Yamamoto T. Proc Natl Acad Sci USA. 1999;96:435–440. doi: 10.1073/pnas.96.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Segal R A, Greengerg M E. Annu Rev Neurosci. 1996;19:463–489. doi: 10.1146/annurev.ne.19.030196.002335. [DOI] [PubMed] [Google Scholar]