Abstract
The X-linked RP3 locus codes for retinitis pigmentosa GTPase regulator (RPGR), a protein of unknown function with sequence homology to the guanine nucleotide exchange factor for Ran GTPase. We created an RPGR-deficient murine model by gene knockout. In the mutant mice, cone photoreceptors exhibit ectopic localization of cone opsins in the cell body and synapses and rod photoreceptors have a reduced level of rhodopsin. Subsequently, both cone and rod photoreceptors degenerate. RPGR was found normally localized to the connecting cilia of rod and cone photoreceptors. These data point to a role for RPGR in maintaining the polarized protein distribution across the connecting cilium by facilitating directional transport or restricting redistribution. The function of RPGR is essential for the long-term maintenance of photoreceptor viability.
Retinitis pigmentosa (RP) is a group of hereditary retinal diseases in which photoreceptor cells degenerate. The genetic defects underlying RP are heterogeneous, with well documented autosomal and X-linked forms (http://www.sph.uth.tmc.edu/RetNet). Many of the genes involved code for well characterized functions essential for the rod phototransduction cascade or for the maintenance of the photoreceptor outer segment structure. Others encode unknown functions. Among the latter is the X-linked locus RP3 (OMIM 312610), which accounts for the majority of X-linked RP families by linkage studies. The retinal disease associated with RP3 appears to have an early age of onset (1) and exhibits early involvement of both cones and rods (2–7). This is in contrast with other types of RP in which rods are primarily affected followed by a secondary degeneration of cones. Because daytime vision and visual acuity depend on cone function, patients with RP3 would experience visual handicaps earlier and with greater severity than the average RP patients. The clinical expression of RP3 suggests that the genetic defect affects an essential function for both rods and cones.
The gene at the RP3 locus was recently isolated and found to encode retinitis pigmentosa GTPase regulator (RPGR) (8, 9), so named because of its similarity to the regulator of chromatin condensation (RCC1), a nuclear protein that catalyzes guanine nucleotide exchange for the small GTPase Ran. The N-terminal half of RPGR consists of six complete tandem repeats of 52–54 amino acids with homology to the repeat structure of RCC1. RCC1 has an essential role in nuclear import and export through its regulation of Ran (10). The presence of an RCC1 homology domain thus raises the possibility that RPGR might participate in some type of intracellular transport process in photoreceptors. The function of RPGR remains unknown (11, 12). To investigate the in vivo function of RPGR and to understand the disease mechanism associated with RPGR mutations, we generated an RPGR-deficient mouse model and examined its phenotype. We also determined the normal subcellular localization of RPGR in photoreceptors. Our results are consistent with RPGR being involved in the maintenance of a polarized distribution of outer segment-specific protein(s).
Materials and Methods
cDNA and Genomic DNA Cloning, Embryonic Stem (ES) Cells, and Generation of RPGR Knockout (KO) Mice.
Using the published human RPGR cDNA sequence (8), we searched the National Center for Biotechnology Information mouse expressed sequence tag database and identified several mouse expressed sequence tags sharing a high degree of homology with the human sequence, indicating that they represented mouse orthologs of RPGR. Oligonucleotide primers were designed based on the expressed sequence tag sequences and were used for PCR amplification of full length mouse RPGR cDNA from mouse retinas. The compiled sequence was subsequently found to be identical to that published by others (12). The X chromosomal origin of this cDNA was confirmed by a PCR-based mapping method using an interspecific backcross mouse DNA panel obtained from The Jackson Laboratory (13). Genomic fragments spanning exons 1–4 and exons 6–8 were amplified by PCR from 129/Sv mouse genomic DNA. These fragments were cloned into the vector pGT1.8IRESβgeo (14) to generate the targeting vector shown in Fig. 1a. Linearized vector DNA was electroporated into J1 ES cells, and neomycin-resistant colonies were selected. Homologous recombinant ES clones were identified by genomic Southern blotting. Two independent targeted clones were microinjected into C57BL/6 blastocysts to generate chimeras. Male chimeric founders were crossed with C57BL/6 females. Nullizygous offspring were obtained through intercross over several generations. Males carrying a single mutant allele and females carrying two mutant alleles were considered to be of the same genotype for the purpose of phenotype analyses and were referred to as nullizygous or KO mice. Subsequent generations of mice were genotyped by PCR using a primer pair spanning the inserted marker. Sequences of the upstream (exon 4) and downstream (exon 6) primers were 5′-TGAGAAGGTGAAACTTGCTGCCTGTGGAAGGAAC and 5′-CCAATCTGCCCTTCAGAATTGTCACCCCACATAA. The sizes of the PCR products were 3.5 kb [wild type (WT)] and 8 kb (KO), respectively.
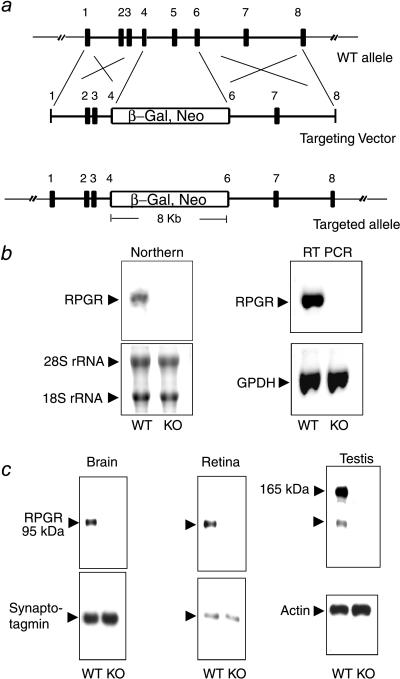
Figure 1.
Targeted disruption of the RPGR gene. (a) The targeting construct. Partial structure of the WT allele is shown (exons 1–8; filled boxes). The targeting vector contains a selectable marker (β-Gal, Neo) that replaces part of exon 4 through part of exon 6. (b) Absence of the normal RPGR transcript in the KO mice. (Left) Northern blot analysis with a full-length RPGR cDNA probe could detect a 3-kb transcript in brain RNA from WT but not from KO mice. Fluorescent dye-stained rRNA bands are shown as a loading control. (Right) Reverse transcription–PCR analysis confirms the absence of the normal RPGR transcript in the KO mice. PCR amplification of GPDH from the same cDNA templates is shown as a control. (c) RPGR protein expression was ablated in the KO mice. Total proteins from brain, retina, or testis were probed with anti-RPGR antibodies on immunoblots. The same blots were reprobed with either synaptotagmin (abundant in neural tissues) or actin antibodies as loading controls.
RNA Analysis.
Total RNA was extracted from mouse tissues. Northern blotting was carried out under high stringency conditions according to standard protocols. For reverse transcription–PCR, cDNA templates were prepared from total RNA by reverse-transcription with the Superscript II preamplification system (GIBCO/BRL) primed with random hexamers. PCR was carried out by using a pair of primers in exon 3 (5′-GTAACAACTGGGGTCAGTTA) and exon 5 (5′-CCTTCATTATTTCCACCAGCTG).
Antibodies, Immunoblotting, and Immunocytochemistry.
A cDNA fragment coding for the C-terminal 250-aa residues of murine RPGR was subcloned into the expression vector pET28 (Novagen). Purified recombinant RPGR was used to immunize both chickens and rabbits. Antisera and/or egg yolk with high titer specific antibodies were collected. Total IgY was extracted from egg yolks. Rabbit antisera and IgY were used, with or without affinity-purification, for immunoblotting and immunocytochemistry. In these experiments, preimmune serum, unrelated total rabbit IgG, or preimmune IgY were included, as appropriate, for negative controls. Immunocytochemistry was performed on aldehyde-fixed frozen sections (10 μm), except that unfixed frozen sections were found necessary and therefore were used for RPGR staining. Frozen sections were cut along the superior to inferior axis. Dissociated photoreceptor cells were prepared as described (15). Both the chicken and rabbit RPGR antibodies gave similar staining patterns on immunoblots and on retinal sections, although the rabbit antibodies gave a better signal to noise ratio. Antibodies specific for the blue and green cone opsin were used as described (16). Anti-glial fibrillary acidic protein (GFAP) antibodies were obtained from Sigma.
Histology and Electroretinogram (ERG) Analyses.
Histology procedures and full-field rod ERG recordings were carried out as described (17). Cone ERGs were elicited by full-field flashes and were recorded under light-adapted conditions based on the method of Peachey et al. (18).We also recorded dark-adapted full-field ERGs in response to bright white flashes (19) to fit the initial segment of the a-wave to a mathematical “component” model that estimates parameters involved in rod photoreceptor transduction (20).
Rhodopsin Quantification.
Rhodopsin was determined spectrophotometrically by using total eye extracts as described (21).
Results
Targeted Disruption of the RPGR Gene.
We designed a knockout vector in which exons 4–6 were replaced by a selectable marker resulting in early truncation of the RPGR coding sequence. The selectable marker was a fusion gene for both neomycin resistance and for the Escherichia coli β-galactosidase. It contained no promoter and depended on the endogenous RPGR gene promoter for expression (Fig. 1a). Transfection of this vector into mouse ES cells gave rise to multiple clones with targeted disruption of the RPGR gene as identified by genomic Southern blotting. Because the ES cells were male-derived and carried a single X chromosome, RPGR expression was ablated in all targeted clones (data not shown). Chimeric founders, derived from two independent ES clones, were crossed with C57BL/6 mice. WT, heterozygous, and nullizygous KO offspring were produced at the expected Mendelian ratio. KO mice were fertile and indistinguishable in growth characteristics from WT and heterozygous littermates.
Northern blotting did not detect any RPGR transcript in tissues from KO mice (Fig. 1b). With the more sensitive reverse transcription–PCR assay using a number of primer combinations, there were either little or no products (Fig. 1b), indicating the mutant transcript was either unstable and/or portions of the RPGR coding sequence were removed by aberrant splicing due to the large insertion. Importantly, in the rare transcript in which the inserted marker had been spliced out, sequencing of PCR-amplified products showed that the coding sequence was out of frame beyond the insertion site. RPGR expression was also examined at the protein level by immunoblotting with RPGR antibodies. A 95-kDa protein was detected in the brain and retina (Fig. 1c). This 95-kDa protein was also detected in a variety of somatic tissues, including liver, lung, kidney, and heart (not shown). An additional larger variant of 165 kDa was found only in testis. Both isoforms were absent in tissues from KO mice (Fig. 1c), confirming ablation of RPGR expression in KO mice. The splicing pattern of the RPGR transcript was reported to be very complex, with numerous splice variants identified (12, 22). However, the immunoblot data here indicate that a 95-kDa polypeptide represents the major RPGR gene product.
Localization of RPGR to Photoreceptor Connecting Cilium.
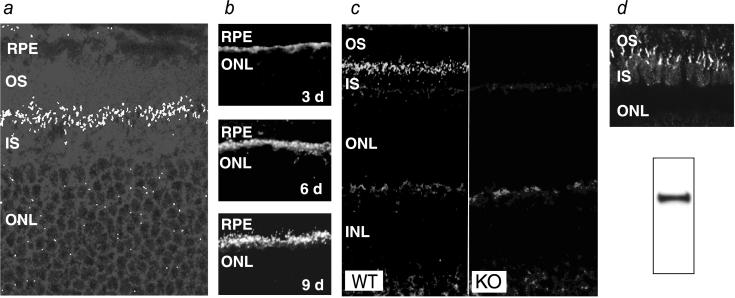
Tissue staining in RPGR-deficient mice for the β-galactosidase reporter, whose transcription was driven by the endogenous RPGR promoter, showed widespread expression in brain, where the particularly high expression in the CA1-CA2 region of the hippocampus stood in contrast to the lack of expression in the CA3 and the dentate gyrus. In the retina, expression was found in the ganglion and photoreceptor cell layers (not shown). We then performed immunocytochemistry to determine the subcellular localization of RPGR in normal retinas. On retinal sections, prominent staining was found at the junction between the inner and outer segments of photoreceptors, indicating staining of the connecting cilia (Fig. 2a). The appearance of RPGR in developing retina was also consistent with the timing and location of the emerging connecting cilia. Thus, RPGR was detectable at postnatal day 3 at the apices of the developing photoreceptor layer and grew in intensity to adult levels by postnatal day 9 (Fig. 2b). RPGR in other layers of the retina was likely to be of low abundance because, by immunofluorescence, the RPGR-deficient retinas differed from WT retinas only in the absence of connecting cilium staining (Fig. 2c). Because mouse photoreceptors are overwhelmingly rods, we turned to the cone dominant ground squirrel retina (23) to ascertain whether RPGR was also localized in the cone connecting cilia. Western blotting confirmed that the RPGR antibodies raised against mouse sequence cross-reacted with the squirrel RPGR (Fig. 2d Lower). On retinal sections, the staining pattern in this cone rich retina indeed indicated presence of RPGR in connecting cilia (Fig. 2d Upper).
Figure 2.
Localization of RPGR in the connecting cilium by immunofluorescence. (a) Confocal image of a normal retina probed with the RPGR antibodies. A band of punctate staining is seen between the inner and outer segments, indicative of connecting cilia. (×500.) (b) Developmental time course of RPGR staining in the mouse retina. Staining first appears around postnatal day 3 as a band at the apex of the photoreceptor layer and increases in intensity and definition through day 9. (c) The connecting cilium staining for RPGR is absent in KO mice. (×250.) (d) RPGR is also localized in the connecting cilia of cone photoreceptors. Shown is RPGR immunostaining of the cone-dominant ground squirrel retina (Upper). Western blotting of ground squirrel tissue homogenates confirms that the RPGR antibodies made against mouse sequence cross-reacts with squirrel RPGR (Lower). RPE, retinal pigment epithelium; OS, outer segment; IS, inner segment; ONL, outer (photoreceptor) nuclear layer.
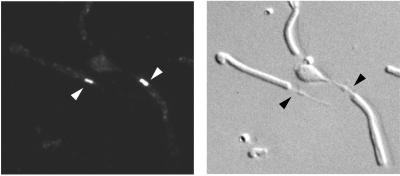
To gain better resolution, we immunostained photoreceptors that had been mechanically dissociated. These preparations contained mostly shaken-off rod outer segments attached to connecting cilia and often with a portion of the inner segments at the proximal end of the connecting cilia. Comparison of the fluorescence and Nomarski images confirmed staining of connecting cilia (Fig. 3). This staining was absent in RPGR-deficient photoreceptors processed identically (not shown). We conclude that RPGR is concentrated in the connecting cilia of both rod and cone photoreceptors.
Figure 3.
Immunostaining of dissociated photoreceptors provides higher spatial resolution. Shown are epifluorescence (Left) and Nomarski (Right) images of the same field of view. Arrows point to connecting cilia of rod photoreceptors. (×500.)
Retinal Disease in RPGR-Deficient Mice.
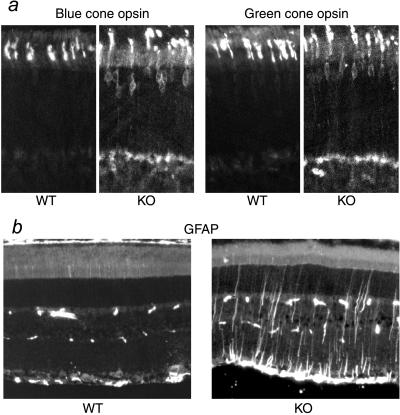
The retinal morphology of RPGR-deficient mice was comparable to that of WT and heterozygous littermates at the completion of retinal development (≈30 postnatal days). Retinal function, as measured by ERG, was also within the normal limits at the early age. However, both cone and rod photoreceptors exhibited subtle abnormalities. Blue and green cone opsins, normally localized in the outer segments, were found to partially mislocalize to the inner segment, perinuclear, and synaptic regions (Fig. 4a). This defect was more pronounced at the retinal quadrants where the respective cone densities were the highest (i.e., superior retina for green cones and inferior retina for blue cones) (24). Mislocalization of cone opsins was found as early as postnatal day 20, the earliest time point examined. Rhodopsin, the photopigment of rod photoreceptors, was not appreciably mislocalized by immunofluorescence. Quantification of rhodopsin by photobleaching difference spectra, however, showed that rhodopsin content in the outer segments was reduced in the RPGR-deficient mice (Table 1). Immunostaining for a number of rod outer segment proteins, including cGMP phosphodiesterase, cGMP-gated cation channel, peripherin/RDS, and arrestin (under both dark- and light-adapted conditions) did not reveal an appreciable difference in the localization pattern between RPGR-deficient and WT littermates.
Figure 4.
Histopathology in KO retinas revealed by immunocytochemistry. (a) Mislocalization of blue and green cone opsins in the KO retinas. (b) GFAP is up-regulated in the KO retinas. (Left) GFAP immunoreactivity is normally found only in the astrocytes in the ganglion cell layer of the retina (near the bottom of figure). The sporadic speckles in the middle of the figure are blood vessel profiles recognized by the anti-mouse secondary antibody. (Right) Muller cell processes are prominently stained for GFAP in a 2-month-old KO retina.
Table 1.
Rhodopsin quantification in RPGR-deficient (KO) and control (WT) mice
| Genotype | Rhodopsin, nmol/eye | n |
|---|---|---|
| KO | 0.44 ± 0.01 | 6 |
| WT | 0.51 ± 0.02 | 6 |
Mice were at 1 month of age. Values are mean ± S.E.M. n = number of mice. P < 0.01 (t test).
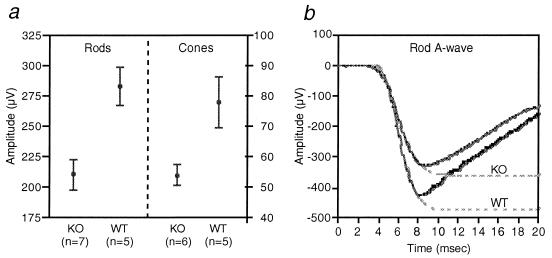
Although retinal cell loss was not apparent in young RPGR-deficient retinas, staining for GFAP, which is up-regulated during retinal degeneration, indicated degenerative changes were already underway at two months of age (Fig. 4b). Photoreceptor cell loss became substantial by 6 months of age, when a reduction of outer segment length and photoreceptor nuclear layer thickness (loss of two rows of nuclei on average) were noted (Fig. 5). Ultrastructurally, the newly formed disk membranes at the base of photoreceptor outer segments were notably disorganized in the mutant mice. The structure of the connecting cilia appeared well maintained (Fig. 6). Thus, RPGR does not appear to be required for the development or maintenance of the connecting cilium axoneme. Concomitant with morphologically apparent degeneration, ERG measurements also began to show abnormalities. Mean rod a-wave amplitude was reduced by 25% in the mutant mice (P = 0.005) (Fig. 7a Left). Cone a-waves in mice are too small to quantitate with these techniques. Cone ERGs showed b-wave amplitudes reduced on average by 31% (P = 0.019) (Fig. 7a Right), consistent with cone cell loss. Component analyses based on responses to bright flashes of light (20) indicated a reduction in the mean slope of the rod a-wave response in the mutant mice (n = 7) compared with WT mice (n = 7) (Fig. 7b), which appeared to be attributable to reductions in rod photoreceptor sensitivity (P = 0.054) and maximal amplitude (P = 0.083). Both are consistent with the observation of loss of outer segment membrane (i.e., fewer cells and shorter outer segments). Ocular findings in the two lines of RPGR-deficient mice were indistinguishable. Heterozygous females, being mosaic for WT and mutant photoreceptors and hence expected to develop a milder form of the disease, were not analyzed in detail.
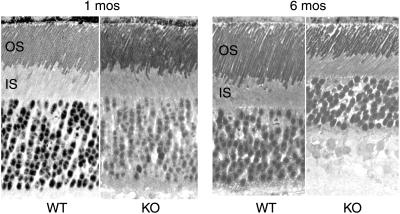
Figure 5.
Light micrographs of retinal sections from WT and KO littermates at 1 and 6 months of age. Note the reduced outer segment length and reduced thickness of the outer nuclear layer in the KO retina at 6 months of age. (×300.)
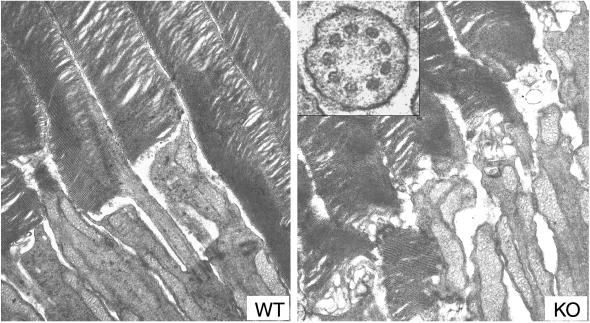
Figure 6.
Electron micrographs of photoreceptors from WT (Left) and KO (Right) mice. Both have well packed disks in the outer segments, but the newly added disk membranes at the base of the outer segments appear disorganized in the KO photoreceptors. (×16,000.) (Inset) Cross section through a connecting cilium showing the normal organization of 9 + 0 microtubule arrangement in the mutant photoreceptor. (×80,000.)
Figure 7.
Rod and cone ERGs from WT and KO mice at 6.5 months of age. (a) Means and SEM of rod ERG a-wave amplitudes (Left) and cone ERG b-wave amplitudes (Right). (b) Representative dark-adapted ERG a-waves in response to a bright flash of white light (28 foot Lambert-seconds) from WT and KO mice. Dashed lines are fitted functions.
Discussion
An important finding of this work is the subcellular localization of RPGR in the photoreceptor connecting cilia. Photoreceptor cells achieve efficient transduction of light to neural signals in part by the elaboration of a specialized organelle known as the outer segment, which is densely packed with stacked disk membranes containing abundant photopigment rhodopsin (in rods) or cone opsins (in cones). The outer segment is linked to the cell body (inner segment) via a connecting cilium, a thin structure analogous to the transition zone of motile cilia (25). The outer segment is continually renewed by shedding the older portion at the tip and adding new membranes at the base. Therefore the connecting cilium is critically important for polarized transport of nascent proteins, including opsin (26), from the inner to the outer segment. The outer segment has a vastly different protein composition, relative to the inner segment, that is geared toward efficient phototransduction. Rhodopsin, for example, is barely detectable in the inner segments but accounts for 85% of outer segment membrane proteins (27). As such, the connecting cilium must also function as a diffusion barrier against redistribution (28). Therefore, our finding that RPGR localizes in the connecting cilia is likely to be of major functional significance. If the ciliary fraction of RPGR alone carries out nonredundant functions in photoreceptors, it would help explain why the disease expression is largely limited to photoreceptors even although RPGR is widely expressed (8).
In the absence of RPGR, the polarized distribution of cone opsins was compromised. Although we were unable to obtain direct evidence that a similar compromise also occurred in rod photoreceptors, the reduced rhodopsin content in the rod outer segments may have resulted from the same underlying defect. Unlike cones, rod outer segment disk membranes are pinched off from plasma membranes as they mature. Therefore, the bulk of rhodopsin is not available for lamellar diffusion back along the connecting cilium. Only the fractions of rhodopsin present in the plasma membrane of rod outer segments (27) and the growing disk membranes would be available for redistribution. Therefore, rods would be expected to have a milder phenotype than cones if the diffusion barrier is compromised. The reported interaction between cone opsin, but not rod opsin, with Ran binding protein 2 suggests the possibility of an additional pathway requiring RPGR in cones (29).
Without RPGR, photoreceptor cells develop normal morphology initially, are able to carry out phototransduction, and remain viable in the first few months of life. Thus, the ciliary function of RPGR does not appear to be central, but facilitative instead. However, over time, a partial mislocalization of opsins apparently reduces the viability of photoreceptors. In this sense, RPGR may be best described as a longevity gene required for the long-term maintenance of photoreceptors. The relatively more pronounced cone phenotype is in agreement with the clinical expression of RPGR mutations, which often manifest as rod-cone or cone-rod dystrophies. Interestingly, it has long been known that some families with X-linked RP manifest an association between retinal disease and motile ciliary abnormalities (30, 31), and at least one such family has been shown to be carrying RP3 (32). This suggests that RPGR may have a general ciliary role, albeit redundant, and that certain alleles of RPGR may result in an interfering mutant protein. Patients carrying these alleles might present with RP in conjunction with an immotile cilia defect.
The presence of an RCC1 homology domain in RPGR, the localization of RPGR in the photoreceptor connecting cilium, and the phenotype of RPGR-deficient mice suggest a potential RanGTP-dependent mechanism that controls the directional movement of opsins against a steep concentration gradient across the cilia. As the guanine nucleotide exchange factor for Ran, RCC1 promotes the generation of RanGTP. Ran mediates a number of cellular processes, including nuclear import and export (10). RanGTP serves both a signaling role and as an energy source for molecular motors that move cargo through the nuclear pore complex. A high local RanGTP concentration is necessary for its functioning. Cells normally maintain a steep gradient of high RanGDP in the cytoplasm and high RanGTP in the nucleus by virtue of sequestration of the RanGTPase-activating protein and RCC1 in the cytosol and nuclei, respectively. Assuming the RCC1 homology domain of RPGR also catalyzes RanGDP to GTP exchange, RPGR will generate a high local concentration of RanGTP in the connecting cilium relative to the inner segment. This provides the conditions for a putative RanGTP-dependent process that drives the unidirectional movement of opsins, and a disruption of this process may be the basis of photoreceptor disease caused by RPGR mutations.
Acknowledgments
We thank Drs. Igal Gery, Robert Molday, Jeremy Nathans, Dorothy Roof, and Gabriel Travis for antibodies, Michael Adamian for electron microscopy, and the Massachusetts General Hospital ES Core Facility for assisting with knockout mouse generation. This work was supported by National Institutes of Health Grant EY10309, the Foundation Fighting Blindness (Baltimore, MD), and the Chatlos Foundation (Longwood, FL). T.L. is a recipient of a career development award from Research to Prevent Blindness.
Abbreviations
- RPGR
retinitis pigmentosa GTPase regulator
- RP
retinitis pigmentosa
- RCC1
regulator of chromatin condensation
- KO
knockout
- WT
wild type
- ES cell
embryonic stem cell
- GFAP
glial fibrillary acidic protein
- ERG
electroretinogram
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.060037497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.060037497
References
- 1.Berson E L, Rosner B, Simonoff E. Am J Ophthalmol. 1980;89:763–775. doi: 10.1016/0002-9394(80)90163-4. [DOI] [PubMed] [Google Scholar]
- 2.Beron E L, Gouras P, Gunkel R D, Myrianthopoulos N C. Arch Ophthalmol. 1969;81:215–225. doi: 10.1001/archopht.1969.00990010217012. [DOI] [PubMed] [Google Scholar]
- 3.Jacobson S G, Buraczynska M, Milam A H, Chen C, Jarvalainen M, Fujita R, Wu W, Huang Y, Cideciyan A V, Swaroop A. Invest Ophthalmol Visual Sci. 1997;38:1983–1997. [PubMed] [Google Scholar]
- 4.Weleber R G, Butler N S, Murphey W H, Sheffield V C, Stone E M. Arch Ophthalmol. 1997;115:1429–1435. doi: 10.1001/archopht.1997.01100160599013. [DOI] [PubMed] [Google Scholar]
- 5.Buraczynska M, Wu W, Fujita R, Buraczynska K, Phelps E, Andreasson S, Bennett J, Birch D G, Fishman G A, Hoffman D R, et al. Am J Hum Genet. 1997;61:1287–1292. doi: 10.1086/301646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer S, Fujita R, Buraczynska M, Abrahamson M, Ehinger B, Wu W, Falls T J, Andreasson S, Swaroop A. Invest Ophthalmol Visual Sci. 1998;39:2470–2474. [PubMed] [Google Scholar]
- 7.Fishman G A, Grover S, Jacobson S G, Alexander K R, Derlacki D J, Wu W, Buraczynska M, Swaroop A. Ophthalmology. 1998;105:2286–2296. doi: 10.1016/S0161-6420(98)91231-3. [DOI] [PubMed] [Google Scholar]
- 8.Meindl A, Dry K, Herrmann K, Manson F, Ciccodicola A, Edgar A, Carvalho M R, Achatz H, Hellebrand H, Lennon A, et al. Nat Genet. 1996;13:35–42. doi: 10.1038/ng0596-35. [DOI] [PubMed] [Google Scholar]
- 9.Roepman R, van Duijnhoven G, Rosenberg T, Pinckers A J, Bleeker-Wagemakers L M, Bergen A A, Post J, Beck A, Reinhardt R, Ropers H H, et al. Hum Mol Genet. 1996;5:1035–1041. doi: 10.1093/hmg/5.7.1035. [DOI] [PubMed] [Google Scholar]
- 10.Gorlich D, Mattaj I W. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 11.Linari M, Ueffing M, Manson F, Wright A, Meitinger T, Becker J. Proc Natl Acad Sci. 1999;96:1315–1320. doi: 10.1073/pnas.96.4.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan D, Swain P K, Breuer D, Tucker R M, Wu W, Fujita R, Rehemtulla A, Burke D, Swaroop A. J Biol Chem. 1998;273:19656–19663. doi: 10.1074/jbc.273.31.19656. [DOI] [PubMed] [Google Scholar]
- 13.Rowe L B, Nadeau J H, Turner R, Frankel W N, Letts V A, Eppig J T, Ko M S, Thurston S J, Birkenmeier E H. Mamm Genome. 1994;5:253–274. doi: 10.1007/BF00389540. [DOI] [PubMed] [Google Scholar]
- 14.Mountford P, Zevnik B, Duwel A, Nichols J, Li M, Dani C, Robertson M, Chambers I, Smith A. Proc Natl Acad Sci. 1994;91:4303–4307. doi: 10.1073/pnas.91.10.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muresan V, Joshi H C, Besharse J C. J Cell Sci. 1993;104:1229–1237. doi: 10.1242/jcs.104.4.1229. [DOI] [PubMed] [Google Scholar]
- 16.Chiu M I, Nathans J. Visual Neurosci. 1994;11:773–780. doi: 10.1017/s0952523800003072. [DOI] [PubMed] [Google Scholar]
- 17.Li T, Sandberg M A, Pawlyk B S, Rosner B, Hayes K C, Dryja T P, Berson E L. Proc Natl Acad Sci. 1998;95:11933–11938. doi: 10.1073/pnas.95.20.11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peachey N S, Goto Y, al-Ubaidi M R, Naash M I. Neurosci Lett. 1993;162:9–11. doi: 10.1016/0304-3940(93)90547-x. [DOI] [PubMed] [Google Scholar]
- 19.Sandberg M A, Lee H, Matthews G P, Gaudio A R. Invest Ophthalmol Visual Sci. 1991;32:1508–1516. [PubMed] [Google Scholar]
- 20.Hood D C, Birch D G. Invest Ophthalmol Visual Sci. 1994;35:2948–2961. [PubMed] [Google Scholar]
- 21.Joseph R M, Li T. Invest Ophthalmol Visual Sci. 1996;37:2434–2446. [PubMed] [Google Scholar]
- 22.Kirschner R, Rosenberg T, Schultz-Heienbrok R, Lenzner S, Feil S, Roepman R, Cremers F P M, Ropers H H, Berger W. Hum Mol Genet. 1999;8:1571–1578. doi: 10.1093/hmg/8.8.1571. [DOI] [PubMed] [Google Scholar]
- 23.von Schantz M, Szel A, van Veen T, Farber D B. Invest Ophthalmol Visual Sci. 1994;35:2558–2566. [PubMed] [Google Scholar]
- 24.Szel A, Rohlich P, Caffe A R, Juliusson B, Aguirre G, Van Veen T. J Comp Neurol. 1992;325:327–342. doi: 10.1002/cne.903250302. [DOI] [PubMed] [Google Scholar]
- 25.Besharse J C, Horst C J. In: Ciliary and Flagellar Membranes. Bloodgood R A, editor. New York: Plenum; 1990. pp. 389–417. [Google Scholar]
- 26.Liu X, Udovichenko I P, Brown S D, Steel K P, Williams D S. J Neurosci. 1999;19:6267–6274. doi: 10.1523/JNEUROSCI.19-15-06267.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molday R S. Invest Ophthalmol Visual Sci. 1998;39:2491–2513. [PubMed] [Google Scholar]
- 28.Spencer M, Detwiler P B, Bunt-Milam A H. Invest Ophthalmol Visual Sci. 1988;29:1012–1020. [PubMed] [Google Scholar]
- 29.Ferreira P A, Nakayama T A, Pak W L, Travis G H. Nature (London) 1996;383:637–640. doi: 10.1038/383637a0. [DOI] [PubMed] [Google Scholar]
- 30.Arden G B, Fox B. Nature (London) 1979;279:534–536. doi: 10.1038/279534a0. [DOI] [PubMed] [Google Scholar]
- 31.van Dorp D B, Wright A F, Carothers A D, Bleeker-Wagemakers E M. Hum Genet. 1992;88:331–334. doi: 10.1007/BF00197269. [DOI] [PubMed] [Google Scholar]
- 32.Dry K L, Manson F D, Lennon A, Bergen A A, Van Dorp D B, Wright A F. Hum Mutat. 1999;13:141–145. doi: 10.1002/(SICI)1098-1004(1999)13:2<141::AID-HUMU6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]