Abstract
Cryo-electron microscopy three-dimensional reconstructions have been made of mature and of emptied bacteriophage φ29 particles without making symmetry assumptions. Comparisons of these structures with each other and with the φ29 prohead indicate how conformational changes might initiate successive steps of assembly and infection. The 12 adsorption capable ‘appendages' were found to have a structure homologous to the bacteriophage P22 tailspikes. Two of the appendages are extended radially outwards, away from the long axis of the virus, whereas the others are around and parallel to the phage axis. The appendage orientations are correlated with the symmetry-mismatched positions of the five-fold related head fibers, suggesting a mechanism for partial cell wall digestion upon rotation of the head about the tail when initiating infection. The narrow end of the head–tail connector is expanded in the mature virus. Gene product 3, bound to the 5′ ends of the genome, appears to be positioned within the expanded connector, which may potentiate the release of DNA-packaging machine components, creating a binding site for attachment of the tail.
Keywords: cryo-electron microscopy, DNA ejection, DNA packaging, φ29 structure
Introduction
Bacteriophages are the most abundant group of naturally occurring organisms in the biosphere (Wommack and Colwell, 2000; Hendrix, 2002). Approximately 96% of bacteriophages belong to the order Caudovirales (Ackermann, 2003) and typically have either an isometric or a prolate icosahedral head attached to a ‘tail'. The tail, which usually has at least six-fold symmetry, is a highly efficient and specialized machine for infecting the host. Whereas most eukaryotic viruses require numerous particles to initiate a successful infection, tailed bacteriophages are usually successful with only a single particle attaching to a host cell. Infection of a host by tailed bacteriophages requires binding to the host cell surface via a specific receptor, penetrating the host's cell wall and membranes, releasing the double-stranded DNA (dsDNA) genome from the capsid, and delivering the genome into the host's cytoplasm. Before DNA release, a signal must be transmitted from the receptor-binding site to trigger DNA ejection. After new viral components have been synthesized in the infected cell, an isometric or prolate icosahedral prohead (the first particle to be assembled during morphogenesis) assembles with a DNA-packaging machine occupying one of the five-fold symmetric vertices. On completion of DNA packaging, part of the machine is discarded, allowing sequential attachment of the tail components (as in φ29) or of an independently assembled tail.
Tailed bacteriophages can be divided into three families: Myoviridae, Siphoviridae, and Podoviridae, which are characterized by contractile, long noncontractile, and short noncontractile tails, respectively (Ackermann, 2003). φ29, which infects Bacillus subtilis, is a member of Podoviridae and is one of the smallest known tailed phages (Anderson and Reilly, 1993). Previous investigations of φ29 have established the sequence of the genome (Vlcek and Paces, 1986), the assembly pathway (Figure 1), and a highly efficient in vitro DNA-packaging system (Guo et al, 1986; Grimes et al, 2002). The 19.3 kb genome codes for about 20 proteins and has the virus-encoded protein gene product 3 (gp3) covalently linked to both 5′ ends. Eight gene products and a 174-base prohead RNA (pRNA) have been identified as essential structural components for producing mature φ29 virions (Méndez et al, 1971; Guo et al, 1986). Proheads (Figure 1) consist of the major capsid protein (gp8), the scaffolding protein (gp7), the head fiber protein (gp8.5), the head–tail connector (gp10), and a pRNA oligomer. Packaging of the DNA–gp3 complex into the prohead is powered by a pRNA-dependent virus-encoded ATPase (gp16) and is accompanied by the exit of the scaffolding protein (Bjornsti et al, 1983; Grimes and Anderson, 1990). After packaging, the pRNA–ATPase complex is released and the lower collar (gp11), knob (gp9), and appendages (gp12*, the cleavage product of gp12) are subsequently attached to the DNA-filled head (Figure 1) to form the mature phage particle. Mature φ29 heads are 530 Å long by 430 Å wide, and the tail is 380 Å long. Incubation with sodium perchlorate causes DNA-gp3 to be released from mature particles, resulting in emptied particles (Tao et al, 1998). The resultant conformational changes might be approximations of the in vivo processes.
Figure 1.
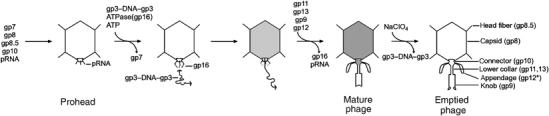
Assembly pathway of φ29. The empty prohead is assembled from the scaffolding protein (gp7), the major capsid protein (gp8), the head–tail connector (gp10), the head fibers (gp8.5), and pRNA. Packaging of DNA–gp3 requires the ATPase gp16 and ATP. The scaffolding protein is lost during DNA packaging. When the DNA–gp3 has been packaged, the pRNA and gp16 components of the packaging machine come off the packaged prohead and are replaced sequentially by the lower collar (gp11 and gp13) and knob (gp9), which, together with the appendages (gp12*), make up the tail. During cell infection, the DNA–gp3 is ejected through the tail, a process that can be mimicked in vitro by treatment with NaClO4.
Previous cryo-electron microscopy (cryoEM) reconstructions of the mature virus (Tao et al, 1998) assumed five-fold symmetry and were only to about 35 Å resolution. The prohead and the mature virus were found to be prolate icosahedrons with T=3 and Q=5 triangulation numbers. X-ray crystallography (HK97 (Wikoff et al, 2000) and T4 (Fokine et al, 2005)) and cryoEM (φ29 (Morais et al, 2005), P22 (Jiang et al, 2003), and ɛ15 (Jiang et al, 2006)) have been used to show that the capsid proteins of the tailed phages HK97, T4, φ29, P22, and ɛ15 have similar folds and associate into similar hexamers and pentamers, indicating a common evolutionary origin of the capsid.
X-ray crystallographic studies of the φ29 connector showed that it is a cone-shaped dodecamer with a central channel. It has three approximately cylindrical regions: the narrow end, the central part, and the wide end (Simpson et al, 2000). The 12-fold symmetric φ29 head–tail connector is located in the unique five-fold symmetric vertex of the capsid and is attached to the tail (Tao et al, 1998). Similarly, symmetry-mismatched head–tail connectors have been found in other tailed phages, including T4 (Leiman et al, 2004), P22 (Tang et al, 2005), ɛ15 (Jiang et al, 2006), T7 (Agirrezabala et al, 2005), and SPP1 (Orlova et al, 2003). In the mature φ29 virion, the narrow end of the connector protrudes out of the capsid and attaches to a spherical bulge in the lower collar (gp11). The bulge has a diameter of about 130 Å and is surrounded by 12 appendages that function to adsorb the virion on host cells (Anderson et al, 1966; Tosi and Anderson, 1973). A thin, 160 Å-long tube, with an outer diameter of 60 Å, leads away from the bulge and was thought to be a part of the lower collar (Hagen et al, 1976; Carazo et al, 1985). The tail knob (gp9) has a cylindrical shape and is attached to the distal end of the lower collar.
Here, we report the three-dimensional cryoEM structures of full and emptied φ29 phage particles to a resolution of approximately 16 and 20 Å, respectively. The use of asymmetric image reconstruction techniques (Morais et al, 2001; Jiang et al, 2006) established that the appendages, which were found to resemble the tailspike structure of bacteriophage P22, can have two alternate conformations, suggesting a mechanism for the partial digestion of the bacterial cell wall during infection. Structural differences between the prohead, the mature virus, and the emptied virus particles suggest how the DNA-packaging process might be completed when gp3 appears to block premature ejection of the genome and how the conformational change of the connector might potentiate detachment of part of the packaging machinery and facilitate tail assembly.
Results and discussion
The head
The quality of the five-fold symmetric reconstruction of the full and emptied head was assessed by analyzing the symmetry of the hexameric capsomer most distal from the tail. This hexamer was previously found to have the least distortion from true six-fold symmetry (Morais et al, 2005), whereas all other hexamers have some distortion. The correlation coefficient between this hexamer density and its rotated density showed correlations of 1.00, 0.88, 0.92, 0.86, 0.92, and 0.88 for rotations of 0, 60, 120, 180, 240, and 300°, respectively, indicating the good quality of the mature virus map. Similar results were obtained for the emptied virus particles.
The cryoEM densities of packaging-competent empty proheads (Morais et al, 2005), full mature virions, and emptied viruses of φ29 were superimposed when calculated at the same resolution, assuming the same EM magnification (Figure 2A and B). This showed that these structures are closely similar in size and in their length-to-width ratios. This ratio is independent of the assumed magnification, although the latter could have some error. Rather large differences in head shape and capsomer structure have been observed in HK97 (Lata et al, 2000) and P22 (Zhang et al, 2000). In particular, the proheads of HK97 and P22 have a smaller volume than the mature viruses and have distorted (‘skewed') hexameric capsomers. In φ29, the degree of skewing for the four kinds of hexamers remains the same in the prohead and in mature particles, implying that either there is no change in head volume during maturation or that the φ29 proheads examined by cryoEM (Tao et al, 1998; Morais et al, 2005) had already undergone conformational changes before DNA packaging. It may be relevant that bacteriophage T4 can package DNA into either immature-shaped proheads or mature-shaped proheads that have already changed their shape to that of the mature virus (Rao and Black, 1985).
Figure 2.
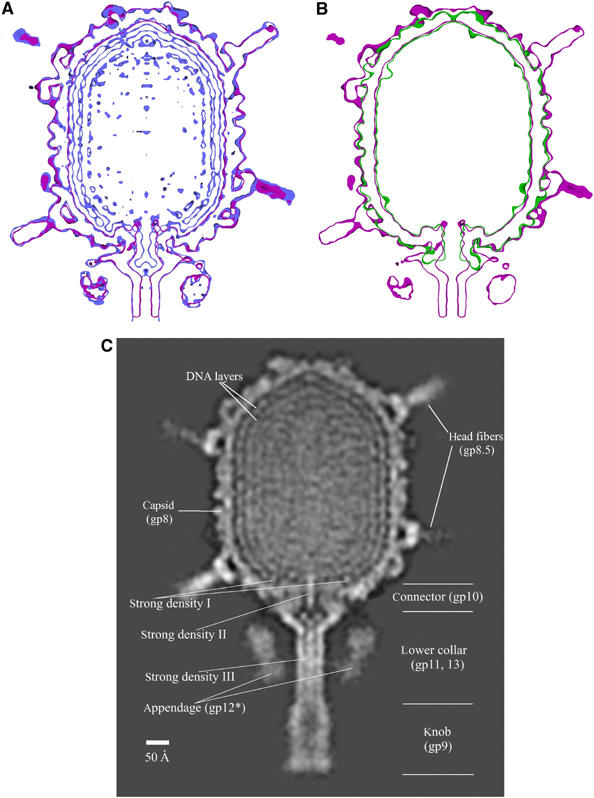
The φ29 structure during the virus' life cycle. Comparison of the cryoEM maps of (A) full (blue) with emptied (red) particles and (B) empty prohead (green) with emptied virion (purple) particles. The width-to-length ratios are the same for all three particles. The maps shown in (A) and (B) are 15 Å-thick slabs showing only the head and proximal parts of the tail. Contours are shown in 2σ intervals. (C) Organization of DNA in mature particles. At least three layers of the packaged genome are resolved in the central cross-section. High densities are white, low densities are black. Various structural components of the phage are labeled, including the dominant, well-defined densities I, II, and III. Horizontal lines designate the approximate boundaries of the different tail components.
Genome organization
Sections through the φ29 reconstruction of the full head show at least three concentric layers inside the capsid separated by a radial distance of about 23 Å (Figure 2A and C), similar to the density attributed to dsDNA in other phages (Cerritelli et al, 1997; Zhang et al, 2000; Fokine et al, 2004; Jiang et al, 2006). This spacing corresponds to hexagonally close-packed parallel dsDNA molecules separated by about 27 Å. The outermost layer, closest to the capsid shell, is better resolved than the innermost layer. Various models have been proposed for the DNA structure inside phage heads (Earnshaw et al, 1978; Harrison, 1983; Black et al, 1985; Lepault et al, 1987; Hud, 1995). Electron microscopy has shown that most of the encapsidated DNA in various bacteriophages is organized into concentric rings as a spool around or along the axis of the phage tail (Cerritelli et al, 1997; Fokine et al, 2004; Chang et al, 2006; Jiang et al, 2006; Lander et al, 2006).
There are three well-resolved regions of the DNA-related density whose average height is about equal to that of the capsid protein, whereas the rest of the DNA density is less than 0.7 of the height of the protein. One of these regions (strong density I on Figure 2C) is a strand of DNA running around the central viral axis above and co-axial with the connector at a radius of 90 Å (Figures 3 and 4A). Similar DNA density has been observed in the structure of the tailed phages ɛ15 (Jiang et al, 2006) and P22 (Chang et al, 2006; Lander et al, 2006), although in these viruses the connector is larger and extends further into the center of the viral capsid, suggesting that in P22 (but not in φ29) this circular DNA structure wraps around the wide end of the connector inside the capsid. Earlier results had shown that supercoiled DNA can wrap around free φ29 connectors (Turnquist et al, 1992), whereas the well-defined circular DNA density in φ29 mature particles is ‘above' and has a larger radius than the wide end of the connector. Nevertheless, this density appears to be functionally important both because it is much better resolved than the other regions of the DNA and because the same ring of DNA occurs in ɛ15 and P22. Lander et al (2006) propose that this circular DNA is related to signaling the terminase to cleave the concatemeric DNA when the P22 head is full. However, that function is not required in φ29, which packages one isolated DNA genome.
Figure 3.

The DNA strong density I (see Figure 2C) near the portal vertex. (A) Stereo view of circular DNA (green) near the connector density (lilac). The capsid, head fibers, lower collar, and appendages are shown in lime green. (B) Side view of circular DNA (green) in the vicinity of the fitted head–tail connector structure (red). The crystal structure of the dodecameric connector is shown as a ribbon diagram.
Figure 4.
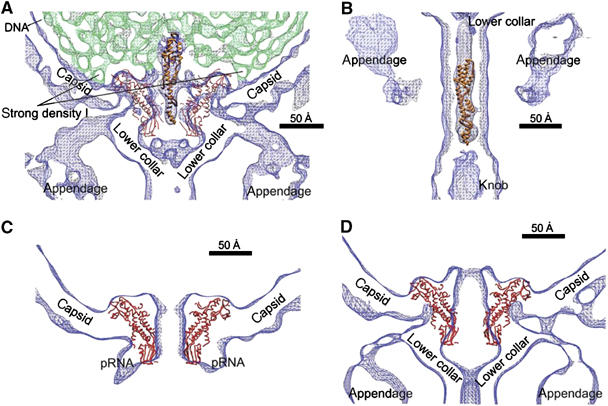
Changes in the connector structure upon phage maturation. (A) Fit of the modified gp3 crystal structure (ribbon representation in orange) into the strong density II (see Figure 2C) in the center of the connector as visualized in the asymmetric cryoEM reconstruction. The structure of the head–tail connector (Cα backbone trace, red) is shown fitted into the density (blue) of the mature phage contoured at 2σ intervals. The DNA density is shown in green. (B) Fit of the modified gp3 crystal structure (orange) into strong density III (see Figure 2C) in the tail's lower collar as visualized in the asymmetric reconstruction. (C) Fit of the connector crystal structure (Cα trace in red) into the cryoEM density of the five-fold averaged reconstruction of the prohead and (D) the cryoEM density of the asymmetric reconstruction of the emptied particles. The narrow end of the connector would have to increase its radius in order to fit into the density (blue) of the emptied particle.
The portal protein of φ29, which assembles to form the head–tail connector, has roughly half the molecular weight of the portal protein of other bacteriophages (Carrascosa and Valpuesta, 1999). In P22, the additional mass of the connector extends further into the capsid and is surrounded by the ring of DNA discussed above (Lander et al, 2006). However, phages other than φ29 package a DNA concatemer that requires a terminase to cleave it when the head is full (Black et al, 1994). In contrast, no signal to terminate DNA packaging is required in φ29, as the genome is of unit length, ending with the gp3 at the right end of the DNA (see below). Thus, it is possible that the extra size of the portal protein in tailed phages other than φ29 may be required for transmitting the ‘head full' signal, presumably when the DNA around the expanded wide end of the connector is placed into position at the end of the packaging process (Orlova et al, 1999; Lander et al, 2006). Hence, the presence of the ring of DNA around and ‘above' the portal protein of φ29 might be a remnant of a function no longer required or of a function in all these tailed bacteriophages that has not yet been identified.
The other two regions that have especially well-defined DNA structure are rod-like densities of about 25 Å diameter, both situated on the central axis of the virus. Density II (Figure 2C) is about 100 Å long, passes through the 75 Å-long connector, and merges with weaker density in the bulge of the lower collar. Density III (Figure 2C) is about 190 Å long and is situated in the axial portion of the lower collar. One interpretation could be that both densities represent the final portion of the packaged DNA, leaving gp3, covalently bound to the 5′ end of DNA, at the distal end of density III. The structure of gp3 (Kamtekar et al, 2006) can be fitted into the distal half of density III after changing the two hinge angles between the three domains (Figure 4B). If density III were the end of the DNA genome with its attached gp3 molecule, it would require that the DNA would start exiting the head and fill the lower collar after the tail has been assembled onto the filled head as has been suggested for phage lambda (Saigo and Uchida, 1974; Thomas, 1974). An additional question that arises in light of the above interpretation of densities II and III is why there is a discontinuity of the DNA density in the bulge of the lower collar. It is possible that the path of DNA in this region is different in different particles, resulting in weak, diffuse density upon averaging between different particles.
Alternatively, density II could be the terminal gp3 with density III being perhaps free gp3 that is purported to be present in φ29 and has been shown to have muralytic activity (Moak and Molineux, 2004). Indeed, density II has the correct size for fitting the structure of gp3 after changing the two hinge angles (Figure 4A). In contrast to the gp3 at the left end of the DNA that is packaged first, the gp3 at the right end would remain in the same position within each capsid and, thus, would be enhanced by the averaging process between different images. Similar density as density II has been observed in the connectors of mature P22 (Chang et al, 2006; Lander et al, 2006) and ɛ15 (Jiang et al, 2006) tailed bacteriophages. In P22, this density was assigned to be a pilot injection protein, but in ɛ15 it was interpreted as the DNA terminus.
The connector
The asymmetric reconstructions of the mature and of the emptied virus show 12 well-resolved densities (Supplementary Figure S1A) representing the connector, consistent with the size and shape of the crystallographically determined structure (Simpson et al, 2000, 2001). The connector is surrounded by the capsid at its wide end and is attached to the lower collar at its narrow end. The symmetry mismatch between the head and the connector results in each of the 12 subunits being associated with a different structural environment. Although most of the 12 monomers of the connector are approximately related by 12-fold symmetry, some have considerable deviation, as is also the case for the connector of ɛ15 (Jiang et al, 2006). A plot (Supplementary Figure S1B) of the height of the portal protein densities at the wide end of the connector shows the largest densities are six subunits apart. It will be shown in the discussion of the appendages (below) that the relationship between the first and sixth position of a 12-fold symmetric object (the appendages) and a five-fold symmetric object (the head capsid) is similar. Thus, the 12 subunits in the connector have some plasticity (as also observed crystallographically, Simpson et al, 2001) and are sensitive to their particular environment. This property is likely to be important for the functioning of the packaging motor, which has five ATPase complexes at 72° intervals (Tao et al, 1998; Simpson et al, 2000; Morais et al, 2001), providing the power for DNA packaging.
The crystal structure of the head–tail connector (Simpson et al, 2000, 2001; Morais et al, 2005) fits well into the cryoEM density of the prohead. The additional lobes seen in the density that create the internal end of the portal channel (Figure 4C and D) correspond to poorly ordered polypeptide in the crystal structure that can be visualized only at low resolution in the cryoEM density (Simpson et al, 2000). However, only the wide end of the connector structure fits into the cryoEM density of the full and emptied virions (Figure 4D). The narrow end of the connector is surrounded by the pRNA in the prohead, but is expanded in the mature and emptied particles. Whatever is the identity of the high density within the connector channel (gp3 or DNA) of the mature φ29 particles (see above), it could be the agent that causes the narrow end of the connector to be expanded in virions relative to the prohead. This expansion might trigger the release of the pRNA and gp16 components of the packaging motor, thus permitting collar attachment to the filled head (Nelson et al, 1976; Camacho et al, 1979). Furthermore, the gp3 might function as a temporary plug to stop the packaged DNA from exiting until the lower collar (gp11) has been able to attach (Carazo et al, 1985).
The appendages
Micrographs of negatively stained φ29 virus had shown that there are 12 appendages radiating out from the lower collar (Anderson et al, 1966). The appendages were better resolved in a cryoEM reconstruction, showing that they attached to the bulge of the lower collar, close to the capsid, and that they were surrounded by a ring of high density (Morais et al, 2001). The current 12-fold averaged results show that each of the appendages consists of a roughly radial, slightly bent rib emanating from the bulge of the lower collar and ending in a tassel-like structure that runs roughly parallel to the length of the virus. These tassel-like structures are best resolved in the asymmetric reconstruction described here, but were unresolved in the earlier reconstructions. Overall, the structure of the appendages is reminiscent of an umbrella with 12 ribs that end in these ‘tassels'. The umbrella pole is formed by the axial region of the lower collar, and the umbrella handle by the tail knob (Figure 5A).
Figure 5.

Structure of appendages labeled 1–12. Surface-shaded views showing (A) an angled side view contoured at 4.5σ and (B) a top view contoured at 3σ. Appendages 1 and 6 are in the ‘up' position, whereas the other appendages are in the ‘down' position. (C) Diagram showing the relationship between the position of the appendages and the position of the head fibers. The appendages in the ‘up' position are shown as green ellipses, whereas those in the ‘down' position are shown as green circles. The lower tier of five fibers (A1–A5) is shown in pink. The more distant upper layer of 10 fibers (B1–B10) is shown in yellow. Note that appendages in the ‘up' position are least hindered by the head fibers.
The asymmetric reconstructions of the mature virus and of the emptied particles both show that two of the 12 appendages are extended radially outwards (the ‘up' position), whereas the other 10 have their tassels ‘hanging' roughly parallel to the length of the virus (the ‘down' position) (Figure 5A and B). Thus, the 12-fold symmetry of the appendages is only approximate, explaining why the asymmetric reconstruction appears to be significantly better than the 12-fold averaged map as judged by the resolution of the individual appendages from each other and by the amount of apparent structural detail.
Most of the appendages have some degree of partial occupancy in both the ‘up' and the ‘down' conformations (Table I) that is correlated to the position of the head fibers. There are two rings of head fibers that can interfere with the position of the appendages. The five head fibers closest to the head portal (A1–A5 in Figure 5C) point ‘downwards', and thus, have the greatest potential for steric hindrance with the appendages. The next set of 10 head fibers (B1–B10 in Figure 5C) also point ‘downwards' but to a lesser extent and, thus, have less interference with the appendages. The symmetry mismatch between the ring of five A and 10 B fibers with the 12 appendages creates a unique environment of head fibers for each appendage (Figure 5). The most occupied ‘up' positions of the appendages occur where there is the least steric interference with the surrounding head fibers (appendages 1 and 6 in Figure 5C). Similarly, the appendages that have no apparent occupancy in the ‘up' position are closer to the downward-pointing head fibers. The correlation between the measured occupancies of the appendages in the ‘down' position and the angular distance from their nearest head fiber was 0.82 (Table I). Thus, the positions of the appendages are dictated by the positions of the head fibers.
Table 1.
Occupancy of appendages in the ‘down' and ‘up' positions, showing the negative correlation with the position of the nearest head fiber (see Figure 5 for appendage number nomenclature)
| Observed relative occupancy (h) |
Nearest head fiber | Distance (x) from nearest head fiber (deg) | Calculateda occupancy ‘down' | ||
|---|---|---|---|---|---|
| Appendage | ‘Down' | ‘Up' | |||
| 1 | 0.27 | 0.52 | A1 & A2 | ±36 | 0.51 |
| 2 | 1.00 | A2 | −6 | 0.91 | |
| 3 | 0.86 | A2 | +24 | 0.67 | |
| 4 | 0.72 | A3 | −28 | 0.60 | |
| 5 | 0.78 | A3 | +12 | 0.83 | |
| 6 | 0.49 | 0.42 | A3 & A4 | ±30 | 0.59 |
| 7 | 0.87 | A4 | 0 | 0.99 | |
| 8 | 0.65 | A4 | +30 | 0.59 | |
| 9 | 0.76 | A5 | −12 | 0.83 | |
| 10 | 0.69 | A5 | +18 | 0.75 | |
| 11 | 0.80 | A1 | −24 | 0.67 | |
| 12 | 0.96 | A1 | +6 | 0.91 | |
| Calculated from h=−0.013∣x∣+0.99. | |||||
A BLAST search showed a number of polysaccharide-binding proteins that had a low level of sequence similarity to φ29 gp12. Of these, rhamnogalacturonase (RGase), present in dicotyledonous plants, had the highest level of sequence similarity (15% amino-acid identity between residues 91–443) for a protein that also had its structure determined (PDB accession number 1RMG). The crystal structure of the RGase monomer (Petersen et al, 1997) is primarily a 12-turn, right-handed β-helix with about 23 residues per turn. A DALI (Holm and Sander, 1998) search for similar structures found more than a dozen other glycosidases and lyases with a Z-score greater than 10. All these structures bind polysaccharides along the length of the β-helix (Jenkins et al, 1998). An approximately 18 kDa C-terminal fragment of φ29 gp12 is cleaved off to give gp12* (Peterson et al, 2001) during maturation. It has been shown that φ29 adsorption is dependent on binding to glucosylated teichoic acid, present on the surface of the B. subtilis cell wall (Young, 1967; Yasbin et al, 1976). Similarly, the P22 tailspike binds and cleaves the host's cell surface lipopolysaccharide (McConnell et al, 1979).
The central domain of the trimeric P22 tailspike (residues 143–540) consists of three parallel β-helices, each with 13 turns (Steinbacher et al, 1994). The β-helix of RGase can be superimposed onto each of the P22 tailspike β-helices with a 1.7 Å rms distance between equivalenced Cα atoms. The superposition also shows that the site of polysaccharide binding is on the outside of each of the monomers in the P22 tailspike trimer (Steinbacher et al, 1997), consistent with the RGase alignment. Given the sequence similarity of φ29 gp12 to RGase and the structural similarity of RGase to the P22 tailspike, it is possible to align these three protein sequences (Figure 6). Residues 91–443 of gp12 correspond to the β-helical component of the P22 tailspike. The approximately 178 carboxy-terminal residues of gp12* are likely to be homologous to the 99 residue carboxy-terminal section of the P22 tailspike, although there is no easily recognizable sequence similarity. This is confirmed by the good fit of the P22 tailspike domain structure into the tassel component of the appendages of the φ29 cryoEM maps (Figure 7). This fit places the three amino termini of the P22 tailspike domain at the proximal end of the tassel, leaving approximately 90 residues to span the distance from the collar to the tassel, formed by the ribs of the umbrella. Secondary structural predictions show that the amino-terminal region of gp12* likely forms an α-helix. Although sequence analysis of the residues in the amino-terminal region of gp12* only weakly indicate that the three amino-terminal polypeptides form a trimeric coiled coil, the 15 Å-radius, 75 Å-long umbrella ribs in the cryoEM density would accommodate such a structure. Bacteriophage P22 also has ‘ribs' connecting the virus neck to the tailspikes (Tang et al, 2005), although P22 has only six ribs and their structure is not a coiled coil (Steinbacher et al, 1997). These observations establish that there are 3 × 12 or 36 copies of gp12* in the mature virus, rather less than the 60 copies suggested earlier (Peterson et al, 2001). This contrasts with the six trimeric tailspikes of ɛ15 (Jiang et al, 2006) and P22 (Tang et al, 2005).
Figure 6.
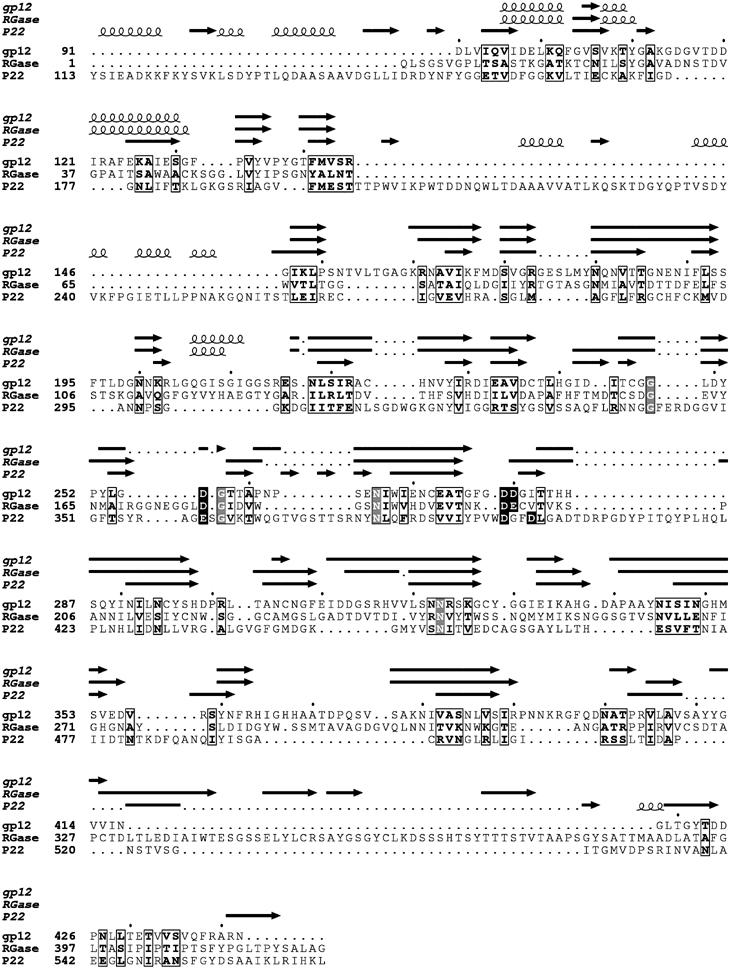
Sequence and structural alignment of φ29 gp12, plant RGase, and the P22 tailspike protein. The predicted secondary structure of gp12, and the observed secondary structures of RGase and the P22 tailspike are shown above the alignments. Conserved residues are boxed. Residues involved in substrate binding and catalysis in RGase and the P22 tailspike are shown in white on a black background. Completely conserved residues are shown in white on a gray background. The program JPred was used for making the secondary structure prediction of gp12 (Cuff and Barton, 2000).
Figure 7.
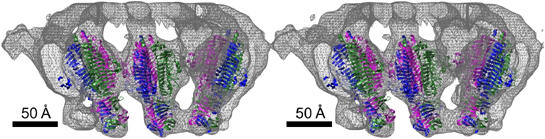
Stereo diagram showing the fit of the P22 tailspike structure into the cryoEM density map as determined by the asymmetric reconstruction for the mature φ29 phage. The trimeric P22 tailspikes are shown as ribbon diagrams using red, green, and blue for the three different monomers. The cryoEM density is shown in gray.
Given the homology between gp12, the tailspike protein of phage P22, and the RGase, the φ29 appendages may cleave the teichoic acid in the B. subtilis cell wall. It may be necessary for the appendages to continually disengage and relocate, allowing the tail knob that may house gp3 muralytic activity (Moak and Molineux, 2004) to digest the underlying peptidoglycan layer of the cell wall as it seeks contact with the cytoplasmic membrane. This movement could be achieved by virtue of the two alternate positions of the appendages. It has been hypothesized (Hendrix, 1978) that the head rotates relative to the connector while the DNA is being packaged, potentiated by the mismatch between the five-fold symmetry of the capsid portal vertex and the hydrophobic exterior of the 12-fold symmetric connector (Simpson et al, 2000). Although the connector rotation may not be necessary for DNA packaging (Baumann et al, 2006), the symmetry mismatch implies that there is no strongly preferred orientation of the connector around the long axis of the phage, thus possibly permitting it to rotate or oscillate in a random manner without having to overcome any large energy barriers. The rotation or oscillation of the head relative to the connector might occur during infection as a result of Brownian motion and would alter the position of the head fibers relative to the appendages, causing the appendages to oscillate between ‘up' and ‘down' positions. Hence, as the head rotates or oscillates, an appendage which moves from ‘down' to ‘up' would cause the virus to move further through the teichoic acid polymers and the cell wall, whereas an appendage that moves from ‘up' to ‘down' would be readied for further cleavage of the teichoic acid. A somewhat similar mechanism has been observed for bacteriophage T4 in which the tail lysozyme rotates as the result of sheath contraction (Kostyuchenko et al, 2003). Although earlier data had indicated that the φ29 head fibers were not essential for phage infection (Reilly et al, 1977), these studies did not determine the rate of adsorption or penetration for wild-type versus fiberless particles. Thus, one of the functions of the fibers might be to increase the speed with which φ29 is able to puncture a hole in the cell wall of B. subtilis.
The lower collar and the tail knob
The lower collar is attached to the expanded narrow end of the head–tail connector and consists of a bulge region and a thin axial tube that extends about 160 Å to the tail knob. The tube has outer and inner diameters of 60 and 31 Å, respectively, in both the mature virus and in emptied particles. Slightly protruding features on the internal surface of the bulge, related approximately by 12-fold symmetry, indicate that the lower collar bulge probably consists of 12 copies of gp11, consistent with a determination of the gp11 copy number (Peterson et al, 2001).
Earlier studies (Hagen et al, 1976; Carazo et al, 1985) had shown that gp11 binds to the connector. As the molecular weight of gp11 (34 kDa) is similar to that of the portal protein gp10 (36 kDa), and considering that the volume of density for the connector and the bulb are similar, it would seem probable that the bulb of the lower collar might be an assembly of 12 gp11 molecules. Although previous studies indicated that gp13 functioned as a morphogenetic factor during phage maturation (Garcia et al, 1983), unpublished results (D Cohen and D Anderson, personal communication) indicate that gp13 is likely a part of the tail.
The distal portion of the tail consists of the tail knob, which extends from the end of the lower collar as a large cylindrical structure about 150 Å long and has an outer diameter of 82 Å in the full particles. The knob was missing in the reconstruction of the ‘new' data set of emptied particles (see Materials and methods), but in an ‘old' data set (Tao et al, 1998) the distal end of the knob was extended by a cone-like structure relative to full particles (Figure 8). The conical-shaped density is 40 Å long and 70 Å wide at the distal end and 60 Å wide at the proximal end. Apparently, this cone is located within the knob in full particles (Figure 8A), but is exposed during or subsequent to DNA ejection (Figure 8B). The cone might span the cytoplasmic membrane to provide a passage for DNA entry, and trigger DNA ejection from the tail.
Figure 8.
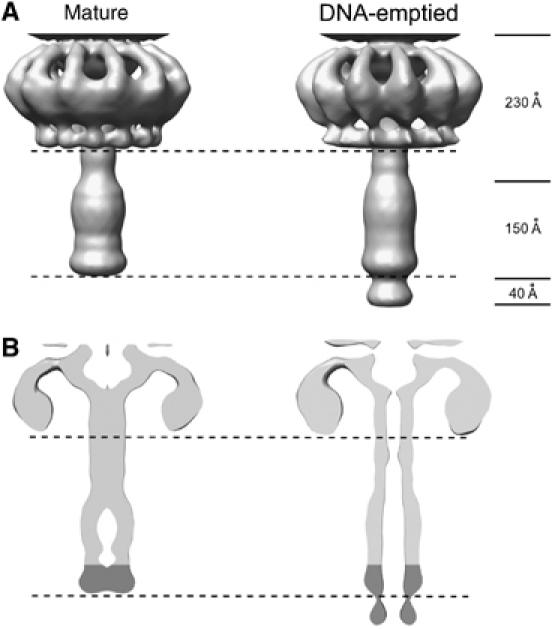
Comparison of the knob and lower collar cryoEM density of the mature and DNA-emptied particles. The DNA-emptied particles have an additional cone-shaped density at their distal end as seen in (A) a surface shaded view and (B) a cross-section of the cryoEM densities. The mass of the cone-shaped density appears to form the end of the knob in the mature virus.
Evolution
The evolution of viruses is dependent not only on gradual adaptation to the environment by spontaneous mutation of the genome but also on the adoption of host genes. Nevertheless, many of the structural components of viruses appear to have had common origins that pre-date specific adaptation. It has long been evident that three-dimensional structure has been conserved over a much longer period of time than amino-acid or nucleotide sequences (Rossmann et al, 1974). For this reason, a number of techniques for finding similar folds in order to discern a common function (Holm and Sander, 1993; Murzin et al, 1995) are now available.
Many eukaryotic viruses and some phages use the ‘jelly-roll' fold in their major capsid protein, indicating a common origin for their capsid fold and assembly (Rossmann et al, 1985; Nandhagopal et al, 2002; Benson et al, 2004). However, all tailed dsDNA phages for which there is sufficient information use the ‘HK97' capsid fold for their capsid proteins, indicating a common origin for tailed phages different to the viruses that utilize the ‘jelly-roll' fold. Therefore, it should not be surprising that the scaffolding protein, required to assemble the phage heads, also has a common structure between φ29 (Morais et al, 2003, 2005) and P22. Nor should it be surprising that the structure of the head–tail connector is common among phages and is similar to that found in φ29 (Simpson et al, 2000), as the capsid protein, the scaffolding protein, and the portal protein are all related to head assembly.
Now, it appears that there is also a common structure for the tailspike of P22 and φ29. However, the comparison with RGase (Petersen et al, 1997) shows that this gene may have been derived from a eukaryotic host. Alternatively, the evolution of these proteins may have diverged long ago from a common ancestor. If the first scenario is true, then the tailspike gene might have been a more recent acquisition, consistent with the different type of tailspike structures found for the K1F (Stummeyer et al, 2004) and the p2 (Spinelli et al, 2006) bacteriophage. Nevertheless, because prokaryotes have had a one billion year head start on eukaryotes, it is to be anticipated that in the evolution of the efficient infection machinery embodied in the tail of most phages, it is likely that some of the tail components will have had a common origin.
Materials and methods
Image reconstruction
Production and purification of full and empty particles was described previously (Tao et al, 1998). Electron micrographs were recorded at a magnification of about 33 000 using a CM300 FEG microscope and 38 000 using a CM200 microscope under low-dose conditions (∼20 electrons/Å2). Micrographs were digitized using a Zeiss SCAI scanner with a step size of 14 μm (Table II).
Table 2.
CryoEM data and image reconstruction
| DNA-filled head | Entire DNA-filled particle | Old data | New data |
||
|---|---|---|---|---|---|
| Emptied taila | Emptied head | Entire emptied particle | |||
| Underfocus (μm)b | 6.4–1.4 | 6.4–1.4 | 6.0–2.6 | 5.8–1.7 | 5.8–1.7 |
| Number of boxed particlesc | 12 184 (10157) | 12 184 (10866) | 3197 (2994) | 7272 (6193) | 7272 (6132) |
| Resolution (Å) | |||||
| Entire box | 16 | 23 | 30 | 20 | 27 |
| Head | 16 | 21 | NA | 20 | 25 |
| Tail | NA | 27 | 30 | NA | 30 |
| Symmetry imposed | 5 | 1 | 12 | 5 | 1 |
| Program used | EMAN | Python | EMAN | EMAN | Python |
| Images were low-pass filtered to 27 Å. | |||||
| Determined from the contrast transfer function of the microscope. | |||||
| The total number of particles boxed for each reconstruction. The number of particles used in each 3D map calculation is given in parentheses. | |||||
| NA=not applicable. | |||||
Individual particle images were selected and boxed with the program ROBEM (http://cryoem.ucsd.edu/programs.shtm). The boxed images were preprocessed to normalize mean intensities and variances and to remove linear background gradients. The contrast transfer function (CTF) parameters were determined with the ‘ctfit' routine in the EMAN package (Ludtke et al, 1999), and improved by using the available pseudo-atomic model of the φ29 capsid (Morais et al, 2005). Both phases and amplitudes of the observed image data were corrected using the CTF parameters.
The initial reconstruction of the mature particles used the previously determined φ29 prohead (Morais et al, 2005) to start a model-based classification as implemented in the program EMAN (Ludtke et al, 1999), assuming five-fold symmetry. This reconstruction was based on images that had been boxed to remove the tail as much as possible. These images were used to produce a reconstruction of the head and a small portion of the top of the tail using five-fold symmetry. In the subsequent asymmetric reconstruction that included the tail portion of the virus, a python script (Jiang et al, 2006) was used to differentiate between the five possible orientations around the long axis of the phage (Morais et al, 2001, 2003; Jiang et al, 2006). Finally, this second reconstruction was 12-fold averaged and then iterated to obtain a 12-fold symmetric structure. Visual inspection showed that the asymmetric reconstruction was considerably superior to the 12-fold averaged density.
The ‘old' data of the emptied particles (Tao et al, 1998) was used for a reconstruction of the tail using 12-fold averaging. Subsequently, ‘new' data were collected from a new sample of emptied phage. The ‘new' data set was both far more plentiful and of better quality. Therefore, it was used to make an asymmetric reconstruction of the complete emptied phage using the same two-step procedure described above for the full particles (Table II). The resultant reconstruction of the ‘new' (but not the ‘old') data set showed that the knob (gp9) was missing from the phage. Protein analysis of the sample established that gp9 was present in solution. Careful inspection of cryoEM and negatively stained micrographs indicated that many particles had shortened tails, suggesting that the knob can be labile in emptied particles. For both full and emptied particle types, centers of the images with only the heads boxed were set to be approximately at the center of the head. Centers of the images containing the whole virus were obtained by shifting the box centers to the head–tail junction.
The resolutions of the various reconstructions were determined by the Fourier shell correlation method (van Heel, 1987). A correlation coefficient of 0.5 between independent half-data sets was used as the cut-off criterion. The head and tail were masked separately to determine their respective resolutions (Table II).
Fitting of the P22 tailspike structure
The program EMfit (Rossmann et al, 2001) was used to fit the structure of the P22 tailspike (Steinbacher et al, 1994) into the cryoEM density of the appendages in the reconstruction of the φ29 mature virus. The tailspike structure was fitted to each of the 12 ‘down' positions, but was reliable only for appendages 1 and 6 in the ‘up' position. The relative occupancies of the 12 appendages were determined by comparing the average Cα densities for each fitted tailspike, normalized to the appendage with the greatest occupancy (A2 in Figure 5C) (Table I). The degree of steric interference of each appendage by the surrounding head fibers was assessed by the angular distance of the appendage from the nearest head fiber (x). The −0.82 correlation (the greatest occupancies correspond to the smallest angular distance of the nearest head fiber) suggested that it would be possible to make a crude prediction of the appendage occupancies. A least-squares fit was made between the experimentally observed occupancies in the ‘down' position (z) and the angular distance x by assuming a linear relation, giving the result
![]()
The predicted occupancies based on this assumption have a 14% ‘R' factor compared to the observed occupancies (Table I).
Supplementary Material
Supplementary Figure S1A
Acknowledgments
We thank Wen Jiang, Kay Choi, Petr Leiman, and Chuan Xiao for many stimulating discussions. We are grateful to Cheryl Towell, Sheryl Kelly, and Sharon Wilder for help in preparing the manuscript. The work was supported by a grant to MGR from the NSF (Grant # MCB-0443899) and to DLA and MGR from the NIH (Grant # DE03606). The work was also supported by the Keck Foundation with a grant to MGR for the purchase of a CM300 electron microscope.
References
- Ackermann HW (2003) Bacteriophage observations and evolution. Res Microbiol 154: 245–251 [DOI] [PubMed] [Google Scholar]
- Agirrezabala X, Martin-Benito J, Castón JR, Miranda R, Valpuesta JM, Carrascosa JL (2005) Maturation of phage T7 involves structural modification of both shell and inner core components. EMBO J 24: 3820–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D, Reilly B (1993) Morphogenesis of bacteriophage φ29. In Bacillus subtilis and Other Gram-Positive Bacteria: Biochemistry, Physiology, and Molecular Genetics, Sonenshein AL, Hoch JA, Losick R (eds), pp 859–867. Washington, DC: American Society for Microbiology [Google Scholar]
- Anderson DL, Hickman DD, Reilly BE (1966) Structure of Bacillus subtilis bacteriophage φ29 and the length of φ29 deoxyribonucleic acid. J Bacteriol 91: 2081–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann RG, Mullaney J, Black LW (2006) Portal fusion protein constraints on function in DNA packaging of bacteriophage T4. Mol Microbiol 61: 16–32 [DOI] [PubMed] [Google Scholar]
- Benson SD, Bamford JKH, Bamford DH, Burnett RM (2004) Does common architecture reveal a viral lineage spanning all three domains of life? Mol Cell 16: 673–685 [DOI] [PubMed] [Google Scholar]
- Bjornsti MA, Reilly BE, Anderson DL (1983) Morphogenesis of bacteriophage φ29 of Bacillus subtilis: oriented and quantized in vitro packaging of DNA–protein gp3. J Virol 45: 383–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black LW, Newcomb WW, Boring JW, Brown JC (1985) Ion etching of bacteriophage T4: support for a spiral-fold model of packaged DNA. Proc Natl Acad Sci USA 82: 7960–7964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black LW, Showe MK, Steven AC (1994) Morphogenesis of the T4 head. In Molecular Biology of Bacteriophage T4, Karam JD (ed), pp 218–258. Washington, DC: American Society for Microbiology [Google Scholar]
- Camacho A, Jiménez F, Viñuela E, Salas M (1979) Order of assembly of the lower collar and the tail proteins of Bacillus subtilis bacteriophage phi 29. J Virol 29: 540–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo JM, Santisteban A, Carrascosa JL (1985) Three-dimensional reconstruction of bacteriophage φ29 neck particles at 2.2 nm resolution. J Mol Biol 183: 79–88 [DOI] [PubMed] [Google Scholar]
- Carrascosa JL, Valpuesta JM (1999) Bacteriophage connectors: structural features of a DNA translocating motor. Recent Res Devel Virol 1: 449–465 [Google Scholar]
- Cerritelli ME, Cheng N, Rosenberg AH, McPherson CE, Booy FP, Steven AC (1997) Encapsidated conformation of bacteriophage T7 DNA. Cell 91: 271–280 [DOI] [PubMed] [Google Scholar]
- Chang J, Weigele P, King J, Chiu W, Jiang W (2006) Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure 14: 1073–1082 [DOI] [PubMed] [Google Scholar]
- Cuff JA, Barton GJ (2000) Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins: Struct Funct Genet 40: 502–511 [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, King J, Harrison SC, Eiserling FA (1978) The structural organization of the DNA packaged within the heads of T4 wild type, isometric and giant bacteriophages. Cell 14: 559–568 [DOI] [PubMed] [Google Scholar]
- Fokine A, Chipman PR, Leiman PG, Mesyanzhinov VV, Rao VB, Rossmann MG (2004) Molecular architecture of the prolate head of bacteriophage T4. Proc Natl Acad Sci USA 101: 6003–6008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokine A, Leiman PG, Shneider MM, Ahvaze B, Boeshans KM, Steven AC, Black LW, Mesyanzhinov VV, Rossmann MG (2005) Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc Natl Acad Sci USA 102: 7163–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia JA, Carrascosa JL, Salas M (1983) Assembly of the tail protein of the Bacillus subtilis phage φ29. Virology 125: 18–30 [DOI] [PubMed] [Google Scholar]
- Grimes S, Anderson D (1990) RNA dependence of the bacteriophage φ29 DNA packaging ATPase. J Mol Biol 215: 559–566 [DOI] [PubMed] [Google Scholar]
- Grimes S, Jardine PJ, Anderson DL (2002) Bacteriophage φ29 DNA packaging. Adv Virus Res 58: 255–294 [DOI] [PubMed] [Google Scholar]
- Guo P, Grimes S, Anderson D (1986) A defined system for in vitro packaging of DNA–gp3 of the Bacillus subtilis bacteriophage φ29. Proc Natl Acad Sci USA 83: 3505–3509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen EW, Reilly BE, Tosi ME, Anderson DL (1976) Analysis of gene function of bacteriophage φ29 of Bacillus subtilis: identification of cistrons essential for viral assembly. J Virol 19: 501–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SC (1983) Packaging of DNA into bacteriophage heads: a model. J Mol Biol 171: 577–580 [DOI] [PubMed] [Google Scholar]
- Hendrix RW (1978) Symmetry mismatch and DNA packaging in large bacteriophages. Proc Natl Acad Sci USA 75: 4779–4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix RW (2002) Bacteriophages: evolution of the majority. Theor Popul Biol 61: 471–480 [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C (1993) Protein structure comparison by alignment of distance matrices. J Mol Biol 233: 123–138 [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C (1998) Touring protein fold space with Dali/FSSP. Nucl Acids Res 26: 316–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hud NV (1995) Double-stranded DNA organization in bacteriophage heads: an alternative toroid-based model. Biophysics J 69: 1355–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins J, Mayans O, Pickersgill R (1998) Structure and evolution of parallel β-helix proteins. J Struct Biol 122: 236–246 [DOI] [PubMed] [Google Scholar]
- Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W (2006) Structure of epsilon 15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature (London) 439: 612–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Li Z, Zhang Z, Baker ML, Prevelige PE Jr, Chiu W (2003) Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nat Struct Biol 10: 131–135 [DOI] [PubMed] [Google Scholar]
- Kamtekar S, Berman AJ, Wang J, Lázaro JM, de Vega M, Blanco L, Salas M, Steitz TA (2006) The φ29 DNA polymerase: protein-primer structure suggests a model for the initiation to elongation transition. EMBO J 25: 1335–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyuchenko VA, Leiman PG, Chipman PR, Kanamaru S, van Raaij MJ, Arisaka F, Mesyanzhinov VV, Rossmann MG (2003) Three-dimensional structure of bacteriophage T4 baseplate. Nat Struct Biol 10: 688–693 [DOI] [PubMed] [Google Scholar]
- Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige PE Jr., Poliakov A, Potter CS, Carragher B, Johnson JE (2006) The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science 312: 1791–1795 [DOI] [PubMed] [Google Scholar]
- Lata R, Conway JF, Cheng N, Duda RL, Hendrix RW, Wikoff WR, Johnson JE, Tsuruta H, Steven AC (2000) Maturation dynamics of a viral capsid: visualization of transitional intermediate states. Cell 100: 253–263 [DOI] [PubMed] [Google Scholar]
- Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG (2004) Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell 118: 419–429 [DOI] [PubMed] [Google Scholar]
- Lepault J, Dubochet J, Baschong W, Kellenberger E (1987) Organization of double-stranded DNA in bacteriophages: a study by cryo-electron microscopy of vitrified samples. EMBO J 6: 1507–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128: 82–97 [DOI] [PubMed] [Google Scholar]
- McConnell M, Reznick A, Wright A (1979) Studies on the initial interactions of bacteriophage ɛ15 with its host cell, Salmonella anatum. Virology 94: 10–23 [DOI] [PubMed] [Google Scholar]
- Méndez E, Ramírez G, Salas M, Viñuela E (1971) Structural proteins of bacteriophage φ29. Virology 45: 567–576 [DOI] [PubMed] [Google Scholar]
- Moak M, Molineux IJ (2004) Peptidoglycan hydrolytic activities associated with bacteriophage virions. Mol Microbiol 51: 1169–1183 [DOI] [PubMed] [Google Scholar]
- Morais MC, Choi KH, Koti JS, Chipman PR, Anderson DL, Rossmann MG (2005) Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of φ29. Mol Cell 18: 149–159 [DOI] [PubMed] [Google Scholar]
- Morais MC, Kanamaru S, Badasso MO, Koti JS, Owen BAL, McMurray CT, Anderson DL, Rossmann MG (2003) Bacteriophage φ29 scaffolding protein gp7 before and after prohead assembly. Nat Struct Biol 10: 572–576 [DOI] [PubMed] [Google Scholar]
- Morais MC, Tao Y, Olson NH, Grimes S, Jardine PJ, Anderson DL, Baker TS, Rossmann MG (2001) Cryoelectron-microscopy image reconstruction of symmetry mismatches in bacteriophage φ29. J Struct Biol 135: 38–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin AG, Brenner SE, Hubbard T, Chothia C (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol 247: 536–540 [DOI] [PubMed] [Google Scholar]
- Nandhagopal N, Simpson AA, Gurnon JR, Yan X, Baker TS, Graves MV, Van Etten JL, Rossmann MG (2002) The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc Natl Acad Sci USA 99: 14758–14763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RA, Reilly BE, Anderson DL (1976) Morphogenesis of bacteriophage φ29 of Bacillus subtilis: preliminary isolation and characterization of intermediate particles of the assembly pathway. J Virol 19: 518–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova EV, Dube P, Beckmann E, Zemlin F, Lurz R, Trautner TA, Tavares P, van Heel M (1999) Structure of the 13-fold symmetric portal protein of bacteriophage SPP1. Nat Struct Biol 6: 842–846 [DOI] [PubMed] [Google Scholar]
- Orlova EV, Gowen B, Dröge A, Stiege A, Weise F, Lurz R, van Heel M, Tavares P (2003) Structure of a viral DNA gatekeeper at 10 Å resolution by cryo-electron microscopy. EMBO J 22: 1255–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Kauppinen S, Larsen S (1997) The crystal structure of rhamnogalacturonase A from Aspergillus aculeatus: a right-handed parallel β helix. Structure 5: 533–544 [DOI] [PubMed] [Google Scholar]
- Peterson C, Simon M, Hodges J, Mertens P, Higgins L, Egelman E, Anderson D (2001) Composition and mass of the bacteriophage φ29 prohead and virion. J Struct Biol 135: 18–25 [DOI] [PubMed] [Google Scholar]
- Rao VB, Black LW (1985) DNA packaging of bacteriophage T4 proheads in vitro evidence that prohead expansion is not coupled to DNA packaging. J Mol Biol 185: 565–578 [DOI] [PubMed] [Google Scholar]
- Reilly BE, Nelson RA, Anderson DL (1977) Morphogenesis of bacteriophage φ29 of Bacillus subtilis: mapping and functional analysis of the head fiber gene. J Virol 24: 363–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann MG, Arnold E, Erickson JW, Frankenberger EA, Griffith JP, Hecht HJ, Johnson JE, Kamer G, Luo M, Mosser AG, Rueckert RR, Sherry B, Vriend G (1985) Structure of a human common cold virus and functional relationship to other picornaviruses. Nature (London) 317: 145–153 [DOI] [PubMed] [Google Scholar]
- Rossmann MG, Bernal R, Pletnev SV (2001) Combining electron microscopic with X-ray crystallographic structures. J Struct Biol 136: 190–200 [DOI] [PubMed] [Google Scholar]
- Rossmann MG, Moras D, Olsen KW (1974) Chemical and biological evolution of a nucleotide-binding protein. Nature (London) 250: 194–199 [DOI] [PubMed] [Google Scholar]
- Saigo K, Uchida H (1974) Connection of the right-hand terminus of DNA to the proximal end of the tail in bacteriophage lambda. Virology 61: 524–536 [DOI] [PubMed] [Google Scholar]
- Simpson AA, Leiman PG, Tao Y, He Y, Badasso MO, Jardine PJ, Anderson DL, Rossmann MG (2001) Structure determination of the head–tail connector of bacteriophage φ29. Acta Crystallogr D 57: 1260–1269 [DOI] [PubMed] [Google Scholar]
- Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG (2000) Structure of the bacteriophage φ29 DNA packaging motor. Nature (London) 408: 745–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli S, Desmyter A, Verrips CT, de Haard HJW, Moineau S, Cambillau C (2006) Lactococcal bacteriophage p2 receptor-binding protein structure suggests a common ancestor gene with bacterial and mammalian viruses. Nat Struct Mol Biol 13: 85–89 [DOI] [PubMed] [Google Scholar]
- Steinbacher S, Miller S, Baxa U, Budisa N, Weintraub A, Seckler R, Huber R (1997) Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 Å, fully refined structure of the endorhamnosidase at 1.56 Å resolution, and the molecular basis of O-antigen recognition and cleavage. J Mol Biol 267: 865–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbacher S, Seckler R, Miller S, Steipe B, Huber R, Reinemer P (1994) Crystal structure of P22 tailspike protein: interdigitated subunits in a thermostable trimer. Science 265: 383–386 [DOI] [PubMed] [Google Scholar]
- Stummeyer K, Dickmanns A, Mühlenhoff M, Gerardy-Schahn R, Ficner R (2004) Crystal structure of the polysialic acid-degrading endosialidase of bacteriophage K1F. Nat Struct Mol Biol 12: 90–96 [DOI] [PubMed] [Google Scholar]
- Tang L, Marion WR, Cingolani G, Prevelige PE Jr, Johnson JE (2005) Three-dimensional structure of the bacteriophage P22 tail machine. EMBO J 24: 2087–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Olson NH, Xu W, Anderson DL, Rossmann MG, Baker TS (1998) Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell 95: 431–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JO (1974) Chemical linkage of the tail to the right-hand end of bacteriophage lambda DNA. J Mol Biol 87: 1–9 [DOI] [PubMed] [Google Scholar]
- Tosi M, Anderson DL (1973) Antigenic properties of bacteriophage φ29 structural proteins. J Virol 12: 1548–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnquist S, Simon M, Egelman E, Anderson D (1992) Supercoiled DNA wraps around the bacteriophage φ29 head–tail connector. Proc Natl Acad Sci USA 89: 10479–10483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M (1987) Similarity measures between images. Ultramicroscopy 21: 95–100 [Google Scholar]
- Vlcek C, Paces V (1986) Nucleotide sequence of the late region of Bacillus φ29 completes the 19285-bp sequence of φ29 genome. Comparison with the homologous sequence of phage PZA. Gene 46: 215–225 [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE (2000) Topologically linked protein rings in the bacteriophage HK97 capsid. Science 289: 2129–2133 [DOI] [PubMed] [Google Scholar]
- Wommack KE, Colwell RR (2000) Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev 64: 69–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasbin RE, Maino VC, Young FE (1976) Bacteriophage resistance in Bacillus subtilis 168, W23, and interstrain transformants. J Bacteriol 125: 1120–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young FE (1967) Requirement of glucosylated teichoic acid for adsorption of phage in Bacillus subtilis 168. Proc Natl Acad Sci USA 58: 2377–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Greene B, Thuman-Commike PA, Jakana J, Prevelige PE Jr, King J, Chiu W (2000) Visualization of the maturation transition in bacteriophage P22 by electron cryomicroscopy. J Mol Biol 297: 615–626 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1A


