Abstract
Protein modification by SUMO conjugation is an important regulatory event. Sumoylation usually takes place on a lysine residue embedded in the core consensus motif ψKxE. However, this motif confers limited specificity on the sumoylation process. Here, we have probed the roles of clusters of acidic residues located downstream from the core SUMO modification sites in proteins such as the transcription factor Elk-1. We demonstrate that these are functionally important in SUMO-dependent transcriptional repression of Elk-1 transcriptional activity. Mechanistically, the acidic residues are important in enhancing the efficiency of Elk-1 sumoylation by Ubc9. Similar mechanisms operate in other transcription factors and phosphorylation sites can functionally substitute for acidic residues. Thus, an extended sumoylation motif, termed the NDSM (negatively charged amino acid-dependent sumoylation motif), helps define functional SUMO targets. We demonstrate that this extended motif can be used to correctly predict novel targets for SUMO modification.
Keywords: Elk-1, SUMO, transcription repression, Ubc9
Introduction
The modification of proteins by SUMO conjugation is an important regulatory post-translational event (reviewed by Johnson, 2004; Gill, 2005; Hay, 2005). Sumoylation can control various facets of protein function, including subcellular localisation, stability and interactions with DNA and other proteins. Many of the functions of SUMO can be attributed to nuclear activities, including important roles in controlling DNA replication/genome stability and gene transcription, although several non-nuclear proteins have been shown to be SUMO modified (reviewed by Watts, 2004). The number of functionally verified mammalian SUMO substrates is rapidly increasing, and recent proteomic screening approaches have uncovered more new targets (Li et al, 2004; Vertegaal et al, 2004; Zhao et al, 2004; Guo et al, 2005). However, we still do not know the number of SUMO substrates that exist in cells, and attempts to address this question are hindered by the low abundance of many SUMO substrates such as transcription factors. This problem is exacerbated by the low stoichiometry of sumoylation, which is usually observed. Furthermore, transient regulated SUMO modification is difficult to detect. Potential SUMO substrates can be identified by searching with the core consensus motif required for SUMO conjugation, ψKxE (where ψ is a bulky hydrophobic residue). However, this motif is found in over a third of all characterised proteins, suggesting that numerous false positives likely exist. Thus, the identification of further determinants would facilitate the prediction of potential SUMO substrates. In addition, several substrates have been identified which are modified on sites which do not conform to this minimal core motif (reviewed by Johnson, 2004).
An extended SUMO consensus motif has been suggested by several groups. For example, the synergy control motif (SC) is defined by the presence of proline residues flanking the core SUMO motif (Subramanian et al, 2003). In addition, an extended motif based on the R-motif repression domain of the transcription factor Elk-1 and the CRD1 domain of p300 was proposed, in which clusters of acidic residues are found downstream from the core SUMO motif (Yang et al, 2003). However, to date, the functional significance of these extended motifs has not been investigated. Recently, the PDSM (phosphorylation-dependent sumoylation motif) has been identified in a subset of substrates that conforms to the extended consensus ψKxExxSP (Hietakangas et al, 2006). Phosphorylation of the SP motif within this consensus sequence plays an important role in promoting sumoylation of several substrates (Gregoire et al, 2006; Hietakangas et al, 2006; Shalizi et al, 2006).
The core SUMO consensus motif ψKxE is recognised by the E2-conjugating enzyme Ubc9, which makes key interactions with this motif (Lin et al, 2002; Bernier-Villamor et al, 2002; Reverter and Lima, 2005) and transfers SUMO onto the lysine residue. Ubc9 is itself activated by the transfer of SUMO from the E1 SUMO-activating enzyme, SAE1/2. E3 ligases such as PIAS proteins and RanBP2 can facilitate substrate sumoylation, but at least in vitro, Ubc9 is sufficient to promote substrate sumoylation (reviewed by Johnson, 2004; Hay, 2005).
Here, we have investigated the functional significance of the acidic residues found within the R-motif sequence. Using Elk-1 as a model, we demonstrate that these acidic residues play functionally important roles through promoting substrate sumoylation. A reciprocal basic patch is identified on the E2-conjugating enzyme Ubc9 that is also important for substrate binding and sumoylation. The extended SUMO consensus motif, termed the NDSM (negatively charged amino acid-dependent sumoylation motif), is also found to play an analogous role in other substrates and can be used to correctly predict SUMO targets.
Results
Functional importance of acidic groups located downstream from the core SUMO site
We previously noted the similarities between the R-motif repression domain in Elk-1 and the CRD1 repression domain in p300. In addition to the functionally important core SUMO motif (ψKxE), these regions contain several acidic residues located in the immediate downstream ‘tail' region (Yang et al, 2002, 2003). We extended this analysis to a larger data set containing all currently validated SUMO sites found in human substrates with a role in transcriptional control (65 proteins, with 94 sites; Figure 1 and Supplementary Table S1). Interestingly, there is a marked over-representation of acidic residues in the tail region of these proteins compared to randomly selected proteins containing putative core SUMO motifs (Figure 1; Supplementary Figure S1A, C and D; see Supplementary Tables S1 and S2 for complete data sets). Conversely, there is an under-representation of basic residues, leading to an overall net negative charge in the tail region (Figure 1; Supplementary Figure S1A and B). This net negative charge is most apparent in a patch between positions 3 and 6, and is characterized by a marked over-representation of acidic residues at positions 3, 4, 5 and 6, and a lack of basic residues at position 4 (Figure 1; Supplementary Figure S1A). This analysis indicates that the SUMO consensus site can be portrayed by a longer sequence, consisting of the core motif and the acidic tail region, which potentially form a functional module. In particular, the conserved acidic nature of the tail region suggests functional importance.
Figure 1.
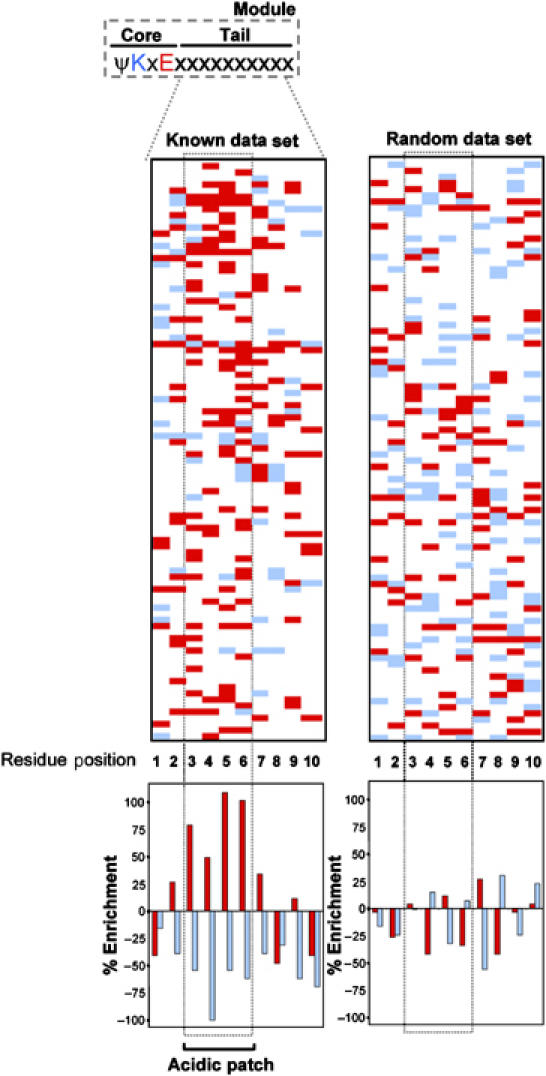
The region downstream from the core SUMO motif is rich in acidic residues. The left panel shows an alignment of the 10-amino-acid tail region located downstream from known characterised SUMO sites found in human transcriptional regulatory proteins (see Supplementary Table S1 for more details). Only acidic (shaded red) and basic (shaded blue) residues are depicted. The graph shown below indicates the percentage enrichment of acidic (red) and basic (blue) residues at each position, relative to the average occurrence of each type of amino acid in the downstream region of a control data set derived from shuffled ψKxE motifs. The panel on the right portrays the same information, but is a random selection from the total core consensus ψKxE data set (see Supplementary Table S2 for details).
To assess the potential functional importance of the acidic residues in the tail region of the extended SUMO module, we first focused on the SUMO sites located in the Elk-1 R-motif. The R-motif has previously been shown to function as a SUMO-dependent repression domain (Yang et al, 2003). As an initial approach, we therefore examined whether mutation of the tail region affected the activity of the Elk-1 R-motif in a transrepression reporter assay (Figure 2A). The Elk-1 R-motif contains two distinct 14-amino-acid modules, each containing one of the reported functional core SUMO modification sites (Figure 2A; Yang et al, 2003). As the SUMO site located at K249 is the most important for transcriptional repression, we focused on module 2. First, we mutated the three acidic residues (E255, E256 and E258) located downstream from K249 to alanine. This mutant, E3A, showed a four-fold increase in activity, similar to that observed when mutating K249 itself (Figure 2A). To enable us to focus directly on module 2, we disabled module 1 by mutating K230, and then probed the role of downstream acidic residues in transcriptional repression mediated by the SUMO site at K249. In this case, in the K230R/E3A mutant, the mutation of the three acidic residues synergised with the K230R mutation in enhancing Elk-1 transactivation capacity (Figure 2A). However, in the absence of the two major SUMO sites in the K230,249R/E3A mutant, the removal of the three acidic residues had no further effect. We also tested the effect of the E3A mutant in the context of full-length Elk-1 on the egr-1 promoter (Figure 2B). Consistent with a repressive effect of sumoylation, mutation of the two SUMO motifs caused an increase in Elk-1 transactivation activity. Importantly, mutation of the downstream acidic residues in the E3A mutant also caused an increase in Elk-1-mediated reporter activation. Thus, functionally, mutating three acidic residues downstream from K249 gave a similar effect to mutating the SUMO modification sites themselves and caused transcriptional derepression.
Figure 2.
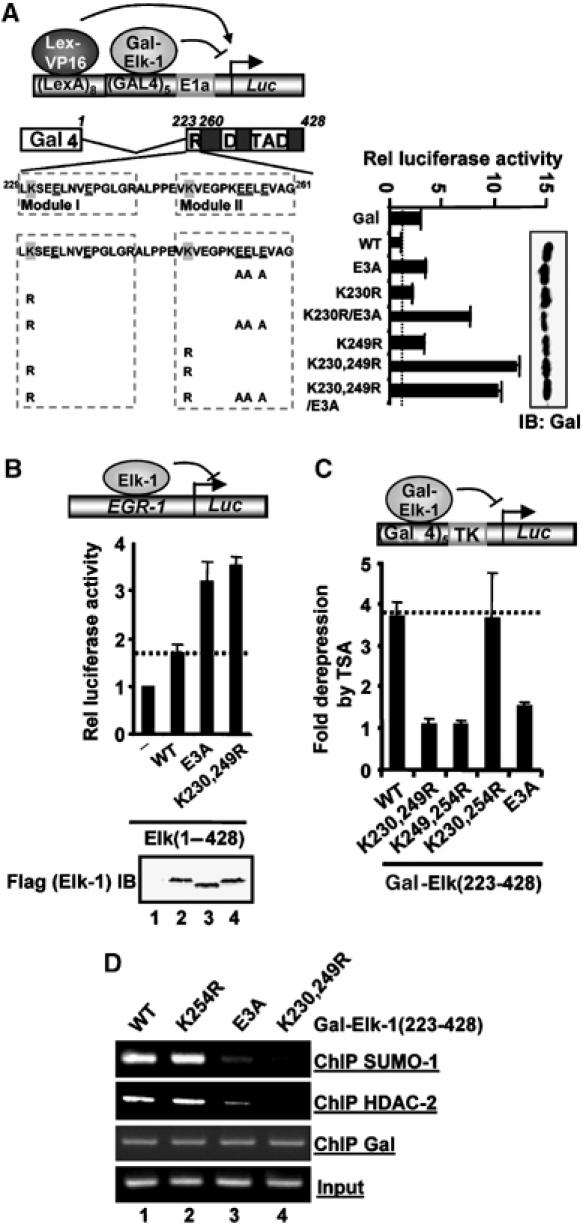
Functional importance of acidic groups located downstream from the core SUMO site. (A) Reporter gene analyses of the activities of the indicated mutant versions of Gal-Elk-1(233–428) fusion proteins in 293 cells. The structure of the LexA-Gal4-driven luciferase reporter used and the domain structure of the Elk-1 moiety is diagrammed and the sequence of the R-motif shown. The SUMO acceptor lysines are shaded grey and downstream acidic residues underlined. The boxes represent 14-amino-acid modules consisting of the core SUMO consensus site ψKXE followed by 10 downstream amino acids (tail). Assays were performed on the reporter in the presence of co-transfected LexA-VP16. The identity of the mutations in Elk-1 are indicated, with E3A representing the simultaneous mutation of E255, E256 and E258 to alanine. Western blots (IB) of Gal-Elk-1 protein levels are shown on the right. Data are shown relative to the activity of wild-type (WT) GAL-Elk-1(223–428) (taken as 1). Luciferase assays are representative of at least two independent experiments (s.e.'s are shown; n=2). (B) Reporter gene analyses of the activities of the indicated WT and mutant versions of full-length Elk-1 proteins in 293 cells. Assays were performed on an egr-1 promoter-driven luciferase reporter (shown schematically). A Western blotting showing the relative expression levels of the Flag-tagged Elk-1 proteins is show below the graph. Data are representative of at least two independent experiments (s.e.'s are shown; n=2). (C) Reporter gene analyses of the activities of the indicated WT and mutant versions of Gal-Elk-1(223–428) fusion proteins in 293 cells in the presence of TSA. Assays were performed on a Gal4-driven luciferase reporter (shown schematically). Data are shown as fold derepression by TSA relative to the activity of each construct in the absence of TSA. Luciferase assays are representative of at least two independent experiments (s.e.'s are shown; n=2). (D) ChIP analysis in the presence of the indicated WT and mutant versions of Gal-Elk-1(223–428) fusion proteins in 293 cells on the promoter of a Gal4-driven E1b luciferase reporter. ChIPs were performed with the indicated antibodies.
The repressive activity of the Elk-1 R-motif relies on its ability to recruit the histone deacetylase HDAC-2 in a SUMO-dependent manner and hence influence local histone acetylation levels (Yang and Sharrocks, 2004). We therefore investigated whether the acidic residues in module 2 had any role in this process. First, we analysed the ability of TSA to derepress the Elk-1 R-motif. Consistent with previous results (Yang et al, 2002), mutational analysis revealed that K249 plays a major role in determining the TSA sensitivity (Figure 2C). In contrast, neither K230 nor K254 appeared to play an important role in this process. Indeed, while K254 has been implicated as a SUMO target (Salinas et al, 2004), it appears to have little role in mediating transcriptional repression (data not shown). Next, we compared the ability of wild-type and mutant forms of Elk-1 to recruit HDAC-2 to promoters by chromatin immunoprecipitation (ChIP) analysis. As expected, the K230,249R mutant exhibited severely reduced HDAC-2 recruitment (Figure 2D, lane 4; Yang and Sharrocks, 2004). The activity of the K254R mutant was again unaffected, but the E3A mutant exhibited greatly reduced HDAC-2 recruitment (Figure 2D, lanes 3 and 4). Moreover, ChIP analysis revealed greatly reduced SUMO occupancy at this promoter in the presence of the E3A mutant version of Elk-1. Thus, the downstream acidic residues appear important for recruiting HDAC activity to Elk-1-regulated promoters.
Collectively, these data therefore demonstrate that acidic residues located downstream from the core motif play an important functional role in determining the repressive properties of the key SUMO module in Elk-1.
Role of downstream acidic groups in determining the efficiency of Elk-1 sumoylation
The function of the acidic residues within the SUMO modules in Elk-1 in transcriptional repression might be either through a direct effect or indirectly by helping to determine the sumoylation status of each module. Indeed, the latter possibility is suggested by the observation that SUMO levels, most likely of Elk-1 itself, are reduced at promoters upon mutation of these residues. To further probe this possible mechanism of action, we directly tested whether mutation of the acidic residues in module 2 had any effect on the levels of SUMO-modified Elk-1. First, we expressed wild-type and mutant versions of Elk-1 in 293 cells in the presence of HA-tagged SUMO and determined their sumoylation status. As observed previously (Yang et al, 2003), mutation of K230 and 249 virtually abolished Elk-1 sumoylation (Figure 3A, lane 4). Similarly, mutation of the acidic residues in the E3A mutant also caused a severe reduction in sumoylation levels (Figure 3A, lane 3). Importantly, the K254R mutant did not show significantly reduced sumoylation levels, indicating that mutation of the acidic residues was affecting the modification of SUMO sites other than K254. These results suggest that the acidic residues might form part of an extended motif that determines the efficiency of substrate sumoylation. To test this directly, we compared the sumoylation efficiency of wild-type and mutant Elk-1 peptides in an in vitro assay. In comparison to the wild-type peptide, the sumoylation efficiency of the mutant peptide was severely compromised (Figure 3B). Thus, both in vitro and in vivo assays demonstrate the importance of the acidic residues in determining the efficiency of Elk-1 sumoylation.
Figure 3.
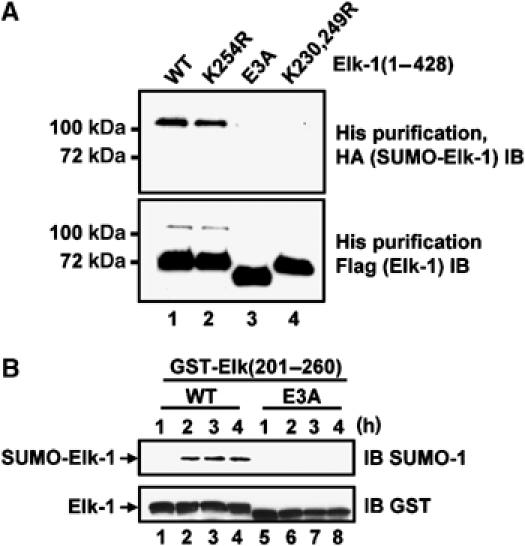
Role of acidic residues in Elk-1 sumoylation. (A) Sumoylation of Elk-1 in vivo. The indicated mutant versions of His-Flag-tagged Elk-1(1–428) were coexpressed with HA-tagged SUMO-1 and His-tagged Elk-1 was precipitated from cell lysates. Sumoylated Elk-1 was detected by immunoblotting (IB) with an anti-HA antibody and total Elk-1 with an anti-Flag antibody. (B) Elk-1 sumoylation in vitro. Purified wild-type (WT) and E3A mutant GST-Elk-1(201–260) (2 μg) were sumoylated in vitro with 500 ng Ubc9 for the indicated times. Sumoylated Elk-1 was detected by IB with an anti-SUMO-1 antibody and total GST-Elk-1 with an anti-GST antibody.
The E2 ubiquitin-conjugating enzyme, Ubc9, is responsible for catalysing the transfer of SUMO onto substrates (reviewed by Johnson, 2004). We therefore investigated whether the acidic residues played a role in stabilising Ubc9–Elk-1 interactions. First, we used GST pulldowns to analyse the binding of purified Ubc9 with GST-Elk-1 fusion proteins in vitro. The mutation of the acidic residues in the mutant Elk-1(E3A) led to a decrease in Ubc9–Elk-1 interactions (Figure 4A). To further corroborate this finding, we investigated Elk-1–Ubc9 interactions in vivo by co-immunoprecipitation analysis. In comparison to wild-type Elk-1, mutation of either the SUMO conjugation motifs (K230,249R) or the acidic residues in the second SUMO module (E3A) caused a large decrease in Ubc9 binding to Elk-1 (Figure 4B).
Figure 4.
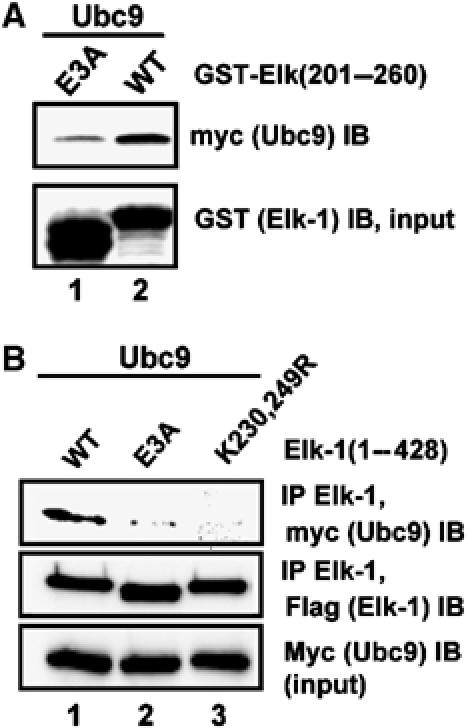
The acidic residues are important in Ubc9–Elk-1 interactions. (A) GST pulldown assay of myc-tagged Ubc9 by the indicated GST-Elk-1 fusion proteins. (B) Co-immunoprecipitation analysis of Ubc9 with Elk-1 from 293 cells co-transfected with expression vectors encoding myc-tagged Ubc9 and the indicated Flag-tagged Elk-1 proteins. Immunoprecipitation (IP) was performed with an anti-Elk-1 antibody, and precipitated Elk-1 and Ubc9 were detected by immunoblotting (IB) with the indicated antibodies.
Recently, an acidic tail located downstream from a hydrophobic patch has been shown to be an important determinant of noncovalent interactions between SUMO and other proteins (Hecker et al, 2006). We therefore tested whether Elk-1 bound to SUMO and if the acidic tail played a role in this binding. However, noncovalent binding of neither wild-type nor E3A mutant Elk-1 could be detected with SUMO-1 (Supplementary Figure S2).
Together, these data therefore demonstrate that the downstream acidic residues play an important role in determining the efficiency of Elk-1–Ubc9 interactions, which lead to reduced Elk-1 sumoylation within the core SUMO motifs.
Role of acidic residues and phosphorylation motifs in other SUMO substrates
To test whether the acidic residues found downstream from core SUMO motifs have similar functional roles in SUMO substrates other than Elk-1, we first investigated MEF2A. MEF2A and other related MEF2 proteins have been shown to be SUMO modified (Gregoire and Yang, 2005; Zhao et al, 2005) and, more recently, a downstream Cdk5 phosphorylation site was shown to play an important role in basal level MEF2 sumoylation (Gregoire et al, 2006; Hietakangas et al, 2006; Shalizi et al, 2006). Importantly, a negatively charged phosphomimicking glutamate residue was able to rescue MEF2 sumoylation, suggesting that it is the negative charge rather than the presence of a phosphate group that is the salient feature. We therefore introduced additional mutations into MEF2A to further probe the importance of acidic residues downstream from the core SUMO motif. First, we established that the core SUMO consensus motif conferring repressive properties on MEF2A contained K395 (data not shown; Riquelme et al, 2006). We then introduced mutations into the extended SUMO module extending 10 amino acids further downstream (Figure 5A and B). Mutation of the Cdk5 phosphoacceptor motif, S400A, also caused an enhancement of MEF2A activity. However, the introduction of an acidic residue further downstream, in the absence of the phosphoacceptor residue in the S400A/R403E mutant, reduced the activity back to near basal levels. Similarly, the introduction of a negatively charged amino acid in place of the phosphoacceptor residue [MEF2A(S400E)] also maintained the activity of MEF2A at a low level. Thus, it appears that it is the presence of a negatively charged residue in this downstream region that is important for maintaining MEF2A in a repressed state rather than a precisely positioned phosphorylated amino acid. To establish whether the phosphoacceptor site and acidic nature of the downstream region was important for MEF2A sumoylation, we first analysed the sumoylation status of wild-type MEF2A and a form that lacked both the phosphoacceptor serine and a downstream acidic residue (S400A/D404A). In comparison to the wild-type protein, MEF2A(S400A/D404A) exhibited much reduced levels of sumoylation in vivo (Figure 5D, lane 3). This is consistent with the observation that mutation of S400 alone substantially reduces the sumoylation of MEF2 proteins (Gregoire et al, 2006; Hietakangas et al, 2006; Shalizi et al, 2006). Importantly, however, we were able to restore substantial levels of sumoylation in the absence of S400 by introducing a negatively charged acidic residue in place of R403 (Figure 5D, lane 2). Thus, the presence of negatively charged phosphate groups or acidic residues downstream from the core SUMO motif is important for maintaining MEF2A sumoylation levels and the resulting transcriptional repressive properties conferred by SUMO modification.
Figure 5.
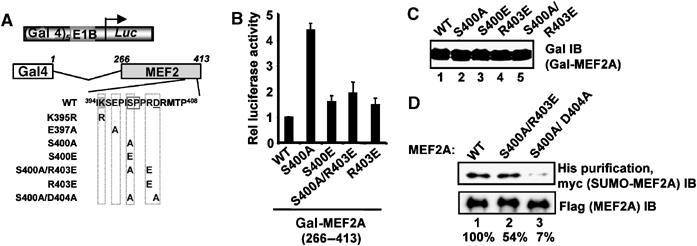
Role of acidic residues in sumoylation of MEF2A. (A) Schematic illustration of the Gal4-driven luciferase reporter and the Gal4-MEF2A(266–413) fusion protein. The sequence of the module containing the core SUMO consensus site ψKXE followed by 10 downstream amino acids is shown. The SUMO acceptor lysines are shaded grey and downstream acidic residues underlined. The residues altered in each mutant protein are indicated. (B) Reporter gene analyses of the activities of the indicated mutant versions of GAL-MEF2A(266–413) fusion proteins in 293 cells. Data are presented relative to the activity of the wild-type (WT) protein (taken as 1). (C) Western blots (IB) of Gal-MEF2A protein levels. (D) Sumoylation of MEF2A in vivo. WT or mutant versions (S400A/D404A or S400A/R403E) of His-Flag-tagged MEF2A(1–486) were coexpressed in 293 cells with myc-tagged SUMO-1 and His-tagged MEF2A was precipitated from cell lysates. Sumoylated MEF2A was detected by immunoblotting (IB) with an anti-myc antibody and total MEF2A with an anti-Flag antibody. The numbers under each lane represent the average binding in three independent experiments, compared to WT MEF2A (taken as 100%).
Next, we investigated the role of acidic residues in determining the sumoylation levels of the orphan nuclear hormone receptor LRH-1 (Chalkiadaki and Talianidis, 2005). As expected, mutation of the target lysine residue, K224, reduced LRH-1 sumoylation (Figure 6A, lanes 3 and 6). However, the mutation of two downstream acidic residues in LRH-1(D229A/E236A) also lead to a large decrease in sumoylation levels. This decrease was also apparent in the presence of the E3 ligase PIASxα, although higher levels of sumoylation were retained (Figure 6A, lane 5). Importantly, sumoylation of LRH-1 in vitro was also compromised by mutation of D229 and E236 (Figure 6B). The significance of these results for LRH-1 activity was demonstrated using a reporter gene assay. Mutation of either the SUMO-modified lysine residue (K224) or the two downstream acidic residues caused a loss of repressive activity and enhanced transactivation by LRH-1 (Figure 6C).
Figure 6.
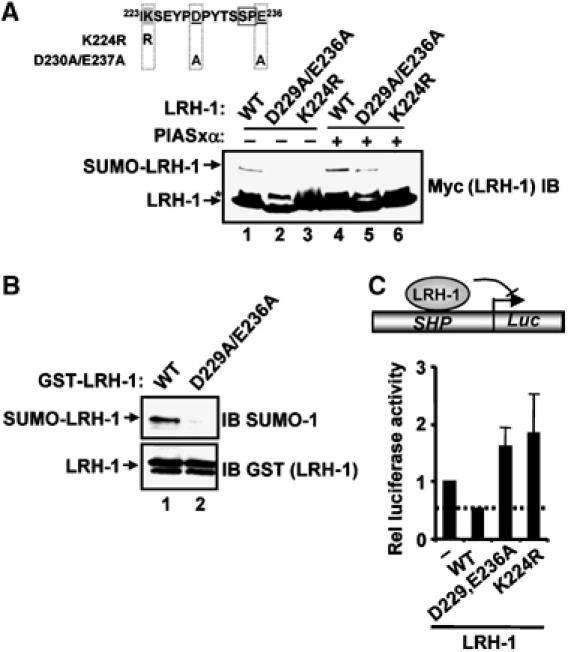
Role of acidic residues in sumoylation of LRH-1. (A) Sumoylation of LRH-1 in vivo. The sequence of the module in LRH-1 containing the core SUMO consensus site ψKXE followed by 10 downstream amino acids is shown. Annotation is as in Figure 5A. The indicated wild-type and mutant myc-tagged LRH-1 proteins were coexpressed in 293 cells with HA-tagged SUMO-1 and PIASxα where indicated. LRH-1- and SUMO-conjugated species were detected by immunoblotting (IB) with an anti-myc antibody. (B) LRH-1 sumoylation in vitro. Purified wild-type (WT) and D229A/E336A mutant GST-LRH-1(134–257) (2 μg) were sumoylated in vitro with 20 ng Ubc9 for 1 h. Sumoylated LRH-1 was detected by immunoblotting. (C) Reporter gene analyses of the activities of the indicated WT and mutant versions of full-length LRH-1 proteins in His-SUMO-HeLa cells. Assays were performed on an SHP promoter-driven luciferase reporter (shown schematically). Data are representative of at least two independent experiments (s.e.'s are shown; n=2).
Collectively, these data therefore demonstrate that in addition to Elk-1, acidic residues located downstream from other substrates play a key role in determining the efficiency of their sumoylation and hence transcriptional regulatory properties. We have therefore termed this extended SUMO module the negatively charged amino acid-dependent sumoylation motif (NDSM) due to the requirement for acidic residues in the downstream region for efficient sumoylation of substrates.
Identification of a basic patch on Ubc9 required for efficient sumoylation of an NDSM-containing substrate
The identification of a negatively charged patch within the NDSM of substrate proteins that is required for their efficient interaction with Ubc9 and their subsequent sumoylation suggests that a cognate positively charged binding surface should exist on Ubc9. We therefore mutated closely spaced pairs of basic residues in Ubc9 and tested their requirement for efficient substrate sumoylation. We used Elk-1 as a model NDSM-containing substrate and RanGAP-1 as a control, as its SUMO modification site does not contain appropriately positioned downstream acidic residues (Figure 7A). Four Ubc9 mutants exhibited reduced activity towards Elk-1 (Figure 7A). However, of these, only the Ubc9(K59E/K61E) mutant did not show a decrease in sumoylation of RanGAP-1 (Figure 7A, lane 5). The selectivity of the defects in Ubc9(K59E/K61E) towards the NDSM-containing substrate Elk-1 were further confirmed by analysing the rates of SUMO conjugation (Figure 7B). Basic residues in the N-terminal α-helix of Ubc9 (including R13, K14, R17 and K18) have previously been shown to be important in binding to SUMO and thioester bond formation, explaining the defects seen in the Ubc9(R13E/K14E) and Ubc9(R17E/K18E) mutants (Figure 7A; Tatham et al, 2003). We confirmed that SUMO loading of Ubc9 was not defective by the mutations K59E/K61E (Supplementary Figure S3, data not shown), although it remains possible that there are subtle kinetic defects in this process.
Figure 7.
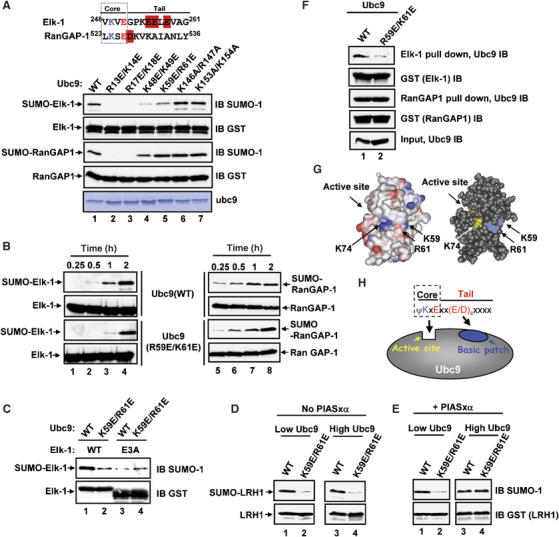
Identification of a basic patch on Ubc9 required for efficient sumoylation of an NDSM-containing substrate. (A) In vitro sumoylation assay of GST-Elk-1(201–260) (top panel) and GST-RanGAP-1(418–587) (third panel) (2 μg) using the indicated Ubc9 mutants (500 ng). Inputs of the GST fusion proteins (second and fourth panels; anti-GST immunoblot (IB)) and Ubc9 mutants (bottom panel; Coomassie stained) are shown. Sumoylated substrates are detected using anti-SUMO-1 IB. The sequences immediately downstream from the major SUMO motifs found in Elk-1 and RanGAP-1 are shown at the top. Downstream acidic residues are highlighted in red. (B) In vitro time-course sumoylation assay of GST-Elk-1(201–260) (left) and GST-RanGAP-1(418–587) (right) using the indicated Ubc9 mutants for the indicated times. Proteins levels and detection were as in (A). (C) In vitro sumoylation assay of wild-type and E3A mutant GST-Elk-1(201–260) (top panel) (2 μg) using the indicated wild-type and mutant Ubc9 (500 ng). Inputs of the GST fusion proteins (bottom panel; anti-GST IB) are shown. Sumoylated substrates are detected using anti-SUMO-1 IB. (D, E) In vitro sumoylation assay of wild-type and E3A mutant GST-LRH-1(134–257) (2 μg) (top panel) using either low (10 ng) or high (50 ng) amounts of the indicated wild-type and mutant Ubc9. Sumoylation assays were carried out in the absence (D) or presence (E) of PIASxα (62.5 ng). Inputs of the GST fusion proteins (bottom panel; anti-GST IB) are shown. Sumoylated substrates are detected using anti-SUMO-1 IB. (F) GST pulldown assay of Ubc9 (3 μg) by GST-Elk-1(201–260) (top panel) and GST-RanGAP-1(418–587) (third panel) (3 μg each). Ubc9 and inputs of the GST fusion proteins (second and fourth panels) were detected using IB with anti-Ubc9 or anti-GST antibodies, respectively. (G) Ubc9 structural models showing the charged surface area (left-hand panel, created using WebLab ViewerPro 4.0) and a space filling model (right-hand panel, created using Cn3D version 4.1). The left-hand model shows basic areas as blue and acidic areas as red. The locations of the basic patch created by K59 and R61 (shown as blue on the right-hand model), K74 (yellow) and the active site (yellow) are indicated. (H) Schematic model depicting the interactions between Ubc9 and the NDSM. The preponderance of acidic residues within positions 3–6 is denoted by (E/D)4 within the tail. The ψKxE core engages with the active site, whereas the tail region binds to the basic patch.
A key prediction from these data is that mutation of the basic patch on Ubc9 should not further compromise its ability to sumoylate the Elk-1 E3A mutant, which has lost the acidic patch. We therefore compared the activity of wild-type Ubc9 and Ubc9(K59E/R61E) towards wild-type and E3A mutant forms of Elk-1 (Figure 7C). In contrast to wild-type Elk-1 (lanes 1 and 2), there was no decrease in the sumoylation activity of Ubc9 towards the Elk-1(E3A) mutant upon removal of the basic patch in the Ubc9(K59E/R61E) mutant (lanes 3 and 4). This result therefore strongly implies that interactions between the basic patch on Ubc9 and the acidic patch on Elk-1 occur, as independent roles for the two regions would combine to further compromise the ability of Ubc9 to sumoylate Elk-1.
We also compared the activity of wild-type Ubc9 and Ubc9(K59E/R61E) towards another protein containing an NDSM, LRH-1, to further generalize our findings. As observed for Elk-1, the Ubc9(K59E/R61E) mutant exhibited reduced activity towards LRH-1 (Figure 7D). LRH-1 sumoylation can be enhanced by the E3 ligase PIASxα (Chalkiadaki and Talianidis, 2005; see Figure 6A). In the presence of PIASxα and low levels of Ubc9, reduced sumoylation was again seen with the Ubc9(K59E/R61E) mutant (Figure 7E, lane 2). However, with higher levels of Ubc9, PIASxα was able to enhance sumoylation of LRH-1 by Ubc9(K59E/R61E). This suggests that by acting as an E3 ligase, PIASxα can overcome the requirement for interactions between the acidic patch in the NDSM and the basic region on Ubc9.
As mutation of the NDSM reduced interaction between substrates and Ubc9 (Figure 4), we predicted that loss of the basic region in Ubc9 would have similar consequences. Indeed, Ubc9(K59E/K61E) exhibited reduced binding to Elk-1 in a GST pulldown assay (Figure 7F).
Collectively, these data therefore identify an important basic patch on the surface of Ubc9 (Figure 7G) that is required for efficient binding and sumoylation of NDSM-containing substrate proteins.
Extended SUMO module allows prediction of novel SUMO substrates
The total number of SUMO substrates is currently unknown, and simple searches with the motif ψKxE using the SUMOplot programme (http://www.abgent.com/doc/sumoplot) return nearly half of all characterised human proteins in the SWISSPROT database. Other approaches have introduced inherent biases into the search protocols such as the SSP programme, which uses phylogenetic conservation as a parameter but also assumes that proteins must contain an NLS (Zhou et al, 2005). We therefore investigated whether our extended SUMO module (NDSM) could be used to identify known or novel potential substrates. A sequence consisting of the core SUMO motif ψKxE followed by at least two further acidic residues in the C-terminal tail, one of which is located between amino acids 3 and 6, was used to search the human proteins within the SWISSPROT database. This search identified 2139 (15%) of these proteins as potential SUMO substrates (Figure 8A; Supplementary Table S3). Importantly, the NDSM shows further discriminatory power when looking at the total numbers of sites identified. On average, the core ψKxE predicts that there are 8398 sites (1.7 sites per protein identified), whereas the NDSM predicts only 2847 (1.3 sites per protein), and the NDSM also produces less false negatives (see Discussion). Candidate proteins found using the extended module showed significant enrichment for a variety of gene ontology categories when compared to a random data set (Figure 8B; Supplementary Table S4). Many of these categories were associated with nuclear function and in particular transcription and chromatin, which are processes with which SUMO is known to be associated with.
Figure 8.
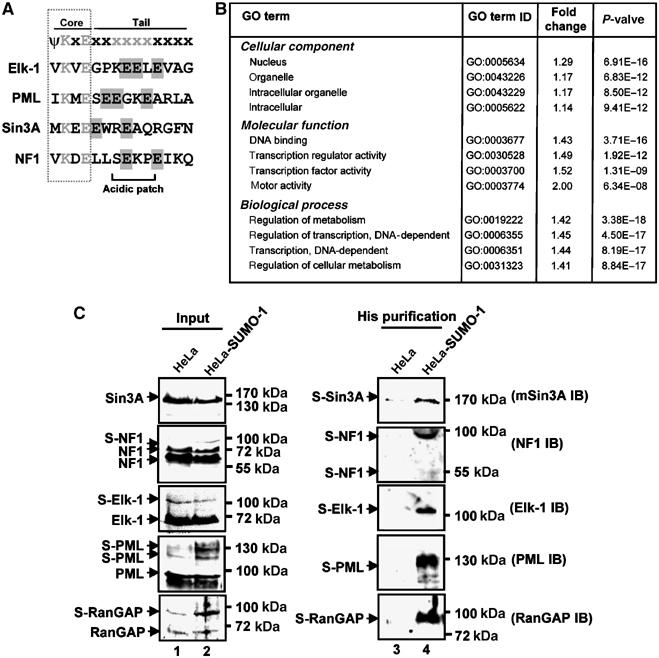
Identification of novel SUMO substrates using an extended SUMO consensus motif. (A) Sequences of the SUMO sites within known and novel substrates predicted using NDSM motif (see Supplementary Table S3 for complete data set). (B) Gene ontologies showing high levels of over-representation in the NDSM-predicted data set (see Supplementary Table S4 for complete data set). Data are represented as fold increase over a control data set generated with a shuffled ψKxE motif with downstream acidic residues. P-values are generated using a χ2 test. (C) Identification of SUMO-conjugated proteins. Proteins conjugated to His-tagged SUMO were precipitated from normal HeLa or His-SUMO-HeLa cells and identified by Western analysis (IB) with the indicated antibodies (right-hand panels). The input proteins are shown on the left. Bands corresponding to sumoylated proteins are indicated by the suffix ‘S-'. A colour version of this figure is available at The EMBO Journal Online.
Recently, several substrates including RBP1 (ARI4A) (Binda et al, 2006) and KLF8 (Wei et al, 2006) have been shown to be modified by SUMO as predicted by searching with the NDSM. To further verify whether the potential SUMO substrates represented bona fide in vivo targets for sumoylation, we randomly selected additional candidates and asked whether the endogenous protein was modified in HeLa cells (Figure 8C). As expected, the known substrates, RanGAP-1, PML and Elk-1 were all sumoylated. In addition, sumoylation of the novel substrates Sin3A and NF1 was detected. Thus, the extended SUMO module can be used to correctly predict novel substrates.
Discussion
The majority of lysine residues that have currently been verified as sites for SUMO conjugation conform to the core consensus motif ψKxE. However, this sequence is still somewhat limited in conferring specificity. Here we have identified an extended SUMO consensus motif, which we have termed the NDSM. In addition to the core consensus motif, this extended sequence encompasses several acidic residues clustered within the 10-amino-acid region located immediately downstream. Within the tail region, the acidic residues tend to occupy a hotspot encompassing residues 3–6. Functionally, these acidic residues play an important role in determining the efficiency of substrate sumoylation.
Recently, the PDSM has been identified in a subset of SUMO substrates that conforms to the extended consensus ψKxExxSP (Hietakangas et al, 2006). Phosphorylation of the SP motif within this consensus sequence plays an important role in promoting sumoylation of several substrates including MEF2A (Gregoire et al, 2006; Hietakangas et al, 2006; Shalizi et al, 2006). Interestingly, the location of the phosphoacceptor residue in the PDSM at position 3 is located within the middle of the acidic patch found in the tail region of known substrates (Supplementary Figure S1A), suggesting that it is the negative character of this residue that is important. Indeed, insertion of an acidic residue in place of the phosphorylated serine residue retained efficient substrate sumoylation and hence imparted transcriptional repressive properties on MEF2 proteins (Gregoire et al, 2006; Hietakangas et al, 2006; Shalizi et al, 2006). However, here we demonstrate that these SUMO-dependent repression properties can also be maintained in the absence of MEF2A phosphorylation at S400, by insertion of an acidic residue further downstream. Thus, it appears that it is the negative charge character of the tail region downstream from the core SUMO motif that is the important parameter in promoting SUMO-dependent transcriptional repression. Our data also imply that phosphorylation at other positions within the tail region other than position 3 might also promote substrate sumoylation, thereby further extending the number of potential SUMO substrates beyond those already defined by the PDSM and NDSM. As potential ‘SP' phosphorylation motifs are often associated with core SUMO consensus motifs (Hietakangas et al, 2006; Shalizi et al, 2006; Supplementary Table S1), then this represents an attractive way in which substrate sumoylation can be controlled in an inducible manner by the introduction of increased negative charge through the deposition of phosphate groups. Furthermore, by altering the sequence of the tail region, it might be possible to weaken interactions with Ubc9 and hence introduce a further regulatory step through requiring E3 ligases as adapter proteins to enhance substrate–Ubc9 interactions. Indeed, we demonstrate that the requirement for the basic patch on Ubc9 for efficient LRH-1 sumoylation can be overcome by the E3 ligase PIASxα (Figures 6 and 7). Thus, the importance of the NDSM might vary depending on the level of core SUMO pathway components and substrate-specific E3 ligases in the cell.
While the PDSM and NDSM represent useful motifs to help identify bona fide sumoylation targets, it should be emphasised that there are likely to be a number of substrates that do not conform to these motifs. Indeed, several substrates do not even match the core ψKxE motif (reviewed by Johnson, 2004). In addition, structural constraints might also contribute to the specificity of sumoylation as recently observed in HSF-1 and HSF-2, where their conserved SUMO sites conform to the core ψKxE motif, but are presented on a structurally constrained loop, and it is the loop length that is critical in determining their differential sumoylation (Anckar et al, 2006). Indeed, the sites we have studied in Elk-1 and LRH-1 fall within unstructured regions, which do not correspond to known domains. Thus, the NDSM might represent a motif that is only presented in the context of extended peptide motifs. Finally, it is possible that other sequence determinants are present in the regions surrounding the SUMO motifs. Indeed, our analysis of known sites identifies serine and proline residues as over-represented in the downstream tail region (Supplementary Figure S1D). The latter observation is consistent with an alternative consensus site based on the SC, which is defined by the presence of proline residues flanking the core SUMO motif (Subramanian et al, 2003). The over-representation of serine residues might reflect further opportunities for control by phosphorylation, but further studies are needed to probe this possibility.
One mechanism through which substrate sumoylation is enhanced by the presence of negatively charged clusters of amino acids appears to be via increased binding to the E2 SUMO-conjugating enzyme Ubc9 (Figure 4). This increased binding is presumably mediated by electrostatic interactions, and consistent with this hypothesis, we have identified an important basic patch on the surface of Ubc9 (Figure 7). While the catalytic cleft and immediate surrounding area in Ubc9 have been shown to interact with the core SUMO motif (Bernier-Villamor et al, 2002; Lin et al, 2002; Reverter and Lima, 2005), it appears that this basic patch acts as a secondary binding site via the acidic region of the NDSM. Indeed, the position of the basic patch is consistent with the trajectory of a peptide binding to the active site of Ubc9, where the glutamate residue in the core ψKxE motif interacts with K74 and the downstream acidic residues would be ideally positioned to interact with K59 and R61 in Ubc9 (Figure 7G). Thus, the NDSM can make two contacts with Ubc9: one between the core ψKxE motif and the active site and one between the acidic tail and the basic patch on the surface of Ubc9 (Figure 7H). Interestingly, a similar mechanism has been shown to operate in MAP kinase–substrate interactions, but in this case, a basic patch on the substrates interacts with an acidic region on the kinase (reviewed by Tanoue and Nishida, 2003). In vivo, it is also possible that the E3 ligases might recognise this region, and hence promote the action of Ubc9. However, as the acidic residues are clearly important for the sumoylation of Elk-1 in vitro in the absence of any E3 ligase, the likely target is Ubc9 itself. Another possibility is that Elk-1 might interact with SUMO in a noncovalent manner, and the acidic tail might facilitate this process. Indeed, an acidic tail located downstream from a hydrophobic patch has recently been shown to be an important determinant of noncovalent interactions between SUMO and other proteins (Hecker et al, 2006). We could not detect noncovalent binding of Elk-1 and SUMO, suggesting that it is unlikely that the acidic tail serves this function, although it remains possible that weak intramolecular interactions between the conjugated SUMO molecule and Elk-1 might occur through this region.
It is important to emphasise that while our data are consistent with the NDSM being a key determinant for Ubc9 binding, we cannot rule out a subsidiary role for either the NDSM in substrates or the basic residues in Ubc9. Indeed, the effects we see in vivo on both sumoylation levels and protein activity are much greater than in vitro. While this might reflect suboptimal conditions in vitro, it equally might reflect a more complex role in vivo, where as yet unknown accessory proteins might act through these motifs to enhance SUMO-dependent effects. Indeed, it is currently unknown whether the acidic residues in the NDSM play a direct role in mediating transcriptional repression, in addition to promoting substrate sumoylation. In Elk-1, they are clearly important for its repressive properties and ability to recruit HDAC-2 to promoters (Figure 2). Moreover, the NDSM shows over-representation in a number of gene ontology categories related to transcription (Figure 8B; Supplementary Table S4). However, as sumoylation is required for imparting repressive properties on proteins like Elk-1, it is currently difficult to dissect these two functions. Further studies are therefore needed to address whether the acidic residues might also play a role in corepressor recruitment.
Finally, an important finding from our studies is that the NDSM can be used to predict novel SUMO substrates. By using this motif to gain more confidence in substrate prediction, it will become easier to identify and verify new targets. Indeed, using our new extended consensus motif, we estimate that 15% of human proteins might be SUMO modified, compared to 35% if searches are carried out with the ψKxE site alone. Here we have verified our approach by showing that the transcription factor NF1 and the corepressor Sin3A are both SUMO modified (Figure 8). In the latter case, sumoylation is likely to confer repressive properties as observed for many SUMO substrates (reviewed by Gill, 2005; Hay, 2005). In common with other transcription factors, NF1 can act as either a transcriptional activator or transcriptional repressor in different contexts (reviewed by Gronostajski, 2000), thus sumoylation might help it switch to a repressive role. Further studies are required to probe the role of sumoylation in these proteins. Importantly, the predictive power of the NDSM is further emphasised when looking at the total number of sites predicted rather than just the numbers of proteins. Indeed, the NDSM outperforms the core ψKxE motif in identifying real targets. By searching the human proteins in the SWISSPROT database with the ψKxE core motif, there are 8398 sites identified, but with the NDSM only 2842 (a three-fold enrichment). This compares with the numbers of proteins identified as 4906 (ψKxE core motif) and 2139 (NDSM) (a 2.3-fold enrichment). Importantly, the average numbers of potential sites per protein are 1.7 (ψKxE core motif) and 1.3 (NDSM). Thus, as this value approaches ‘1', the NDSM does not only highlight particular proteins but also enables the identification of sites within proteins. Furthermore, the predictive power of the NDSM among the multiple sites found within proteins (rather than for the proteins themselves) can be determined by analysing the ‘known target set'—see Supplementary Table S1). Within these proteins, there are sites that are not modified and yet contain the ψKxE core motif. Thus, the success rate of predictions based on the presence of a SUMO motif that is known to be modified can be obtained. Using this parameter, 81% of NDSM sites within these proteins are modified by SUMO. However, only 63% of sites containing the ψKxE core motif are actually modified (note if one takes into account possible PDSMs that are related to the NDSM, then this ratio goes down to 50%). We have also investigated the predictive power on proteins that have been identified as SUMO substrates since we generated the data set in this study. Within these proteins, the NDSM correlates with a modified site in 78% of cases, whereas the ψKxE core motif is only modified in 53% of cases (46% if PDSMs are omitted). These ratios are very similar to the initial data set, demonstrate and very low false-negative rate (20%) and underline the strong predictive power of the NDSM. Once an NDSM has been identified, it is possible to then use other features to provide further confidence that a protein is a true SUMO substrate. For example, searching databases such as HPRD or BIND can reveal interactions with SUMO pathway components such as Ubc9 or E3 ligases.
In summary, we have identified an important additional determinant of SUMO modification, enabling the derivation of an extended SUMO consensus motif termed the NDSM. This has important implications for both our understanding of how substrate sumoylation is achieved and for facilitating the identification of novel SUMO substrates.
Materials and methods
Plasmid constructs
Details of plasmid constructs can be found in Supplementary data.
Tissue culture, cell transfection and reporter gene assays
The 293, HeLa and HeLa-SUMO-1 (kindly provided by P O'Hare) and HeLa-myc-His-SUMO-1 (kindly provided by Ron Hay) cells were grown and transfection experiments were carried out as described previously (Yang et al, 2003; Yang and Sharrocks, 2005). For TSA stimulation, cells were treated with 330 nM TSA 16 h before harvesting. For Gal4 fusion-driven luciferase reporter gene assays, 0.25 μg of reporter plasmid and 0.125 μg of pCH110 were co-transfected with 0.1 μg of GAL4-fusion expression plasmids. Cell extracts were prepared and luciferase and β-galactosidase assays were carried out as described previously (Yang et al, 1998).
SUMO assays, Western blotting, immunoprecipitation analysis and GST pulldown assays
In vivo sumoylation assays with His-tagged Elk-1 and MEF2A were performed as described previously (Yang et al, 2003). LRH-1 sumoylation levels were assessed in total cell lysates made in SUMO lysis buffer as described previously (Chalkiadaki and Talianidis, 2005). In vitro sumoylation assays were performed as described previously (Yang et al, 2003), except that the E1 enzyme (140 ng/assay) was from Alexis Biochemicals corporation. Assays were performed in 15–20 μl containing 5 mM Tris (pH 8.0), 0.5 mM MgCl2, 0.2 mM ATP, 1 mM creatine phosphate, 3.5 U/ml creatine kinase, 60 μU/ml inorganic pryrophosphatase and typically 3 μg SUMO-1, 10–500 ng Ubc9 and 2 μg substrate. GST-Elk-1, GST-LRH-1, GST-PIASxα and GST-Ubc9 fusion proteins were purified (Shore and Sharrocks, 1994), as described previously.
Western blotting was carried out with the primary antibodies: Flag (Sigma), HA (Roche), Elk-1, Gal4(DBD), myc, GST, Sin3A, NF1, SUMO-1(D-11) and PML (Santa Cruz Biotech), RanGAP (Abcam), Ubc9 (BD Transduction Lab.) and HRP-conjugated secondary antibodies (BD Transduction Lab.), as described previously (Yang and Sharrocks, 2005).
GST pulldown assays were carried out essentially as described previously (Shore and Sharrocks, 1994). Ubc9 or SUMO-1 was cleaved from GST by treatment with thrombin before performing the pulldown experiments with GST-Elk-1 or GST-RanGAP-1. Residual uncleaved Ubc9 or SUMO-1 was removed prior to binding to GST fusions by preclearing with glutathione agarose beads. Co-immunoprecipitation analysis was performed from cells lysed in 0.5 × lysis buffer as described previously (Yang and Sharrocks, 2005).
SUMO loading assays were performed as described previously (Pichler et al, 2002), using 150 ng E1, 100 ng Ubc9 (E2) and 100 ng SUMO-1 for 45 min at 37°C.
ChIP assays and quantitative real-time PCR ChIP assay
ChIP assays using antisera specific to SUMO-1 (Santa Cruz) and HDAC-2 (Abcam) were performed as described previously (Yang and Sharrocks, 2005), except that crosslinking was performed with 1% formaldehyde for 2–5 min. Bound promoters were detected by standard PCR as described previously (Yang and Sharrocks 2005), using the primer pairs for the Gal-driven luciferase reporter promoter described previously (Yang et al, 2003).
Bioinformatics approaches
Details of approaches used are provided in the Supplementary data.
Supplementary Material
Supplementary Data
Figure 8
Data Sets 1
Data Sets 2
Data Sets 3
Data Sets 4
Acknowledgments
We thank Anne Clancy for excellent technical assistance; Stefan Roberts, Alan Whitmarsh and members of our laboratory for comments on the manuscript and stimulating discussions, Roland Ennos for statistical advice; and Peter O'Hare, Iannis Talianidis, Alan Whitmarsh and Ron Hay for reagents. This work was supported by grants from the Wellcome Trust and a BBSRC studentship to JW.
References
- Anckar J, Hietakangas V, Denessiouk K, Thiele DJ, Johnson MS, Sistonen L (2006) Inhibition of DNA binding by differential sumoylation of heat shock factors. Mol Cell Biol 26: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD (2002) Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108: 345–356 [DOI] [PubMed] [Google Scholar]
- Binda O, Roy JS, Branton PE (2006) RBP1 family proteins exhibit SUMOylation-dependent transcriptional repression and induce cell growth inhibition reminiscent of senescence. Mol Cell Biol 26: 1917–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalkiadaki A, Talianidis I (2005) SUMO-dependent compartmentalization in promyelocytic leukemia protein nuclear bodies prevents the access of LRH-1 to chromatin. Mol Cell Biol 25: 5095–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G (2005) Something about SUMO inhibits transcription. Curr Opin Genet Dev 15: 536–541 [DOI] [PubMed] [Google Scholar]
- Gregoire S, Tremblay AM, Xiao L, Yang Q, Ma K, Nie J, Mao Z, Wu Z, Giguere V, Yang XJ (2006) Control of MEF2 transcriptional activity by coordinated phosphorylation and sumoylation. J Biol Chem 281: 4423–4433 [DOI] [PubMed] [Google Scholar]
- Gregoire S, Yang XJ (2005) Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors. Mol Cell Biol 25: 2273–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronostajski RM (2000) Roles of the NFI/CTF gene family in transcription and development. Gene 249: 31–45 [DOI] [PubMed] [Google Scholar]
- Guo D, Han J, Adam BL, Colburn NH, Wang MH, Dong Z, Eizirik DL, She JX, Wang CY (2005) Proteomic analysis of SUMO4 substrates in HEK293 cells under serum starvation-induced stress. Biochem Biophys Res Commun 337: 1308–1318 [DOI] [PubMed] [Google Scholar]
- Hay RT (2005) SUMO: a history of modification. Mol Cell 18: 1–12 [DOI] [PubMed] [Google Scholar]
- Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I (2006) Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem 281: 16117–16127 [DOI] [PubMed] [Google Scholar]
- Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L (2006) PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci USA 103: 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES (2004) Protein modification by SUMO. Annu Rev Biochem 73: 355–382 [DOI] [PubMed] [Google Scholar]
- Li T, Evdokimov E, Shen RF, Chao CC, Tekle E, Wang T, Stadtman ER, Yang DC, Chock PB (2004) Sumoylation of heterogeneous nuclear ribonucleoproteins, zinc finger proteins, and nuclear pore complex proteins: a proteomic analysis. Proc Natl Acad Sci USA 101: 8551–8556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Tatham MH, Yu B, Kim S, Hay RT, Chen Y (2002) Identification of a substrate recognition site on Ubc9. J Biol Chem 277: 21740–21748 [DOI] [PubMed] [Google Scholar]
- Pichler A, Gast A, Seeler JS, Dejean A, Melchior F (2002) The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108: 109–120 [DOI] [PubMed] [Google Scholar]
- Reverter D, Lima CD (2005) Insights into E3 ligase activity revealed by a SUMO–RanGAP1–Ubc9–Nup358 complex. Nature 435: 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme C, Barthel KK, Liu X (2006) SUMO-1 modification of MEF2A regulates its transcriptional activity. J Cell Mol Med 10: 132–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas S, Briancon-Marjollet A, Bossis G, Lopez MA, Piechaczyk M, Jariel-Encontre I, Debant A, Hipskind RA (2004) SUMOylation regulates nucleo-cytoplasmic shuttling of Elk-1. J Cell Biol 165: 767–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalizi A, Gaudilliere B, Yuan Z, Stegmuller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A (2006) A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science 311: 1012–1017 [DOI] [PubMed] [Google Scholar]
- Shore P, Sharrocks AD (1994) The transcription factors Elk-1 and serum response factor interact by direct protein–protein contacts mediated by a short region of Elk-1. Mol Cell Biol 14: 3283–3291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian L, Benson MD, Iniguez-Lluhi JA (2003) A synergy control motif within the attenuator domain of CCAAT/enhancer-binding protein alpha inhibits transcriptional synergy through its PIASy-enhanced modification by SUMO-1 or SUMO-3. J Biol Chem 278: 9134–9141 [DOI] [PubMed] [Google Scholar]
- Tanoue T, Nishida E (2003) Molecular recognitions in the MAP kinase cascades. Cell Signal 15: 455–462 [DOI] [PubMed] [Google Scholar]
- Tatham MH, Kim S, Yu B, Jaffray E, Song J, Zheng J, Rodriguez MS, Hay RT, Chen Y (2003) Role of an N-terminal site of Ubc9 in SUMO-1, -2, and -3 binding and conjugation. Biochemistry 42: 9959–9969 [DOI] [PubMed] [Google Scholar]
- Vertegaal AC, Ogg SC, Jaffray E, Rodriguez MS, Hay RT, Andersen JS, Mann M, Lamond AI (2004) A proteomic study of SUMO-2 target proteins. J Biol Chem 279: 33791–33798 [DOI] [PubMed] [Google Scholar]
- Watts FZ (2004) SUMO modification of proteins other than transcription factors. Semin Cell Dev Biol 15: 211–220 [DOI] [PubMed] [Google Scholar]
- Wei H, Wang X, Gan B, Urvalek AM, Melkoumian ZK, Guan JL, Zhao J (2006) Sumoylation delimits KLF8 transcriptional activity associated with the cell cycle regulation. J Biol Chem 281: 16664–16671 [DOI] [PubMed] [Google Scholar]
- Yang SH, Bumpass DC, Perkins ND, Sharrocks AD (2002) The ETS domain transcription factor Elk-1 contains a novel class of repression domain. Mol Cell Biol 22: 5036–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Jaffray E, Hay RT, Sharrocks AD (2003) Dynamic interplay of the SUMO and ERK pathways in regulating Elk-1 transcriptional activity. Mol Cell 12: 63–74 [DOI] [PubMed] [Google Scholar]
- Yang SH, Sharrocks AD (2004) SUMO promotes HDAC-mediated transcriptional repression. Mol Cell 13: 611–617 [DOI] [PubMed] [Google Scholar]
- Yang SH, Sharrocks AD (2005) PIASx acts as an Elk-1 coactivator by facilitating derepression. EMBO J 24: 2161–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Yates PR, Whitmarsh AJ, Davis RJ, Sharrocks AD (1998) The Elk-1 ETS-domain transcription factor contains a MAP kinase targeting motif. Mol Cell Biol 18: 710–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Sternsdorf T, Bolger TA, Evans RM, Yao TP (2005) Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol Cell Biol 25: 8456–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Kwon SW, Anselmo A, Kaur K, White MA (2004) Broad spectrum identification of cellular small ubiquitin-related modifier (SUMO) substrate proteins. J Biol Chem 279: 20999–21002 [DOI] [PubMed] [Google Scholar]
- Zhou F, Xue Y, Lu H, Chen G, Yao X (2005) A genome-wide analysis of sumoylation-related biological processes and functions in human nucleus. FEBS Lett 579: 3369–3375 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Figure 8
Data Sets 1
Data Sets 2
Data Sets 3
Data Sets 4


