Abstract
The computer and the cell both use information embedded in simple coding, the binary software code and the quadruple genomic code, respectively, to support system operations. A comparative examination of their system architecture as well as their information storage and utilization schemes is performed. On top of the code, both systems display a modular, multi-layered architecture, which, in the case of a computer, arises from human engineering efforts through a combination of hardware implementation and software abstraction. Using the computer as a reference system, a simplistic mapping of the architectural components between the two is easily detected. This comparison also reveals that a cell abolishes the software–hardware barrier through genomic encoding for the constituents of the biochemical network, a cell's ‘hardware’ equivalent to the computer central processing unit (CPU). The information loading (gene expression) process acts as a major determinant of the encoded constituent's abundance, which, in turn, often determines the ‘bandwidth’ of a biochemical pathway. Cellular processes are implemented in biochemical pathways in parallel manners. In a computer, on the other hand, the software provides only instructions and data for the CPU. A process represents just sequentially ordered actions by the CPU and only virtual parallelism can be implemented through CPU time-sharing. Whereas process management in a computer may simply mean job scheduling, coordinating pathway bandwidth through the gene expression machinery represents a major process management scheme in a cell. In summary, a cell can be viewed as a super-parallel computer, which computes through controlled hardware composition. While we have, at best, a very fragmented understanding of cellular operation, we have a thorough understanding of the computer throughout the engineering process. The potential utilization of this knowledge to the benefit of systems biology is discussed.
Keywords: information storage, information retrieval, system architecture, computation process, process management
1. Introduction
The complexity of a cell, the basic unit of an organism, is attracting the attention of investigators from a diverse array of scientific disciplines. This has arisen, at least in part, owing to the pioneering work of Adleman (1994), which used biological materials to solve a computational problem. Recently, a complex computer composed of biological materials was successfully assembled to implement the Turing machine, the computing model underlying all silicon-based computers (Benenson et al. 2004). These developments, however, do not reflect how a cell operates as a molecular system (Ji 1999). Research into DNA computing is now chiefly concerned with investigating biochemical processes that can be viewed as logical computations and then using them to our advantage (Parker 2003). It was suggested that a cell be studied as a DNA-based molecular computer (Ji 1999), the direction this paper adopts.
Meanwhile, the paradigm in experimental molecular and cellular biology, which used to be much more reductionism oriented, is shifting towards a synthetic one. This shifting is facilitated and necessitated by genomic sequencing efforts and the routine use of high-throughput research methodologies. As such, a new synthetic theoretical framework for interpreting systems operations will help. However, it is still premature for us theoretically to comprehend the enormous complexity of the cell as a molecular system. Instead, a comparative study with a simpler but comparable system can be illuminating and conducive to integrative thinking, as molecular and cellular biological research has historically used simpler model species to shed light on issues encountered in studying higher species. Such a reference system always shares similar system architecture and operation with the target system to ensure that a comparative study is helpful. Additionally, in order to make reliable theoretical projections we must have a complete (or at least a much better) understanding of the model system.
Artificial complex systems, and the knowledge accumulated in the engineering process, represent rich resources for such comparative study. Commonality between biological systems and artificial complex systems does exist. Exploring this commonality has proved fruitful. For example, the scale-free network concept was developed based on common phenomenon shared by biochemical network with non-biological networks (Barabási & Oltvai 2004).
The computer, arguably the most sophisticated system produced by human engineering, is the ideal choice of reference system for this comparative study. First, both a computer and a cell are systems with enormous intrinsic complexity. Intriguingly, both use seemingly very simple code, the computer binary code and the quadruple genetic code (A, C, G, T), respectively, to regulate complex system operation. Second, we have a complete understanding of a computer, since it is the product of human engineering efforts. On the other hand, we have at best a very fragmented understanding of any cellular system.
Therefore, a comparative study between a cell and a computer will be illuminating for systems biology, of which one goal is to understand the system architecture and operation of a cell. In this paper, we perform a comparative examination of the system architectures, the computing processes and the process management schemes of the two systems. Many interesting parallels between the two seemingly completely different systems are discussed. We also point out major differences that make a cell a much more efficient system than an electrical computer.
2. The prototypes and the scope
Cell and computer are abstract concepts, each referring to a highly heterogeneous group, i.e. there are many types of computers and cells. We need representative, and well-defined, prototype systems for this comparative study. It is also necessary to limit the scope of this study. As the two systems are tremendously complex, it is not feasible to cover all aspects of them in one article.
Even though there are many types of computer, the underlying computing theory is shared by all. System-level functionality is supported by a complex hierarchical architecture, the result of a combination of hardware implementation and software abstraction at various levels. We follow the description of this computer architecture by Tanenbaum (1999). Each computer type represents a unique way of implementing the computing theory, i.e. a unique combination of hardware implementation and software abstraction. Identical functions can be carried out by a single-central processing unit (CPU) computer and also by a faster, multi-processor machine. Parallel and distributed computing can be considered as an extension of a single-CPU model, as the effect can be virtually implemented through software abstraction in a single-CPU computer (Tanenbaum 1999). For the sake of simplicity, a single-CPU model is adopted in this paper.
The cell is the smallest structural unit of an organism that is capable of autonomous system integrity and functionality in a changing environment, with no dependence on an external source for architectural components. There are numerous types of cells. All invariantly rely on information embedded in the quadruple genomic DNA code for system operation. Unless otherwise specified, this discussion is best suited for the yeast Saccharomyces cerevisiae, a popular model system in biological research. Most topics, however, are also applicable to other cell types.
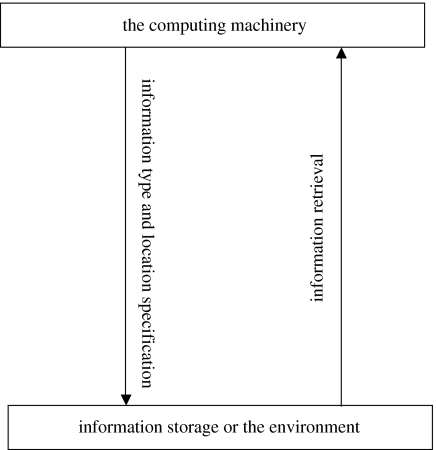
We restrict our focus to how a cell utilizes the genetic code to specify adaptive actions in response to environmental signals, a scheme universal for all cell types. Many important aspects of cellular operation are considered peripheral and are not discussed. Aspects that are specific to the cell and therefore lack comparative values, such as cell replication, are also excluded. We will compare the information organization schemes and the information utilization processes, which are central to both cells and computers. The information utilization process is described as an operation cycle of three steps: information specification, information retrieval and the computing process (figure 1). Information specification means specifying the information (instruction or data) type expected by the computing machinery and the information's location, often a specific address inside the information storage whose specification interlaces intricately with the information organization scheme. In the information retrieval step, the specified instruction/data are transmitted to the computing machinery, which executes the action and then specifies information required for the next cycle. This information utilization process also incorporates environmental signals. For example, in the case of a computer, information specification can be requesting specific user keyboard actions. Information retrieved will then be what the user keyboards back. As another example, the retrieved information can be a direct user input, such as a signal to terminate an executing program. Many details are black-boxed into the three steps and will be analysed below.
Figure 1.
A schematic of an operation cycle.
3. The cell as a multi-layered complex system
Let us begin the comparison with a very simplistic overview of a cell as a multi-layered system, using a computer as the reference system. Molecular and cellular biology research methods, which enabled us to break open a cellular system and see into its internal structure and mechanisms of operation, helped us to gain a better understanding of a variety of distinct regulatory processes. A regulatory process can be divided into multiple steps. One or more gene products, usually proteins, carry out the regulatory events at each step. The information corresponding to a gene product and its temporal–spatial expression profile is embedded inside the seemingly simplistic, one-dimensional genomic sequence. However, at the cellular level, genes and their products are organized into a hierarchical multi-dimensional structure (Barabási & Oltvai 2004), sometimes termed genetic circuit. This structure shows some striking similarities to the multi-layered computer architecture (Tanenbaum 1999), as discussed below. Just as a computer register works as an information (instruction or data) storage unit (Tanenbaum 1999), each individual gene serves as a single genetic unit that stores information corresponding to a regulatory event. Cellular functionality, on the other hand, can be interpreted as the overall properties or behaviours displayed by the cellular system as a whole. Just as any function of a computer is supported by a set of instructions organized into processes (Tanenbaum 1999), a cellular function is generally not a result of the action of just a single gene. There are multiple layers of actions before the primary genetic code gets translated into cellular behaviour. First, the gene set of a cell is organized into biochemical pathways, such as metabolism and signal transduction pathways. Individual pathways are then organized into functional sectors through their interconnectivity and intercorrelation. Examples of the functional sectors include the metabolism network, signal transduction network, cell cycle control network and others. These functional sectors are then joined together to form a dynamic hierarchical network or circuit that works as a cellular operating system, on which a cell executes sets of programmes to maintain its systematic integrity and to make necessary adjustments in response to its environment. Nevertheless, our knowledge is very fragmentary. We are still at an early stage of the journey to a systematic view of how the genetic information governs the cellular machinery's operation to support the overall properties or functions of a cell.
Central to the issue is the information utilization process (outlined in figure 1), of which we have a complete understanding in a computer. A striking similarity exists between a computer and a cell. In both systems, the code provides instruction for an operation. The state of the system, on the other hand, determines how the code will be interpreted and/or used. In a cell, the state of the cellular machinery determines which gene should be expressed or shut down. In a computer, this means where in the memory the CPU will go to fetch data or instructions. We therefore look inside the architecture of a computer for potential theoretical insight.
We discuss a comparative dissection of the multi-layered architecture in a computer and that in a cell, in an attempt to generate a simplistic synthetic view of cellular architecture and process management. We find a striking similarity in system operation, in many cases the same principle is differentially implemented in the two systems. One theme of difference is a clear hardware–software barrier in a computer but a very blurred one in a cell. Another one is the virtual parallelism in a computer and the maxi-parallelism implemented at hardware level in a cell.
4. A comparative study of architectural components
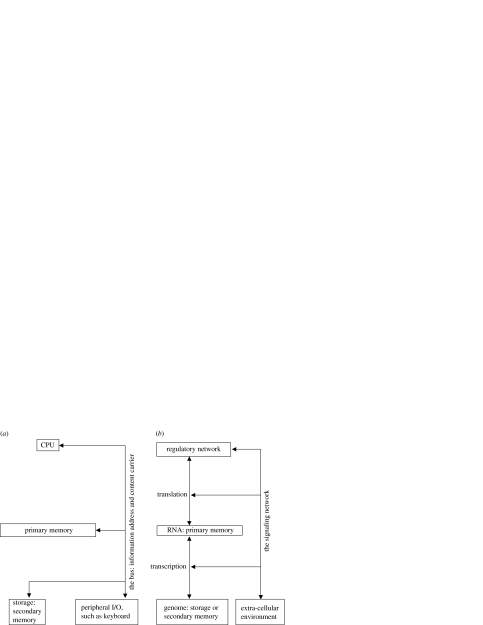
Let us briefly outline the architecture of a computer. The major components of a computer are the CPU, the memory and the input/output (I/O) system (figure 2a). A computer uses a two-layered memory system, the primary and the secondary memory. The secondary memory is the storage for CPU data and instructions. At any moment, parts of the data and instructions are loaded into the primary memory. The loaded information determines what the CPU will do. The computer bus, major components of the I/O system, connects the CPU with the memory and other peripheral I/O apparatus. It sometimes serves as the carrier of the information being loaded or stored. Sometimes, the binary electronic signals in the wires of a bus denote a numeric memory address, from which the CPU receives input or to which it sends output (figure 2a).
Figure 2.
A simplistic schematic comparison of the architecture of a computer (a) and a cell (b). Note that production of components of the signalling network through gene expression is neglected in (b) to simplify the figure, since kinase regulation is predominantly post-translational.
Functional equivalents can be identified for these components in a cell (figure 2b). A cell seems to use the two-layered memory system as well. The genome stores all the data and instructions. However, it is the information loaded into the RNA space that determines what a cell does at any moment. The information stored in the RNA space gives rise to the components of a dynamic biochemical network, a cell's equivalent to computer hardware. Computer hardware, as described above, can be divided into the bus and the CPU. The cellular hardware can be arbitrarily divided in a similar fashion as well. The kinase signal transduction network and the gene expression (transcription and translation) machinery interconnect other parts of the biochemical network, the memory system and the extracellular environment (figure 2b). They therefore function as a cellular bus. Other parts of the biochemical network act as the cellular equivalent to the computer CPU (figure 2b). Next, we compare the computing process in the two systems, and the analogy becomes more appealing.
5. The computing process
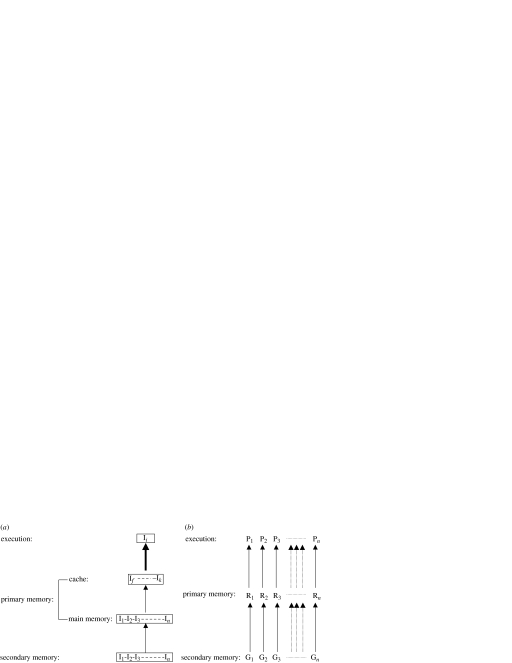
We consider the computing process composed of the information loading from the storage to the primary memory, the upload of loaded information to the computing machinery, and the execution of the encoded actions. A striking parallel between the computing processes of a computer and those of a cell also exist (figure 3 and table 1).
Figure 3.
A comparative snapshot of the serial computing process in a computer (a) and the parallel computing process in a cell (b). In a computer (a), the instructions (I) are loaded through addressing in a serial manner from storage to main memory. Instructions with high priority are then stored in the cache, which has a higher bus bandwidth with the CPU owing to reduced access time. Instructions are executed serially, with only one instruction of a program being executed by the CPU at any moment. In a cell (b), genes (G) are transcribed into RNAs (R) through transcription factor-binding process and then translated into proteins (P) in a parallel manner. All steps of a pathway also proceed in a parallel manner.
Table 1.
The computing process.
| computer | cell | |
|---|---|---|
| form of computation | controlled electrical flux | controlled chemical flux |
| flux-control mechanism | transistor conductivity control | catalysis |
| flux-control agent | transistors organized into circuit | mostly protein |
| function of an instruction | binary electrical signal transformation | chemical reaction |
| input to an instruction | binary electrical signal | reactants of a chemical reaction |
| output from an instruction | binary electrical signal | products of a chemical reaction |
| instruction implementation | data-path wiring control | production of the catalyst |
| multiple instruction execution | sequential data-path control | parallel production of the catalysts |
Memory loading in a computer and transcription in a cell parallel each other, in that they both transfer information needed by the computation machinery from the storage to the primary memory. The information is located through memory addressing in a computer (figure 3a) and through transcription factor-binding process in a cell (figure 3b). A transcription factor complex is a set of proteins that identify a specific DNA location—they therefore represent a mechanism to retrieve information from a specific DNA address.
A computer uses a common bus to specify memory location and to retrieve (or store) specified information (figure 3a). A cell mainly uses the signalling network to regulate the biochemical activities of the transcription factors that determine when and where a gene is activated, analogous to address specification. The retrieval of the information is through a different channel, the transcription machinery. The transcription process loads information from the genome to the RNA space, the ribonome. Transcription factor binding often occurs at multiple genes simultaneously, so that multiple genes can be transcribed in a parallel manner. In other words, the bandwidth of the bus system increases through implementing more hardware, enabling parallel retrieval of multiple fragments of information (figure 3b).
Loaded processes can then be executed. In both systems, a process is divided into many steps. Each step of a computer represents an instruction. An instruction usually is a mechanism, through which an input is transformed to an output in a CPU (or data path) cycle. The computing in a computer is through electrical process: turning on/off a connection. The computing inside a cell is through a chemical process: turning on/off a chemical reaction by catalysis (table 1). A protein usually is, or is part of, an enabling force for a particular chemical reaction. Therefore, a protein is equivalent to the arithmetic logic unit (ALU) of a data path inside a computer CPU.
In a computer, computation is achieved through changing what the CPU does: the CPU executes different instructions at different steps (figure 3a). However, in a cell, computation is achieved through implementing instruction execution at each step as a protein or a protein complex, with each step receiving input from the upstream step (figure 3b). The input is in the format of chemical molecules instead of the electronic signals retrieved from a register in a computer (table 1). The translation machinery plays a major role. The translation process loads information from the RNA to the protein, generating the computing hardware to enable a chemical reaction that is analogous to the execution of a computer CPU instruction. The translation process in a cell is therefore equivalent to the CPU instruction-fetching process in a computer: a computer instruction informs the CPU what to do, while the translation process in a cell produces the enabling factor for the chemical reaction at each step.
In a computer, accessing memory causes CPU delay. This is because the main memory is outside the CPU and connected with the CPU through the bus. Accessing main memory is always slower than CPU execution. Thus, to reduce the CPU delay, a hierarchy of primary memory exists in a computer. High priority CPU data or instructions are stored inside the faster cache, which sometimes locates inside the CPU to completely abolish the delay and which is more expensive to build than the main memory (figure 3a). The cells minimize this delay by two mechanisms. First, analogous to the caching of high priority data or instructions, cells constitutively express many proteins, whose activities are often controlled allosterically or by post-translational modification. Second, the delay is reduced through enhanced bus bandwidth. Multiple RNAs can be translated in a parallel manner into proteins (figure 3b), whereas instructions in a computer program are retrieved sequentially through the bus.
6. Multi-layered architecture revisited
Let us revisit the system architecture in greater detail now in order to see how a complex task is achieved. The theme is fixed hardware and CPU time-sharing in a computer in contrast to a dynamic hardware composite and parallel processing in a cell (table 2). Owing to the high cost of building hardware, the layers in a computer are implemented through hardware implementation and software abstraction. The hardware implementation ends in the middle of the instruction set architecture (ISA) level. In the cells, on the other hand, each element of the biochemical network, including the cellular bus system, is usually implemented directly in hardware, in the form of a gene product dynamically produced through the transcription–translation channel.
Table 2.
Multi-layered architecture from ground up.
| level | function | implementation in computer | implementation in cell |
|---|---|---|---|
| digital logic | processing engine | gate (organized into circuit) as basic element | amino acid as basic element |
| primary memory | gate (organized into register) as basic element | ribonucleotide as basic element | |
| secondary memory | miscellaneous, depending on media | deoxyribonucleotide as basic element | |
| micro-architecture | basic computing structure | data path (the ALU, a circuit, connected with a number of registers) | protein |
| ISA | instruction specification | predetermined formatting | the transcribed region of a gene |
| instruction function | controlling the data path | enabling a chemical reaction | |
| data specification | type and format | substrate specificity | |
| memory addressing | numerical | promoter and enhancer binding | |
| operating system | memory management | swapping information in and out of primary memory from secondary memory | gene transcription and RNA degradation |
| process management | shared resource such as semaphore | shared route by pathways | |
| interprocess signal | pathway cross-talk | ||
| CPU time-sharing | pathway bandwidth management | ||
| file system | continuous or indexed | discontinuous indexed (TF binding) |
Let us briefly, and simplistically, overview the layers in the computer from the transistor/gate level up to the higher programming language level, following the definition and description by Tanenbaum (1999). We will see how a complex computation is achieved through sequential execution of very simple instructions. The gate, a number of transistors wired together to perform a basic Boolean calculation, is the unit of computer CPU and primary memory. At the digital logic level, a unique Boolean function is implemented through the unique way these transistors are wired together. One layer up is the micro-architecture level. In the memory, they are wired into groups to form a register, the unit of memory. In the CPU, gates are combined to form a data path, the main component of the computing engine and in which conductivity of some transistors is dynamically controlled. Controlled transistor conductivity results in dynamically regulated electricity flow. A data path can perform different types of calculation if its internal electricity flow pattern is changed. The ISA level defines the set of instructions available to the software programmer as well as their predetermined function and format. Each instruction actually specifies a specific control of one to several data-path cycles; in other words, how transistor conductivity is controlled for the data path to perform a specific calculation in each cycle. The instruction can be either a mathematic calculation or a memory manipulation. Information in memory is organized through its numeric location (addressing) and predetermined format. Instructions and data have predetermined formats. Consequently, the CPU engages different protocols when dealing with different types of instructions or data in the memory. Currently, the ISA level is the end of hardware implementation. From this level up, everything is through software abstraction. The operating system level defines additional instructions (system calls), implemented through software abstraction, for virtual memory management, I/O operation and process management. The benefit of this level is convenience, relieving the computer programmer of these tedious tasks through automation, which is achieved through abstracting many instructions into one system call. The other layers up are programming levels, either assembly language or higher-level languages, such as C or Java. A computer program is essentially a collection of instructions (ISA level or operating system level) in a predefined order. Essentially, it is the dissection of a complex calculation into many sequential steps of simple calculations that are directly executable by the CPU. The same task can usually be accomplished by multiple, very different algorithms. When the algorithm is efficient, minimum numbers of CPU cycles are used. While tremendous efforts have been made to generate parallel execution effect, it is still mostly sequential computing owing to the limited hardware resources. Parallel computing effect is simulated through CPU time-sharing, either among multiple processes or among multiple fragments, when not mutually dependent, of the same process.
In a cell, the functionality of all these layers is implemented at hardware level. The unit of the computing machinery is the amino acid. The unit of primary and secondary memory is the nucleotide in RNA and DNA, respectively. Instead of using one single data path executing an instruction set through sequential control, a cell implements each instruction through the production of a protein from mRNA, the primary memory. The primary memory is organized through a predetermined format (an open reading frame) and signals embedded in the associated non-translated regions of mRNA. These embedded signals determine when and where the translation process is initiated, analogous to the numerical location (address) of computer memory. Information in mRNA is in turn loaded out of genomic DNA, the secondary memory (or information storage). Here, the signals embedded in the promoter and the enhancer regions are analogous to computer addressing of secondary memory, in that both are used by the information retrieval machinery to locate a linear fragment of information. The format is the start and the end of transcription. In eukaryotic cells, especially in multi-cellular species, it is more complicated owing to the occurrence of RNA splicing and editing, which blurred the distinction between addressing and formatting in memory organization. The gene expression machinery dynamically interprets a transcribed genomic region. In other words, if we refer to the gene expression machinery as the information channel, a gene exhibits different meanings under different channel conditions (Wang in press).
The set of catalysed chemical reactions essentially constitutes the ISA level of a cell. The catalysing agents, mostly proteins, represent the computing hardware of a cell. Contrary to using one data path executing a set of instructions in a computer, a cell implements each instruction in a unique enabling agent, usually as a protein produced from mRNA, as discussed earlier. This results in a blurred hardware–software barrier in a cell. Consequently, the evolution process, the designer of the cell, changes the software through genetic mutations, which in turn, codes for the hardware components of cellular computing and memory organization. In a computer, on the other hand, a programmer changes only the software. The new functionality is generated through changing what the hardware will do. In other words, the difference is that the cell is a computer that constructs itself.
7. Memory management: from storage to main memory
In a computer, memory management means keeping track of the secondary and primary memory and managing the loading of necessary information to the main memory to meet the demand of the CPU, which sends the instruction through the computer bus. The information retrieval in a computer uses a binary scheme via a memory management unit (MMU). The MMU keeps track of the content of the primary memory and provides mechanisms for locating (through a translation table between numerical memory addresses) a fragment of the secondary memory in order to load it into the primary memory. Fragments with low priority are purged from the primary memory. Each file is either not retrieved or retrieved as a single copy. A process represents just a sequentially ordered action by the CPU. In other words, the ‘bandwidth’ of each process contains just one execution thread. However, in a cell, it is far more complicated. The gene expression machinery receives input from the signalling network and outputs requested genes to the RNA space. The scheme used is far more complicated than a binary scheme. The input to the gene expression machinery determines how many copies of a gene product, either RNA or protein, are produced. Similarly to purging low-priority information from the primary memory in a computer, RNA is also being degraded. The balance between production and degradation determines abundance of each RNA species, which is a major determinant of the abundance of the corresponding protein. The abundance of a gene product can have a wide range. The abundance of the constituents of a biochemical pathway in turn determines the number of concurrently executing threads, a major determinant of the bandwidth of the pathway. This gives rise to a complex biochemical network, which is reconfigurable through controlled information loading by the gene expression process and which is at the core of the cellular computing machinery. Coordinating the bandwidth of multiple biochemical pathways thus becomes an important issue, which will be re-visited when we discuss cellular process management later.
8. The file system in a cell: the storage
Efficient information retrieval from the secondary memory to the primary memory, as discussed above, relies on good organization of the secondary memory. Computer memory is organized into files. Computer files can be classified in many ways. By information type, some contain just data, while others, the executables, contain instructions. By way of information storage, some are just ordered byte-stream. Others contain lists of logical record, each with a well-defined structure. By way of information retrieval, some are sequentially retrieved. Some are indexed. However, in a cell, most genes code for proteins corresponding to instructions. If you analogise the gene set of a cellular process to a computer file, the majority of files stored in a genome are executable files. The genes of a process are located through indexes, the promoters and the enhancers. Prokaryotic operons are similar to a computer process file in that the instructions of a biochemical pathway are organized together in a sequential order to form a contiguous unit. However, in eukaryotic cells, genes of a process are dispersed throughout the genome although they can be commonly addressed through transcription factors. Parallel retrieval of multiple instructions often occurs through the occurrence of the same set of transcription factor-binding sites in the regulatory regions of these genes. A transcription factor becomes a one-to-many memory pointer or an index. Extreme examples include the locus control region, where no genes are coded but pointers to multiple genes are provided.
9. Process management in a cell: from primary memory to CPU
At the operating level in a cell, process management is necessary but there is no need for managing CPU time owing to the parallel computing nature. In a computer, each loaded process represents just one execution thread. The bandwidth of the computing machinery determines how many CPU cycles can be achieved in a unit time. Process management is achieved through scheduling, whether and when to assign CPU time to a process. However, process management becomes much more complicated in a cell.
The operating system is implemented at hardware level in a cell. Unlike a computer process, a cellular process is inherently multi-threaded at hardware level. As discussed earlier, each biochemical pathway has a manageable bandwidth, a major determinant of which is the gene expression process. Process management therefore has to be achieved through coordinating the bandwidth of biochemical pathways. The abundance (copy number) and the biochemical activity (kinetic parameters) of the proteins in the process are major determinants of the bandwidth of the process. The constituent's abundance determines the number of parallel executing threads. The constituent's biochemical activity determines the execution speed of each thread. Therefore, two mechanisms are extensively used in process bandwidth management.
First, process control is achieved by post-translational effects. One way is modulating the activity of the proteins through allosteric interaction or covalent modifications. One such mechanism is protein phosphorylation. For example, glycogen synthase, a key enzyme in glucose metabolism, is regulated through phosphorylation by the kinase GSK-3 (Cohen 1986). Another way is for multiple processes to share common junction points or routes, commonly termed cross-talk in molecular and cellular biology. These cross-talks are analogous to the interprocess signals and shared resources (such as semaphore) used in computers to orchestrate multiple processes.
Second, process control is achieved through modulating the abundance of the proteins in a pathway through control of gene expression as well as RNA or protein degradation. Essentially, the information loading process becomes part of the process management scheme. The best example for this process management mechanism is the prokaryotic polycistronic operon, which is more analogous to computer process management through organizing instructions of a process into one modular unit of execution and management. In eukaryotic cells, all transcription units are single genes. However, micro-array analysis revealed that genes of a biochemical pathway are more likely to be coregulated at transcription level (Hughes et al. 2000). The other determinant of the abundance of a gene product is the degradation process. Not surprisingly, RNA degradation is regulated in a similar manner (Wang et al. 2002). The ubiquitin protein degradation system also has proved to be highly regulated, even though new high-throughput technology is yet to be developed to measure the process.
10. System-level properties: the goal of the computation process
All aspects at various levels of the two systems support interdependent properties manifested at the system level. System integrity, for example, results from coordinating activities of different parts of the system. System functionality, as another example, means responding to environmental signals. A computer responds to external signals such as user input to perform requested tasks. A cell actively detects environmental signals, such as the presence of stress or changes in nutrient availability, and makes relevant adjustments. Both systems are capable of executing programs in response to environmental factors and, at the same time, maintaining system integrity.
11. Discussion
The computer is an increasingly important biological research tool for data storage and analysis as well as for in silico modelling. In this paper, we propose that it may serve yet another role, a model system for systems biology. This is consistent with the use of model systems in the history of biology, such as the use of yeast as a model for the study of higher eukaryotic species.
We have compared the computing architecture in a computer with that in a cell. These systems are made of different materials and at different scales (macro- versus micro-scales) and seemingly have nothing in common, but they display a surprising parallel in many aspects of their system architecture and operation control. We think it is possible to derive a unifying abstract computational model. This model can be divided into an information-processing module, a control module and a communication module that interconnects the processing module, the control module and the environment. Modern computers implement the model in a highly serial (or virtual parallel through time-sharing) manner and with clear distinction between hardware and software, performing computations through controlled electric flux. A cell is an implementation of such a model in a highly parallel manner, and with a blurred software–hardware barrier, performing computations through controlled catalysis of chemical reactions.
One of the major differences between a computer and a cell, as discussed above, is the embedding of code for dynamic construction of hardware components inside the genome, blurring the distinction between hardware and software. This enables biochemical pathways to have a bandwidth manageable through controlling the abundance of individual components of the hardware, i.e. the hardware composition. Consequently, while a computer performs computation through serial CPU control, in a cell, the computation is performed through managing hardware composition. Molecular and cell biology has elucidated mechanisms at the micro-architecture and the ISA levels. However, at the operating system level, pathway bandwidth management represents the next target of research. Tremendous challenges lie ahead, since as yet we do not have a systematic understanding of gene expression regulation.
The other major difference is the maxi-parallel computing nature of a cell. Whereas parallelism is virtually implemented through CPU job scheduling in computers, it is directly implemented at the hardware level in cells. Essentially, all elements of the biochemical network are implemented in corresponding gene products dynamically produced through the gene expression channel. In contrast to a centralized CPU in a computer, gene products are loosely distributed in the cells. The computation is distributed to multiple cellular network modules, which in eukaryotic cells often reside in different organelles (Srere 1987; Hartwell et al. 1999; Norris et al. 1999).
It should be pointed out that a computer entirely lacks one cellular functional domain: the replication process during which some genomic mutations occur. The evolution process, the designer of the cell, selects advantageous changes to the genomic sequences, embedded in which are the codes for the hardware components of cellular computing and memory organization. This evolutionary process has been mimicked by evolutionary computing technique (Koza et al. 1999). In one experiment, each self-replicating computer program improved itself by introducing mutations in its code during replication to better compete for the host computer's CPU time, analogous to the selection pressure in biological evolution. Emergence of complex features was observed (Lenski et al. 2003). Currently computers have no self-replication capability. However, a computer can be interpreted as a cell in a non-replicating state (as most human cells are) to perform some functionality, in which case, the cell cycle machinery can be neglected. A cell, on the other hand, is a computer with the capability for self-replication, in which the genome serves as the carrier for information transmission between generations.
Even though we have focused on potential benefits for biologists, this comparative study should also prove useful for computer scientists. Computer engineering can benefit in at least two aspects. First, parallel computing is currently a major research topic in computer sciences. Looking into cells should provide ample insights for parallel computing principles and their implementation. Second, the cells exhibit much better error tolerance capability, resulting in improved system robustness. Redundancy plays a major role. The cells have multiple mechanisms to process the same information. If one fails, there are other ways to ensure that crucial cellular processes, such as programmed cell death, are completed. Computer scientists should look into cells for potential principles in redundancy design and in balancing redundancy and economical issues in system design.
Another benefit of this comparative study is in molecular and cellular biology education, which is facing increasing challenges in the face of genomic sequence availability and the routine application of high-throughput research technologies (Bialek & Botstein 2004). This comparison suggests that principles of computer architecture design and system operation may serve as a potential framework (although simplistic) for organizing molecular and cellular biological knowledge, which currently is highly fragmentary and often lacking in general design principles. This approach will effectively break the barrier between life sciences and other branches of sciences. Such an integrative approach should equip a student with a better, organically organized, knowledge set to face the daunting challenges in the post-genomic era.
In summary, besides being a research tool, the computer may potentially serve as a simple model system for systems biology. It may prove helpful for the current integrative approaches in biological research. It may also be useful in molecular and cellular biological education, which increasingly demands interdisciplinary approaches (Bialek & Botstein 2004), and which is inundated with huge amounts of details—often lacking an organizational framework.
References
- Adleman L.M. Molecular computation of solutions to combinatorial problems. Science. 1994;266:1021–1024. doi: 10.1126/science.7973651. [DOI] [PubMed] [Google Scholar]
- Barabási A.L, Oltvai Z.N. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Benenson Y, Gil B, Ben-Dor U, Adar R, Shapiro E. An autonomous molecular computer for logical control of gene expression. Nature. 2004;429:423–429. doi: 10.1038/nature02551. [DOI] [PubMed] [Google Scholar]
- Bialek W, Botstein D. Introductory science and mathematics education for 21st-century biologists. Science. 2004;303:788–790. doi: 10.1126/science.1095480. [DOI] [PubMed] [Google Scholar]
- Cohen P. Muscle glycogen synthase. In: Boyer P, Krebs E, editors. The enzymes. Academic Press; New York: 1986. pp. 461–497. [Google Scholar]
- Hartwell L.H, Hopfiled J.J, Leibler S, Murray A.W. From molecular to modular cell biology. Nature. 1999;402(Suppl. 6761):C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- Hughes T.R, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Ji S. The cell as the smallest DNA-based molecular computer. BioSystems. 1999;52:123–133. doi: 10.1016/s0303-2647(99)00039-8. [DOI] [PubMed] [Google Scholar]
- Koza J.R, Bennett F.H, III, Andre D, Keane M.A. Morgan Kaufmann; San Francisco, CA: 1999. Genetic programming III: Darwinian invention and problem solving. [Google Scholar]
- Lenski R.E, Ofria C, Pennock R.T, Adami C. The evolutionary origin of complex features. Nature. 2003;423:139–144. doi: 10.1038/nature01568. [DOI] [PubMed] [Google Scholar]
- Norris V, et al. Hypothesis: hyperstructures regulate bacterial structure and the cell cycle. Biochimie. 1999;81:915–920. doi: 10.1016/s0300-9084(99)00203-5. [DOI] [PubMed] [Google Scholar]
- Parker J. Computing with DNA. EMBO Rep. 2003;4:7–10. doi: 10.1038/sj.embor.embor719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srere P.A. Complexes of sequential metabolic enzymes. Annu. Rev. Biochem. 1987;56:89–124. doi: 10.1146/annurev.bi.56.070187.000513. [DOI] [PubMed] [Google Scholar]
- Tanenbaum A.S. 4th edn. Prentice Hall; Upper Saddle River, NJ: 1999. Structured computer organization. [Google Scholar]
- Wang, D. In press ‘Molecular gene’: interpretation in the right context. Biol. Phil.20 (2).
- Wang Y, Liu C.L, Storey J.D, Tibshirani R.J, Herschlag D, Brown P.O. Precision and functional specificity in mRNA decay. Proc. Natl Acad. Sci. USA. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]