Abstract
SMG6 and SMG5 are essential factors in nonsense-mediated mRNA decay, a conserved pathway that degrades mRNAs with premature translation termination codons. Both SMG5 and SMG6 have been predicted to contain a C-terminal PIN (PilT N-terminus) domain, present in proteins with ribonuclease activity. We have determined the structures of human SMG5 and SMG6 PIN domains. Although they share a similar overall fold related to ribonucleases of the RNase H family, they have local differences at the putative active site. SMG6 has the canonical triad of acidic residues that are crucial in RNase H for nuclease activity, while SMG5 lacks key catalytic residues. The structural differences are reflected at the functional level. Only the PIN domain of SMG6 has degradation activity on single-stranded RNA in vitro. This difference in catalytic activity is conserved in Drosophila, where an SMG6 with an inactive PIN domain inhibits NMD in a dominant-negative manner. Our findings suggest that the NMD machinery has intrinsic nuclease activity that is likely to contribute to the rapid decay of mRNAs that terminate translation prematurely.
Keywords: decay, EST1A, NMD, P-bodies, RNA degradation
Introduction
Nonsense-mediated mRNA decay (NMD) is a post-transcriptional regulatory pathway that recognizes and rapidly degrades mRNAs containing premature translation termination codons (PTCs or nonsense codons) (reviewed in Baker and Parker, 2004; Conti and Izaurralde, 2005; Lejeune and Maquat, 2005). PTC-containing mRNAs may arise from alternative splicing events, from errors in transcription or mRNA processing or by transcription of mutated alleles. In addition, genome-wide identification of NMD targets in yeast, fruitfly and human cells have shown that NMD regulates post-transcriptionally about 3–10% of the transcriptome (reviewed in Conti and Izaurralde, 2005).
The NMD pathway is conserved from yeast to human, although the molecular machinery increases in complexity in metazoans (reviewed in Conti and Izaurralde, 2005). UPF1 (also known as SMG2) is an RNA helicase present in all species and is a key factor in NMD. It associates with UPF2 and UPF3 (also known as SMG3 and SMG4, respectively) to form a core complex that is assembled on mRNAs as a result of an improper translation termination event when the ribosomes encounter a PTC. UPF1 activity is regulated by cycles of phosphorylation and dephosphorylation (Page et al, 1999). Current models for mammalian NMD converge on a mechanism whereby UPF1 is recruited to PTC-containing mRNAs by prematurely terminating ribosomes. The interaction with components of the NMD machinery including UPF2, UPF3 and the SMG1 kinase leads to the phosphorylation of UPF1 and subsequent recruitment of degradative enzymes (reviewed in Conti and Izaurralde, 2005; Lejeune and Maquat, 2005; Behm-Ansmant and Izaurralde, 2006). Both 5′–3′ RNases and 3′–5′ RNases have been implicated in the degradation of NMD targets (reviewed in Parker and Song, 2004).
Phosphorylated UPF1 is directly recognized by SMG5, SMG6 and SMG7 via a conserved 14-3-3 like domain present in all three proteins (Fukuhara et al, 2005). These proteins are thought to serve as adaptors between phosphorylated UPF1 and protein phosphatase 2A (PP2A), thereby triggering UPF1 dephosphorylation (Anders et al, 2003; Chiu et al, 2003; Ohnishi et al, 2003). However, the molecular mechanism of PP2A recruitment is unknown. Co-immunoprecipitation data suggest the involvement, either direct or indirect, of the C-terminus of SMG5, which contains a PIN domain (Ohnishi et al, 2003). A PIN domain is also present in the C-terminus of SMG6, while the C-terminus of SMG7 contains a low-complexity region that functions in marking bound mRNAs for decay (Unterholzner and Izaurralde, 2004). SMG7 localizes to P-bodies (large cytoplasmic domains where mRNA degradation enzymes localize), and promotes the recruitment of SMG5 and UPF1 to these bodies (Unterholzner and Izaurralde, 2004; Fukuhara et al, 2005). In contrast to the SMG5–SMG7 complex, SMG6 is not found in P-bodies (Unterholzner and Izaurralde, 2004).
PIN domains were first identified in a bacterial protein involved in the biosynthesis of type IV pili (PilT N-terminus, from which the name PIN originates) (Wall and Kaiser, 1999). PIN-containing proteins are present in all three domains of life. In bacteria, PIN domains are typically found in toxin–antitoxin systems, where their toxic effect is thought to arise from ribonuclease activity (Anantharaman and Aravind, 2003). A nuclease activity of PIN domains was originally suggested by bioinformatic analysis, which predicted a similarity to nucleases of the FLAP family of proteins, such as T4 RNase H or Taq polymerase (Clissold and Ponting, 2000). This prediction was confirmed recently, with the first crystal structures of archaeal PIN domains showing a similar fold to the nuclease domain of T4 RNase H (Arcus et al, 2004; Levin et al, 2004). Despite the overall low sequence similarity, conserved acidic residues at the active site of T4 RNase H (Bhagwat et al, 1997) are present at the same structural positions in archaeal PIN domains, suggesting a similar role in coordinating a metal ion for catalysis. The expectation for eukaryotic PIN domain-containing proteins such as SMG5, SMG6 or the ribosomal processing protein Nob1p (Fatica et al, 2004) is of a similar arrangement of active site residues fulfilling a similar function. Here we show that despite a similar overall fold, the structures of the PIN domain of SMG5 and SMG6 have differences at the active site resulting in different nuclease activities in vitro and in vivo. Our results identify an intrinsic ribonuclease activity within the mRNA surveillance complex.
Results and discussion
Structure determination of human SMG6 and SMG5 PIN domains
The structure of the PIN domain of human SMG6 (hSMG6-PIN, residues 1239–1421, Figure 1A) was solved by single-wavelength anomalous dispersion (SAD) using crystals of the E1282C mutant protein (that had an alternative crystal packing to the wild-type crystals, see Materials and methods). The structure of wild-type hSMG6-PIN crystals was determined by molecular replacement using the coordinates of the E1282C mutant and refined at 1.9 Å resolution with Rfree of 26.7% and R factor of 22.2%. The crystals contain three molecules per asymmetric unit. The model discussed in the text (molecule A; Figure 1B) includes residues 1239–1418, with the exception of short disordered loop regions (residues 1290–1295, 1343–1350, 1373–1379). The structure has good stereochemistry, with more than 90% of the residues lying in the most favored regions of the Ramachandran plot (see Table I for phasing and refinement statistics).
Figure 1.
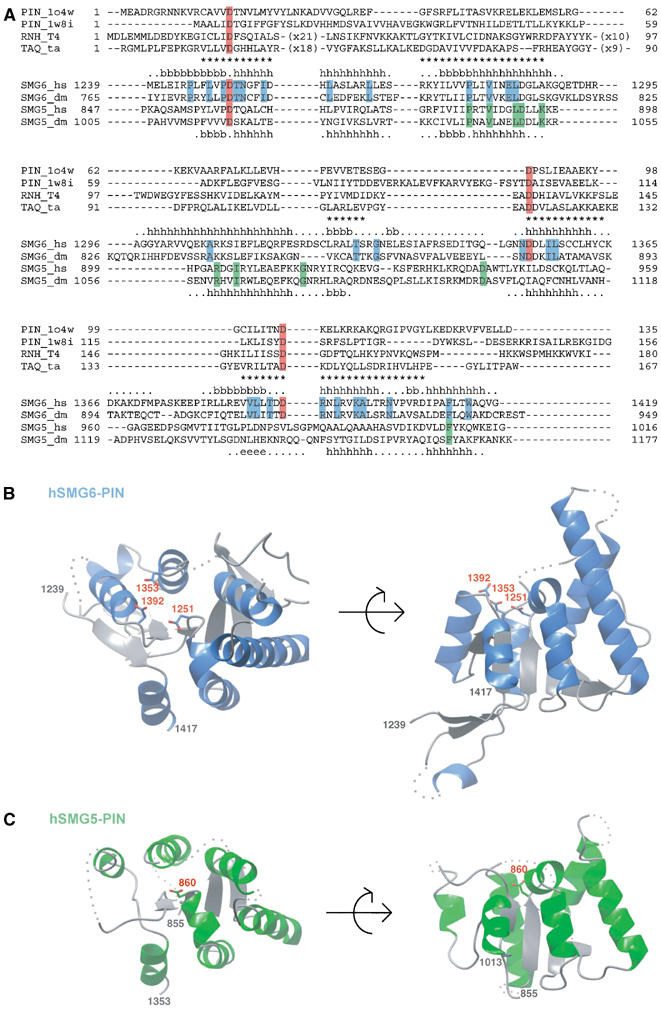
Overall sequence conservation and structure of the PIN domain of hSMG6 (also known as EST1A) and hSMG5 (also known as EST1B). (A) Structure-based sequence alignment of SMG6-PIN and SMG5-PIN with related proteins of known structure: the archaeal PIN domains (pdb code 1o4w and 1w8i) and the nuclease domains of T4 RNase H (RNH_T4) and Thermus aquaticus TAQ polymerase (TAQ_ta). The secondary structure elements of human hSMG6 and human hSMG5 are indicated (above and below the sequences, respectively) with the symbols h for α-helices, b for β-strands, dots for extended portions and no symbol for residues that are disordered and are not present in the final model. Asterisks indicate residues of human hSMG6 present at equivalent structural position in the PIN domains of known structure (i.e. r.m.s.d. less than 2.5 Å between corresponding Cα atoms after optimal superposition). The three aspartic-acid residues at the active site of nucleases are highlighted in red. Highlighted in blue are residues that are conserved in SMG6 based on a sequence alignment including SMG6 from Homo sapiens (hs) and Drosophilla melanogaster (dm) (shown) and also Caenorhabditis elegans and Danio rerio (not shown). Highlighted in green are conserved residues of SMG5 as of a sequence alignment using SMG5 sequences from the same species. (B) Structure of human SMG6-PIN (residues 1239–1417) in two orientations related by a 90° rotation along a horizontal axis. Helices are shown in blue, β-strands in gray and disordered parts of the polypeptide chain are indicated with dots. In sticks are the three conserved aspartic acids that are highlighted in red in (A). The N- and C-termini as well as the aspartates are labeled. All ribbon drawings in the figures were prepared using CCP4 mg (Potterton et al, 2002). (C) Structure of human SMG5-PIN (residues 855–1013). The structure is shown in similar orientations to the hSMG6-PIN structure in (B), with analogous labeling and color coding (with exception of the helices that are shown in green).
Table 1.
Crystallographic data collection, phasing, and refinement statistics
| SMG6 E1282Ca | Iodide-SADb | SMG6 wild typea | SMG5 wild typea | |
|---|---|---|---|---|
| Data collection and phasing statistics | ||||
| X-ray source | Rigaku MicroMax | Rigaku MicroMax | SLS | SLS |
| Wavelength (Å) | 1.542 | 1.542 | 0.978 | 0.979 |
| Space group | P21212 | P212121 | P21 | |
| a (Å) | 69.36 | 69.29 | 36.84 | 45.69 |
| b (Å) | 73.34 | 73.38 | 71.18 | 42.56 |
| c (Å) | 37.31 | 37.36 | 181.21 | 65.31 |
| α, β, γ (deg) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 96.06, 90 |
| Resolution (Å)c | 30–1.9 (2.0–1.9) | 30–2.45 (2.58–2.45) | 30–1.8 (1.9–1.8) | 30–2.75 (2.9–2.75) |
| Unique reflections | 15 634 | 13 823 | 45 496 | 6552 |
| Completeness (%)c | 99.9 (100.0) | 99.4 (98.9) | 99.7 (100.0) | 98.8 (98.8) |
| Multiplicityc | 6.9 (6.9) | 22.1 (22.1) | 5.2 (3.8) | 3.2 (3.2) |
| I/σc | 13.3 (6.5) | 45.3 (24.5) | 6.4 (1.9) | 8.8 (2.0) |
| Rsym (%)c | 4.0 (11.0) | 6.3 (14.3) | 7.5 (34.4) | 6.5 (32.7) |
| FOM (acentric)d | 0.57 | |||
| Phasing power (acentric)d | 2.8 | |||
| FOM after solvent flipping | 0.79 | |||
| Refinement and model statisticse | ||||
| Rfree (%) | 23.9 | 26.7 | 32.9 | |
| Rwork (%) | 19.7 | 22.2 | 27.1 | |
| R.m.s.d. bond lengths (Å) | 0.011 | 0.018 | 0.011 | |
| R.m.s.d. bond angles (deg) | 1.3 | 1.8 | 1.4 | |
| Ramachandran φ, ψ angles (non Pro/Gly residues) (%) | ||||
| Most favored | 94.5 | 90.9 | 93.5 | |
| Additionally allowed | 5.5 | 8.9 | 6.5 | |
| Generously allowed | 0 | 0.2 | 0.0 | |
| Disallowed | 0 | 0 | 0 | |
| Protein residues | 165 | 461 | 227 | |
| Water molecules | 179 | 221 | 0 | |
| aData processed with MOSFLM. | ||||
| bData processed with XDS. | ||||
| cValues in parentheses correspond to the highest resolution shell. | ||||
| dAs determined with AutoSHARP. | ||||
| eAs determined with REFMAC5 and PROCHECK. FOM, figure of merit. | ||||
The structure of the PIN domain of human SMG5 (hSMG5-PIN, residues 853–1016) was determined by molecular replacement using the coordinates of hSMG6-PIN and refined to 2.8 Å resolution. There are two independent molecules in the asymmetric unit of the crystals (molecules A and B). The model discussed in the text (molecule A, Figure 1C) includes residues 855–896, 899–923, 944–959, 975–986 and 990–1013, while chain B is less well ordered. The R factor is 27.1% and the Rfree 32.9%. Given that the electron density maps show a good agreement with the model (shown for the active site region discussed in the text; Supplementary Figure 1), the relatively high Rfree is likely due to the significant proportion of poorly disordered residues (particularly in chain B, where only 38 residues in the core have defined density).
SMG6 and SMG5 have similar folds with different active sites
hSMG6-PIN folds into a five-stranded parallel β-sheet that is highly twisted and is flanked by α-helices on both sides (Figure 1B). The structure has a groove at the C-terminal end of the β-strands. As predicted by bioinformatic analysis (Clissold and Ponting, 2000), the groove is lined by conserved aspartic-acid residues (D1251, D1353 and D1392, highlighted in red in Figure 1A), in much the same way as observed in archaeal PIN domains (Arcus et al, 2004; Levin et al, 2004) and in the FLAP family of 5′ nucleases (Mueser et al, 1996; Urs et al, 1999).
Superposition of hSMG6-PIN with archaeal PIN domains (pdb codes 1w8i and 1o4w) using the program DALI (Holm and Sander, 1993) results in an root-mean-square deviations (r.m.s.d.) of 2.5 Å in the α-carbon positions of at least 100 residues with sequence identity lower than 14% (Figure 2A and B). The sequence and structural similarity is lower to FLAP nuclease domains, such as those in T4 RNase H or Taq polymerase. Superposition of hSMG6-PIN onto the nuclease domain of T4 RNase H results in an r.m.s.d. of 3.5 Å over 106 residues (overall sequence identity of 6%). Despite being distant homologs, the active-site residues of T4 RNase H are strictly conserved in hSMG6-PIN (Figure 2C and D). In FLAP nucleases, the acidic residues coordinate a metal ion (Mg2+ in T4 RNase H) that is believed to activate water for the nucleophilic attack on the phosphodiester bond of the nucleic acid. At the corresponding structural region of hSMG6-PIN, we find electron density that can be interpreted either as an ion or as a water molecule, or as a mixture of both.
Figure 2.
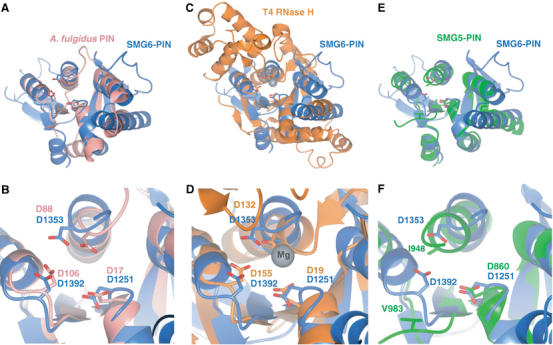
The PIN domains of human SMG6 and SMG5 have a core structure similar to 5′ nucleases. (A, B) Structural superposition of hSMG6-PIN (blue) to an archaeal PIN domain from Archaeoglobus fulgidus (pdb code 1o4w, in pink). The structures are viewed in the same orientation as the left panels in Figure 1B. (B) A detailed view of the active site region. The conserved aspartic acid residues (in red in Figure 1A) are shown in stick format and labeled with residue numbers from both proteins. (C, D) Structural superposition of hSMG6-PIN (blue) to T4 RNase H (in orange), in a similar representation to panels (A, B). The gray sphere in the right panel represent an Mg ion bound in the T4 RNase H structure. (E, F) Structural superposition of hSMG6-PIN (blue) to hSMG5-PIN (green), in a similar representation to panels (A, B). The superpositions show that hSMG6-PIN has a similar active site as compared to structurally homologous nucleases, while hSMG5-PIN shares only partial conservation.
hSMG6-PIN and hSMG5-PIN are more similar to each other, both in terms of sequence (25% identity, Figure 1A) and in structure (Figure 2E and F), than they are to archaeal PIN domains or to FLAP nucleases. The structures of hSMG5-PIN and hSMG6-PIN superpose with a low r.m.s.d. (1.7 Å) in the Cα position of 111 residues. Despite their overall similarity, hSMG6-PIN and hSMG5-PIN have significant differences that are localized at the putative active site. Only one of the three active-site aspartates is conserved at the equivalent structural position in hSMG5-PIN: D860 in human SMG5 corresponding to D1251 in human SMG6 (Figure 2C and D). Thus, the structural analysis shows an intact active site similar to that of nucleases in hSMG6-PIN and an impaired active site in hSMG5-PIN.
In vitro RNase degradation activity of SMG6 and SMG5 PIN domains
The structural results imply that the PIN domains of SMG5 and SMG6 might harbor different catalytic properties. To test this, we performed in vitro degradation assays using a 5′[32P]-end-labeled single-stranded (ss) RNA oligo, (U)30. In the assay, hSMG6-PIN degrades the (U)30 RNA oligo in the presence of manganese and to a much lesser extent magnesium (Figure 3A, lanes 4 and 5). The purified protein is inactive most likely because the purification was performed without adding metals to the buffers. The activity is specific of hSMG6-PIN and not due to Escherichia coli contaminants, since the nuclease activity is strongly impaired upon mutation of one of the three active-site aspartates (D1353A Figure 3A, lane 6). In agreement with this result, hSMG5-PIN, which also lacks two of the three active-site aspartates, exhibits a nearly equivalent activity to that of the hSMG6-PIN mutant (Figure 3A, lanes 8 versus 6).
Figure 3.
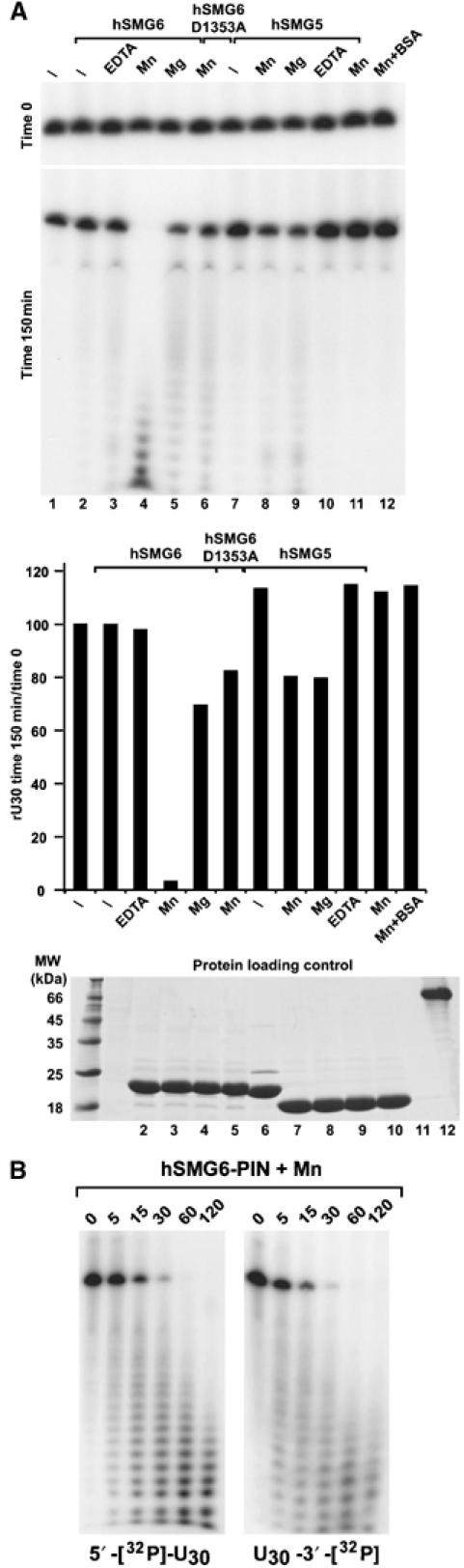
The PIN domain of SMG6 has nuclease activity on single-stranded RNA. (A) hSMG6-PIN and hSMG5-PIN (either wild-type or mutant) were incubated with a 32P-labelled (U)30 RNA with or without addition of 3 mM MgCl2, 3 mM MnCl2 or 2.5 mM EDTA as indicated. One eight of the input was loaded immediately after mixing (time 0, RNA loading control) and one eight after incubation at room temperature for 150 min on 10% polyacrylamide gel containing 8 M urea. RNA signals were visualized by autoradiography and quantified using the Fuji FLA-2000 phosphoimager system. The lower panel shows the RNA signals measured following the 150-min incubation normalized to those at time 0. Proteins used in (A) were analyzed by SDS–PAGE, followed by Coomassie staining (protein loading control). (B) Pattern of decay intermediates. 600 ng of cold (U)30 RNA (10.8 pmol) were mixed with either 6 ng of 5′-[32P]-end-labelled (U)30 RNA (0.1 pmol) or 6 ng of 3′-[32P]-end-labelled (U)30 RNA (0.1 pmol) and incubated with 5 μg of purified proteins in 60 μl of buffer containing 20 mM HEPES pH 7.5, 150 mM NaCl, 10% glycerol and 1 mM DTT. Samples were analyzed at the indicated time points. The extent of RNA degradation was evaluated by analyzing the reaction mixtures on 12% denaturing polyacrylamide gels.
These results are consistent with the structural data: significant RNA degradation is detected for the PIN domain with 3 aspartates at the active site, while only low level of RNA degradation is detected for the PIN domain where one of the aspartates is not present. A similar pattern of decay intermediates was observed independently of whether the radioactive label was incorporated at the 5′ or the 3′ end of the ribo-oligonucleotide, suggesting that the hSMG6-PIN domain may have endonuclease activity (Figure 3B). The activity appears to be specific to ssRNA, since ssDNA or double-stranded (ds) RNA is not degraded (Supplementary Figure 2). Interestingly, ssDNA acts as an inhibitor of RNA degradation (Supplementary Figure 2), suggesting that deoxyribonucleotides might be able to bind hSMG6-PIN, but that the 2′OH of ribonucleotides is needed for degradation.
The catalytic activity of SMG6 is conserved
We next asked whether the activity of the PIN domain of SMG6 is conserved and can be observed in vivo. For this, we made use of a tethering assay previously described for Drosophila proteins (Rehwinkel et al, 2005). This assay involves the expression of a firefly luciferase reporter mRNA harboring five BoxB sites (5BoxB) inserted in the 3′UTR (F-Luc-5BoxB reporter). BoxB sites are bound with high affinity by a peptide derived from the N protein of bacteriophage λ (λN-peptide). λN-fusions of full-length Drosophila SMG5, SMG6 (dSMG5 or dSMG6), or of the corresponding PIN domains were coexpressed with the F-Luc-5BoxB reporter in Drosophila Schneider cells (S2 cells). A plasmid encoding Renilla luciferase (R-Luc) served as a transfection control. In the experiments described below, firefly luciferase activities and mRNA levels were normalized to those of Renilla luciferase to compensate for possible differences in transfection efficiencies.
When λN-dSMG6 or the PIN domain was tethered to the reporter transcript, firefly luciferase expression was reduced relative to that in cells expressing the λN-peptide alone or fused to maltose binding protein (λN-MBP; Figure 4A and B). The reduction in firefly luciferase expression is in part explained by a reduction in the steady-state levels of the reporter mRNA, as shown by Northern blot (Figure 4C). In cells expressing λN-dSMG5 or λN-dSMG5-PIN, no reduction of luciferase expression was observed (Figure 4A), even though these proteins were expressed at levels comparable to those of dSMG6 (data not shown).
Figure 4.
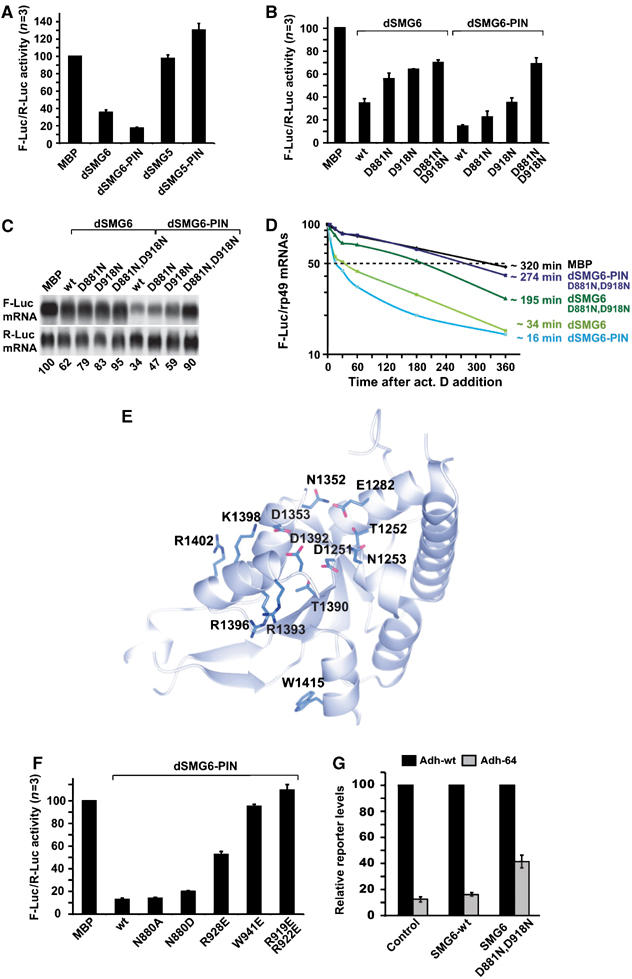
In vivo analysis of SMG6 and SMG5 PIN domains. (A–C) S2 cells were transfected with the F-Luc-5BoxB reporter, a plasmid expressing Renilla luciferase, and vectors expressing the λN-protein fusions. Firefly luciferase activity was normalized to that of Renilla and set to 100 in cells expressing the λN-MBP. Mean values±standard deviations from three independent experiments (n=3) are shown. In (C), representative RNA samples corresponding to (B) were analyzed by Northern blot. (D) Decay of the F-Luc-5BoxB mRNA was monitored in cells expressing the different λN-protein fusions. The levels of the F-Luc-5BoxB mRNA normalized to rp49 are plotted against time. mRNA half-lives (t1/2) calculated from the decay curves are indicated. (E, F) Conserved solvent-exposed residues in the hSMG6-PIN are required for RNA degradation. Residues discussed in the text are shown on the structure and indicated (E). In particular, R1393, R1396, R1402 and W1415 correspond to residues of Drosophila SMG6-PIN (R919, R922, R928 and W942, respectively) tested in the tethering assays in (F). (G) S2 cells were transfected with vectors expressing adh-wt or adh-64 (which carries a PTC at codon 64). A truncated version of adh (adhΔ) served as a transfection control. Vectors expressing dSMG6 wild type, dSMG6 mutant (D881N, D918N) or the corresponding empty vector (control) were included in the transfection mixtures as indicated. Total RNA samples were isolated and analyzed by Northern blot using a probe specific for adh mRNA (not shown). The levels of the adh mRNA reporters were quantitated, normalized to the levels of adhΔ mRNA. These levels were set to 100% for adh-wt in each experimental condition. Mean values±standard deviations of four independent experiments are shown.
Due to the high conservation between Drosophila and human sequences (Figure 1A), mutants based on the structure of the human SMG6 and SMG5 proteins can be confidently mapped onto the Drosophila orthologues. When two active-site aspartates were mutated in Drosophila SMG6, luciferase expression was restored (Figure 4B). Thus, the results using tethering assays with Drosophila SMG5 and SMG6 are consistent with the results using in vitro degradation assays with human orthologues.
We next investigated whether the decreased steady-state levels of the mRNA reporter are due to increased mRNA degradation rates. To this end, the levels of F-Luc-5BoxB mRNA were analyzed over time after inhibition of transcription by actinomycin D and normalized to those of the long-lived (half-life>8 h) endogenous rp49 mRNA (which encodes the ribosomal protein L32). In control cells expressing λN-MBP, the half-life of F-Luc-5BoxB mRNA was ca. 320 min, while in cells expressing λN-dSMG6 or λN-dSMG6-PIN, the half-life of this mRNA was reduced to ca. 34 min and 16 min respectively (Figure 4D). The half-life of the reporter was restored when the tethered proteins were mutated at the active site (Figure 4D). Thus, tethering of dSMG6 or of dSMG6-PIN causes a reduction of the steady-state levels of bound mRNAs by increasing their degradation rate.
Evolutionary conserved surface residues required for SMG6-PIN activity
The finding that RNA degradation activity is conserved in human and Drosophila SMG6, prompted us to investigate whether SMG6-PIN presents surface residues in addition to the aspartate triad that contribute to the nuclease activity. Several residues are strictly conserved in the PIN domains of SMG6 orthologues (Figure 1A, highlighted in blue). A subset of the invariant residues is conserved for structural reasons. These include not only hydrophobic residues that are buried in the inner core but also polar residues that are next to the active-site aspartates (T1252 and N1352, in hSMG6; Figures 1A and 4E) and that are involved in hydrogen-bonding interactions with the polypeptide backbone (likely to be important for maintaining the active site in a proper conformation for catalysis).
Another subset of invariant residues is exposed to solvent without any apparent structural role. A set of well-conserved solvent-exposed residues is present in the PIN domain of SMG6 orthologues but not of SMG5, including R1393, R1396, R1402 and W1415 (Figure 4E). Given their chemical properties and location in proximity of the active site, these residues are good candidates for contributing to RNA binding. To test this hypothesis, we substituted the corresponding residues in the PIN domain of Drosophila SMG6 (R919, R922, R928 and W941) to glutamic acid and tested the mutants in the tethering assay. The R928E mutant partially impaired the activity of the PIN domain, while the W941E mutant and the double-mutant R919,922E prevented the PIN domain from inhibiting the expression of the luciferase reporter (Figure 4F). In contrast, no effect on activity in the tethering assay is observed upon mutation of N880 in Drosophila SMG6 (structurally equivalent to human N1253, that is also positioned near the active site) (Figure 4F). These results indicate that a subset of conserved surface residues of SMG6-PIN is important for activity.
Overexpression of a catalytic mutant of SMG6 inhibits NMD
The experiments described above clearly establish that the PIN domain of SMG6 is active in vivo and elicits degradation of bound mRNAs. However, these experiments do not address whether the nuclease activity of this domain is required for NMD. To begin to investigate the role of this domain in NMD, we coexpressed wild-type dSMG6 or a dSMG6 mutant together with an NMD reporter based on the Drosophila alcohol dehydrogenase (adh) gene, which carries a PTC at codon 64 (adh-64; Gatfield et al, 2003). The SMG6 mutant carries substitutions of two active-site aspartates to asparagines in the catalytic site (D881N, D918N). Wild-type adh mRNA (adh-wt) is expressed at 10-fold higher levels than adh-64 (Figure 4G), which is degraded by NMD as reported before (Gatfield et al, 2003). Overexpression of wild-type dSMG6 has no effect on reporter expression levels. In contrast, overexpression of the dSMG6 mutant inhibits NMD, and consequently leads to a four-fold increase in the steady-state levels of the PTC-containing mRNA (Figure 4G). Thus, a dSMG6 protein with an inactive PIN domain inhibits NMD in a dominant-negative manner, suggesting that this activity is required for efficient degradation of NMD substrates in vivo.
Concluding remarks
The structure of SMG6-PIN shows a similar fold and conserved active site to that of T4 RNase H. Consistently, SMG6-PIN degrades single-stranded RNA in vitro and the reaction depends on the presence of metal ions. In vivo, using a reporter assay for mRNA decay in Drosophila cells, we find that the PIN domain of Drosophila SMG6 increases the rate of degradation of bound transcripts. The effect is abolished upon mutation of active-site aspartate residues. In contrast, both human and Drosophila SMG5 shows very low RNase activity in vitro and in vivo. The structure provides an explanation for these functional differences. Although SMG5 has the expected PIN-like fold and extensive sequence similarity with SMG6, the active site is impaired, with only one of the three active-site acidic residues conserved. In agreement with this, mutation of acidic residues in the active site of SMG6 abolishes nuclease activity.
What is the relevance of these results for NMD? A rationale for the presence of nuclease activity in SMG6 but not in SMG5 comes from their localization. Transiently expressed SMG5 and SMG7 localize to P-bodies together with enzymes involved in general mRNA degradation (Unterholzner and Izaurralde, 2004). If SMG7 targets mRNAs associated with phosphorylated UPF1 to P-bodies, there is no evolutionary pressure to maintain a functional nuclease active site in the SMG7–SMG5 complex. On the other hand, SMG6 does not localize to P-bodies, and has maintained nuclease activity. This nuclease activity is required for NMD because overexpression of a nuclease inactive SMG6 mutant partially inhibits NMD in a dominant-negative manner.
These results point to the presence of either alternative degradative pathways or of consecutive steps in NMD. The possibility of alternative NMD pathways in mammalian cells has already been postulated (Gehring et al, 2005). If alternative SMG6-dependent and SMG5–SMG7-dependent pathways were at play, the expectation is that different PTC-containing mRNAs might be degraded preferentially via one and not the other. However, microarray analysis indicates that SMG5 and SMG6 regulate common targets at least in Drosophila, suggesting that in this organism they act along the same pathway (Rehwinkel et al, 2005). If SMG6 and SMG5 function at consecutive steps in the same pathway, an attractive hypothesis is that SMG6 might be the endonuclease that initiates NMD in Drosophila (Gatfield and Izaurralde, 2004). These findings on the presence of a nuclease activity within the mRNA surveillance complex have to be reconciled with the observation that SMG5, SMG6 and SMG7 are all involved in UPF1 dephosphorylation (Anders et al, 2003; Chiu et al, 2003; Ohnishi et al, 2003). One possibility is that UPF1 dephosphorylation is linked to mRNA degradation. More generally, given that recent studies have implicated human SMG6 (also known as EST1A) in telomere maintenance (Reichenbach et al, 2003; Snow et al, 2003), the question arises as to whether the nuclease activity we identified in the PIN domain of SMG6 has functions beyond NMD.
Materials and methods
Protein expression, purification and crystallization
hSMG6-PIN (residues 1239–1421) was expressed as a TEV-cleavable GST-fusion protein in E. coli. The protein was purified by affinity chromatography, TEV protease was added to cleave the GST tag and hSMG6-PIN was purified to homogeneity by size exclusion chromatography in 20 mM HEPES pH 7.5, 150 mM NaCl, 1 mM DTT. All mutants used for structure solution or for functional analysis were constructed according to a modified Stratagene QuikChange protocol and the mutations verified by DNA sequencing. The mutant proteins were purified with a similar protocol to that used for the wild type and displayed a similar biochemical behavior. hSMG5-PIN (residues 853–1016) was expressed and purified with a similar protocol.
hSMG6-PIN was concentrated to 10 mg/ml and crystallized using sitting-drop vapor diffusion against 30% Jeffamine 2000, 100 mM HEPES pH 7.6 at 18°C. The E1282C mutant was concentrated to 6 mg/ml and crystallized in similar conditions to the wild-type protein. The best crystals of the hSMG6-PIN E1282C mutant were obtained at 18°C in 20% Jeffamine 2000, 100 mM HEPES pH 7.6 using microseeding. hSMG5-PIN was crystallized with a protein concentration of 30 mg/ml against a well buffer containing 20% PEG 4000 100 mM citrate pH 5.5 using sitting drop vapor diffusion at 4°C. Crystals were cryoprotected using Paratone-N and flash-cooled in liquid nitrogen.
To overcome difficulties in binding heavy atoms to wild-type crystals, we engineered point mutations based on secondary-structure predictions. Serendipitously, the E1282C mutant yielded a different crystal form where heavy-atom sites could be located. Wild-type hSMG6-PIN crystals contain three molecules per asymmetric unit, two of which are related by translational symmetry giving rise to a peak in the native Patterson map. The E1282C hSMG6-PIN mutant crystallizes in an orthorhombic space group with one molecule per asymmetric unit. Both crystal forms diffract beyond 1.9 Å resolution using synchrotron radiation (Table I). hSMG5-PIN crystallizes as thin needles that diffract to 2.8 Å resolution and contain two molecules per asymmetric unit.
Structure determination
The structure of the human SMG6 E1282C mutant was determined by SAD on a crystal soaked in a stabilizing solution supplemented with 500 mM NaI for 40 s. SAD data were collected to 2.45 Å resolution using an in-house X-ray source (Rigaku 007 equipped with Xenocs mirrors). For high multiplicity, 1080 frames (1° oscillation each) were collected. Data were processed with XDS (Kabsch, 1993).
Iodide sites were located using the SHELX package (Sheldrick, 1998). Heavy-atom refinement, phasing and density modification were performed using AutoSHARP (Bricogne et al, 2003), resulting in a readily interpretable electron density map. Approximately 50% of the protein molecule was built automatically using ARP/wARP (Morris et al, 2003). The remainder of the atomic model was built manually in COOT (Emsley and Cowtan, 2004) and refined against a 1.9 Å resolution native data set of E1282C using REFMAC5 (Murshudov et al, 1997). The structure of wild-type SMG6 protein was solved by molecular replacement with Phaser (Storoni et al, 2004; McCoy et al, 2005) using the coordinates of the refined E1282C structure. The structures of wild-type and E1282C mutant proteins are very similar, with pair-wise α-carbon r.m.s.d. of less than 0.6 Å. The structure of human SMG5 was also solved by molecular replacement with Phaser and refined using TLS refinement in REFMAC5. Data collection, phasing and refinement statistics are shown in Table I.
In vitro nuclease assays
For the degradation assay in Figure 3A, 100 ng of cold (U)30 RNA (10.8 pmol) were mixed with 1 ng of 5′-[32P]-end-labelled (U)30 RNA (0.1 pmol) and incubated with 500 ng of purified proteins in 10 μl of buffer containing 20 mM HEPES pH 7.5, 150 mM NaCl, 10% glycerol and 1 mM DTT. The extent of RNA degradation was evaluated by analyzing the reaction mixtures on 12% denaturing polyacrylamide gels.
Tethering assay in S2 cells and RNA analysis
For the expression of λN-HA-peptide fusions, cDNAs encoding full-length Drosophila SMG5, SMG6 or PIN domains were amplified with primers containing appropriate restriction sites, using a (dT)15-primed S2 cDNA library as template. The amplified cDNAs were cloned into a vector allowing the expression of λN-HA-peptide fusions (pAc5.1-λN-HA). The adh-wt and adh-64 reporters were described by Gatfield et al (2003). The F-Luc-5BoxB and R-Luc plasmids were described by Rehwinkel et al (2005).
Transfections were performed in six-well dishes using Effectene transfection reagent (Qiagen). For the tethering assay the following plasmids were cotransfected: 0.15 μg reporter plasmid (F-Luc-5BoxB), 0.4 μg pAc5.1-R-Luc as transfection control and 1 μg pAc5.1 λN-HA construct for the expression of λN-HA-fusions. At 24 h after transfection, firefly and Renilla luciferase activities were measured using the Dual-Luciferase reporter assay system (Promega), and total RNA was isolated using TriFast (Peqlab biotechnologies). For the measurement of mRNA half-lives, transfected cells were treated with actinomycin D (5 μg/ml final concentration) 24 h after transfection, and harvested at the time points indicated.
For the assay shown in Figure 4G cells were transfected with 0.25 μg of adh-wt or adh-64, 0.25 μg of a truncated version of the adh gene that served as transfection control (adh-Δ), and 0.1 μg of a vector expressing dSMG6 or the SMG6 mutant. RNA samples were collected 24 h after transfection and analyzed as described (Rehwinkel et al, 2005).
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Acknowledgments
We are grateful to beamline scientists at SLS for assistance during data collection and Doris Lindner for skilled technical support. We also thank Atlanta Cook, Martin Jinek, Esben Lorentzen and Peter Brick for help in various crystallographic stages and for critical reading of the manuscript. This study was supported by the European Molecular Biology Organization (EMBO), the Human Frontier Science Program Organization (HFSPO) and the American Cystic Fibrosis Foundation. IB-A is a recipient of a fellowship from the European Molecular Biology Organization (EMBO).
References
- Anantharaman V, Aravind L (2003) New connections in the prokaryotic toxin–antitoxin network: relationship with the eukaryotic nonsense-mediated RNA decay system. Genome Biol 4: R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders KR, Grimson A, Anderson P (2003) SMG-5, required for C. elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J 22: 641–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcus VL, Backbro K, Roos A, Daniel EL, Baker EN (2004) Distant structural homology leads to the functional characterization of an archaeal PIN domain as an exonuclease. J Biol Chem 279: 16471–16478 [DOI] [PubMed] [Google Scholar]
- Baker KE, Parker R (2004) Nonsense-mediated mRNA decay: terminating erroneous gene expression. Curr Opin Cell Biol 16: 293–299 [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Izaurralde E (2006) Quality control of gene expression: a stepwise assembly pathway for the surveillance complex that triggers nonsense-mediated mRNA decay. Genes Dev 20: 391–398 [DOI] [PubMed] [Google Scholar]
- Bhagwat M, Meara D, Nossal NG (1997) Identification of residues of T4 RNase H required for catalysis and DNA binding. J Biol Chem 272: 28531–28538 [DOI] [PubMed] [Google Scholar]
- Bricogne G, Vonrheim C, Flensburg C, Schiltz M, Paciorek W (2003) Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr D 59: 2023–2030 [DOI] [PubMed] [Google Scholar]
- Chiu SY, Serin G, Ohara O, Maquat LE (2003) Characterization of human Smg5/7a: a protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA 9: 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clissold PM, Ponting CP (2000) PIN domains in nonsense-mediated mRNA decay and RNAi. Curr Biol 10: R888–R890 [DOI] [PubMed] [Google Scholar]
- Conti E, Izaurralde E (2005) Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr Opin Cell Biol 17: 316–325 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fatica A, Tollervey D, Dlakic M (2004) PIN domain of Nob1p is required for D-site cleavage in 20S pre-rRNA. RNA 10: 1698–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E (2005) SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell 17: 537–547 [DOI] [PubMed] [Google Scholar]
- Gatfield D, Izaurralde E (2004) Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 429: 575–578 [DOI] [PubMed] [Google Scholar]
- Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E (2003) Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J 22: 3960–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring NH, Kunz JB, Neu-Yilik G, Breit S, Viegas MH, Hentze MW, Kulozik AE (2005) Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol Cell 20: 65–75 [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C (1993) Protein structure comparison by alignment of distance matrices. J Mol Biol 233: 123–138 [DOI] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 26: 795–800 [Google Scholar]
- Lejeune F, Maquat LE (2005) Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr Opin Cell Biol 17: 309–315 [DOI] [PubMed] [Google Scholar]
- Levin I, Schwarzenbacher R, Page R, Abdubek P, Ambing E, Biorac T, Brinen LS, Campbell J, Canaves JM, Chiu HJ, Dai X, Deacon AM, DiDonato M, Elsliger MA, Floyd R, Godzik A, Grittini C, Grzechnik SK, Hampton E, Jaroszewski L, Karlak C, Klock HE, Koesema E, Kovarik JS, Kreusch A, Kuhn P, Lesley SA, McMullan D, McPhillips TM, Miller MD, Morse A, Moy K, Ouyang J, Quijano K, Reyes R, Rezezadeh F, Robb A, Sims E, Spraggon G, Stevens RC, van den Bedem H, Velasquez J, Vincent J, von Delft F, Wang X, West B, Wolf G, Xu Q, Hodgson KO, Wooley J, Wilson IA (2004) Crystal structure of a PIN (PilT N-terminus) domain (AF0591) from Archaeoglobus fulgidus at 1.90 A resolution. Proteins 56: 404–408 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Storoni LC, J RR (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr D 61: 458–464 [DOI] [PubMed] [Google Scholar]
- Morris RJ, Perrakis A, Lamzin VS (2003) ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol 374: 229–244 [DOI] [PubMed] [Google Scholar]
- Mueser TC, Nossal NG, Hyde CC (1996) Structure of bacteriophage T4 RNase H, a 5′ to 3′ RNA–DNA and DNA–DNA exonuclease with sequence similarity to the RAD2 family of eukaryotic proteins. Cell 85: 1101–1112 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin A A, Dodson E J (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, Ohno S (2003) Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell 12: 1187–1200 [DOI] [PubMed] [Google Scholar]
- Page MF, Carr B, Anders KR, Grimson A, Anderson P (1999) SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol Cell Biol 19: 5943–5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Song H (2004) The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol 11: 121–127 [DOI] [PubMed] [Google Scholar]
- Potterton E, McNicholas S, Krissinel E, Cowtan K, Noble M (2002) The CCP4 molecular-graphics project. Acta Crystallogr D 58: 1955–1957 [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E (2005) Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA 11: 1530–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach P, Hoss M, Azzalin CM, Nabholz M, Bucher P, Lingner J (2003) A human homolog of yeast Est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr Biol 13: 568–574 [DOI] [PubMed] [Google Scholar]
- Sheldrick GM (1998) SHELX: applications to macromolecules. In Direct Methods for Solving Macromolecular Structures, Fortier S (ed) pp 401–411. Dordrecht: Kluwer Academic Publishers [Google Scholar]
- Snow BE, Erdmann N, Cruickshank J, Goldman H, Gill RM, Robinson MO, Harrington L (2003) Functional conservation of the telomerase protein Est1p in humans. Curr Biol 13: 698–704 [DOI] [PubMed] [Google Scholar]
- Storoni LC, McCoy AJ, Read RJ (2004) Likelihood-enhanced fast rotation functions. Acta Crystallogr D 60: 432–438 [DOI] [PubMed] [Google Scholar]
- Unterholzner L, Izaurralde E (2004) SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell 16: 587–596 [DOI] [PubMed] [Google Scholar]
- Urs UK, Murali R, Krishna Murthy HM (1999) Structure of taq DNA polymerase shows a new orientation for the structure-specific nuclease domain. Acta Crystallogr D 55: 1971–1977 [DOI] [PubMed] [Google Scholar]
- Wall D, Kaiser D (1999) Type IV pili and cell motility. Mol Microbiol 32: 1–10 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2


