Abstract
POT1 (protection of telomere 1) is a highly conserved single-stranded telomeric binding protein that is essential for telomere end protection. Here, we report the cloning and characterization of a second member of the mouse POT family. POT1b binds telomeric DNA via conserved DNA binding oligonucleotide/oligosaccharide (OB) folds. Compared to POT1a, POT1b OB-folds possess less sequence specificity for telomeres. In contrast to POT1a, truncated POT1b possessing only the OB-folds can efficiently localize to telomeres in vivo. Overexpression of a mutant Pot1b allele that cannot bind telomeric DNA initiated a DNA damage response at telomeres that led to p53-dependent senescence. Furthermore, a reduction of the 3′ G-rich overhang, increased chromosomal fusions and elevated homologous recombination (HR) were observed at telomeres. shRNA mediated depletion of endogenous Pot1b in Pot1a deficient cells resulted in increased chromosomal aberrations. Our results indicate that POT1b plays important protective functions at telomeres and that proper maintenance of chromosomal stability requires both POT proteins.
Keywords: genomic instability, POT1b, telomere, telomerase
Introduction
Telomeres are specialized protein–DNA complexes that cap the ends of linear chromosomes and maintain genome stability by providing both end protection and a mechanism for complete replication of chromosomal termini. In mammals, telomeres consist of TTAGGG repetitive sequences that terminate in single-stranded (ss) 3′ G-rich overhangs that can be sequestered into lariat-like structures termed the t-loop (Griffith et al, 1999; Murti and Prescott, 1999; Nikitina and Woodcock, 2004). A model has been proposed in which this altered telomeric conformation is required to protect telomeres from being recognized as double-stranded DNA breaks (DSBs) that would otherwise activate DNA damage checkpoint responses (reviewed in de Lange, 2005). Failure to properly maintain telomere end capping functions results in dysfunctional telomeres that are processed by the DNA damage response pathways to initiate the onset of replicative senescence or fuel genomic instability associated with cancer cells (Karlseder et al, 1999; Celli and de Lange, 2005; Wong et al, 2006).
The telomeric G-rich overhang is evolutionarily conserved and is a substrate for ss DNA binding proteins TEBPαβ (telomere end binding protein) found in the ciliate Oxytricha nova (Price and Cech, 1987), Cdc13p from Saccharomyces cerevisiae (Garvik et al, 1996; Chandra et al, 2001) and protection of telomere 1 (POT1), first identified in S. pombe and are present in diverse organisms including plant, chicken, mouse and human (Baumann and Cech, 2001; Baumann et al, 2002; Wei and Price, 2004; Shakirov et al, 2005; Wu et al, 2006). All three proteins bind ss telomeric DNA through oligosaccharide/oligonucleotide binding motifs (OB-fold), a structurally conserved protein domain capable of binding to ss DNA with both high affinity and specificity (Mitton-Fry et al, 2002; Lei et al, 2003, 2004). While POT1, Cdc13p and TEBPαβ all use OB-folds to bind ss telomeric DNA, considerable diversity exists between the DNA binding domains of these proteins. Cdc13p uses only one OB-fold to bind DNA, while POT1 and TEBPαβ use two or more OB-folds for DNA binding (Price and Cech, 1987; Horvath et al, 1998; Lei et al, 2002, 2004; Trujillo et al, 2005). The importance of POT1 in mediating telomere end protection is further revealed by observations that deletion of SpPot1 results in catastrophic loss of telomeric DNA and cell death (Baumann and Cech, 2001). Survivors emerge with circularized chromosomes, obviating the need for the telomeric protective function of Pot1a. In addition, conditional knockout of the mouse Pot1a gene leads to chromosomal fusions and initiation of replicative senescence (Wu et al, 2006). These results indicate that POT1 is an integral telomere end protection protein, and that loss of POT1 results in dysfunctional telomeres that are targeted for chromosomal end-joining reactions.
POT1 is part of a complex of six core telomere-binding proteins termed the telosome (Liu et al, 2004b) or shelterin (de Lange, 2005). Both human and mouse POT1 interacts with the TPP1 protein through its C-terminal domain, which in turn interacts with the protein TIN2 (Kim et al, 2004; Liu et al, 2004a, 2004b; Ye et al, 2004a, 2004b; Wu et al, 2006). This protein complex can then interact with TRF1 or TRF2/Rap1 to form subcomplexes, although additional studies are required to determine the specific stochiometry of protein subunits (Kim et al, 2004; Liu et al, 2004b; Ye et al, 2004b; Yang et al, 2005). As the only ss telomere binding protein in the complex, POT1 serves a critically important role by transducing telomere length information to the TRF1 complex (Loayza and de Lange, 2003). Indeed, reduction of endogenous POT1 levels by siRNA knockdown (Liu et al, 2004a; Ye et al, 2004a) or elimination of POT1 protein by a conditional knockout approach (Wu et al, 2006) results in telomere length elongation. In addition, biochemical analyses using purified POT1 and telomerase demonstrated that POT1 negatively regulates telomerase activity in vitro by limiting its access to the terminal G residue of telomeres (Kelleher et al, 2005; Lei et al, 2005). These results are consistent with POT1's function as a negative regulator of telomere length.
Several organisms possess more than one Pot gene, and recent data in plants suggest that different POT proteins may perform different functions in telomere maintenance (Shakirov et al, 2005). In this study, we report the identification and characterization of POT1b, a second member of the mouse POT family. POT1b is highly homologous to POT1a and efficiently binds telomeric DNA in vitro via its conserved OB-folds. However, in contrast to POT1a, a truncated POT1b protein possessing only the OB-folds can efficiently localize to telomeres in vivo. Overexpression of a mutant allele of POT1b resulted in a potent DNA damage response at telomeres and the onset of a p53-dependent senescence phenotype. shRNA mediated depletion of Pot1b in the setting of Pot1 deficiency resulted in increased chromosomal aberrations. Our results indicate that POT1b plays important protective functions at telomeres and that proper maintenance of chromosomal stability requires both POT proteins.
Results
Cloning and sequence analysis of mouse Pot1b
While the human genome contains a single Pot1 gene, we identified two Pot1 orthologs in the mouse and rat genomes (Supplementary Figure 1; Wu et al, 2006). In this report, we focus on the characterization of mouse Pot1b. Pot1b encodes a protein 640 amino acids in length, is located on murine chromosome 17 and is highly homologous to mouse Pot1a and human Pot1 (Supplementary Figure 1A). Like POT1a, POT1b also possesses two OB-folds: OB1 is 74% identical to the corresponding regions in human POT1 and mouse POT1a, while OB2 is 73 and 81% identical to the corresponding regions in mouse POT1a and human POT1, respectively (Supplementary Figure 1A). Importantly, aromatic residues required for stacking interactions of telomeric ss DNA residues, including Phe31, Phe62, Tyr89 and Tyr223 (Lei et al, 2004), are evolutionarily conserved across mouse, rat and human POT proteins (Supplementary Figure 1B). In addition, the C-terminal portion of POT1b is 71 and 73% identical to the corresponding region in mouse POT1a and human POT1, respectively. RT–PCR analysis revealed that like Pot1a, Pot1b is ubiquitously expressed, detected in early embryonic stages and in all adult mouse tissues examined (Supplementary Figure 1C).
POT1b efficiently binds to 12 bp telomeric DNA
Biochemical and structural analyses have revealed that the POT1 OB-folds are critical for binding to telomeric substrates in vitro (Baumann and Cech, 2001; Baumann et al, 2002; Lei et al, 2003, 2004; Loayza and de Lange, 2003; Loayza et al, 2004; Wei and Price, 2004; Wu et al, 2006). Efficient binding of human POT1 requires ss telomeric DNA of 9–10 nucleotides that included the core telomeric sequence (T)TAGGGTTAG (Loayza et al, 2004; Lei et al, 2004). To determine the length of ss telomeric DNA substrate required to efficiently bind POT1b, we generated N-terminal Flag-tagged POT1b (POT1b N) containing both OB-folds (residues 1–341; Figure 2A). Pot1b N was translated in vitro, and electrophoretic mobility shift assays (EMSA) were performed with radiolabeled ss telomeric DNA substrates of 9–13 nucleotides containing the indicated permutations (Figure 1A). While some complex formation was observed when POT1b N was incubated with the 10-nucleotide core telomeric sequence, efficient binding of POT1b N to telomeric DNA required a minimum of 12 nucleotides (Figure 1A). In addition to the 10 bp core sequence, an additional two 5′ G residues greatly promoted binding, while the addition of three 5′ G residues diminished binding by ∼50% compared to the optimal 12-mer GGTTAGGGTTAG substrate (Figure 1A).
Figure 2.
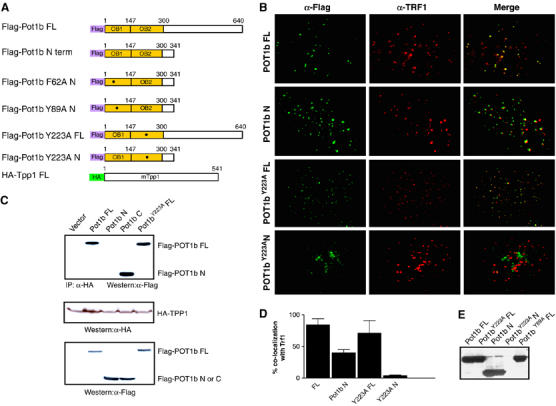
Biological properties of POT1b. (A) Schematic of N-terminal Flag-tagged full-length (FL) POT1B, POT1B N and derivative point mutants. (B) Co-localization of Flag-tagged FL POT1B, POUT1B N, FL POT1BY223A and POT1BY223A N with TRF1 in p53−/− MEFs. Growing cells were transiently infected with the indicated retroviral constructs, fixed and stained with mouse anti-Flag and rabbit anti-TRF1. Representative images are shown. (C) POT1b interacts with TPP1 via its C-terminal domain. Flag-tagged POT1B constructs were transfected into 293 T cells along with HA-tagged TPP1. IP-Western: IP pulldown and Western analysis were performed with the indicated antibodies. Western blots with anti-Flag and anti-HA antibodies were used to quantitate protein expression levels. (D) Quantitation of POT1b co-localization with TRF1 at telomeres. Error bars represent standard error of the mean (s.e.m.). (E) Anti-Flag Western blot demonstrating that MEF cell lines used in this report expressed the indicated transgenes at approximately equal levels.
Figure 1.
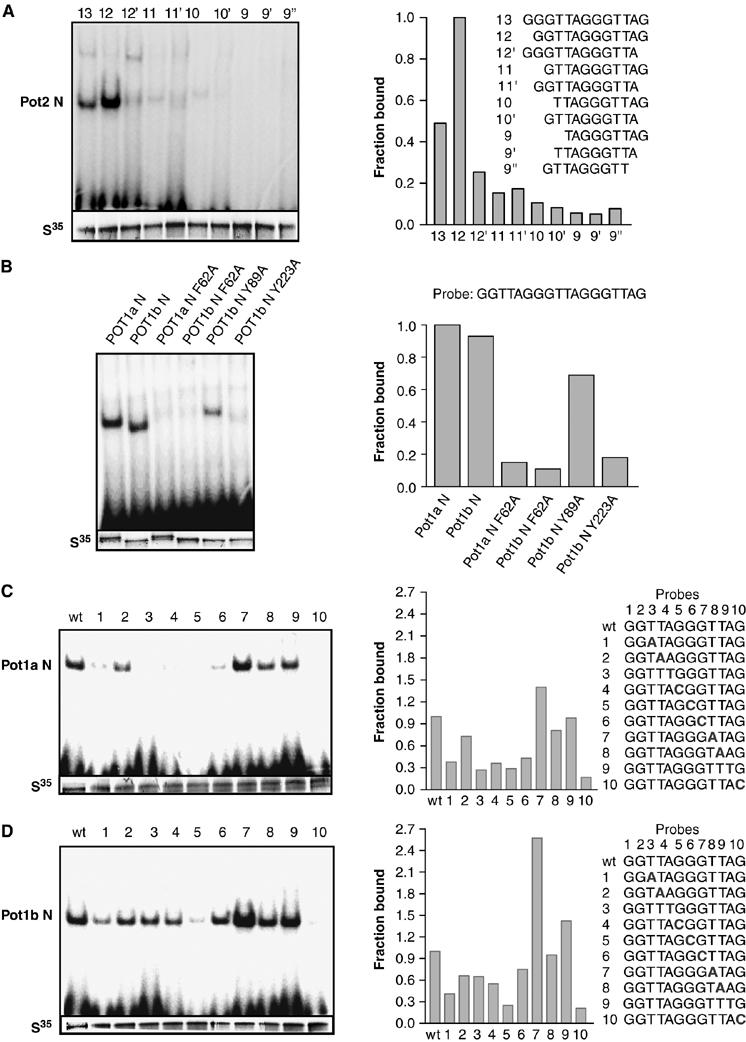
Characterization of POT1b interaction with ss telomeric DNA in vitro. (A) POT1b efficiently binds to a 12 base ss telomeric DNA sequence GGTTAGGGTTAG. In vitro translated POT1b (S35) was incubated with 32P-labeled telomeric oligonucleotides with the indicated nucleotide permutations and complexes were resolved by native PAGE. The fraction of POT1b bound to various telomeric substrates is plotted on the right, with POT1b binding to the 12-mer telomeric sequence set at 1.0. (B) Mutations with the OB-folds abolish POT1a and POT1b binding to ss telomeric DNA. POT1a N and POT1b N constructs bearing wild-type OB-folds or the indicated point mutations were incubated with the12-mer ss telomeric DNA sequence and subjected to gel mobility shift assays. The fraction of complex formed is plotted to the right. (C) Specificity of ss telomeric DNA binding to POT1a N and (D) POT1b N. Numbers indicate nucleotide position within the 12-base core tight-binding telomeric DNA sequence, in which one nucleotide at a time (in bold) was substituted with its complement. The fraction of complex formed is plotted on the right, with binding to wild-type telomeric sequence set at 1.0.
Crystal structural analysis of both S. pombe and human POT1 proteins identified a series of highly conserved aromatic residues within the OB-folds predicted to be important for binding to telomeric sequences (Lei et al, 2003, 2004). To test whether these amino-acid residues are required for POT1b binding to ss telomeric DNA, we generated Pot1b N constructs containing either wild-type OB-folds or a series of Pot1b N mutants possessing specific point mutations within OB1 (Phe62Ala and Tyr89Ala) and OB2 (Tyr223Ala) using site-directed mutagenesis (Figure 2A). Structural and genetic analyses predicted that these mutations should disrupt POT1b binding to ss telomeric DNA (Lei et al, 2003). Amino-acid residue Y223 within OB2 is of particular interest, because it is predicted from structural analysis to sequester the terminal G-residue of telomeric DNA, and access of this particular G-residue by telomerase appears to be critical for telomere elongation (Lei et al, 2004, 2005; Kelleher et al, 2005). EMSA performed with the 12-mer GGTTAGGGTTAG ss telomeric DNA confirmed that POT1a N and POT1b N possessing wild-type OB-folds efficiently bound ss telomeric DNA with similar affinities to form protein–DNA complexes (Figure 1B). In contrast, the F62A and Y223A substitutions within both POT1a N and POT1b N diminished complex formation by ∼80% (Figure 1B and data not shown), confirming their importance for binding to ss telomeric DNA. Interestingly, POT1bY89A N was able to bind ss telomeric DNA at reduced affinity, even though structural analysis predicted that Y89 is important for stacking interactions involving telomeric residues A3 and G4 (Lei et al, 2004). Taken together, these results support data from human POT1 crystal structural studies that evolutionarily conserved aromatic residues within the POT1b OB-folds are important for binding to ss telomeric DNA.
To identify nucleotides within the telomeric repeat that is critical for stable interaction with both mouse POT1a and POT1b, a series of 12 nucleotide ss telomeric DNAs containing single-nucleotide substitutions within the core telomeric sequence was evaluated for POT1a N and POT1b N binding via EMSA. Comparison of the mobility shift profiles between these two proteins revealed that POT1a N displayed greater specificity for telomeric sequences (Figures 1C and 2D). POT1b N was able to bind with at least 30% efficiency to almost all of the substituted ss telomeric DNA substrates, with the exception of a G to C substitution at residue 5, which almost completely abolished binding (Figure 1D). In contrast, substitutions at positions 1, 3, 4, 5 and 6 eliminated POT1a N binding to ss telomeric DNAs, a result reminiscent of the human POT1pN–ss telomeric DNA complex, where sequence specificity was greatest for nucleotides 2–5 (Figure 1C; Lei et al, 2004). A G to C substitution at position 10 of the telomeric substrate abolished ss telomeric DNA binding to both POT1a N and POT1b N, supporting previous results indicating that this 3′ terminal G10 residue is of critical importance for human POT1 binding to telomeres (Lei et al, 2004).
POT1b binds telomeres in vivo
Localization of human POT1 to telomeres in vivo is independent of its OB-folds but does require an intact C-terminus (Loayza and de Lange, 2003; Liu et al, 2004a). To determine whether POT1b localizes to telomeres in vivo, Flag-tagged Pot1b cDNA constructs encoding either full-length Pot1b, Pot1b N and derivative point mutants (Figure 2A) were transiently expressed in mouse embryo fibroblasts (MEFs) at approximately equal levels (Figure 2E). Indirect immunofluorescence revealed that cells expressing full-length Pot1b with wild-type OB-folds displayed a punctate staining pattern, in which ∼84% of POT1b containing foci co-localized with the telomere binding protein TRF1 (Figure 2B and D). In addition, full-length Pot1b constructs possessing either the F62A, Y89A or the Y223A point mutations also localized to telomeres. These results suggest that like mouse POT1a, functional OB-folds are not required for telomeric localization of POT1b in vivo (Figure 2B; Wu et al, 2006). Surprisingly, overexpression of Pot1b N in MEFs resulted in the formation of punctuate nuclear foci, ∼40% of which co-localized with TRF1 (Figure 2B and D). This result suggests that POT1b can localize to telomeres in vivo via only its OB-folds, in marked contrast to human POT1 and mouse POT1a, which requires an intact C-terminus for telomeric localization (Loayza and de Lange, 2003; Liu et al, 2004a; Wu et al, 2006). The telomere binding specificity of the POT1b OB-folds was further confirmed by the observation that the mutant POT1bY223A N protein cannot localize to telomeres in vivo (Figure 2B and D).
The protein TPP1 interacts with the C-terminus of POT1 to recruit it to telomeres (Houghtaling et al, 2004; Liu et al, 2004a; Ye et al, 2004a). To investigate whether POT1b also interacts with TPP1, we conducted co-immunoprecipitation experiments with full-length HA-tagged mouse Tpp1 and several Flag-tagged Pot1b constructs. Full-length POT1b, POT1bF62A, POT1bY89A and POT1bY223A mutant proteins possessing intact C-termini readily interacted with TPP1, while POT1b N did not (Figure 2C and data not shown). Direct interaction between full-length POT1b and TPP1was also detected with a yeast two-hybrid system (Table I). Finally, we demonstrate that both mouse POT1a and POT1b form a complex with TPP1 at telomeres in vivo (Supplementary Figure 2A and B). Taken together, these results suggest that while efficient recruitment of POT1b to telomeres in vivo requires an intact C-terminus, the OB-folds alone can target POT1b to telomeres.
Table 1.
Two-hybrid interaction of TPP1 with POT1b WT and POT1b mutants
| TPP1∷GAL4 activation domain (AD) | PPC86∷GAL4 AD | |
|---|---|---|
| Pot1b FL∷GAL4 DNA binding (DB) | Positive | |
| Pot1b F62A FL∷GAL4 DB | Positive | |
| Pot1b N∷GAL4 DB | Negative | |
| PDEST32 vector only | Negative | |
| PCL1GAL4 DB | Positive | |
| Interactions are determined qualitatively by comparing intensity of β-galactosidase staining against the PCL1Gal4-PPC86 positive control. |
MEFs expressing mutant Pot1b alleles exhibit p53-dependent senescence
To examine the role of POT1b at telomeres, we expressed full-length wild-type Pot1b, Pot1bY89A and Pot1bY223A mutant alleles in early passage MEFs. Expression of full-length Pot1b in p53+/+ MEFs minimally perturbed cellular proliferation. In contrast, overexpression of mutant Pot1b alleles resulted in rapid entry (within 2 days) into a phenotype resembling replicative senescence (enlarged cells with a highly vacuolated cytoplasm) that was accompanied by a four- to six-fold induction in the number of senescence-associated (SA)-β-galactosidase positive cells and an ∼4-fold decline in the number of BrdU positive cells (Figure 3A and B and data not shown). This premature entry into senescence was rescued in a p53−/− background (Figure 3C), suggesting that it may be due to increased DNA damage initiated by the mutant Pot1b alleles. We noticed that p53−/− MEFs expressing full-length Pot1bY223A exhibited a slower growth rate compared to cells expressing other mutant Pot1b alleles, with an ∼2-fold reduction in the number of BrdU positive cells (Figure 3D). This decline in cellular growth correlated with an ∼15-fold increase in the number of anaphase bridges observed in DAPI-stained cells expressing full-length Pot1bY223A over vector expressing controls (Figure 3E). Anaphase bridges, a hallmark of chromosomal end-to-end fusions, were also elevated in MEFs expressing full-length Pot1bY89A (Figure 3E). In contrast, anaphase bridges were rare in cells expressing full-length Pot1b, Pot1b N or Pot1b N mutants (Figure 3E). Finally, overexpression of mutant Pot1b alleles did not appear to adversely influence overall telomere length (see below). Taken together, these data suggest that mutant Pot1b alleles perturb telomere capping function to initiate the formation of altered chromosomes, which promotes growth arrest in the setting of an intact p53 pathway.
Figure 3.
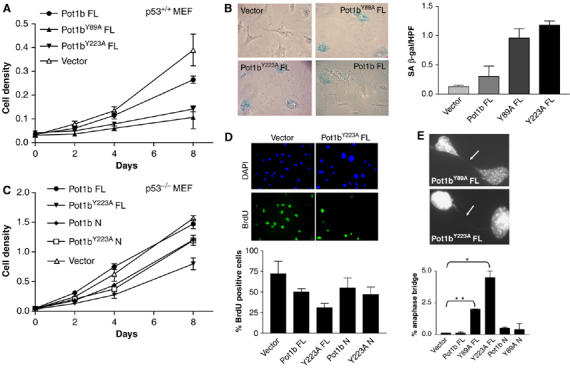
Expression of mutant Pot1b alleles in p53+/+ MEFs results in premature senescence. (A) Growth curve of p53+/+ MEFs expressing FL Pot1b, FL Pot1bY89A, FL Pot1bY223A and vector control. Error bars represent s.e.m. (B) Prominent SA-β-gal staining observed in MEFs expressing FL Pot1b mutant alleles. (C) Growth curve of p53−/− MEFs expressing the indicated constructs. Error bars represent s.e.m. (D) BrdU incorporation of p53−/− MEFs expressing indicated Pot1b constructs. Representative images are shown. Error bars represent s.e.m. (E) Elevated number of anaphase bridges (arrows) observed in p53−/− MEFs expressing FL Pot1bY89A and FL Pot1bY223A. Anaphase bridges are expressed as a per cent of total anaphases observed. Error bars represent s.e.m. *P<0.01; **P<0.03.
Elevated DNA damage response and chromosomal aberrations in MEFs expressing full-length Pot1bY223A
The induction of a phenotype resembling replicative senescence in p53+/+ MEFs expressing mutant Pot1b alleles prompted us to examine whether a DNA damage response is activated at telomeres to initiate the onset of replicative senescence (d'Adda di Fagagna et al, 2003; Takai et al, 2003). Dysfunctional telomeres are detectable by the telomere dysfunction induced foci (TIF) assay that monitors telomeric association of DNA damage proteins such as γH2AX and 53BP1 (d'Adda di Fagagna et al, 2003; Takai et al, 2003; Celli and de Lange, 2005). 53BP1 associated TIFs were minimal in p53−/− MEFs expressing full-length Pot1b and Pot1bY223A N (Figure 4A and B). In contrast, expression of full-length Pot1bY223A and Pot1b N induced at least eight TIFs in ∼50% and ∼20% of cells examined, respectively, suggesting a robust induction of the DNA damage response at telomeres by these mutant proteins (Figure 4A and B). TIF formation was accompanied by the accumulation of other DNA damage markers, including ATM phospho-Ser 1981 and phospho-Chk2 in cells expressing full-length Pot1bY223A, and to a lesser extent in Pot1b N expressing cells (Figure 4C).
Figure 4.
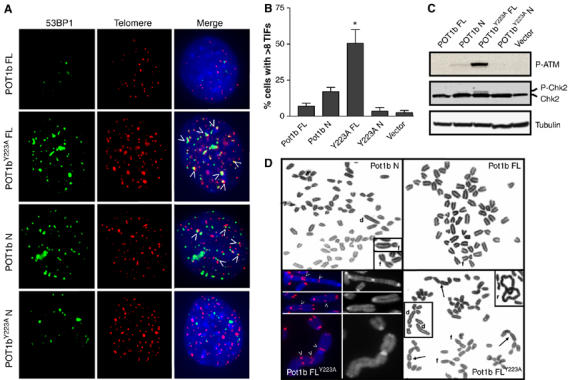
Elevated DNA damage response and chromosomal aberrations in MEFs expressing mutant Pot1b alleles. (A) Co-localization of DNA damage response foci to telomeres in p53−/− MEFs stably expressing the indicated constructs. 53BP1 was detected with anti-53BP1 antibody (FITC, green) and telomeres were detected by telomere PNA-FISH (TRITC, red). (B) Quantitation of co-localization of 53BP1 with telomeres in p53−/− MEFs stably expressing the indicated constructs. Percentage of cells possessing >8 TIFs are indicated. A total of 100 nuclei were scored in two separate experiments. Error bars represent s.e.m. *P<0.01 (Student's t-test). (C) Immunoblots detecting the phosphorylation status of ATM and Chk2 in MEF cells expressing the indicated constructs. (D) Chromosomal aberrations are prominent in p53−/− MEFs expressing FL Pot1bY223A and Pot1b N. Arrows point to tandem chromosomal translocations, and telomere PNA-FISH characterized the majority of these fusions as possessing TTAGGG-repeats at the sites of fusion (arrowheads). r: ring chromosomes; f: p–p arm chromosomal fusions, d: dicentric chromosomes.
In accord with the presence of dysfunctional telomeres in cells expressing full-length Pot1bY223A, multiple chromosomal aberrations including p–p and p--g chromosomal arm fusions, dicentrics, fragments and rings were observed in cells expressing this mutant allele (Figure 4D; Supplementary Table 1). On average, there were 1.8 fusions per metaphase, and several cells showed tandem fusions of 4–5 chromosomes that were never observed in MEFs expressing full-length wild-type Pot1b or Pot1b N (Figure 4D; Supplementary Table 1). Telomere PNA FISH characterized the majority of these fusions (82%) as possessing abundant TTAGGG repeats at the site of fusion. Overexpression of Pot1b N also generated fused chromosomes, although at a reduced level (Figure 4D; Supplementary Figure 3 and Supplementary Table 1). These results are consistent with the notion that a subset of telomeres is rendered dysfunctional by overexpression of mutant POT1b alleles, resulting in end-to-end chromosomal fusions (Figure 4D; Supplementary Table 1).
Reduction of 3′ G-rich telomere overhang in MEFs expressing mutant Pot1b alleles
Previous studies have indicated that the extensive end-to-end chromosomal fusions seen in cells expressing mutant TRF2 are due to loss of the terminal 3′ G-rich overhang (van Steensel et al, 1998; Celli and de Lange, 2005). Since POT1b readily binds to ss telomeric DNA in vitro, we ascertained the status of the 3′ G-rich overhang in MEFs expressing wild-type or mutant Pot1b alleles. A quantitative 3′ overhang-specific hybridization protection assay (HPA) revealed that MEFs expressing full-length Pot1bY223A exhibited ∼60% reduction in the amount of G-rich overhang (Figure 5A; Tahara et al, 2005). The G-rich overhang was also slightly reduced (∼18%) in MEFs expressing Pot1b N. In contrast, overhang length did not change appreciably in MEFs expressing full-length Pot1b and Pot1bY223A N. The G-strand overhang was almost completely removed following treatment with Exonuclease I, demonstrating that the ss G-rich repeats originated from the 3′ end of telomeres (Figure 5A). Consistent with the robust telomeric signals detected by PNA FISH in MEFs expressing all Pot1b constructs (Figure 4A), HPA revealed that overall telomere length remained unchanged in cells expressing mutant Pot1b alleles (Figure 5B). Taken together, these results suggest that expression of full-length Pot1bY223A (and to a lesser extent Pot1b N) resulted in a reduction of the 3′ G-overhang at telomeres, leading to telomere deprotection and end-to-end chromosomal fusions that are most likely mediated by the non-homologous end-joining (NHEJ) pathway (Smogorzewska et al, 2002; Celli and de Lange, 2005).
Figure 5.
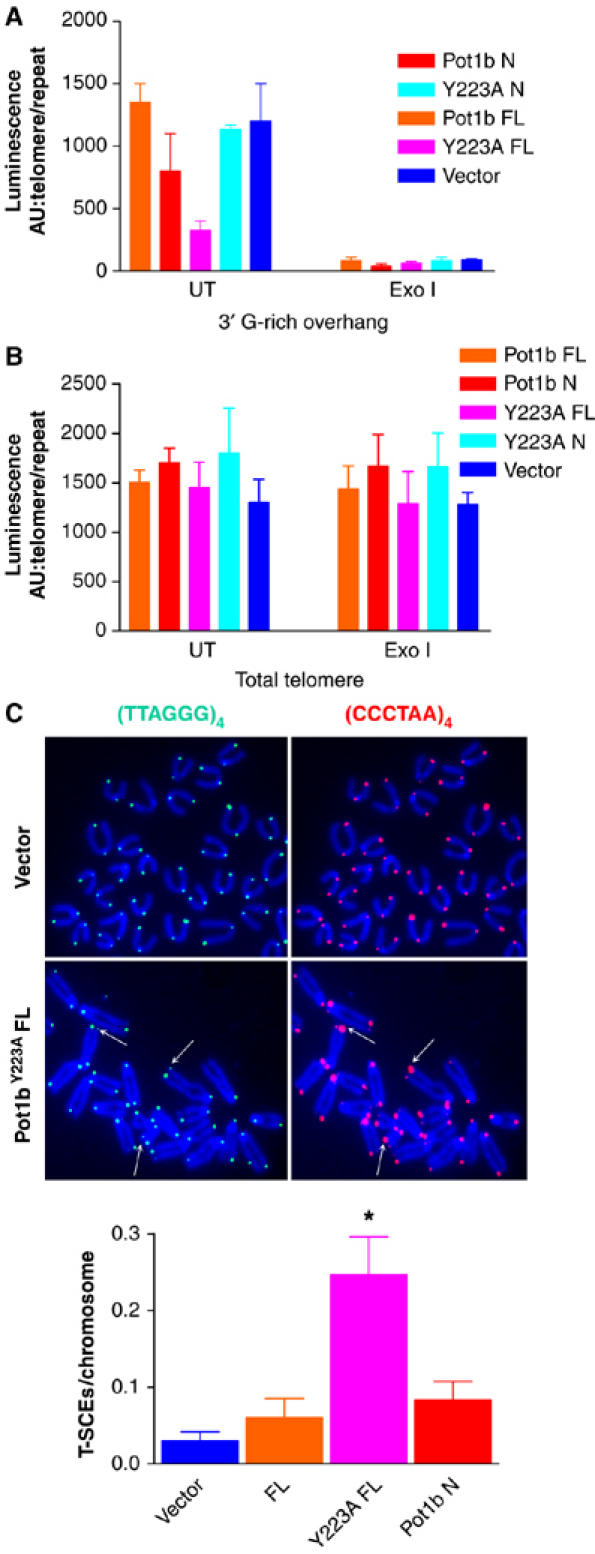
Reduction of 3′ G-rich telomere overhang and elevated T-SCEs in MEFs expressing mutant Pot1b alleles. (A) HPA using telomeric probes was performed on genomic DNA isolated from p53−/− MEFs (PD 4) expressing the indicated constructs to assess the length of the 3′ G-rich overhang and total telomeric DNA (B). UT: untreated; ExoI: genomic DNA were treated with ExoI to remove the 3′ G-rich overhang. Luminescence intensity in arbitrary units (AU) was normalized against mouse repetitive DNA A1a. (C) CO-FISH revealed elevated T-SCE in p53−/− MEFs expressing full-length Pot1bY223A (arrows). Telomeres were labeled with Tam-OO-[CCCTAA]4 PNA probe (red) to detect lagging strand or FITC-OO-[TTAGGG]4 probe (green) to detect leading strand telomeres. (D) Quantification of T-SCE in p53−/− MEFs of the indicated genotypes. A minimum of 750 chromosomes was scored per genotype. Error bars represent s.e.m. *P<0.01 (Student's t-test).
POT1b represses homologous recombination (HR) at telomeres
Formation of the t-loop is postulated to prevent engagement of telomeric termini by the NHEJ pathway by sequestering the 3′ G-rich overhang. However, a t-loop resembles a Holiday junction (HJ), a structure characteristic of substrates undergoing HR. Inappropriate HR at telomeres can promote rapid deletion of telomeres, resulting in telomere shortening and premature entry into senescence (Lustig, 2003; Wang et al, 2004). The elevated number of chromosomal aberrations characteristic of telomere HR in Pot1 deficient MEFs (Wu et al, 2006) prompted us to assess whether POT1b has a role in repressing HR at telomeres. We utilized Chromosome Orientation (CO)-FISH to assess the frequency of HR between telomeres of sister chromatids (telomere sister chromatid exchanges, T-SCEs) (Bailey et al, 2001). CO-FISH typically yields a characteristic pattern of two telomere signals, one on each end of the chromatid marking either lagging or leading strands, depending on the probe used for hybridization. However, a T-SCE event within telomeric DNA results in a three- (or four-) telomere hybridization signals of unequal intensity. T-SCEs were rare in MEFs expressing full-length Pot1b and Pot1b N (Figure 5C). In contrast, MEFs expressing full-length Pot1bY223A exhibited ∼5-fold increase in both leading- and lagging-strand T-SCE over controls (Figure 5C and D). In addition, telomere PNA FISH also detected additional cytogenetic features characteristic of aberrant telomere HR, including the presence of telomeric DNA-containing Double Minute (TDM) chromosomes (Supplementary Figure 3A and Supplementary Table 1). TDMs are proposed to result from aberrant HR events between telomeric sequences and intrachromosomal telomeric repeats (Zhu et al, 2003; Laud et al, 2005). These results suggest that POT1b plays a role in repressing aberrant HR at telomeres.
Both POT1a and POT1b are required to maintain chromosomal stability
Conditional deletion of Pot1a results in elevated chromosomal instability and aberrant HR at telomeres (Wu et al, 2006). To determine whether depletion of Pot1b affected chromosomal stability, we utilized retroviral delivery of Pot1b shRNAs into MEFs. We identified two Pot1b shRNAs, which reduced Pot1b expression by >90% without affecting Pot1a levels (Figure 6A and data not shown). Compared to vector controls, shRNA-Pot1b-1 and 3 depleted cells exhibited ∼15-fold increase in cytogenetic aberrations, including chromosomal fusions without telomeres at the sites of fusion, isochromatid rings resulting from fusions of sister chromatids and TDMs (Figure 6B and D; Supplementary Table 1 and data not shown). Comparison of these chromosomal aberrations to those detected in MEFs expressing full-length Pot1bY223A revealed that the number of fused chromosomes possessing TTAGGG repeats at the site of fusion is ∼3-fold higher in MEFs expressing the mutant Pot1b allele (Figure 6D). Finally, we asked whether both POT1a and POT1b are required to maintain chromosomal stability. shRNA-Pot1b mediated depletion of Pot1b in Pot1aΔ/Δ, p53−/− MEFs resulted in an increase in the number of TDMs (1.8 per metaphase) and isochromatid rings (3.1 per metaphase) observed over shRNA-Pot1b depleted MEFs (1.3 TDMs and 1.9 isochromatid rings per metaphase) and Pot1aΔ/Δ, p53−/− MEFs (1.1 TDM and 0.45 isochromatid rings per metaphase) (Figure 6D; Wu et al, 2006). These results suggest that both POT1a and POT1b are required to maintain chromosomal stability.
Figure 6.
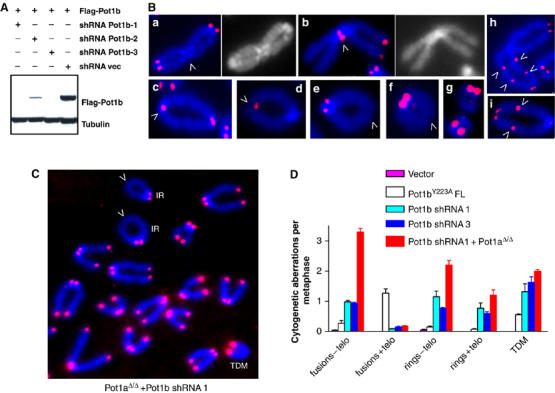
Depletion of Pot1b results in multiple cytogenetic aberrations. (A) shRNA mediated knockdown of Pot1b in p53−/− MEFs. p53−/− MEFs expressing Flag-tagged Pot1b were transfected with pSUPER control (shRNA vec) or pSUPER-Pot1b shRNAs 1, 2 and 3 (shRNA Pot1b 1, 2, 3). Flag-POT1b was detected by Western. (B) Knockdown of Pot1b induced multiple chromosomal aberrations, including p–p arm fusions without TTAGGG repeats at fusion sites (a, arrowhead); p–p arm fusions with TTAGGG repeats at fusion sites (b, arrowhead); isochromatid rings with TTAGGG repeats at the sites of fusion (c, d, arrowhead indicates site of fusion); isochromatid rings without TTAGGG repeats at sites of fusion (e, f, arrowheads); TDMs (g); and dicentric chromosomes with TTAGGG repeats at multiple sites of fusion (h, j, arrowheads). (C) Depletion of both Pot1 and Pot1b result in increased HR at telomeres. TDM: telomere double minute chromosomes, IR: isochromatid rings. (D) Quantification of cytogenetic aberrations of the indicated genotypes. N=164 total metaphases scored. Error bars represent s.e.m.
Discussion
The data presented here indicate that POT1b, ss telomere binding protein, plays a role in repressing both NHEJ and HR at telomeres. The presence of two POT proteins in the mouse genome suggests that they may mediate distinct roles at telomeres. Our data suggest that POT1a negatively regulates telomere length (Wu et al, 2006), while POT1b plays a role in maintaining the 3′ G-rich overhang.
Role of POT1b in telomere end protection
Inappropriate repair of dysfunctional telomeres could adversely impact cell survival by eliciting apoptosis or cell cycle arrest in p53 competent cells (Karlseder et al, 1999; Celli and de Lange, 2005). Dysfunctional telomeres can be processed as double-stranded DNA breaks by the NHEJ machinery, in which Ligase IV-mediated joining of ends create fused chromosomes with TTAGGG sequences at the site of fusion (van Steensel et al, 1998; Smogorzewska et al, 2002; Celli and de Lange, 2005). NHEJ-mediated chromosomal fusions are inhibited by the presence of the 3′ G-rich overhang, and its removal promotes chromosomal end-joining (Zhu et al, 2003; Celli and de Lange, 2005). Repression of inappropriate NHEJ at telomeres requires formation of the telosome/shelterin complex, since deletion of specific components in this complex (TRF2 and POT1) results in NHEJ-mediated chromosomal fusions (van Steensel et al, 1998; Celli and de Lange, 2005; Wu et al, 2006). Since POT1 sequesters the terminal G10 residue of 3′ G-rich overhang into a deep binding pocket, it is predicted to be essential for telomere end protection (Lei et al, 2004). Additional evidence that POT1 is required for the formation of a protective telomeric structure comes from a recent study indicating that telomeres become transiently deprotected during the G2 phase of the cell cycle after partial loss of telomeric POT1 (Verdun et al, 2005).
Our data are consistent with the notion that a second member of the mouse POT family, POT1b, is also required for telomere end protection. POT1b preferentially binds to the 5′GGTTAGGGTTAG3′ sequence and its OB-folds are sufficient to target it to telomeres. Overexpression of mutant Pot1b alleles results in the activation of a robust ATM-dependent DNA damage response at telomeres that initiates a p53-dependent cell cycle arrest despite the fact that double-stranded telomeric repeats remain relatively constant. In the absence of p53, this senescent phenotype is bypassed, with elevated anaphase bridge formation and accumulation of end-to-end chromosomal fusions with TTAGGG sequence at the site of fusion. Mutant POT1b proteins might exert their effect by directly perturbing endogenous POT1b function at telomeres. For example, POT1b N could potentially compete with endogenous POT1b for access to the 3′ G-rich overhang. On the other hand, full-length POT1bY223A could inhibit access of endogenous POT1b to the telosome/shelterin complex by binding TPP1. Another possibility is that overexpression of mutant POT1b alleles may disrupt POT1b's interaction with other members of the telosome/shelterin complex. The long tandemly fused chromosomes observed in cells expressing the mutant Pot1bY223A allele is reminiscent of the type of chromosomal fusions observed in human cells expressing Trf2ΔBΔM and in MEFs null for Trf2 (van Steensel et al, 1998; Celli and de Lange, 2005), suggesting that full-length POT1bY223A may disrupt TRF2-POT1b interaction at telomeres. While a functional interaction between TRF2 and POT1b remains to be determined, TRF2 interacts with human POT1 at telomeres (Yang et al, 2005).
Although POT1b represses NHEJ at telomeres, this activity is minor compared to TRF2's role in preventing NHEJ-mediated chromosomal fusions (Celli and de Lange, 2005). This result suggests that POT1b may have additional functions at telomeres. We speculate one way that cells can inhibit inappropriate NHEJ at telomeres is to adopt a t-loop structure at the ends of chromosomes to sequester the 3′ ss overhang. However, a t-loop also resembles a HR substrate. In normal cells, inappropriate HR at telomeres must be repressed, since branch migration of the 3′ overhang in a t-loop can create an HJ that can be cleaved by HJ resolvase to initiate telomere deletion (Lustig, 2003; Wang et al, 2004). Recently, we demonstrated that POT1a plays a role in repressing inappropriate HR at telomeres (Wu et al, 2006). In cells deficient in Pot1a, multiple chromosomal aberrations characteristic of telomere HR were present. Here, we extend this observation to POT1b, and show that shRNA-mediated depletion of Pot1b also resulted in elevated telomere HR. Cytogenetic abnormalities characteristic of telomere HR, including TDMs, isochromatid rings and elevated T-SCEs are elevated in Pot1b depleted cells and in cells expressing mutant Pot1b alleles. In addition, depletion of both Pot1a and Pot1b led to increased chromosomal aberrations, suggesting that both POT proteins are required to repress inappropriate NHEJ and HR at telomeres.
Separation of function in mouse POT proteins
The observation that there are two POT proteins encoded by the mouse genome while there is only one in human suggests that mouse POT proteins may perform separate functions at telomeres. This notion is supported from recent data indicating that Arabidopsis has two POT1-like proteins, one which is involved in telomere length regulation and the other involved in chromosome end protection (Shakirov et al, 2005). We recently demonstrated that mouse POT1a is required for telomere length homeostasis, since conditional deletion of Pot1a in MEFs resulted in telomere length elongation. In addition, deletion of Pot1a correlated with an increase in the 3′ G-rich overhang, suggesting that it may regulate a C-strand nuclease (Wu et al, 2006). In contrast, data presented here suggest that POT1b is required for maintenance of the 3′ G-rich overhang and has no appreciable role in telomere length regulation. Both proteins are required to repress NHEJ and telomere HR. Taken together, these results suggest that the two mouse POT proteins may have both distinct and overlapping functions in mediating telomere end protection. This observation is interesting in light of a recent report suggesting that telomeres exist in extendible and non extendible states, and that telomerase acts preferentially on the shortest telomeres (Teixeira et al, 2004). POT1a may negatively regulate telomere length by inhibiting telomerase access to the terminal G-residue in long telomeric substrates while promoting telomere extension by enabling telomerase to access short telomeric ends (Kelleher et al, 2005; Lei et al, 2005). POT1b would serve to protect the unfolded 3′ overhang from exposure to degrading nucleases, and given that its OB-folds can directly bind to telomeres in vivo, it is possible that this end protective function may not require other components of the telosome. It will be of interest to determine whether subcomplexes containing varying ratios of the POT proteins exist on telomeres at different phases of the cell cycle to modulate telomere elongation and protection.
Materials and methods
Cloning of murine Pot1b, site-directed mutagenesis and expression analysis
Based on cDNA XM_355022, we used a PCR strategy to clone mouse Pot1b cDNA in frame with a 5′ Flag epitope tag, inserted it into the KS plasmid (Stratagene) and fully sequenced. Pot1b deletion constructs were generated by PCR and the sequenced inserts subcloned into pcDNA3.1 (Clonetech) and pQCXIP (Novex) vectors. N-terminal POT1b consisted of aa 1–341. Site-directed mutagenesis was performed according to the manufacturer's instructions (Strategene), in which phenylalanine at position 62 and tyrosine at position 89 and tyrosine at position 223 were individually mutated into alanines. First-strand cDNA panels (Clontech) were used to detect Pot1b expression. Transcripts were amplified using AdvanTaq 2 PCR Kit (Clontech) and G3PDH was amplified under the same conditions following manufacturer's recommendations.
Cell culture, immunolocalization studies and microscopy
p53+/+ and p53−/− MEFs (passage 1–3) and 293 T cells were maintained in Dulbecco's modified Eagle's medium and cultured at 3% oxygen to minimize premature entry into senescence (Parrinello et al, 2003). For immunolocalization studies, Pot1b constructs subcloned in the retroviral vector pQCXIP were infected into rapidly growing early passage MEFs, either wild type or null for p53. Infected cells were plated onto eight-well chambers and harvested 24–48 h after infection. Immunofluorescence was performed as described (Laud et al, 2005) and POT1b was localized with anti-Flag antibody (Sigma) at 1:10 000. Polyclonal antibody against mTRF1 (a generous gift from Jan Karlseder, Salk Institute) was used at 1:2000. Images were acquired on a Nikon Eclipse 800 with × 63 and × 100 plan-apo objectives and photographed with a cooled CCD camera. CCD chip nonlinearities were removed by taking bias and dark current frames and the optical train flat fielded to eliminate vignetting. Individual raw images were taken through DAPI, Rhodamine and FITC narrowband filters and stacked in MetaMorph and PhotoShop CS were utilized to compose the final images. The same amount of linear histogram stretch was applied to all raw images before stacking to maintain the same degree of contrast enhancement in all images.
In vitro translation, gel shift and immunoprecipitation assays
Pot1b constructs under the control of the T7 expression vector were translated in vitro with the TNT coupled reticulocyte lysate kit (Promega) under the manufacturer's recommended conditions. DNA binding assays were performed essentially as described (Baumann et al, 2002) and fractionated at 4°C on a 4–20% polyacrylamide Tris–borate EDTA gel. The gels were dried, and radiolabeled ssDNA was visualized by exposure to a PhosphorImager. To determine the interaction between mouse POT1b and TPP1, we cloned the Tpp1 cDNA based on cDNA gi:22823923 (Liu et al, 2004a), in frame with a 5′ HA epitope tag and verified the construct by sequencing. Various Flag-tagged Pot1b constructs and full-length HA-Tpp1 were co-transfected into 293T cells, and IP performed as described (Ye et al, 2004a).
Yeast two-hybrid analysis
Yeast two-hybrid was performed using Proquest Two-Hybrid System with Gateway Technology (Invitrogen), according to the manufacturer's protocol. Mouse Tpp1 was cloned into pDESt22 to generate a fusion protein with the yeast GAL4 activation domain, while full-length Pot1b, full-length Pot1bF62A and Pot1b N constructs were cloned into pDEST32 to generate a fusion protein with the GAL4 DNA binding domain using Gateway LR Recombination Systems (Invitrogen). Proteins interactions were tested using β-Gal Filter Assay, as described (Wu et al, 2006).
Western blot analysis, growth analysis, SA β-gal assay and anaphase bridge determination
The antibodies used for Western analyses were phospho-ATM Ser 1981 (Cell Signalling; 1:1000) and Chk2 (BD Biosciences, 1: 500). Anti-mouse γ-tubulin (Sigma, 1:10 000) was used as a loading control. For growth curves, 4 × 104 stably infected MEFs were plated in triplicate into six-well plates. At 0, 2, 4, 8 days post plating, cells were fixed and stained with crystal violet. SA β-gal assay was performed as described (Dimri et al, 1995). BrdU incorporation was performed essentially as described (Smogorzewska et al, 2002). Anaphase bridges were quantitated from a minimum of 200 metaphase plates undergoing anaphase and determining percentage possessing a distinct chromatin bridge after DAPI staining.
G-tail HPA
HPA was performed as described (Tahara et al, 2005) and 1 μg non-denatured genomic DNA and 0.5 μg heat-denatured genomic DNA was used per assay for the detection of telomere 3′ overhangs and total telomere DNA, respectively. To check specificity of G-tail detection, non-denatured genomic DNA was treated with Exonuclease I (30 U/μg DNA) at 37°C overnight, and heat inactivated at 80°C for 20 min, before G-tails were assayed.
CO-FISH analysis for T-SCEs
CO-FISH has been described in detail previously (Bailey et al, 2001). Hybridization of metaphase spreads was performed with TRITC-OO-(TTAGGG)4 or FITC-OO-(CCCTAA)4 peptide nucleic acid probes (Applied Biosystems). For simultaneous visualization of both leading and lagging strands, both probes were used at a concentration of 0.5 μg/ml. For CO-FISH, a minimum of 750 chromosome ends were scored blinded for each genotype, and pairwise comparisons for statistical significance were made by Student's t-test. Differences between genetic backgrounds are considered significant only when P-values were less than 0.01. Images were captured with Metamorph Premiere (Molecular Devices) and processed with Metamorph and Adobe Photoshop CS.
shRNA interference
Three shRNA-Pot1b were generated in pSuper as described (Deng et al, 2003). To generate pRetro-Super constructs, EcoR1- and XhoI-digested insert from pSuper was subcloned into the same site into pRetro-Super vector (Brummelkamp et al, 2002). The target sequence for mouse Pot1b is: shRNA–Pot1b-1: position 372 to 390 (5′CTTCACTGCTCAGGAC TAC3′); shRNA-Pot1b-2: position 623641 (5′GTCACATTCTGCAGCTACA3′); shRNA–Pot1b-3: position 962–980 (5′GCTCTGAATCAGACCTAGT3′)
Note added in proof:
Cloning and characterization of Pot1b was recently reported by Hockemeyer et al (2006) Cell 126: 63–77.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2A, 2B
Supplementary Figure 3
Supplementary Table 1
Acknowledgments
We are grateful to Jan Karlseder for providing TRF1 antibody, and Philip Carpenter for the 53BP1 antibody. We acknowledge the TC Hsu Molecular Cytogenetics Core for outstanding cytogenetic services (NCI No. CA016672). We thank members of the Chang lab for helpful comments. SC acknowledges generous financial support from the Welch Foundation, the Elsa U Pardee Foundation, the Sidney Kimmel Foundation for Cancer Research, the Abraham and Phyllis Katz Foundation, and the Michael Kadoorie Cancer Genetic Research Program.
References
- Bailey SM, Cornforth MN, Kurimasa A, Chen DJ, Goodwin EH (2001) Strand-specific postreplicative processing of mammalian telomeres. Science 293: 2462–2465 [DOI] [PubMed] [Google Scholar]
- Baumann P, Cech TR (2001) Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292: 1171–1175 [DOI] [PubMed] [Google Scholar]
- Baumann P, Podell E, Cech TR (2002) Human Pot1 (protection of telomeres) protein: cytolocalization, gene structure, and alternative splicing. Mol Cell Biol 22: 8079–8087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553 [DOI] [PubMed] [Google Scholar]
- Celli GB, de Lange T (2005) DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol 7: 710–712 [DOI] [PubMed] [Google Scholar]
- Chandra A, Hughes TR, Nugent CI, Lundblad V (2001) Cdc13 both positively and negatively regulates telomere replication. Genes Dev 15: 404–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T (2005) Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19: 2100–2110 [DOI] [PubMed] [Google Scholar]
- d'Adda di Fagagna FD, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, von Zglinicki T, Saretzki G, Carter NP, Jackson SP (2003) A DNA damage checkpoint response in telomere-initiated senescence. Nature 426: 194–198 [DOI] [PubMed] [Google Scholar]
- Deng Y, Ren X, Yang L, Lin Y, Wu X (2003) A JNK-dependent pathway is required for TNFalpha-induced apoptosis. Cell 115: 61–70 [DOI] [PubMed] [Google Scholar]
- Dimri GP, Lee XH, Basile G, Acosta M, Scott C, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereirasmith O, Peacocke M, Campisi J (1995) A biomarker that identifies senescent human-cells in culture and in aging skin in-vivo. Proc Natl Acad Sci USA 92: 9363–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvik B, Carson M, Hartwell L (1996) Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol 15: 6128–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T (1999) Mammalian telomeres end in a large duplex loop. Cell 97: 503–514 [DOI] [PubMed] [Google Scholar]
- Hemann MT, Greider CW (1999) G-strand overhangs on telomeres in telomerase-deficient mouse cells. Nucl Acids Res 27: 3964–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Daniels JP, Takai H, de Lange T (2006) Recent expansion of the telomeric complex in rodents: two distinct POT1 proteins protect mouse telomeres. Cell 126: 63–77 [DOI] [PubMed] [Google Scholar]
- Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC (1998) Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell 95: 963–974 [DOI] [PubMed] [Google Scholar]
- Houghtaling BR, Cuttonaro L, Chang W, Smith S (2004) A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr Biol 14: 1621–1631 [DOI] [PubMed] [Google Scholar]
- Karlseder J, Broccoli D, Dai Y, Hardy S, De Lange T (1999) p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science 283: 1321–1325 [DOI] [PubMed] [Google Scholar]
- Kelleher C, Kurth I, Lingner J (2005) Human protection of telomeres 1 (POT1) is a negative regulator of telomerase activity in vitro. Mol Cell Biol 25: 808–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Beausejour C, Davalos AR, Kaminker P, Heo SJ, Campisi J (2004) TIN2 mediates functions of TRF2 at human telomeres. J Biol Chem 279: 43799–43804 [DOI] [PubMed] [Google Scholar]
- Laud PR, Multani AS, Bailey SM, Wu L, Ma J, Kingsley C, Lebel M, PathakS, DePinho RA, Chang S (2005) Elevated telomere–telomere recombination in WRN-deficient, telomere dysfunctional cells promotes escape from senescence and engagement of the ALT pathway. Genes Dev 19: 2560–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Baumann P, Cech TR (2002) Cooperative binding of single-stranded telomeric DNA by the Pot1 protein of Schizosaccharomyces pombe. Biochemistry 41: 14560–14568 [DOI] [PubMed] [Google Scholar]
- Lei M, Podell ER, Baumann P, Cech TR (2003) DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature 426: 198–203 [DOI] [PubMed] [Google Scholar]
- Lei M, Podell ER, Cech TR (2004) Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol 11: 1223–1229 [DOI] [PubMed] [Google Scholar]
- Lei M, Zaug AJ, Podell ER, Cech TR (2005) Switching human telomerase on and off with hPOT1 protein in vitro. J Biol Chem 280: 20449–20456 [DOI] [PubMed] [Google Scholar]
- Liu D, O'Connor MS, Qin J, Songyang Z (2004b) Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J Biol Chem 279: 51338–51342 [DOI] [PubMed] [Google Scholar]
- Liu D, Safari A, O'Connor MS, Chan DW, Laegeler A, Qin J, Songyang Z (2004a) PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol 6: 673–680 [DOI] [PubMed] [Google Scholar]
- Loayza D, de Lange T (2003) POT1 as a terminal transducer of TRF1 telomere length control. Nature 423: 1013–1018 [DOI] [PubMed] [Google Scholar]
- Loayza D, Parsons H, Donigian J, Hoke K, de Lange T (2004) DNA binding features of human POT1: a nonamer 5′-TAGGGTTAG-3′ minimal binding site, sequence specificity, and internal binding to multimeric sites. J Biol Chem 279: 13241–13248 [DOI] [PubMed] [Google Scholar]
- Lustig AJ (2003) Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nat Rev Genet 4 (11): 916–923 [DOI] [PubMed] [Google Scholar]
- Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke DS (2002) Conserved structure for single-stranded telomeric DNA recognition. Science 96: 145–147 [DOI] [PubMed] [Google Scholar]
- Murti KG, Prescott DM (1999) Telomeres of polytene chromosomes in a ciliated protozoan terminate in duplex DNA loops. Proc Natl Acad Sci USA 96: 14436–14439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitina T, Woodcock CL (2004) Closed chromatin loops at the ends of chromosomes. J Cell Biol 166: 161–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J (2003) Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol 5: 741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CM, Cech TR (1987) Telomeric DNA–protein interactions of Oxytricha macronuclear DNA. Genes Dev 1: 783–793 [DOI] [PubMed] [Google Scholar]
- Shakirov EV, Surovtseva YV, Osbun N, Shippen DE (2005) The Arabidopsis Pot1 and Pot1b proteins function in telomere length homeostasis and chromosome end protection. Mol Cell Biol 25: 7725–7733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Karlseder J, Holtgreve-Grez H, Jauch A, de Lange T (2002) DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr Biol 12: 1635–1644 [DOI] [PubMed] [Google Scholar]
- Tahara H, Kusunoki M, Yamanaka Y, Matsumura S, Ide T (2005) G-tail telomere HPA: simple measurement of human single-stranded telomeric overhangs. Nat Methods 2: 829–831 [DOI] [PubMed] [Google Scholar]
- Takai H, Smogorzewska A, de Lange T (2003) DNA damage foci at dysfunctional telomeres. Curr Biol 13: 1549–1556 [DOI] [PubMed] [Google Scholar]
- Teixeira MT, Arneric M, Sperisen P, Lingner J (2004) Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117: 323–335 [DOI] [PubMed] [Google Scholar]
- Trujillo KM, Bunch JT, Baumann P (2005) Extended DNA binding site in Pot1 broadens sequence specificity to allow recognition of heterogeneous fission yeast telomeres. J Biol Chem 280: 9119–9128 [DOI] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, De Lange T (1998) TRF2 protects human telomeres from end-to-end fusions. Cell 92: 401–413 [DOI] [PubMed] [Google Scholar]
- Verdun RE, Crabbe L, Haggblom C, Karlseder J (2005) Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol Cell 20: 551–561 [DOI] [PubMed] [Google Scholar]
- Wang RC, Smogorzewska A, de Lange T (2004) Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119: 355–368 [DOI] [PubMed] [Google Scholar]
- Wei C, Price CM (2004) Cell cycle localization, dimerization, and binding domain architecture of the telomere protein cPot1. Mol Cell Biol 24: 2091–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KK, Chang S, Depinho RA (2006) Modeling cancer and aging in the telomerase-deficient mouse. In Telomeres, de Lange T, Lundblad V, Blackburn E (eds), pp 109–138. Cold Spring Harbor: Cold Spring Harbor Laboratory Press [Google Scholar]
- Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, Deng Y, Behringer RR, Chang S (2006) Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell 126: 49–62 [DOI] [PubMed] [Google Scholar]
- Yang Q, Zheng YL, Harris CC (2005) POT1 and TRF2 cooperate to maintain telomeric integrity. Mol Cell Biol 25: 1070–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye JZ, Donigian JR, van Overbeek M, Loayza D, Luo Y, Krutchinsky AN, Chait BT, de Lange T (2004b) TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J Biol Chem 279: 47264–47271 [DOI] [PubMed] [Google Scholar]
- Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T (2004a) POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev 18: 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T (2003) ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol Cell 12: 1489–1498 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2A, 2B
Supplementary Figure 3
Supplementary Table 1


