Abstract
VPgs are essential for replication of picornaviruses, which cause diseases such as poliomyelitis, foot and mouth disease, and the common cold. VPg in infected cells is covalently linked to the 5' end of the viral RNA, or, in a uridylylated form, free in the cytoplasm. We show here the first solution structure for a picornaviral VPg, that of the 22-residue peptide from poliovirus serotype 1. VPg in buffer is inherently flexible, but a single conformer was obtained by adding trimethylamine N-oxide (TMAO). TMAO had only minor effects on the TOCSY spectrum. However, it increased the amount of structured peptide, as indicated by more peaks in the NOESY spectrum and an up to 300% increase in the ratio of normalized NOE crosspeak intensities to that in buffer. The data for VPg in TMAO yielded a well defined structure bundle with 0.6Å RMSD (vs. 6.6 Å in buffer alone), with 10–30 unambiguous constraints per residue. The structure consists of a large loop region from residues 1–14, from which the reactive tyrosinate projects outward, and a C-terminal helix from residues 18–21 that aligns the sidechains of conserved residues on one face. The structure has a stable docking position at an area on the poliovirus polymerase crystal structure identified as a VPg binding site by mutagenesis studies. Further, UTP and ATP dock in a base-specific manner to the reactive face of VPg, held in place by residues conserved in all picornavirus VPgs.
Keywords: viral replication, polymerase interaction, picornavirus, circular dichroism, trimethylamine N-oxide (TMAO), solvent stabilization, uridylylation, post-translational modification
1. Introduction
"Viral proteins linked to the genome" (VPgs), first discovered attached to the genomic RNA of poliovirus nearly three decades ago [1, 23, 28, 38, 54], are essential for virus growth and well conserved in all picornaviruses, including those that cause poliomyelitis, hepatitis A, foot and mouth disease and the common cold [10, 20, 29, 45, 51]. VPgs are linked to the 5’ terminal UMP of the genomic RNAs via phosphodiester bonds to the hydroxyl group of Tyr3 [1, 28, 44]. Free VPg peptides, uridylylated at the same reactive tyrosine residue (VPgpU or VPgpUpU), are found in the cytoplasm of infected cells [37, 59–61] [16, 31] and serve as primers for viral replication. Functional studies of VPg mutants have shown that even residues quite distant in the sequence from Tyr3, such as Pro14, Arg17 and Lys20, reduced viral replication and uridylylation in the in vitro assay[35, 36]. This indicated that the peptide must form a compact structure to be uridylylated. Thus we undertook solution studies to determine structural features that could account for the interaction of the VPg of poliovirus serotype 1 with the viral polymerase.
Many peptides are flexible in solution, and thus determining their NMR structures is not straightforward. Typical results for studies of unstabilized peptides show mostly random or undetermined structure, especially in the N- and C-termini. Our initial results, using buffer alone, showed that VPg was inherently flexible in low salt buffer at neutral pH. Addition of TMAO, a solvent known to stabilize other proteins and peptides [5, 47, 53] [58] [26] [15, 25] [14] enabled us to determine a high resolution structure of the VPg peptide of poliovirus serotype 1. The structure is maintained by residues conserved in picornavirus VPgs. Further, by comparing the chemical shifts and peak intensities in the stabilizing solvent with those in buffer alone, we were able to demonstrate that the solvent stabilized the major conformation in buffer, and reduced internal flexibility by interactions with both the solvent and individual sidechains.
Thanks to the resolution of the structure in the stabilizing solvent, we were able to determine the relative position of sidechains of amino acids required for VPg activity and interaction with the polymerase. Our results indicate that these aid in forming a compact structure, with residues shown by mutagenesis studies to be essential for uridylylation and viral replication serving to stabilize the position of the reactive tyrosine. Evidence for the biological significance of the stabilized structure was obtained by molecular docking studies. UTP and ATP dock to the VPg NMR structure in a base-specific manner, held in position by conserved residues that are required for VPg activity. VPg itself docks near residues on the poliovirus polymerase that were previously shown to interact with the peptide during uridylylation by mutagenesis studies[3, 31]. The structure will thus aid in elucidating the role of this essential peptide in picornavirus replication.
2. Materials and Methods
2.1 VPg peptide and solution conditions
For NMR experiments, chemically synthesized VPg of poliovirus serotype 1 (a generous gift from J. H. van Boom [11]) was dissolved to 3.7 mM (5 mg in 0.6 ml final volume) in 10 mM Na phosphate pH 7.0–10%D20. Where indicated, a 4M solution of deuterated TMAO (Cambridge Isotope Lab., DLM-4779), adjusted to pH 7 with HCl and 10 mM Na phosphate and 10% D2O, was added to a final concentration of 1 M. A small amount of DSS (sodium 2,2-dimethyl-2-silapentane-5-sulfonate) in D2O was added as an internal reference. The samples were centrifuged for 10 min in a desktop centrifuge at 10,000 x g before transferring to an NMR tube. They were then degassed with N2 and topped with argon before closing. Samples were stored at 4 ºC and the NMR spectra were collected at 10 ºC.
2. 2 Circular dichroism
The peptide was dissolved in buffer with or without TMAO added as described above to the concentrations indicated in Figure 1. Spectra were measured at room temperature in an AVIV spectrophotometer model 62DS[48].
Figure 1.
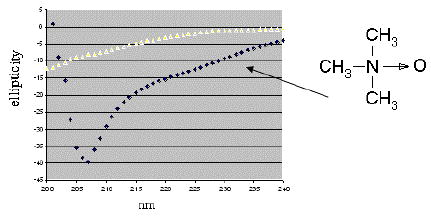
Circular dichroism spectra of chemically synthesized VPg of poliovirus serotype 1 in two solvent conditions. In 10 mM Na phosphate buffer, pH 7 at a concentration of 0.04 mM (top line), the peptide shows little evidence of stable secondary structure. The spectrum of a ~0.2 mM solution in 1M TMAO (bottom line) indicates the peptide is ordered. The minimum at 207 nm and the trough between 220–230 nm suggests a combination of secondary structure elements, with some helical content.
2.3 NMR experiments
All spectra were recorded on Varian UnityPlus 600 MHz or 750 MHz spectrometers equipped with Z-axis gradients. WET-NOESY [50]spectra were collected at 5 and 10 ºC, with 100 and 200 ms mixing times, 7 kHz sweep widths, 1024 complex data points in f2, and 256 to 1024 complex points in f1. TOCSY spectra [41]were recorded at 10 ºC with 60 ms mixing times, 7 kHz sweep widths, and 1024 by 256 complex data points in f2 and f1, respectively. Spectra were analyzed with FELIX (Accelrys) software. Chemical shift assignments were done manually, using data from TOCSY, NOESY, and DQF-COSY [39]experiments.
2.4 Structure calculation with NOAH/DIAMOD
We used our program suite NOAH/DIAMOD[6, 7, 13, 32, 34, 56] to automatically assign the NOESY cross-peaks and calculate bundles of structures[13, 57]. Previous studies demonstrate that the 3D structures and automated assignments generated with these programs are in excellent agreement with results from conventional manual assignments [33, 56].
2.5 %NOE intensity
NOE peak intensities (from FELIX) that were clearly visible in both spectra (i.e., with and without TMAO) were divided by the relevant diagonal crosspeak intensities to give %NOE intensity. The ratio of the normalized intensity in TMAO to that in buffer was then calculated to determine the effect of TMAO on NOE transfer efficiency. As peaks between the amide protons were clearest in the buffer spectrum run on the 600 MHz spectrometer, this was used for the comparisons in the first section of Table 1. For the bottom half of the table, peaks were calculated using a NOESY spectrum in buffer collected on the 750 MHz spectrometer at 5°C.
Table 1.
NMR and refinement statistics for the structure of PV-VPg in 1M TMAO.
| Protein | |
|---|---|
| NMR distance & dihedral constraints | |
| Distance constraints | 266 |
| Total NOE | 471 |
| Intra-residue | 127 |
| Inter-residue | 139 |
| Sequential (|i−j| = 1) | 79 |
| Medium-range (|i−j| < 4) | 240 |
| Long-range (|i−j| >= 5) | 26 |
| Intermolecular | --- |
| Hydrogen bonds | 0 |
| Total dihedral angle restraints | 10 |
| phi | 5 |
| psi | 5 |
| Structure Statistics | |
| Violations (mean and s.d.) | |
| Distance constraints (Å) | 0.041±0.108 |
| Dihedral angle constraints (º) | 1.34±3.51 |
| Max. dihedral angle violation (º) | 15.1 |
| Max. distance constraint violation (Å) | 0.66 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.011 |
| Bond angles (º) | 1.9 |
| Impropers (º) | --- |
| Average pairwise r.m.s.d.* (Å) | |
| Heavy | 0.93 ± 0.21 |
| Backbone | 0.57 ± 0.19 |
Pairwise r.m.s.d. was calculated among 10 refined structures.
2.6 Molecular docking
The ZDOCK program[9, 30], which uses a fast Fourier transform method to evaluate possible binding modes for a ligand on a given receptor protein, was used for all dockings. The ZDOCK score for complexes contains terms for shape complementarity, desolvation energy, and the electrostatic potentials of the molecular surfaces. Drawings were done with MOLMOL[21] or (for Figure 5B) SwissPDBViewer. The poliovirus polymerase structure was taken from PDB file 1RA6[52]. The ATP structure was taken from PDB file 1AOI. Several different UTP ligand files were used for docking, either drawn with Chem3D (to control the protonation state) or with structures taken from PDB files 1GX6 and 1UEI. Figure 5B shows the docking of a UTP molecule with full protonation of the phosphate oxygens.
Figure 5.
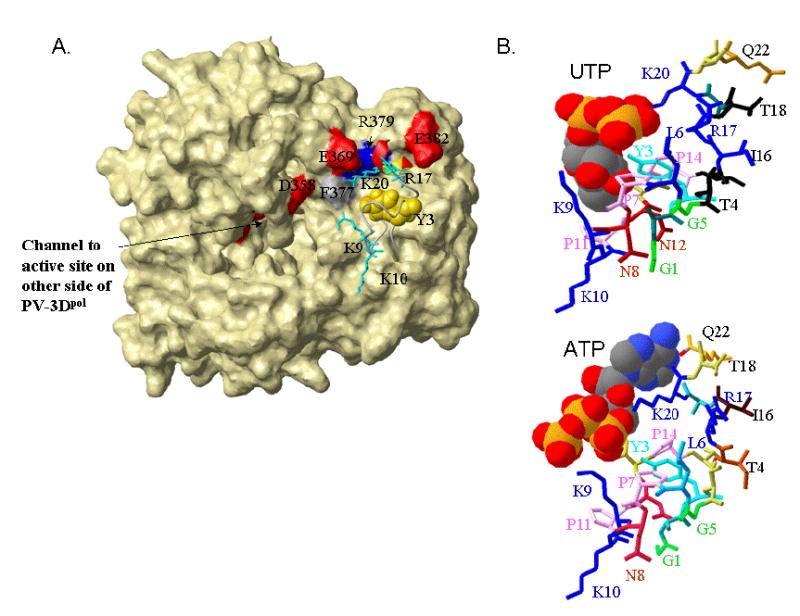
Results of docking studies with the stabilized VPg structure. A) The stabilized VPg structure docks near amino acids known to be in the interaction site (Glu382, red, Arg379 blue, Phe377 gray[31]) on the surface of the poliovirus 3Dpol . The polymerase is shown in surface representation and other nearby acidic residues (Glu369, Asp358, red) are highlighted and labeled. The VPg is in ribbon format, with Tyr3 space filling and colored gold, and the positively charged conserved sidechains (Lys 9,10, 20 and Arg17) shown in neon and colored turquoise. B) Possible binding modes for UTP (top) and ATP on VPg. The VPg structure is shown in ribbon format with the side chains (in neon) differentially colored, and the nucleotides are in CPK format. Consistent with the net positive charge of the adenine base, interactions of ATP with VPg are primarily through its triphosphate tail. The negatively charged uridine base has many interactions with the VPg sidechains, particularly to the Lys9 and the amides of the asparagines residues, in addition to interactions between the phosphate groups and Lys20. As the top dockings varied somewhat in the exact positioning of the nucleotides, the complexes selected for this figure have a short distance between the α-phosphate of the docked nucleotide and the Tyr3-OH group. This was complex 5 for UTP (Z-dock score18.7) and complex 2 for ATP (Zdock score 38.41).
3. Results
3.1 CD and NMR spectra of VPg in different solvent environments
Initial CD results for a solution of chemically synthesized VPg of poliovirus serotype 1 (Figure 1) indicated the peptide was not in a well-defined conformation in phosphate buffered solution. Adding 1M TMAO gave a spectrum suggesting an ordered conformation, with possibly some helical content as indicated by a minimum at 207nM (Figure 1, bottom spectrum). The increase in ordered structure was consistent with other reports that 1–3 M TMAO solution stabilized the active conformation of enzymes and DNA binding factors [5, 12]. The effect of 1M TMAO on proton 2D NMR spectra of VPg was then tested. We first found that chemical shifts, determined from TOCSY experiments, for most protons showed only a slight decrease, of 0.01–0.03 ppm, in the presence of TMAO when compared to buffer alone. Only a few chemical shifts, such as that for Gly5-NH (7.27 vs 7.40 ppm), increased in the presence of TMAO. The lack of significant change in the chemical shifts suggest is consistent with TMAO modifying the population (or ensemble) of structures, rather than structurally altering the peptide.
On the other hand, TMAO greatly enhanced the intensity and number of cross peaks in all areas of the NOESY spectrum of VPg (Figure 2). Several peaks arising from interactions between the exchangeable amide protons of neighboring residues seen in the spectrum in phosphate buffer are much enhanced by the presence of TMAO (Fig.2a). As the expanded fingerprint region of the spectrum shows (Figure 2b), while many peaks remain unchanged in both spectra, TMAO increases the number of NOE peaks. These additional peaks are consistent with the solvent stabilizing the position of the 3 prolines and interactions between the (non-exchanging) proline Hδ protons and the amide protons from the previous residues. Interactions between large sidechains are also stabilized. There is, for example, a small crosspeak, just visible in Figure 2b, between the Gly5NH and the Tyr3BH, in the NOE spectrum in the presence of TMAO.
Figure 2.
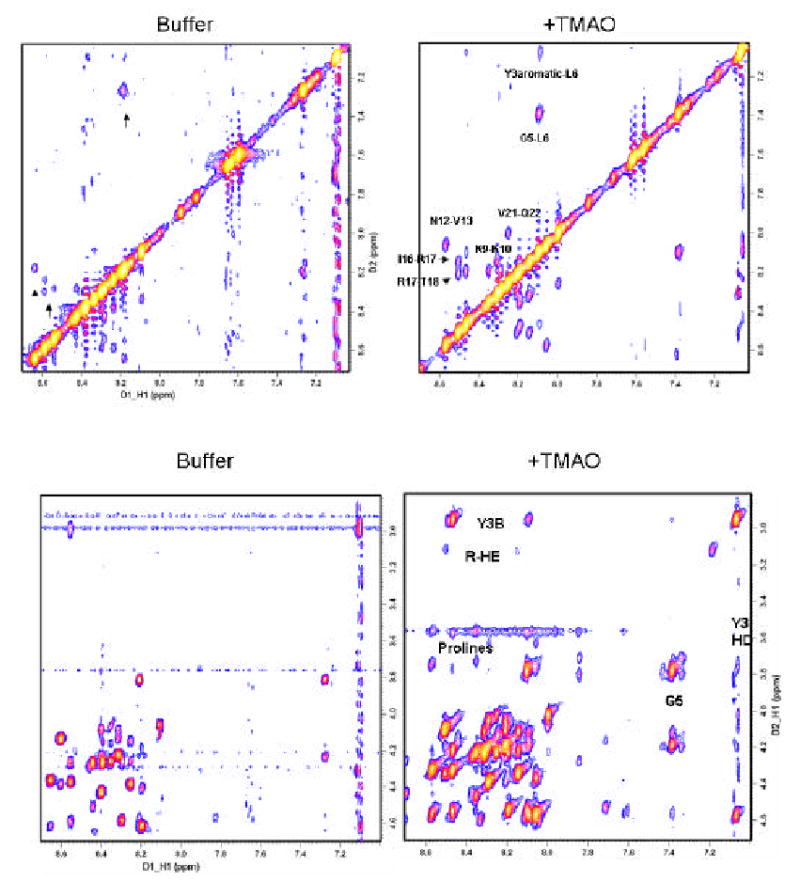
TMAO stabilizes structure based interactions between residues, as indicated by the enhanced size and number of peaks in NOESY spectra. a) The amide proton interaction regions (N-N) of NOESY spectra in buffer without (left) and with 1M TMAO (right) are shown. The presence of several peaks in both spectra (arrows) indicate TMAO stabilizes a peptide configuration that is already present, but at a lower concentration, in buffer not containing TMAO. b) Fingerprint region of NOESY spectra in buffer without (left) and with (right) TMAO. Note: Interresidue crosspeaks clearly visible in the TMAO spectrum (such as between backbone amides and the 3 prolines, Gly5, the Hβ of the reactive tyrosine, and the Hɛ of Arg 17) are seen only at very low contour level in the buffered sample. Thus the buffer spectrum pictures were made at a lower contour level (higher sensitivity but more background peaks) than those for TMAO, to better highlight peaks common to both spectra. Background peaks can be distinguished by their low intensity, repeating patterns and their lack of correlation with any of the chemical shifts for the peptide protons. Table 2 gives a more exact quantitation of the intensity difference between peaks common to the two spectra.
3.2 The structure bundle in TMAO is better defined
Structures calculated by the NOAH-DIAMOD program from the NOESY spectrum in buffer converged but the bundle was relatively undefined (Figure 3a). In contrast, structure calculations from the sample in TMAO converged rapidly to a compact bundle (Figure 3b), after about half of the NOESY peaks had been assigned unambiguously (Table 1 and supplemental information). The primary secondary structure elements of VPg, a large loop formed from residue 1–14, stabilized by the 3 conserved proline residues, and a helix from residues 18–21, are present also in some of the conformations without TMAO (Figure 3a vs 3b). The RMSD of the structure bundle is reduced from 6.6 Å to ~ 0.6 Å in 1M TMAO. There are about 10 unambiguous assignments per residue in the TMAO NOESY peaklist, with many more constraints for key residues, such as Arg17 (more than 30; supplemental information and Figure 3c). Figure 4 shows that hand-assigned long- and short-range interresidue peaks support the presence of a helix at the C-terminus.
Figure 3.
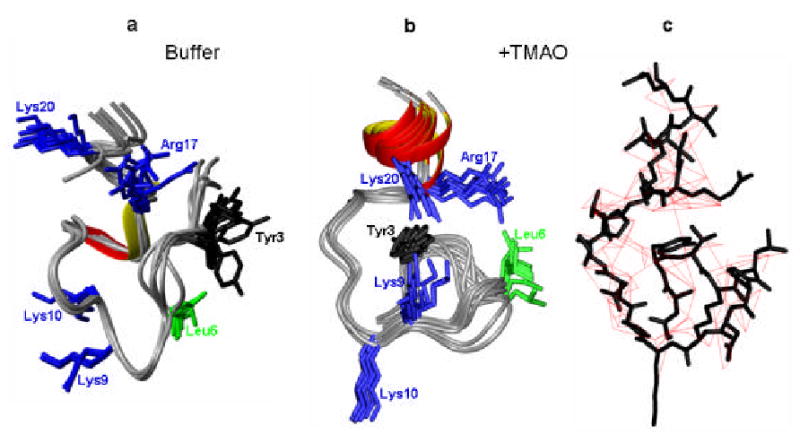
Comparison of the structures calculated for PV-VPg in buffer (a) and in the presence of 1 M TMAO (b) The top 10 structures in buffer alone show much more conformational flexibility, both in the backbone and in the position of the conserved side chains. c) Inter-residue constraints (automatically assigned) used in determining the VPg structure in TMAO.
Figure 4.
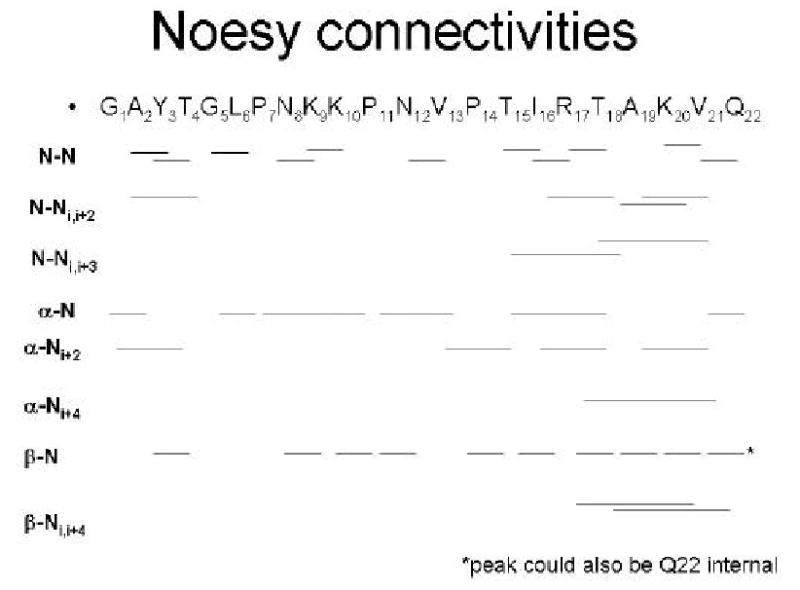
Inter-residue NOE connectivities support the helix at the C-terminus of the structure shown in Figure 3b. (Peaks were hand assigned, from the NOE spectrum in 1M TMAO, described in Figure 2).
The difference between the structure bundles is due to the stabilization of the structure in TMAO that allowed us to obtain sufficient constraints to fix the positions of the large sidechains, and determine how they interact with one another to form a compact structure.
3.3 TMAO increases the amount of structured peptide
While it is understood that membrane and many viral proteins require a hydrophobic environment to fold properly[55], the correct solution environment for maintaining the structure of soluble peptides and proteins is less defined. Adding co-solvents that stabilize proteins against aggregation and deactivation is often necessary to obtain the highly concentrated solutions needed for structural studies[5, 19, 42, 46, 47, 49, 53]. Previous work has shown that TMAO, unlike solvents such as Trifluoroethanol[8], does not particularly stabilize any one secondary structure element, but does stabilize the active structures of enzymes and DNA binding factors[26, 53]. To quantify the position specific effect of TMAO on VPg residues, we compared normalized peak intensities for many different types of proton-proton interactions that were clearly visible in both spectra. To correct for a potential overall increase in signal intensity due to reduced exchange with the solvent[12], peak intensities were normalized by dividing by the diagonal peak intensity to obtain the %NOE. The % NOE's in the spectrum with TMAO are as much as 300% greater than those in buffer alone (Table 2). Because the concentration of the peptide is 33% lower in the TMAO sample, this increase in peak intensities indicates that TMAO acts to restrict the internal movement of the peptide residues, and increases the amount of structured VPg. Were TMAO's effect only to slow hydrogen exchange rates, then the peaks resulting from NOE transfer would increase to the same degree as the diagonal peak intensities.
Table 2.
The ratio of NOE crosspeak intensities to that of the diagonal peaks is enhanced by the presence of 1M TMAO. This indicates that the co-solvent increases the amount of structured peptide, and has other effects besides decreasing the exchange rate with the solvent.
| Crosspeak assignment | % intensity Buffer | % intensity TMAO | Increase in NOE transfer in TMAO |
|---|---|---|---|
| Between exchangeable protons: | |||
| N12-NH/V13-NH | 1.7 | 3.5 | 2.06 |
| R17-NH/ I16-NH | 0.61 | 1.5 | 2.45 |
| R17-NH/T18-NH | 0.5 | 1.7 | 3.4 |
| N8-NH/ K9-NH | 0.46 | 1.1 | 2.4 |
| K10-NH K9-NH | 0.85 | 2.6 | 3.05 |
| L6-NH G5NH | 0.43 | 1.4 | 3.25 |
| Q22-NH/ V21NH | 0.46 | .79 | 1.72 |
| N12HD1 N12HD2 | 11.6 | 27.8 | 2.4 |
| N8HD1 N8HD2 | 13.2 | 30.4 | 2.3 |
| Intra- and inter-sidechain (nonexchanging protons): | |||
| Y3HA toHB3 | 2.3 | 3.88 | 1.68 |
| L6 HG toHD | 5.0 | 5.5 | 1.09 |
| P7 HA/HB | 9.28 | 6.96 | .75 |
| P7 HA/HG1 | 1.03 | 1.51 | 1.46 |
| P7 HA/HG2 | 2.6 | 1.88 | 0.72 |
| N8 HA to HB | 1.0 | 1.34 | 1.34 |
| V13 HB/HG | 2.7 | 4.1 | 1.54 |
| K9 HB2/HB3 | 15. | 19.3 | 1.28 |
| K10 HA/HG | 1.46 | 1.47 | 1.01 |
| K10 HA/P11HD | 11.4 | 14.6 | 1.48 |
| K10 HA/P11 HB | 1.15 | 1.65 | 1.43 |
| V13HA/P14HB2 | 4.0 | 6.4 | 1.6 |
| V13HA/P14HG2 | 2.4 | 4.8 | 2.0 |
| I16 HB/HG1 | 2.5 | 2.64 | 1.06 |
| I16 HB/HG2 | 5.35 | 5.36 | 1.00 |
| I16HG1/HD | 2.8 | 4.3 | 1.55 |
| R17HG2/V21HG | 0.69 | 1.1 | 1.59 |
Further, this enhanced NOE transfer rate was seen for peaks from protons throughout the structure. The highest observed effects were on crosspeaks where both protons were exchangeable (e.g., between backbone amides of neighboring residues). There was also a significant increase in intensity of NOE peaks between inter-residue, non-exchangeable sidechain and Cα protons in the fingerprint regions. While some intra-residue interactions between sidechain protons were relatively unaffected, especially for protons in the proline sidechains (note the intensity decrease represents exactly that of the concentration), others between protons at the end of large sidechains, such as Tyr3 and Ile16, with their Hα/Hβ protons, were also enhanced by TMAO. Peaks between sidechains where both protons were non-exchangeable were also enhanced, as for example between the V13 sidechain and P14.
3.4 Docking studies with the stabilized structure
VPg must interact productively with the poliovirus polymerase and nucleotides, particularly UTP, to be uridylylated. Previous mutagenesis studies have indicated that VPg interacts with the poliovirus polymerse (PV-3Dpol) at a cleft on the face opposite to the active site[31]. As figure 5A shows, the VPg NMR structure has a low energy docking position near the PV-3Dpol residues previously identified, namely Phe377, Arg379 and Glu382.
In addition, it is known that the potyvirus VPg, which is a much larger protein than the VPg of picornaviruses, can bind to nucleotides, and that this activity is required for its uridylyation[40]. We thus determined low energy docking sites for both UTP and ATP on our stabilized structure of PV-VPg. Both of these nucleotides docked to the VPg NMR structure, in a base specific manner (Figure 5B). That is, conserved VPg residues interacted with both the base and the phosphate groups of UTP, while stable contacts with ATP were primarily to the phosphate oxygen atoms. The exact position of UTP varied to some extent with the protonation state. The docking shown (for UTP with the phosphate oxygens protonated) is one in which Tyr3 hydroxyl lies closest to the α-phosphate position. A possible mechanism for uridylylation of VPg, in which the 5' end of an RNA molecule aids in positioning the nucleotide, is discussed in detail elsewhere (Schein et al., 2006, Proteins, in press).
4. Discussion
Flexible peptides present a problem for structure determination. Either one finds many possible structures, such as the bundle shown in Figure 3a, or one can fix the conformation, by crystallizing the peptide or by adding stabilizing solvents. In the first case, a peptide model structure, supported by a few constraints common to most conformations, is often shown. It may be argued that none of these approaches presents a satisfactory picture of the biological situation, where other proteins, small molecules and cofactors provide interactions that induce a specific fold. Further, a flexible structure may be needed for modifications [17], such as the uridylylation reaction that VPg undergoes to perform its biological functions
We have thus chosen to show both the structures of VPg in buffer and in TMAO (Figure 3), to emphasize that while the structure in TMAO is consistent with mutational and docking data (Figure 5), additional conformations are possible depending on solution conditions. Other studies have shown that high concentrations of TMAO can replace one partner in protein complexes[24, 25] and stabilize the active conformation of enzymes[2]. The comparison of the spectra (Table 2 and Figure 2) shows that TMAO enhances NOE cross peak intensity and lowers the internal motion of VPg. Explanations for how TMAO can stabilize proteins include that it reduces proton exchange with the solvent, and raises the barrier for interconversion of dynamic folding intermediates that might be more prone to aggregation than the native state [4, 18]. At high concentrations, the amphipathic structure of TMAO, clear from figure 1, could mimic the reactive groups available in the protein rich environment of the cytosol. We also note that most of the sidechains of VPg, particularly the 3 prolines, one tyrosine, 3 lysines, and one arginine residue would be expected to interact favorably with TMAO according to measurements of their transfer free energy[2].
4.1 Conserved residues interact with the reactive Tyrosine in the structure
The structure in TMAO is consistent with experimental data and molecular docking studies. The residues conserved in VPgs of picornaviruses (Table 3) are all on the same face of the structure as Tyr3, the residue that is attached through its phenolic moiety to the 5' end of the viral RNA (or to UMP in free VPgpU). In addition to the N-terminal Gly and C-terminal Gln of VPg, conserved residues (in PV-VPg numbering) include Gly5, Lys 9 and 10, Pro14 and Arg17; Lys 20 is conserved in the enteroviruses. Mutating any of these positions reduced or prevented the growth of the poliovirus in HeLa cells or yielded quasi-infectious virus (i.e., the progeny virus reverted to wild type) [22, 23, 27, 36, 43]. Most of these conserved residues, as Figure 5B shows, could aid in forming a surface suitable for binding nucleotides near the reactive tyrosinate side chain. Mutating Arg17, which in our structure interacts with nearly every other residue in the C-terminal half of the peptide (Figure 3c), completely eliminated VPg activity. Pro14, which lies at the turning point between the two major secondary structure areas, cannot be mutated without reducing the functionality of VPg. Mutating other conserved residues inhibits interaction of VPg with the polymerase and can alter structure as well. For example, two mutants of PV-VPg, Gly5Pro and Thr3Tyr4 (where the positions of the Thr and Tyr residues are switched) completely eliminate uridylylation and reduce its interaction with the polymerase[36]. Both of these mutants have considerably different TOCSY spectra from the wild type, as indicated by a decrease in the chemical shift of the Tyr3 amide (data not shown). In contrast, adding TMAO to the wild type PV-VPg had a much smaller effect on the TOCSY spectrum, and the relative position of the Tyr3 chemical shifts was not altered.
Table 3.
Picornavirus VPgs have conserved sequence features. Bold capital letters: essential for poliovirus replication according to mutagenesis studies[36].
| Poliovirus | GAYTGL-PNkkPnVPTiRtAKVQ |
| Coxsackievirus B3 | GAYTGv-PNqkPrVPTlRqAKVQ |
| Enterovirus consensus: | GaYtGl-pnxkpkvPtlRqakvQ |
| Foot and mouth disease-1 | GPYAGPLERQKPLKVRAKLPQQE |
| Foot and mouth disease-3 | GPYEGPVKKPVALKVKAKNLIVTE |
| Hepatitis A | GVYHGV-TKPKQVIKLDADPVE |
| Human Rhinovirus 2 | GPYSGE-PKPKTKIPERRVVtQ |
| Human Rhinovirus 14 | GPYSGnpPhnKlKaPtlRpVVvQ |
| Human Rhinovirus 89 | GPYSGE-PKPKsRaPERRVVaQ |
| Rhinovirus consensus: | GPYSGE-PKPKtkvPERrVVaQ |
Molecular docking studies (Figure 5A) also indicate that the lowest target function structure in TMAO docks efficiently to the previously determined interaction site for VPg on the polyvirus polymerase[31]. Docking studies with UTP or ATP also indicated that the reactive face of VPg forms a potential binding surface for nucleotides (Figure 5B). These results are consistent with a mechanism in which VPg plays a role in its own uridylyation, which need not occur within the active site of the polymerase. (Schein et al., Proteins, 2006, in press).
4.2 In conclusion, this first structure of VPg, a peptide essential for picornavirus replication, indicates that residues conserved throughout its sequence contribute to the overall structure. The use of TMAO was critical for determining a highly-refined VPg structure; the similarity of the TOCSY spectra in the two solvent conditions indicated that TMAO stabilized a major configuration of VPg in buffer. TMAO increased the amount of structured peptide, as indicated by the enhanced NOE transfer efficiency (% NOE) as well as the number of peaks visible in NOESY spectra of the peptide. The structure docked efficiently to a predetermined binding site on the PV-polymerase, and the reactive face provides a surface that could play a role in binding of nucleotides. This structure of VPg in solution paves the way for further structural studies with mutants and peptidomimetics, to better understand the interactions of VPg with the poliovirus polymerase.
Acknowledgments
This work was supported by grants from the U.S. Department of Energy (Grant Number DE-FG-00ER63041), the Sealy Center for Vaccine Development, and the National Institutes of Health to Eckard Wimmer (Grant number R37AI015122) and Werner Braun (R21AI55746). The NMR facilities of the Sealy Center for Structural Biology and Molecular Biophysics, which were funded by grants from the Sealy Foundation and Building funds from the NIH (Grant number 1CO6CA59098), were used in this project. We thank Lucy Lee for running the CD experiments.
Footnotes
Competing interests: The authors declare they have no competing financial interest that would be affected by publication of this article.
Supplemental figures: Description of the NMR structure calculation for poliovirus VPg in 1M TMAO. a)convergence of the NOAH-DIAMOD structure calculation b) local RMSD of the final structure bundle, c) the number of unambiguous constraints per residue.
References
- 1.Ambros V, Baltimore D. Protein is linked to the 5' end of poliovirus RNA by a phosphodiester linkage to tyrosine. J Biol Chem. 1978;253:5263–6. [PubMed] [Google Scholar]
- 2.Auton M, Bolen DW. Predicting the energetics of osmolyte-induced protein folding/unfolding. Proc Natl Acad Sci U S A. 2005;102:15065–8. doi: 10.1073/pnas.0507053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boerner J, Lyle JM, Daijogo S, Semler BL, Schultz SC, Kirkegaard K, et al. Allosteric effects of ligands and mutations on poliovirus RNA-dependent RNA polymerase. Journal of Virology. 2005;79:7803–11. doi: 10.1128/JVI.79.12.7803-7811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolen D, Baskakov IV. The osmophobic effect: natural selection of a thermodynamic force in protein folding. J Mol Biol. 2001;310:955–63. doi: 10.1006/jmbi.2001.4819. [DOI] [PubMed] [Google Scholar]
- 5.Bolen DW. Protein stabilization by naturally occurring osmolytes. Methods Mol Biol. 2001;168:17–36. doi: 10.1385/1-59259-193-0:017. [DOI] [PubMed] [Google Scholar]
- 6.Braun W, Go N. Calculation of protein conformations by proton-proton distance constraints - A new efficient algorithm. J Mol Biol. 1985;186:611–26. doi: 10.1016/0022-2836(85)90134-2. [DOI] [PubMed] [Google Scholar]
- 7.Braun W, Vasak M, Robbins AH, Stout CD, Wagner G, Kägi JHR, et al. Comparison of the NMR solution structure and the X-ray crystal structure of rat metallothionein-2. Proc Nat Acad Sci. 1992;89:10124–8. doi: 10.1073/pnas.89.21.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Celinski S, Scholtz JM. Osmolyte effects on helix formation in peptides and the stability of coiled-coils. Protein Sci. 2002;11:2048–51. doi: 10.1110/ps.0211702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen R, Li L, Weng Z. ZDOCK: An Initial-stage Protein Docking Algorithm Proteins Structure. Function and Genetics. 2003;52:80–7. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 10.Cheneau Y, Rweyemamu MM, Astudillo V, Lubroth J. Global Intiatives for the progressive control and eradication of foot and mouth disease. In: Dodet B, Vicari M, editors. Foot and Mouth Disease Control Strategies. Paris, Amsterdam.,etc.: Elsevier; 2003, p. 247-pf.
- 11.Dreef-Tromp CM, van der Elst H, van den Boogart JE, van der Marel GA, van Boom JH. Solid-phase synthesis of an RNA nucleopeptide fragment from the nucleoprotein of poliovirus. Nucleic Acid Res. 1992;20:2435–9. doi: 10.1093/nar/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foord R, Leatherbarrow RJ. Effect of osmolytes on the exchange rates of backbone amide protons in proteins. Biochemistry. 1998;37:2969–78. doi: 10.1021/bi9712798. [DOI] [PubMed] [Google Scholar]
- 13.Garimella R, Xu Y, Schein CH, Rajarathnam K, Nagle GT, Painter SD, et al. NMR solution structure of attractin, a water-borne protein pheromone from the mollusk Aplysia californica. Biochemistry. 2003;42:9970–9. doi: 10.1021/bi0274322. [DOI] [PubMed] [Google Scholar]
- 14.Gursky O. Probing the conformation of a human apolipoprotein C-1 by amino acid substitutions and trimethylamine-N-oxide. Protein Sci. 1999;8:2055–64. doi: 10.1110/ps.8.10.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henkels C, Kurz JC, Fierke CA, Oas TG. Linked folding and anion binding of the Bacillus subtilis ribonuclease P protein. Biochemistry. 2001;40:2777–89. doi: 10.1021/bi002078y. [DOI] [PubMed] [Google Scholar]
- 16.Hope DA, Diamond SE, Kirkegaard K. Genetic dissection of interaction between poliovirus 3D polymerase and viral protein 3AB. J Virol. 1997;71:9490–8. doi: 10.1128/jvi.71.12.9490-9498.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iakoucheva LM, Radivojac P, Brown C, O'Connor T, Sikes J, Obradovic Z, et al. Intrinsic disorder and protein phosphorylation. Nucleic Acids Research. 2004;32:1037–49. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaravine VA, Rathgeb-Szabo K, Alexandrescu AT. Microscopic stability of cold shock protein A examined by NMR native state hydrogen exchange as a function of urea and trimethylamine N-oxide. Protein Science. 2000;9:290–301. doi: 10.1110/ps.9.2.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klibanov A. Why are enzymes less active in organic solvents than water? Trends Biotechnol. 1997;15:97–101. doi: 10.1016/S0167-7799(97)01013-5. [DOI] [PubMed] [Google Scholar]
- 20.Knowles NJ, Samuel AR. Molecular epidemiology of foot and mouth disease virus. Virus Research. 2003;91:65–80. doi: 10.1016/s0168-1702(02)00260-5. [DOI] [PubMed] [Google Scholar]
- 21.Koradi R, Billeter M, Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graphics. 1996;14:51–5. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 22.Kuhn RJ, Tada H, Ypma-Wong MF, Dunn JJ, Semler BL, Wimmer E. Construction of a "mutagenesis cartridge" for poliovirus genome -linked viral protein: isolation and characterization of viable and nonviable mutants. Proc Natl Acad Sci USA. 1988;85:519–23. doi: 10.1073/pnas.85.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuhn RJ, Tada H, Ypma-Wong MF, Semler BL, Wimmer E. Mutational analysis of the genome -linked protein VPg of poliovirus. J Virol. 1988;62:4207–15. doi: 10.1128/jvi.62.11.4207-4215.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar R, Betney R, Li J, Thompson EB, McEwan IJ. Induced alpha-helix structure in AF1 of the androgen receptor upon binding transcription factor TFIIF. Biochemistry. 2004;43:3008–13. doi: 10.1021/bi035934p. [DOI] [PubMed] [Google Scholar]
- 25.Kumar R, Lee JC, Bolen DW, Thompson EB. The conformation of the glucocorticoid receptor af1/tau1 domain induced by osmolyte binds co-regulatory proteins. J Biol Chem. 2001;276:16146–8152. doi: 10.1074/jbc.M100825200. [DOI] [PubMed] [Google Scholar]
- 26.Kumar R, Serrette JM, Thompson EB. Osmolyte-induced folding enhances tryptic enzyme activity. Arch Biochem Biophys. 2005;436:78–82. doi: 10.1016/j.abb.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 27.Lama JSMA, Rodriguez PL. A role for 3AB protein in poliovirus genome replication. Journal of Biological Chemistry. 1995;270:14430–8. doi: 10.1074/jbc.270.24.14430. [DOI] [PubMed] [Google Scholar]
- 28.Lee YF, Nomoto A, Detjen BM, Wimmer E. A protein covalently linked to poliovirus genome RNA. Proc Natl Acad Sci USA. 1977;74:59–63. doi: 10.1073/pnas.74.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leforban Y, Gerbier G. Recent history of foot and mouth disease in Europe. Foot and Mouth Disease Control Strategies. Paris: Elsevier; 2003.
- 30.Li L, Chen R, Weng Z. RDOCK: Refinement of Rigid-body Protein Docking Predictions. PROTEINS: Structure, Function, and Genetics. 2003;53:693–707. doi: 10.1002/prot.10460. [DOI] [PubMed] [Google Scholar]
- 31.Lyle J, Clewell A, Richmond K, Richards O, Hope D, Schultz S, et al. Similar structural basis for membrane localization and protein priming by an RNA-dependent RNA polymerase. J Biol Chem. 2002;277:16324–31. doi: 10.1074/jbc.M112429200. [DOI] [PubMed] [Google Scholar]
- 32.Mumenthaler C, Braun W. Automated Assignment of Simulated and Experimental NOESY Spectra of Proteins by Feedback Filtering and Self-correcting Distance Geometry. Journal of Molecular Biology. 1995;254:465–80. doi: 10.1006/jmbi.1995.0631. [DOI] [PubMed] [Google Scholar]
- 33.Mumenthaler C, Güntert P, Braun W, Wüthrich K. Automated combined assignment of NOESY spectra and three-dimensional protein structure determination. J Biomol NMR. 1997;10:351–62. doi: 10.1023/a:1018383106236. [DOI] [PubMed] [Google Scholar]
- 34.Oezguen N, Adamian L, Xu Y, Rajarathnam K, Braun W. Automated assignment and 3D structure calculations using combinations of 2D homonuclear and 3D heteronuclear NOESY spectra. Journal of Biomolecular NMR. 2002;22:249–63. doi: 10.1023/a:1014925824100. [DOI] [PubMed] [Google Scholar]
- 35.Paul AV. Possible unifying mechanism of picornavirus genome replication. In: Semler BL, Wimmer E, editors. Molecular Biology of Picornaviruses. Washington: ASM press; 2002, p. 227–45.
- 36.Paul AV, Peters J, Mugavero J, Yin J, van Boom JH, Wimmer E. Biochemical and genetic studies of the VPg-uridylylation reaction catalyzed by the RNA polymerase of poliovirus. J Virol. 2003;77:891–904. doi: 10.1128/JVI.77.2.891-904.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paul AV, Rieder E, Kim DW, van Boom JH, Wimmer E. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J Virol. 2000;74:10359–70. doi: 10.1128/jvi.74.22.10359-10370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paul AV, van Boom JH, Filippov D, Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393:280–4. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- 39.Piantini U, Sorenson OW, Ernst RR. Multiple quantum filters for elucidating NMR coupling networks. J Am Chem Soc. 1982;104:6800–1. [Google Scholar]
- 40.Pusstinen P, Makinen K. Uridylylation of the potyvirus VPg by viral replicase NIb correlates with the nucleotide binding capacity of VPg. J Biol Chem. 2004;279:38103–10. doi: 10.1074/jbc.M402910200. [DOI] [PubMed] [Google Scholar]
- 41.Rance M. Improved techniques for homonuclear rotating-frame and isotropic mixing experiments. J Magn Reson. 1987;74:557–64. [Google Scholar]
- 42.Rariy R, Klibanov A. Correct protein folding in glycerol. PNAS, USA. 1997;94:13520–3. doi: 10.1073/pnas.94.25.13520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reuer Q, Kuhn RJ, Wimmer E. Characterization of poliovirus clones containing lethal and nonlethal mutations in the genome-linked protein VPg. J Virol. 1990;64:2967–75. doi: 10.1128/jvi.64.6.2967-2975.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rothberg PG, Harris TJ, Nomoto A, Wimmer E. 04-(5'-uridylyl)tyrosine is the bond between the genome-linked protein and the RNA of poliovirus. Proc Natl Acad Sci USA. 1978;75:4868–72. doi: 10.1073/pnas.75.10.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saiz M. Foot and mouth disease virus:biology and propects for diease control. Microbes and Infection. 2002;4:1183–92. doi: 10.1016/s1286-4579(02)01644-1. [DOI] [PubMed] [Google Scholar]
- 46.Schein CH. Controlling oligomerization of pharmaceutical proteins. Pharm Acta Helv. 1994;69:119–26. doi: 10.1016/0031-6865(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 47.Schein CH. Solubility as a function of protein structure and solvent components. Bio/Technology. 1990;8:308–17. doi: 10.1038/nbt0490-308. [DOI] [PubMed] [Google Scholar]
- 48.Schein CH, Nagle GT, Page JS, Sweedler JV, Xu Y, Painter SD, et al. Aplysia attractin: Biophysical characterization and modeling of a water-borne pheromone. Biophys J. 2001;81:463–72. doi: 10.1016/S0006-3495(01)75714-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh R, Haque I, Ahmad F. Counteracting Osmolyte Trimethylamine N-Oxide Destabilizes Proteins at pH below Its pKa: Measurements of thermodynamic parameters in the presence and absence of Trimethylamine N-oxide. J Biol Chem. 2005;280:11035–42. doi: 10.1074/jbc.M410716200. [DOI] [PubMed] [Google Scholar]
- 50.Smallcombe SH, Patt SL, Keifer PA. WET Solvent Suppression and its application to LC NMR and High-Resolution NMR Spectroscopy. J Magn Reson. 1995;117:295–303. [Google Scholar]
- 51.Sutmoller P, Barteling SS, Olascoaga RC, Sumption KJ. Control and eradication of foot and mouth disease. Virus Res. 2003;91:101–44. doi: 10.1016/s0168-1702(02)00262-9. [DOI] [PubMed] [Google Scholar]
- 52.Thompson AA, Peersen OB. Structural Basis for Proteolysis-Dependent Activation of the Poliovirus RNA-Dependent RNA. Polymerase Embo J. 2004;23:3462–71. doi: 10.1038/sj.emboj.7600357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tulla-Puche J, Getun IV, Woodward C, Barany G. Native-like conformations are sampled by partially folded and disordered variants of bovine pancreatic trypsin inhibitor. Biochemistry. 2004;43:1591–8. doi: 10.1021/bi035301a. [DOI] [PubMed] [Google Scholar]
- 54.Weitz M, Baroudy B, Maloy W, Ticehurst J, Purcell R. Detection of a genome-linked protein (VPg) of hepatitis A virus and its comparison with other picornaviral VPgs. J Virol. 1986;60:124–30. doi: 10.1128/jvi.60.1.124-130.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilkinson T, Tellinghuisen TL, Kuhn RJ, Post CB. Association of sindbis virus capsid protein with phospholipid membranes and the E2 glycoprotein: implications for alphavirus assembly. Biochemistry. 2005;44:2800–10. doi: 10.1021/bi0479961. [DOI] [PubMed] [Google Scholar]
- 56.Xu Y, Jablonsky MJ, Jackson PL, Braun W, Krishna NR. Automatic assignment of NOESY cross peaks and determination of the protein structure of a new world scorpion neurotoxin using NOAH/DIAMOD. J Magn Reson. 2001;148:35–46. doi: 10.1006/jmre.2000.2220. [DOI] [PubMed] [Google Scholar]
- 57.Xu Y, Wu J, Gorenstein D, Braun W. Automated 2D NOESY assignment and structure calculation of crambin (S22–I25) with the self-correcting distance geometry based NOAH/DIAMOD programs. J Magnetic Resonance. 1999;136:76–85. doi: 10.1006/jmre.1998.1616. [DOI] [PubMed] [Google Scholar]
- 58.Yancey P, Siebenaller JF. Trimethylamine oxide stabilizes teleost and mammalian lactate dehydrogenases against inactivation by hydrostatic pressure and trypsinolysis. J Exp Biol. 1999;202:3597–603. doi: 10.1242/jeb.202.24.3597. 1999 Dec;202(Pt 24) [DOI] [PubMed] [Google Scholar]
- 59.Yang Y, Rijnbrand R, Watowich S, Lemon SM. Genetic Evidence for an Interaction between a Picornaviral cis-Acting RNA Replication Element and 3CD Protein. J Biol Chem. 2004;279:12659–67. doi: 10.1074/jbc.M312992200. [DOI] [PubMed] [Google Scholar]
- 60.Yang Y, Rijnbrand R, McKnight KL, Wimmer E, Paul A, Martin A, et al. Sequence requirements for viral RNA replication and VPg uridylylation directed by the internal cis-acting replication element (cre) of human rhinovirus type 14. J Virol. 2002;76:7485–94. doi: 10.1128/JVI.76.15.7485-7494.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yin J, Paul AV, Wimmer E, Rieder E. Functional dissection of a poliovirus cis-acting replication element [PV-cre(2C)]: analysis of single- and dual-cre viral genomes and proteins that bind specifically to PV-cre RNA. J Virol. 2003;77:5152–66. doi: 10.1128/JVI.77.9.5152-5166.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]


