Abstract
Introduction: Given the common use of acronyms and initialisms in the health sciences, searchers may be entering these abbreviated terms rather than full phrases when searching online systems. The purpose of this study is to evaluate how various MEDLINE Medical Subject Headings (MeSH) interfaces map acronyms and initialisms to the MeSH vocabulary.
Methods: The interfaces used in this study were: the PubMed MeSH database, the PubMed Automatic Term Mapping feature, the NLM Gateway Term Finder, and Ovid MEDLINE. Acronyms and initialisms were randomly selected from 2 print sources. The test data set included 415 randomly selected acronyms and initialisms whose related meanings were found to be MeSH terms. Each acronym and initialism was entered into each MEDLINE MeSH interface to determine if it mapped to the corresponding MeSH term. Separately, 46 commonly used acronyms and initialisms were tested.
Results: While performance differed widely, the success rates were low across all interfaces for the randomly selected terms. The common acronyms and initialisms tested at higher success rates across the interfaces, but the differences between the interfaces remained.
Conclusion: Online interfaces do not always map medical acronyms and initialisms to their corresponding MeSH phrases. This may lead to inaccurate results and missed information if acronyms and initialisms are used in search strategies.
Highlights
Acronyms and initialisms are widely used in the health sciences and may be used when searching the literature.
Acronyms and initialisms do not always correctly map to Medical Subject Headings (MeSH).
Mapping accuracy varies depending on the MEDLINE MeSH interface used.
Searching only by acronyms or initialisms could lead to missed or lost information.
Implications for practice
The differences in mapping to MeSH between systems should be considered when selecting an interface and developing search strategies.
Given the common use of these abbreviated terms, it may be beneficial to include information regarding searching by acronyms and initialisms when instructing users.
It may be advisable for system developers to consider use of acronyms and initialisms when creating mechanisms that automatically map terminology to MeSH.
INTRODUCTION
An acronym is a “word formed from the initial letters of other words” [1]. It is derived from a combination of the Greek words akros, meaning top, and onyma, meaning name [2]. Although the word “acronym” is commonly used to refer to any phrase abbreviated by the use of initial letters of each word or syllable, the specific definition is for those initial-letter abbreviations that can be pronounced as words. Some common examples include NATO (North Atlantic Treaty Organization), JAMA (Journal of the American Medical Association), AWOL (absent without leave), and AIDS (acquired immunodeficiency syndrome). An initialism, on the other hand, is “a group of initial letters used as an abbreviation for a name or expression, each letter or part being pronounced separately” [3]. Examples of initialisms include UN (United Nations), TWA (Trans World Airlines), and FDA (Food and Drug Administration).
Some scholars require that a true acronym be pronounceable as noted in the definition above and have a minimum of three letters. In 1962, Baum noted that the definition of acronym was becoming “blurred and confused” [4]. In current usage, both initialisms and acronyms are often referred to as acronyms and two-letter terms are included. While there has been much discussion regarding the definition of acronym in comparison to other shortening devices such as abbreviations, word clippings, and word blends, these topics are beyond the scope of this study. They are, however, well documented in linguistics literature, and several examples are cited here [4–9]. For the purposes of this study, both acronyms and initialisms are included in the test data as well as two-letter terms, such as MS (multiple sclerosis).
The term “acronym” appears to have been coined in 1943 [10]. Although this term was first mentioned in 1943, initial-letter abbreviations have been used for some time. It has been noted that these types of abbreviations occurred during the Roman Empire, with SPQR used for Senatus Populusque Romanus [5]. While such historic initial-letter abbreviations are recorded, it is impossible to know whether readers of the day pronounced them as acronyms or as initialisms or, on seeing the letters, actually spoke the full phrase [9]. Use of acronyms and initialisms increased over time, particularly during and after World War II [5, 7, 11].
In the present day, the use of acronyms and initialisms abounds in the English language throughout all aspects of both written and verbal communications. On any given day, acronyms and initialisms will be encountered in news reports and popular literature. The use of acronyms and initialisms has seeped into everyday conversations as well, for example, ASAP (as soon as possible), ETA (estimated time of arrival), and VW (Volkswagen). During a visit to a chat room, one may come across BRB (be right back), LOL (laughing out loud), or TTYL (talk to you later), among others.
This increase in the use of acronyms is also the case in the health sciences. In 1970, Britton observed that “in science, new words normally emerge in response to the need for novel, precise, and economical communication” and that the acronym is a result of the “need for economy and convenience of expression … usually formed from the initials of a phrase name” [12]. It is now rather unusual when a medical condition or procedure does not come with an associated acronym or initialism, for example, MRI (magnetic resonance imaging), ALS (amyotrophic lateral sclerosis), COPD (chronic obstructive pulmonary disease), HRT (hormone replacement therapy), and SARS (severe acute respiratory syndrome), to name just a few.
For some years, editorials, letters to editors, and articles have been published in the health sciences literature lamenting the use, overuse, misuse, or abuse of acronyms in medical journals [13–19] Baue mentions that in one issue of a clinical journal, fifty-two acronyms are found [13]. Fred and Cheng go further noting that more than ninety undefined acronyms were located in one review article [14].
Given this common use of acronyms and initialisms in the health sciences, searchers may be entering these abbreviated forms rather than full phrases when searching online systems. This may be particularly true when the full phrase is lengthy and/or difficult to spell. In some instances, searchers may know the acronym or initialism but are unsure of its exact meaning. Searchers may also be assuming that online systems will properly translate acronyms and initialisms to their corresponding full phrases.
The confusion over searching by acronyms has been recognized, and several systems have been developed to aid in matching acronyms to their corresponding definitions [20]. The effectiveness of searching by acronyms and initialisms in MEDLINE depends on how successfully they are mapped to their corresponding Medical Subject Headings (MeSH). It has been shown that different MEDLINE interfaces do not map to MeSH in the same way [21]. Federiuk has found that searching MEDLINE with abbreviations and acronyms is not a straightforward process and may require the use of the abbreviation, full phrase, and subject heading to acquire all unique citations [22]. This may lead to different search results and potentially lost or incomplete information. The purpose of the current study is to compare how different interfaces to MEDLINE map acronyms and initialisms to their corresponding MeSH terms and to note variations among systems.
METHODOLOGY
This research is an extension of a pilot study, the results of which were presented in a poster at MLA '04, the 2004 Medical Library Association (MLA) annual meeting [23]. The pilot study examined 114 randomly selected acronyms and initialisms. That research was expanded to include a total of 415 terms in the current study's test data set.
The interfaces used in this study included: the PubMed MeSH database, the PubMed Automatic Term Mapping feature, the NLM Gateway Term Finder, and Ovid MEDLINE. Test terms were randomly selected from two print sources: The Dictionary of Medical Acronyms and Abbreviations by Jablonski [24] and Elsevier's Dictionary of Abbreviations, Acronyms, Synonyms and Symbols Used in Medicine by Tsur [25]. The following procedure was used to select the test terms:
Microsoft Excel was used to generate two lists of random numbers. The first list included numbers randomly selected between 1 and 440 (the number of pages in the Jablonski book). The second list of random numbers was obtained for numbers between 1 and 843 (the number of pages in the Tsur book).
For each randomly generated page number of each book, all acronyms and initialisms were selected for possible inclusion in the test data set. Other types of abbreviations, eponyms, and symbols were excluded.
Each of the selected acronyms and initialisms was included in the test data set if its corresponding full phrase was a MeSH term. The NLM MeSH Browser was used to establish if the full phrases were MeSH terms. Phrases that were main headings or entry terms in the MeSH record were included. For example, if HRT were an acronym found on one of the selected pages from one of the books, then the full meaning, “Hormone Replacement Therapy,” was searched in the NLM MeSH Browser. If the full phrase was a MeSH term, as in this case, then the acronym or initialism was added to the test data set. If the full phrase or meaning was not a MeSH term, it was excluded. The NLM MeSH Browser was selected for this task because most of the MeSH print sources have ceased publication due to the “greater coverage, flexibility, and currency of the MeSH Browser” [26].
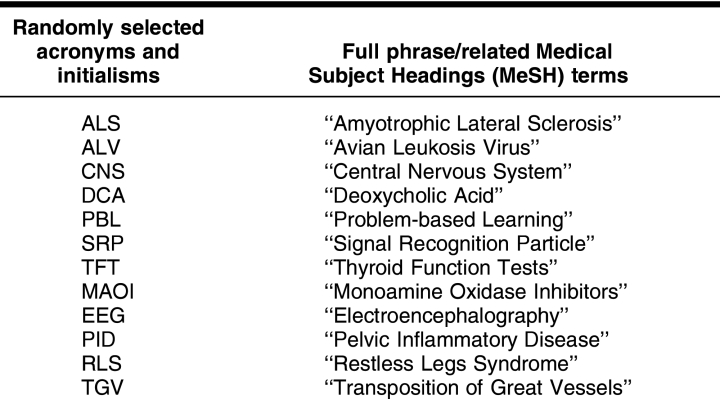
Of the total 415 test terms, 263 were drawn from the Jablonski book, including the 114 test terms from the original pilot study. For the current study, 152 test terms were also obtained from the Tsur book. Table 1 shows examples of randomly selected test terms included in the study along with their corresponding MeSH terms.
Table 1 Examples of randomly selected acronyms
Some acronyms and initialisms have more than one meaning. For example, ALS stands for amyotrophic lateral sclerosis, but it also stands for afferent loop syndrome and antilymphocyte serum. If a test term had multiple meanings that equated to MeSH terms, then that term was added to the database and counted and tested separately for each meaning. Of the 415 terms in the test data set, 176 were unique with only 1 meaning. The remaining test terms were derived from 68 randomly selected acronyms and initialisms that had multiple meanings.
Each test term was entered into each MEDLINE MeSH interface to determine if it mapped to its corresponding MeSH term. For example, “DCL” was entered into each of the selected interfaces to see if it correctly mapped to “Diffuse Cutaneous Leishmaniasis.” The 114 terms from the original pilot study were retested with the expanded set of terms for the current study. The detailed steps for each interface are:
In Ovid MEDLINE, each test term was entered into the query box. The Map Term to Subject Heading option was selected. A search was performed that resulted in a new page with either one specific MeSH term or a list of possible MeSH terms from which to choose. If the MeSH term associated with the entered test term was found on this page, then the mapping was successful. If the corresponding MeSH term was not listed, then the mapping failed.
For PubMed, two features were tested: the PubMed MeSH database and the Automatic Mapping option. For the PubMed MeSH database, this feature was selected from the sidebar menu of PubMed's main screen. Each test term was entered into the query box, and the Go button selected. The results displayed one or more MeSH terms from which to choose. The complete list of results was reviewed to determine if the associated MeSH term was present. If the MeSH term associated with the entered test term was found in the list, then the mapping was considered successful. If the corresponding MeSH term was not listed, then the mapping failed.
For PubMed's Automatic Mapping feature, each test term was entered into the search box on PubMed's main page. The Go button was selected, and a search was performed with citations retrieved. At this point, the Details option was selected to view the Query Translation Box that showed how the entered terms were translated using MeSH terms and PubMed's search rules and syntax [27]. If the corresponding MeSH term appeared in the Query Translation Box, then the mapping was considered successful. If the corresponding MeSH term was not displayed, then the mapping failed.
For the NLM Gateway interface, the Term Finder was selected from the menu bar. Each of the test terms was entered in the query box, and the Go button was selected. The resulting page was either a direct match for the MeSH term or a list of possible MeSH terms from which to choose. If the MeSH term associated with the test term was retrieved, then the mapping was considered successful. If the term was not retrieved, then the mapping failed.
As was done in the pilot study [23], a second narrowly constructed test was performed on common acronyms and initialisms. These terms were not randomly selected but were gathered via an informal survey of library colleagues, who were asked to provide a list of common acronyms they have encountered in their searching. This second set of acronyms and initialisms was developed to compare commonly used terms against the randomly selected set. This set of common acronyms and initialisms was tested using the same method as the randomly selected data. The set of common terms included forty-six acronyms and initialisms. Table 2 includes examples of common terms tested.
Table 2 Examples of common acronyms derived from informal survey
RESULTS
Randomly selected data set
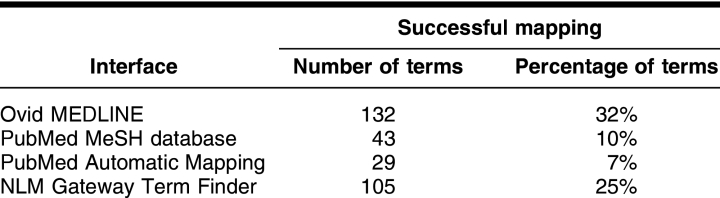
For the randomly selected test terms, all tested interfaces performed with low success rates (Table 3). Ovid MEDLINE performed with the highest success rate of 32%, followed by the NLM Gateway Term Finder with a success rate of 25%. Although all tested interfaces performed poorly, the systems also showed a wide difference in success rates.
Table 3 Frequency of successful mapping of randomly selected acronyms and initialisms to MeSH (n = 415 terms)
Commonly selected terms
The forty-six commonly used tested acronyms and initialisms had substantially higher success rates in mapping to MeSH as shown in Table 4. As in the randomly selected data set, Ovid MEDLINE and the NLM Gateway Term Finder had the highest success rates, both at 89%. There were also wide differences in the success rates of the common acronyms and initialisms.
Table 4 Frequency of successful mapping of common acronyms and initialisms to MeSH (n = 46 terms)
DISCUSSION
The results of this research show that successful mapping of acronyms and initialisms to MeSH is generally low across all interfaces studied. The findings also show wide differences in the rates of mapping when the tested interfaces are compared. The low success rates may be due to the random selection of test data, which includes acronyms and initialisms that may be less commonly known or used.
The differences in mapping rates may be related to how each system translates the search strategy and maps it to MeSH. According to the Help Screens in both PubMed and the NLM Gateway, both the PubMed Automatic Term Mapping feature and the NLM Gateway Term Finder utilize the Unified Medical Language System (UMLS) Metathesaurus [28, 29]. Given the substantial differences in the mapping success rates between these two interfaces, it is clear that they employ different algorithms for mapping. Ovid MEDLINE, which had the highest success rate, uses its own statistical analysis to map a term to the controlled vocabulary [30].
The mapping of commonly used acronyms and initialisms was far more successful across all systems than the randomly selected set. As this portion of the study did not include a randomly selected set of test data, the results should be viewed cautiously. While the results of the commonly used terms were much better than the randomly selected set, there were some surprises. Even some very highly used terms did not map successfully in all interfaces. For example, EMT should lead to the MeSH term, “Emergency Medical Technicians.” In both the PubMed MeSH database and the PubMed Automatic Mapping feature, this did not occur. The same was true for PSA (prostate-specific antigen), EBM (evidence-based medicine), STD (sexually transmitted disease), and TB (tuberculosis).
The PubMed Automatic Term Mapping feature had the lowest success rates for both randomly selected terms and for common terms. UTI (urinary tract infections), HRT (hormone replacement therapy), and PMS (premenstrual syndrome) are some examples of common acronyms that did not map successfully in the Automatic Term Mapping feature of PubMed. In most cases, the entered term was searched in all fields when it did not map to the correct MeSH phrase. In some cases, mapping was incorrect. For example, HRT was mapped to the MeSH term “Heart” instead of “Hormone Replacement Therapy.”
During this study, it was also observed that the PubMed MeSH database often displayed substance names (supplementary concept records) in the mapped lists [31]. While the availability of the supplementary concept records might offer the opportunity for more precise searching, it might also lead to confusion or inefficiency for the average searcher. For example, MS (multiple sclerosis) elicited a list of sixty-nine possible matches in the PubMed MeSH database. The corresponding MeSH term, “Multiple Sclerosis,” was the last item listed at number sixty-nine. Of the sixty-eight terms listed ahead of “Multiple Sclerosis,” sixty-six of them were substance names. This was also true when PSA (prostate-specific antigen) was entered. Of the fifteen items in the retrieved list, “Prostate-Specific Antigen” was not among them, and all fifteen were substance names. While the mapping to substance names may be beneficial, it appears that, in certain circumstances, substance names overshadow the mapping to MeSH terms. This may lead searchers to conclude that appropriate MeSH terms do not exist, when in fact they are available.
CONCLUSION
The results of this study identify differences between interfaces regarding the effectiveness of mapping medical acronyms and initialisms to MeSH terms. These differences have implications for those who use MEDLINE for online searching, medical librarians, and system developers.
Average searchers might not be finding what they need and might not be aware that information is missing from their search results. If searchers enter acronyms or initialisms and search them as keywords rather than using their corresponding MeSH terms, they may retrieve extremely large sets of citations including numerous false hits. This may lead to inefficiencies in the use of time by searchers and, again, to the possibility of lost information, as crucial citations can be overlooked in large retrieval sets. Researchers aiming for exhaustive searches for patient care and human subject research should be cautious in developing search strategies and should avoid using acronyms and initialisms alone, without also incorporating their equivalent full phrases or MeSH terms.
The implications for librarians include the necessity for selecting the most appropriate interface, understanding the limitations of the systems' mapping capabilities, and instructing users on effective search techniques. The MeSH vocabulary is a powerful tool in searching MEDLINE and can be used to increase the comprehensiveness of search results as well as to effectively narrow sets to usable sizes. While the mapping was poor across all interfaces, each system had successful matches. This leads to the conclusion that acronyms can and are being coded to map to MeSH terms in certain cases. Perhaps this type of coding and mapping can be improved to allow for a better performance of mapping medical acronyms and initialisms to MeSH. In the meantime, it is advisable to develop search strategies that utilize full phrases or MeSH terms rather than only acronyms or initialisms. It is hoped that system developers can use the information from this study to refine mapping mechanisms to increase the effectiveness of acronym and initialism searching and decrease the possibility of lost or missed information.
Acknowledgments
A number of colleagues at the Library of the Health Sciences, University of Illinois at Chicago, were kind enough to submit examples of commonly used acronyms. The author thanks: Sandra De Groote, AHIP, Jo Dorsch, AHIP, Amanda Huston, Richard McGowan, Robin Mittenthal, Carol Scherrer, AHIP, Lisa Wallis, AHIP, and Ann Carol Weller.
REFERENCES
- Acronym. In: Oxford English dictionary. 2nd ed. Oxford, UK: Oxford University Press, 1989. [Google Scholar]
- Diringer D. Acronym. In: Shally-Jensen M, ed. Encyclopedia Americana. International ed. Danbury, CT: Scholastic Library Publishing, 2005:120. [Google Scholar]
- Initialism. In: Oxford English dictionary. 2nd ed. Oxford, UK: Oxford University Press, 1989. [Google Scholar]
- Baum SV.. The acronym, pure and impure. Am Speech. 1962;37(1:):48–50. [Google Scholar]
- Cannon G.. Abbreviations and acronyms in English word-formation. Am Speech. 1989;64(2:):99–127. [Google Scholar]
- Baum SV.. From “awol”to “veep”: the growth and specialization of the acronym. Am Speech. 1955;30(2:):103–10. [Google Scholar]
- Baum SV.. Formal dress for initial words. Am Speech. 1957;32(1:):73–5. [Google Scholar]
- Heller LG, Macris J.. A typology of shortening devices. Am Speech. 1968;43(3:):201–8. [Google Scholar]
- Richard GS. An analysis of the acronym [dissertation]. Providence, RI: Brown University, 1968. [Google Scholar]
- Davenport B.. Answers. Am Note Queries. 1943;11(11:):167. [Google Scholar]
- Riordan JL.. Some “G. I. alphabet soup.”. Am Speech. 1947;22(2:):108–14. [Google Scholar]
- Britton WE.. Some effects of science and technology upon our language. Coll Compos Commun. 1970;21(5:):342–6. [Google Scholar]
- Baue AE. It's acronymania all over again: with due reference to YB Yogi Berra. Arch Surg. 2002 Apr; 137(4:):486–9. [DOI] [PubMed] [Google Scholar]
- Fred HL, Cheng TO.. Acronymesis: the exploding misuse of acronyms. Tex Heart Inst J. 2003;30(4:):255–7. [PMC free article] [PubMed] [Google Scholar]
- Summers JB, Kaminski J.. Acronym addiction. Tex Heart Inst J. 2004;31(1:):108–9. [PMC free article] [PubMed] [Google Scholar]
- Cheng TO. Please let every acronym be defined (PLEAD). Catheter Cardiovasc Interv. 2003 Nov; 60(3:):424–5. [DOI] [PubMed] [Google Scholar]
- Cheng TO. Acronyms must always bE defiNed (AMEN). Circulation. 2002 Dec 17; 106(25:):e225. [DOI] [PubMed] [Google Scholar]
- Jaffe BM. Acronymitis. Surgical Rounds. 1990 Jun; 13:11–2. [Google Scholar]
- Brubaker RF, Brubaker JH. Does somebody else out there hate acronyms? Arch Ophthalmol. 1999 May; 117(5:):701–2. [PubMed] [Google Scholar]
- Wren JD, Chang JT, Pustejovsky J, Adar E, Garner HR, and Altman RB. Biomedical term mapping databases. Nucleic Acids Res. 2005 Jan 1; 33(database issue:):D289–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gault LV, Shultz M, and Davies KJ. Variations in medical subject headings (MeSH) mapping: from the natural language of patron terms to the controlled vocabulary of mapped lists. J Med Libr Assoc. 2002 Apr; 90(2:):173–80. [PMC free article] [PubMed] [Google Scholar]
- Federiuk CS. The effect of abbreviations on MEDLINE searching. Acad Emerg Med. 1999 Apr; 6(4:):292–6. [DOI] [PubMed] [Google Scholar]
- Shultz M. Variations in acronym mapping to MeSH. Presented at: MLA '04, 104th Annual Meeting of the Medical Library Association; Washington, DC; 2004. [Google Scholar]
- Jablonski S. Dictionary of medical acronyms and abbreviations. 4th ed. Philadelphia, PA: Hanley & Belfus, 2001. [Google Scholar]
- Tsur SA. Elsevier's dictionary of abbreviations, acronyms, synonyms and symbols used in medicine. 2nd, enlarged ed. Amsterdam, The Netherlands: Elsevier, 2004. [Google Scholar]
- National Library of Medicine. Planned changes to MeSH publications. NLM Tech Bull [serial online]. 2003 Jul– Aug;333. [cited 21 Nov 2005]. <http://www.nlm.nih.gov/pubs/techbull/ja03/ja03_technote.html>. [Google Scholar]
- National Center for Biotechnology Information. PubMed help: displaying the search results. [Web document]. Bethesda, MD: National Library of Medicine, 2005. [cited 20 Nov 2005]. <http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=helppubmed.section.pubmedhelp.Displaying_the_Searc>. [Google Scholar]
- National Library of Medicine. Term finder—help pages. NLM gateway. [Web document]. Bethesda, MD: The Library, 2005. [cited 20 Nov 2005]. <http://gateway.nlm.nih.gov/gw/Cmd?Help.x>. [Google Scholar]
- National Center for Biotechnology Information (NCBI). PubMed help appendices: how PubMed works: automatic term mapping. [Web document]. Bethesda, MD: National Library of Medicine, 2005. [cited 20 Nov 2005]. <http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=helppubmed.section.pubmedhelp.Appendices>. [Google Scholar]
- Vocabulary mapping. OVID help screens. [Web document]. New York, NY: Ovid Technologies, 2005. [cited 20 Nov 2005]. <http://gateway.ut.ovid.com/gw1.ovidweb.cgi>. [Google Scholar]
- Knecht L. Heading mapped-to maintenance: for supplementary concept records' names of substance. NLM Tech Bull [serial online]. 2003 Nov–Dec;335. [cited 29 Dec 2005]. <http://www.nlm.nih.gov/pubs/techbull/nd03/nd03_map_to.html>. [Google Scholar]