Abstract
The difficulty of obtaining pedigrees for wild populations has hampered the possibility of demonstrating inbreeding depression in nature. In a small, naturally restored, wild population of grey wolves in Scandinavia, founded in 1983, we constructed a pedigree for 24 of the 28 breeding pairs established in the period 1983–2002. Ancestry for the breeding animals was determined through a combination of field data (snow tracking and radio telemetry) and DNA microsatellite analysis. The population was founded by only three individuals. The inbreeding coefficient F varied between 0.00 and 0.41 for wolves born during the study period. The number of surviving pups per litter during their first winter after birth was strongly correlated with inbreeding coefficients of pups (R2=0.39, p<0.001). This inbreeding depression was recalculated to match standard estimates of lethal equivalents (2B), corresponding to 6.04 (2.58–9.48, 95% CI) litter-size-reducing equivalents in this wolf population.
Keywords: inbreeding depression, lethal equivalents, pedigree, conservation biology, wolf
1. Introduction
Inbreeding depression is assumed to be a serious problem for the conservation of small populations (Gilpin & Soulé 1986), but has been difficult to demonstrate in nature (Caughley 1994). The main obstacle has been the construction of pedigrees necessary for calculating inbreeding coefficients. Recently, modern molecular techniques have allowed indirect genetic measurement of inbreeding depression in wild animals, including mammals (e.g. Coltman et al. 1999; Slate et al. 2000). Inbreeding measured as genetic similarity between individuals does not directly translate to inbreeding coefficients. This often prevents one from establishing the level of inbreeding responsible for the decreased fitness, as well as direct comparisons with other studies. These problems are circumvented when using pedigree analysis; however, this has rarely been done in wild populations, with some notable exceptions (e.g. Keller 1998; Loeske et al. 2002). By combining DNA techniques with ecological field data, we have constructed a complete pedigree and demonstrated severe inbreeding depression in the wild Scandinavian wolf, Canis lupus, population. The wolf became functionally extinct in Scandinavia (Norway and Sweden) at the end of the 1960s. Around 1980, at least two wolves immigrated and founded a new population in south-central Scandinavia, 900 km from the edge of the large Finnish/Russian source population (Wabakken et al. 2001; Vila et al. 2003). The first reproduction occurred in 1983, and by 2002 the population included approximately 100 wolves.
2. Materials and methods
(a) Field data
The wolf population has been monitored since 1978, based on snow tracking and, from 1998, also on radio telemetry. Territorial pairs were distinguished and the number of animals in packs counted (Wabakken et al. 2001). A ‘pair’ is two breeding adults producing offspring together, while a ‘pack’ is the total number of individuals in a family, for example, the pair and its dependent offspring. The ‘territory’ is the geographical area where the pair is living. As a fitness measure, we used the number of pups per litter surviving until the first winter after birth (‘winter litter size’). We used data for first-born litters of each breeding pair only, because for subsequent litters, tracks from pups of the year could not be separated from those of yearlings and older philopatric siblings (Mech 1970). In darted wolves, ageing was based on the growth zone in the tibia for pups and tooth wear for adults, and in retrieved dead wolves annual tooth cementum layers (C1) were counted.
(b) Genetic analyses
Samples were derived from the blood of captured wolves, the muscle of dead wolves (‘tissue’), from oestrus blood on snow and from scats. Genomic DNA from tissue was isolated using standard phenol/chloroform–isoamylalcohol extraction protocols. Two isolates were extracted from faecal samples with a Qiamp DNA stool mini kit (Qiagen, Valencia, CA, USA). Faecal and oestrus blood samples were extracted in a separate workspace treated with ultraviolet light to avoid contamination (Sarkar & Sommer 1990). Negative extraction controls were used throughout.
We scored tissue samples for allelic variation at 32 autosomal microsatellite loci, and faecal samples on a subset of 16 (for details see Electronic Appendix). To minimize scoring errors associated with low quality DNA (Taberlet et al. 1999), faecal samples were amplified a minimum of four times (twice per isolate). Heterozygotes were accepted if both alleles were present in two amplifications and homozygotes if four positive amplifications showed only one allele. If neither condition was met, samples were re-amplified. Problematic samples were amplified up to 10 times. In the few samples, where an ambiguous result still occurred, we recorded a half-locus (Miller et al. 2002).
The pedigree was determined by parentage analysis. We used material from 163 wolf individuals; 113 of these were based on muscle from dead wolves or blood from anaesthetized wolves, the rest from faeces and/or from oestrus blood found in snow. A missing genotype of one parent was reconstructed from genotypes of the known parent and pups of that pair. Of the 48 breeding wolves in the pedigree used in the analysis, genotypes of 16 were reconstructed, 25 were based on tissue (muscle or blood drawn directly from the animal) and seven were based on faeces/oestrus blood. The three incestuous pairs in the period 1987–1990 were completely reconstructed from genotypes of 10 wolves born during this period. Here several alternatives were possible. We chose the most parsimonious alternative, but tested all possible alternatives, and none changed the results of this study other than marginally. Inbreeding coefficients were calculated with the software Pedigree Viewer 5.0 (© Brian and Sandy Kinghorn).
(c) Statistical analyses
We used parametric statistics (ANCOVA) in the analyses of inbreeding effects, including the interaction terms between the independent variables in the initial model. Ages of breeding females were treated as a two state variable: young (2–3 years) and old (4 years or older). Genetic load is expressed in terms of lethal equivalents, based on viability data (Kalinowski & Hedrick 1998). We calculated an analogous parameter, litter-reducing equivalents, by regressing litter size (Wi) against the inbreeding coefficient (fi) using the relationship ln Wi=ln (W0−Bfi), where W0 is the litter size for outbred litters (f0). Inbreeding effects on population growth rate (λ) were tested using a Leslie matrix with five age classes. We used data from our study population for survival and reproduction, adjusted to give a baseline growth rate similar to the one observed in the period 1991–2000 of λ=1.29 (Wabakken et al. 2001).
For further details on material and methods, see the Electronic Appendix.
3. Results and discussion
(a) Pedigree, inbreeding coefficients and litter sizes
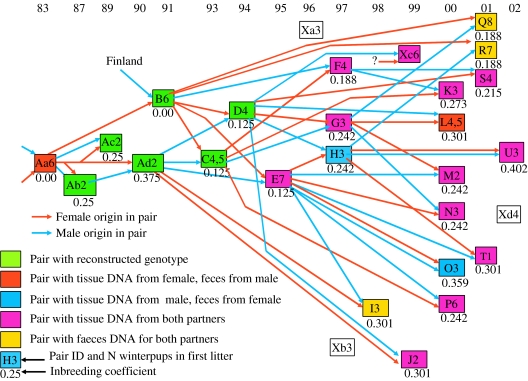
We traced the complete ancestry for both male and female in 24 of the 28 breeding wolf pairs registered during the period 1983–2002, constructing the first complete pedigree back to its founders that has been published for a wild mammal population (Keller & Waller 2002), and calculated inbreeding coefficients (F; figure 1). The first founding wolf pair reproduced for 3 years (1983–1985) until the female was shot in 1985, but offspring from this pair continued to breed within the same territory until 1994 through incestuous matings (figure 1). In 1991, an immigrant male mated with a daughter of the first breeding pair and contributed to the large variation in the inbreeding coefficient F in the population (0.00–0.41). Apart from the early incestuous matings, we recorded only two later cases of full sibling pairings (pairs O and U in figure 1). Nevertheless, most animals born after 1997 have inbreeding coefficients close to or higher than 0.25, a level corresponding to full sibling mating (figure 1).
Figure 1.
Pedigree of the Scandinavian wolf population. Boxes indicate breeding pairs and arrows trace the ancestry of male (blue) and female (red) in each pair. Colours of boxes indicate how the genotype of the wolves in the pair was determined. Unfilled boxes indicate pairs with missing genotype. Pairs are fitted to the time-scale on top according to their year of first reproduction. The number of winter pups in first litters and inbreeding coefficients for offspring, are indicated. If number of pups was determined as a range (e.g. 4–5) we used the mean value (e.g. 4.5).
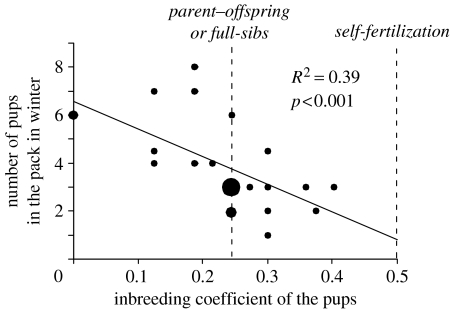
The sizes of winter litters for first breeding pairs were strongly affected by the inbreeding coefficient of the pups (n=24, R2=0.39, p<0.001; figure 2), while the inbreeding coefficients of the mother (partial R2=0.04, p=0.23), of the father (partial R2=0.07, p=0.11), the age of the mother (partial R2=0.09, p=0.076) and time (partial R2=0.10, p=0.058) did not contribute significantly to the same model. After removing offspring inbreeding coefficients, there was indeed an effect of the mother's inbreeding coefficient (n=24, R2=0.27, p=0.01), but not from the father's (partial R2=0.02, p=0.41), nor from age of the mother (partial R2=0.08, p=0.13) or time (partial R2=0.10, p=0.075). Inbreeding of the father (R2=0.06, p=0.25), or time (R2=0.001, p=0.88) had no effect alone. The inbreeding coefficient increased over the years for pups and mothers (r=0.49, p=0.016 and r=0.58, p=0.003).
Figure 2.
The number of pups that survived to winter for first-born litters in relation to the inbreeding coefficient of the pups. Small dots refer to one data point each, medium sized to two data points and large to four data points. Inbreeding levels corresponding to parent–offspring or full-sibling mating, and self-fertilization are indicated.
We are confident that the demonstrated inbreeding effect was not a by-product of association with coincidental trends in the environment, for example weather or food, as time itself had no effect on litter size. Change in prey availability can also be discarded considering that the number of moose (Alces alces), the most important prey for wolves in Scandinavia (Sand et al. in press), stayed high (greater than 1 moose per km2 in all wolf territories) during the study period (Hörnberg 2001). It was well above the threshold (0.5 moose per square kilometre) under which wolf populations are reported to be affected (Messier, 1994).
(b) Effects of inbreeding on demography
The quantitative inbreeding effect was a reduction of 1.15 winter pups per litter for each increase of 0.1 in the F for pups (winter litter size=6.54–11.51F; figure 2). In our population model, an increase of offspring inbreeding coefficient F of 0.1 reduced the growth rate λ from 1.29 to 1.21, assuming all litters were affected equally by inbreeding. Zero population growth (λ=1) would be reached at an average F of 0.48. Our chosen fitness measure, winter litter size, actually represents a combination of fecundity and early survival. It is possible that more fitness components, for example, yearling or adult survival, could be affected, which would make the demographic consequences even more severe. The Scandinavian wolf population thus may have a gloomy future unless it can be purged of its genetic load through natural selection, or receives new genetic variation from outside. However, the effectiveness of purging in small populations has been questioned (Hedrick & Kalinowski 2000), and the probability of natural immigration also seems low, as no new immigrants have appeared in the last 13 years. In an earlier report concerning this population, it was claimed that the male immigrating in 1991 ‘rescued’ the population (Vila et al. 2003). Our interpretation is that before this male arrived there was no population but just a strongly inbred family. The arrival of this newcomer allowed young wolves to find partners outside of their own family, and this sparked off a rapid initial increase, but has not prevented the succeeding inbreeding.
(c) Conservation implications
This study has general implications for the ‘small population paradigm’ (Caughley 1994), and is especially relevant for the conservation of large carnivores. These are charismatic species with large public support, but as powerful predators also highly controversial, they are often forced into small fragmented populations. The wolf could be useful as a model species for this dilemma, in part because there are several studies of inbreeding in captive populations of this species. A captive Swedish wolf population, partly founded from the same source as our study population, also expressed severe inbreeding effects (Laikre 1999), while in two American captive populations of red and Mexican wolf, no effects were noted on demographic parameters (Kalinowski et al. 1999), although effects on body size were noted in the Mexican wolves (Fredrickson & Hedrick 2002). The genetic load of our wild population (6.04±3.44), 95% CI) was substantially heavier than that for the red and Mexican wolves (0.63 and 0.71, respectively), and also clearly higher than the average estimate of 3.14 in a study of 40 captive mammal populations (Ralls et al. 1988). This indicates that impact of inbreeding can vary substantially, even within the same species, depending on the random subset of genes from the source population drawn by the founders, and succeeding random drift. For the famous wild wolf population on Isle Royale in Minnesota, USA, 50 years after founding by only two individuals there still is only some indirect evidence of demographic effects of inbreeding (Wayne 1991; Peterson et al. 1998), but a detailed analysis of inbreeding, of the type demonstrated in this paper, has not been used.
The conservation implication for our study population is that genetic exchange with the source population should be strongly promoted. In the meanwhile, the close demographic and genetic monitoring of the population should be continued. The potential for further exploration of inbreeding effects on more demographic parameters should be pursued.
Acknowledgements
Funding was provided to O.L., H.A. and H.S. by the Swedish Environmental Protection Agency, the World Wide Fund for Nature (Sweden), the Swedish Association for Hunting and Wildlife Management, the private foundation ‘Olle och Signhild Engkvists Stiftelser’, to H-C.P. and P.W. by the Norwegian Research Council, the Norwegian Directorate for Nature Management, the Norwegian Institute for Nature Research, Norwegian Ministry of Environment, and the Hedmark University College and to S.B. by the Swedish Research Council. We thank Philip Hedrick, John Vucetich and Josephine Pemberton and two anonymous referees for insightful comments and suggestions on an earlier version of the manuscript.
Supplementary Material
References
- Caughley G. Directions in conservation biology. J. Anim. Ecol. 1994;63:215–244. [Google Scholar]
- Coltman D.W, Pilkington J.G, Smith J.A, Pemberton J.M. Parasite-mediated selection against inbred Soay sheep in a free-living island population. Evolution. 1999;53:1259–1267. doi: 10.1111/j.1558-5646.1999.tb04538.x. [DOI] [PubMed] [Google Scholar]
- Fredrickson R.J, Hedrick P.W. Body size in endangered Mexican wolves: effects of captivity, inbreeding, and cross-lineage matings. Anim. Cons. 2002;5:39–43. [Google Scholar]
- Gilpin M.E, Soulé M.E. Minimum viable populations: processes and extinction. In: Soulé M.E, editor. Conservation biology. The science of scarcity and diversity. Sinauer Associates; Sunderland, MA: 1986. pp. 19–34. [Google Scholar]
- Hedrick P.W, Kalinowski S.T. Inbreeding depression in conservation biology. Ann. Rev. Ecol. Syst. 2000;31:139–162. [Google Scholar]
- Hörnberg S. Changes in population density of moose (Alces alces) and damage to the forests in Sweden. Forest Ecol. Mngmt. 2001;149:141–151. [Google Scholar]
- Kalinowski S.T, Hedrick P.W. An improved method for estimating inbreeding depression in pedigrees. Zool Biol. 1998;17:481–497. [Google Scholar]
- Kalinowski S.T, Hedrick P.W, Miller P.S. No inbreeding depression observed in Mexican and red wolf captive breeding programs. Conserv. Biol. 1999;13:1371–1377. [Google Scholar]
- Keller L.F. Inbreeding and its fitness effects in an insular population of song sparrows (Melospiza melodia) Evolution. 1998;52:240–250. doi: 10.1111/j.1558-5646.1998.tb05157.x. [DOI] [PubMed] [Google Scholar]
- Keller L.F, Waller D.M. Inbreeding effects in wild populations. Trends Ecol. Evol. 2002;17:230–241. [Google Scholar]
- Laikre L. Conservation genetics of Nordic carnivores: lessons from zoos. Hereditas. 1999;130:203–216. doi: 10.1111/j.1601-5223.1999.00203.x. [DOI] [PubMed] [Google Scholar]
- Loeske E.B, Kruuk L.E, Sheldon B.C, Merilä J. Severe inbreeding depression in collared flycatchers (Ficedula albicollis) Proc. R. Soc. B. 2002;269:1581–1589. doi: 10.1098/rspb.2002.2049. doi:10.1098/rspb.2002.2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mech L.D. The Natural History Press; New York: 1970. The wolf: the ecology and behaviour of an endangered species. [Google Scholar]
- Messier F. Ungulate population models with predation: a case study with the North American moose. Ecology. 1994;75:478–488. [Google Scholar]
- Miller C.R, Joyce P, Waits L.P. Assessing allelic dropout and genotype reliability using maximum likelihood. Genetics. 2002;160:357–366. doi: 10.1093/genetics/160.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson R.O, Thomas N.J, Thurber J.M, Vucetich J.A, Waite J.A, Waite T.A. Population limitation and the wolves of Isle Royale. J. Mammal. 1998;79:828–841. [Google Scholar]
- Ralls K.J, Ballou J.D, Templeton A.D. Estimates of lethal equivalents and the cost of inbreeding in mammals. Conserv. Biol. 1988;2:185–193. [Google Scholar]
- Sand, H., Zimmerman, B., Wabakken, P., Andrén, H. & Pedersen, H. C. In press. GPS-technology and GIS-cluster analyses as tools to estimate kill rates in wolf–ungulate ecosystems. Wildlife Soc. Bull.
- Sarkar G, Sommer S. Shedding light on PCR contamination. Nature. 1990;343:27. doi: 10.1038/343027a0. [DOI] [PubMed] [Google Scholar]
- Slate J, Kruuk L.E.B, Marshall T.C, Pemberton J.M, Clutton-Brock T.H. Inbreeding depression influences lifetime breeding success in a wild population of red deer (Cervus elaphus) Proc. R. Soc. B. 2000;267:1657–1662. doi: 10.1098/rspb.2000.1192. doi:10.1098/rspb.2000.1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlet P, Waits L.P, Luikart G. Noninvasive genetic sampling: look before you leap. Trends Ecol. Evol. 1999;14:323–327. doi: 10.1016/s0169-5347(99)01637-7. [DOI] [PubMed] [Google Scholar]
- Vila C, Sundqvist A.-K, Flagstad Ø, Seddon J, Blörnerfeldt S, Kojola I, Casulli A, Sand H, Wabakken P, Ellegren H. Rescue of a severely bottlenecked wolf (Canis lupus) population by a single immigrant. Proc. R. Soc. B. 2003;270:91–97. doi: 10.1098/rspb.2002.2184. doi:10.1098/rspb.2002.2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wabakken P, Sand H, Liberg O, Bjärvall A. The recovery, distribution, and population dynamics of wolves on the Scandinavian peninsula, 1978–1998. Can. J. Zool. 2001;79:710–725. [Google Scholar]
- Wayne R.K, et al. Conservation genetics of the endangered Isle Royale wolf. Conserv. Biol. 1991;5:41–51. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.