Abstract
Nanodiscs are an example of discoidal nanoscale self-assembled lipid/protein particles similar to nascent high-density lipoproteins, which reduce the risk of coronary artery disease. The major protein component of high-density lipoproteins is human apolipoprotein A-I, and the corresponding protein component of Nanodiscs is membrane scaffold protein 1 (MSP1), a 200-residue lipid-binding domain of human apolipoprotein A-I. Here we present magic-angle spinning (MAS) solid-state NMR studies of uniformly 13C,15N-labeled MSP1 in polyethylene glycol precipitated Nanodiscs. Two-dimensional MAS 13C-13C correlation spectra show excellent microscopic order of MSP1 in precipitated Nanodiscs. Secondary isotropic chemical shifts throughout the protein are consistent with a predominantly helical structure. Moreover, the backbone conformations of prolines derived from their 13C chemical shifts are consistent with the molecular belt model but not the picket fence model of lipid-bound MSP1. Overall comparison of experimental spectra and 13C chemical shifts predicted from several structural models also favors the belt model. Our study thus supports the belt model of Nanodisc structure and demonstrates the utility of MAS NMR to study the structure of high molecular weight lipid-protein complexes.
INTRODUCTION
Human apolipoprotein A-I (apo A-I) constitutes up to 70% of all proteins in high-density lipoprotein (HDL) particles, the amount of which in blood plasma is used to assess risk of coronary artery disease (CAD) (1,2) and atherosclerosis. Apo A-I is a 243-residue polypeptide whose 200-residue C-terminal lipid-binding domain consists of a series of eight 22-mer and two 11-mer amphipathic α-helices, which are interrupted by prolines or glycines. Apo A-I has several functions in the pathway of reverse cholesterol transport, where cholesterol is taken up from peripheral tissues and transported to liver for processing and excretion, and to steroidogenic tissue for hormone biosynthesis (3–6). In its action as an acceptor for cholesterol, apo A-I increases the activity of lecithin-cholesterol acyltransferase (LCAT) (7) and interacts with receptors such as scavenger receptor class B type I (SR-BI) upon transport of cholesteryl esters to the liver (8). During this process, apo A-I is known to undergo sequential structural changes from lipid-free to nascent discoidal HDLs to spherical HDLs upon cholesterol incorporation and esterification (9).
A model lipid/protein complex representative of nascent discoidal HDL, called the Nanodisc, has recently been developed and characterized by multiple biophysical techniques (10,11). The Nanodisc had been engineered as a robust membrane bilayer vehicle for the stoichiometrically controlled incorporation and study of membrane proteins such as cytochrome P450 (12–15), bacteriorhodopsin (bR) (16), and G-protein-coupled receptors (17,18). In addition, the Nanodisc also represents an excellent model system for studying HDL and the atomic-resolution structure of apo A-I. Nanodiscs are soluble, monodisperse, self-assembled particles, 10 nm in diameter, consisting of two molecules of membrane scaffold protein 1 (MSP1) wrapped around the outside edge of a discoidal bilayer fragment consisting of ∼160 saturated lipids. MSP1 is a 25 kDa, 200-residue C-terminal lipid-binding domain of apo A-I protein and has the same predicted secondary structure consisting of a sequence of amphipathic helices.
Apo A-I is a dynamic molecule with multiple functions. The mechanisms underlying these functions cannot be understood in detail without additional structural information about each conformational state (19). A high resolution structure of lipid-free full-length apo A-I has been reported very recently (20). This structure reveals intramolecular antiparallel helix bundles similar to other lipid-free apolipoproteins. It is remarkably different from the structure of Δ(1–43) apo A-I, which contains a ring-shaped four-helix bundle involving four molecules of apo A-I (21). The structure of apo A-I in lipid-bound form, especially in discoidal HDL, has also been examined extensively by both experimental and computational methods (22,23). These studies were motivated by both the fundamental interest in understanding the antiatherogenic function of HDL and the potential value of HDL as therapeutic agents (24). To date, no high resolution structure of apo A-I in discoidal HDL is available, in part because the heterogeneity of lipid/protein complexes presents significant challenges to x-ray crystallography.
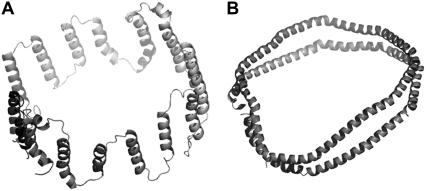
In the absence of single crystal diffraction data, several structural models have been proposed, among which the belt model and the picket fence model are the two most favorable competing models. The main distinguishing feature between these two models is the orientation of the axes of the amphipathic helices with respect to the acyl chains of lipids; they are perpendicular in the belt model and parallel in the picket fence model. Although the low-resolution models were first proposed as early as the 1970s, only recently have all-atom computational models been constructed (25–27). Fig. 1 shows the schematic representations of these two models. The recently proposed helical hairpin model (28,29) is very similar to the belt model except that each apo A-I is bent in the middle to form a hairpin. Various experimental techniques have been employed to gather evidence supporting or disproving these models. The belt model is supported by recent Fourier transform infrared spectroscopy (30), Trp fluorescence quenching (31), and mass spectrometry studies (32). On the other hand, results from limited proteolysis experiments (33) and very recent scanning tunneling microscopy (34) support the picket fence model.
FIGURE 1.
Cartoon representation of (A) the picket fence model and (B) the belt model. Lipid molecules are not shown, to make the orientation of the amphipathic helices of apo A-I surrounding the lipid bilayer clearly visible.
NMR spectroscopy is a complementary technique to x-ray crystallography for high-resolution structure determination. However, the long rotational correlation times associated with the relatively large particle sizes of lipoproteins present significant technical difficulties to solution NMR studies. Although a discoidal lipid/protein complex assembled from an 18 amino acid peptide mimicking apo A-I has been studied by solution NMR very recently (35), extending this study to full length apo A-I in discoidal HDL likely will require further technical developments. Solid-state NMR (SSNMR) has emerged in the past decade as an extremely powerful tool for structural studies of solid proteins and supramolecular assemblies with slow tumbling rates. Historically, SSNMR was only applicable to proteins that could be isotopically labeled at a small number of sites by solid-phase peptide synthesis or mutagenesis and bacterial expression in defined media (amino acid type-specific isotopic labels) (36). This limitation has been overcome in the last decade by the introduction of high magnetic field (>14 Tesla) and high-frequency magic-angle spinning (MAS) in combination with multi-dimensional dipolar recoupling sequences (37). This approach permits the examination of a wide range of protein samples with higher throughput and fidelity than previously possible by employing uniformly 13C, 15N-labeled samples in concert with multi-dimensional methods. For example, chemical shift assignments of several globular proteins have recently been performed using SSNMR methods (38–42). Recent studies (43,44) have also illustrated the potential for applying this approach to membrane proteins, which are difficult to study by x-ray crystallography for similar reasons as lipoproteins. The critical distinguishing feature of SSNMR is that, unlike crystallography, macroscopically ordered samples are not required. Therefore even macroscopically disordered, precipitated samples yield high-resolution spectra, as long as the protein maintains microscopic order and is properly hydrated.
We demonstrate here that the microscopic structural homogeneity of Nanodiscs, precipitated by low molecular weight polyethylene glycol (PEG), is ideal for high-resolution structural studies by SSNMR to determine which model of Nanodisc molecular structure (the belt or the picket fence) agrees best with the experimental data. With modern SSNMR instrumentation, the spectra of uniformly 13C, 15N-labeled MSP1 yield spectral line widths comparable to those observed in microcrystalline globular proteins, such as those we have previously reported (41). We demonstrate that PEG precipitation and resolubilization does not disassemble the Nanodisc samples and that the sample integrity is maintained for several days to weeks under conditions suitable for NMR analysis. The identification of signals by amino acid type and the interpretation of individual resolved signals in the two-dimensional (2D) 13C-13C correlation spectra validate the MSP1 secondary structure on a site-specific basis. These results strongly support the belt model (26) over the picket fence model (25).
MATERIALS AND METHODS
Sample preparation
MSP1 was expressed in Escherichia coli by adopting a protocol described elsewhere (10). To obtain uniformly 13C, 15N-labeled MSP1, 13C,15N-enriched M9 minimal medium was employed: 100 ml of 10× M9 (containing 67.8 g Na2HPO4, 30 g KH2PO4, and 5 g NaCl in 1 L of autoclaved H2O, pH = 7.4, adjusted using 5 M KOH), with 1 g 15NH4Cl, 2 g 13C glucose, 0.011 g of CaCl2·2H2O, 0.246 g MgSO4, 0.02 g thiamine, kanamycin at working concentration of 30 mg/L, and 10 ml of 10× 13C,15N-labeled BioExpress rich growth media (45), filled to 1 L H2O and sterile filtered. Isotopically labeled materials were obtained from Cambridge Isotope Laboratories (Andover, MA). MSP1 purification and Nanodisc preparation were both performed as described previously (10).
For preparation of SSNMR samples, 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) lipids (Avanti Polar Lipids, Alabaster, AL) were used in an 80:1 DMPC/MSP1 preparation ratio. The Nanodiscs were prepared in Tris buffer (100 mM NaCl, 10 mM Tris, 1 mM EDTA, pH = 7.4) and precipitated using 40% PEG 3350. Samples contained ∼5–6 mg/mL of MSP1 in a Nanodisc precipitated with ∼3 volume equivalents of 40% PEG 3350. The mixture was vortexed lightly and left overnight at 4°C. The samples were then centrifuged for ∼5 h at 8000 rpm. The supernatant was removed, and the pellet was transferred into a 3.2 mm thin wall rotor (Varian NMR, Palo Alto, CA) with working volumes of ∼36 μL and confined to active sample region (central 30 μL) of the rotor by Kel-F and rubber spacers as described elsewhere (41). The supernatant contained no observable protein, determined by absorbance at 280 nm. The total mass of the material packed into the rotor was ∼30 mg. Lipid concentration in the rotor was 20–30% by mass, and protein concentration was ∼10–15%, due to the high PEG concentration in the pellet. The amount of protein was determined by comparing the intensity of one-dimensional (1D) 13C and 15N spectra to standard proteins of known quantity.
To test the integrity of the precipitated Nanodiscs, a sample was prepared with natural abundance MSP1 and subjected to the exact precipitation treatment as the isotopically labeled Nanodiscs prepared for SSNMR before placement into a rotor. The PEG supernatant was removed, and the precipitated pellet was resuspended in 125 μl of standard buffer. Water was added further in small aliquots until the sample appeared to be visually clear, resulting in a final volume of 325 μl. The sample was then injected onto the Superdex 200 HR 10/30 column (Amersham Biosciences, Piscataway, NJ), with the column, injection volume, and the flow rate kept constant throughout the experiment. From the first injection of the resuspended sample, fractions 24–27 were collected and pooled together and reinjected onto the column. The sample initially resuspended in 325 μl was diluted further by addition of more water, and an aliquot of this was injected onto the column. With the exception of slight shifts in the retention time attributable to the presence of PEG, the precipitated and resolubilized Nanodisc samples showed the same behavior as freshly prepared Nanodiscs (data in Supplementary Material).
SSNMR spectroscopy
SSNMR experiments were performed on Varian NMR spectrometers. The 750 MHz Inova three-channel spectrometer is equipped with a 3.2 mm 1H-13C-15N BioMAS probe (46) and the 500 MHz InfinityPlus four-channel spectrometer with a 3.2 mm 1H-13C-15N Balun probe. The temperature of the sample cooling gas was maintained at −10.0 ± 0.2°C with 100 scfh flow rate for all the 2D experiments; the effective offset in temperature due to friction, radiofrequency heating (46), and thermocouple calibration results in an absolute sample temperature of ∼10°C ± 5°C in this case, although there is not yet any well-established technique for measuring this absolute temperature internally. Therefore we report the temperature of the cooling gas for comparison throughout this study. The temperature was chosen based on consideration of both the spectral resolution (discussed below) and sample stability. For experiments at 750 MHz, pulse sequences were implemented with tangent ramped cross-polarization (CP) (47) at ∼55 kHz on 1H and ∼45 kHz on 13C and two pulse phase modulation (TPPM) 1H decoupling (48) at 78 kHz. The typical π/2 pulse widths are 2.5 μs on 1H and 3.5 μs on 13C. For experiments at 500 MHz, the 13C field for CP is ∼45 kHz and 1H field for TPPM decoupling is 70 kHz. The typical π/2 pulse widths are 2.5 μs on 1H and 3.0 μs on 13C.
Data were processed with NMRPipe (49) with back linear prediction and polynomial baseline (frequency domain) correction applied to the direct dimension. Zero filling and Lorentzian-to-Gaussian apodization were employed for each dimension before Fourier transformation. Additional acquisition and processing parameters for each spectrum are included in the figure captions. Chemical shifts were referenced externally with adamantane (50).
RESULTS AND DISCUSSION
Sample integrity and stability
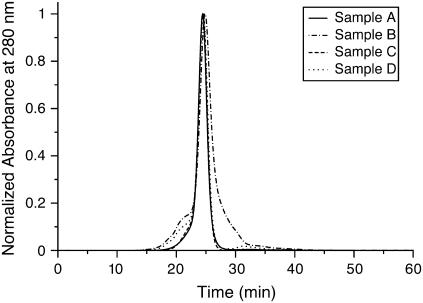
Sample integrity was tested by gel permeation chromatography and phosphate analysis on resuspended Nanodisc samples. Fig. 2 shows the elution profiles from gel permeation chromatography. The elution profile of initially resuspended Nanodiscs (sample B) reveals a sharp peak broadened at the base that is offset compared to the Nanodisc sample before precipitation (sample A). This tail appears to indicate a homogeneous population of slightly smaller sized particles, which we suspected might be due to the interaction of PEG with the column. This was confirmed by calibration of the column with standards mixed with the PEG 3350 (Supplementary Material) and further verified by the fact that additional dilution restores the retention time to the initial position (samples C and D) and minimizes the broadening caused by PEG. Additional confirmation that the samples were assembled as Nanodiscs was found by subjecting fractions from the resuspended sample to phosphate analysis. A stoichiometric ratio of 87 ± 4 lipid/MSP1 was observed in both the precipitated and resuspended samples, whereas the control Nanodisc samples (which were not precipitated) yielded a value of 85 ± 4. These results strongly support the robustness of the Nanodiscs throughout the employed precipitation protocol. The integrity of SSNMR samples as a function of time in the magnet is furthermore confirmed by comparisons of 1D 13C and 15N, and 2D 13C-13C spectra before and after lengthier 2D and three-dimensional (3D) experiments.
FIGURE 2.
Normalized gel filtration chromatograms from the calibrated Superdex 200 HR 10/30 gel filtration column equilibrated in standard buffer (100 mM NaCl, 10 mM Tris/HCl, 1 mM EDTA). The mobile phase flow rate was 0.5 mL/min on a Waters Millennium system, with monitoring of protein absorbance at 280 nm, with one fraction collected per minute. Injection volume was 200 μl for each sample. Sample A: Nanodisc sample before precipitation. Sample B: precipitated Nanodiscs resuspended in 125 μl standard buffer with 200 μl water added. Sample C: reinjection of fractions 24–27 of sample B. Sample D: sample B further diluted with water.
1D CP-MAS 13C spectra
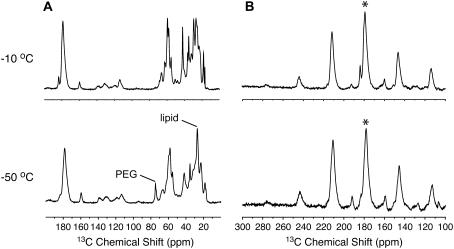
For small globular proteins such as ubiquitin (51) and GB1 (41) and the membrane protein complex LH2 (43), SSNMR spectral resolution is temperature dependent in a system-specific manner that relates both to microscopic homogeneity and molecular dynamics. To find the optimal temperature for multi-dimensional experiments of Nanodiscs, 1D CP-MAS 13C spectra were acquired in the temperature range of −10°C to −50°C. As described in the Methods section, here we refer to the temperature of cooling gas instead of the actual sample temperature, which is generally 15°C–25°C higher. Fig. 3 A shows the 13C 1D spectra at −10°C and −50°C. The trend in signal intensities and spectral resolution agrees with our previous observations in GB1 (41). Protein signals are broadened as temperature is lowered from −10°C to −50°C, with side-chain signals being more sensitive to temperature than backbone signals. In addition to protein, the precipitated Nanodisc sample contains significant amount of lipids and PEG 3350, which are generally more mobile than protein at the higher range of temperatures in this study. Therefore PEG signals are not observed in the −10°C spectrum but appear with significant intensity in the −50°C spectrum. Similarly, the signals at 25–28 ppm, in the expected range of lipid methylene and methyl signals, are significantly enhanced at lower temperatures. Although the lipids are not 13C labeled, the intensity observed is consistent with the large number of lipid molecules at natural 13C isotopic abundance. This observation suggests that 1H-13C heteronuclear couplings of lipids and PEG are motionally averaged at the higher temperature, resulting in the very low CP efficiency for these signals.
FIGURE 3.
750 MHz CP-MAS 1D 13C spectra of MSP1 in Nanodiscs at −10°C and −50°C. 1H-13C CP conditions were optimized at each temperature and other acquisition parameters are the same. Spectra were acquired on ∼150 nmol (∼4 mg) MSP in a 3.2 mm thin wall rotor. (A) Spectra at MAS rate of 12.25 kHz, 20.48 ms acquisition time (2048 × 10 μs), 78 kHz 1H TPPM decoupling (6.4 μs, −10°C), 512 scans, 1 s pulse delay. CP contact time was 0.6 ms at −10°C and 0.9 ms at −50°C (optimized for total signal intensity). The data were processed with 30-Hz net line broadening (Lorentzian-to-Gaussian apodization). (B) Carbonyl region of the spectra at MAS rate of 6.125 kHz. The centerband is marked with an asterisk. The additional sideband manifold centered at ∼160 ppm arises from Tyr Cζ signals.
As an additional probe of the backbone rigidity of MSP1 at different temperatures, the bulk carbonyl chemical shift anisotropy (CSA) was examined. Fig. 3 B shows the carbonyl region of the 1D 13C spectra acquired at a MAS rate of 6.125 kHz. By fitting the sideband intensity to simulated spectra using the averaged asymmetry parameters of different amino acids, the span of CSA was estimated to be 147 ppm at −50°C and 140 ppm at −10°C. The value at −50°C is very close to the rigid lattice limit, and the scaling at −10°C is only 5%. So although small amplitude backbone librations may be present, the Nanodiscs as a whole must be rigid on the NMR timescale to produce the large observed CSA tensor magnitude. Furthermore, signal enhancements in CP experiments are substantial; the enhancement of the 13C signal relative to direct 13C polarization is ∼2-fold, and the (1H)-15N-13C specific CP (52) efficiency in comparison to 1H-13C CP is ∼40%. These values are comparable to the best performance observed in well-behaved globular proteins (41). Therefore Nanodiscs are rigid, ordered particles at the nanoscale and demonstrate the essential sample characteristics required for successful implementation of more sophisticated 2D and 3D MAS experiments.
2D 13C-13C correlation spectra
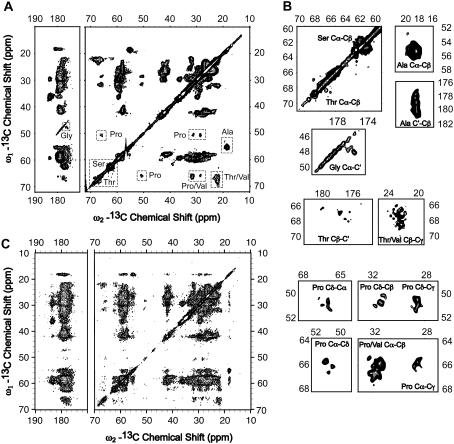
The 2D 13C-13C homonuclear correlation spectra allow for identification of spins in the same residue. Each identified group of spins, i.e., spin system, can be assigned to specific type(s) of amino acid residues according to their characteristic 13C chemical shifts (53). Fig. 4 A shows the 2D 13C-13C homonuclear correlation spectrum of uniformly 13C, 15N-labeled MSP1 in Nanodiscs acquired at 750 MHz 1H frequency with 50 ms dipolar-assisted rotational resonance (DARR) mixing time (54) . For many of the unambiguously identifiable amino acid spin systems in our spectra (Ala, Val, Glu, Pro, Ser, Thr, and Gly), signals with helical secondary chemical shifts (55–57) dominate the overall intensity. Ala, Gly, Ser, and Thr are prominent examples. The Ala chemical shift range could be identified from the Cα-Cβ crosspeaks (Fig. 4, A and B): 53.8–56.5 ppm for Cα and 17.5–19.5 ppm for Cβ. These values are consistent with Ala chemical shifts in a helical secondary structure. A few peaks can be observed that deviate from the rest of the Ala spin systems and have Cα values shifted upfield to 52.5–53.5 ppm. In particular, an individual Ala can be identified at Cα of 52.9 ppm and Cβ of 19.3 ppm. This chemical shift is more characteristic of an Ala in a coiled secondary structure (although its line width is comparable to other individual peaks, indicating a specific preferred conformation). Several Ala residues in the MSP1 sequence—such as Ala-121, Ala-164, and Ala-167—are in a vicinity of a helix break, or possibly a kink, and the observed signal could correspond to one of these Ala not in a helical conformation. In particular, Ala-121 precedes a Pro residue, which would have an effect on its conformation resulting in an upfield shift of Cα (58). Other regions of interest include the Gly Cα-C′ and Thr and Ser Cα-Cβ crosspeaks (Fig. 4, A and B). Several individual Gly residues can be identified, and they all have the Cα chemical shift in the range between 47.1 and 48.5 ppm, and C′ in the range between 174.5 and 177.5 ppm, again consistent with the expected Cα chemical shifts for Gly in a helical secondary structure. The proximity of Cα-Cβ crosspeaks of Ser and Thr to the diagonal is another unambiguous indication of helical secondary structure. Overall, our analysis indicates that MSP1 is predominantly helical, which is consistent with the reported 80% helical content of apo A-I in discoidal HDL (59).
FIGURE 4.
(A) 2D 13C-13C chemical shift correlation spectrum of MSP1 at 750 MHz with 50 ms DARR mixing time. (B) The expansion of the critical regions of the spectrum shown in (A). (C) 2D 13C-13C chemical shift correlation spectrum of MSP1 at 750 MHz with 200 ms DARR mixing time. The data were acquired at −10°C and 12.25 kHz spinning frequency for 38 h. Other acquisition parameters are 0.8 ms 1H-13C CP contact time, 78 kHz 1H TPPM decoupling (6.4 μs, 10°), 10.75 ms t1 acquisition time (512 × 21 μs) with States-TPPI detection, 20.48 ms t2 acquisition time (2048 × 10 μs), 1 s pulse delay. The data were processed with 0.13 ppm (25 Hz) net line broadening (Lorentzian-to-Gaussian apodization), ∼63° phase shifted sine bell functions, and zero filled to 8192 (ω2) × 2048 (ω1) complex points before Fourier transformation. Chemical shifts of the aliphatic 13C signals are diagnostic of the expected helical secondary structure. Expanded regions further exemplify the resolution indicative of a microscopically well-ordered, precipitated Nanodisc formulation.
In the 2D 13C-13C correlation spectrum with 50 ms mixing time (Fig. 4 A), most regions are congested due to a large number of the common amino acid (spin system) types in similar secondary structure environments. This observation is not to be confused with a lack of microscopic order in the sample preparation. Outlying signals in the spectrum indicate that individual line widths are <0.3 ppm. To perform a more detailed analysis of line width, a 2D 13C-13C correlation spectrum with a longer mixing time was acquired, which contains a significant number of interresidue crosspeaks with better chemical shift dispersion. Table 1 shows the line widths of 15 individually resolved peaks randomly picked from the 2D 13C-13C spectrum with 200 ms DARR mixing time (Fig. 4 C). Since the line width in the direct dimension is generally not limited by digital resolution, it is a better indication of the conformational homogeneity of the sample. The line widths (from a spectrum processed with 30 Hz line broadening) range from 61 Hz to 89 Hz with an average of 75 Hz, i.e., 0.4 ppm. This narrow line width suggests the excellent order of the sample on the length scale relevant to SSNMR (∼5–10 Å), comparable to observations for nanocrystalline proteins such as ubiquitin (39,40,51) and GB1 (41) and the membrane protein diacylglycerol kinase recently studied by SSNMR (60). It is well known from previous studies on small globular proteins that residual dipolar (1H-13C) and scalar (13C-13C) couplings and static field inhomogeneities contribute to the majority of the remaining observed line widths. Therefore, the underlying natural line widths in the spectra are clearly indicative of a microscopically well-ordered sample formulation, which is especially favorable for multi-dimensional SSNMR studies. Beyond this technical detail of the NMR data acquisition, the observed line widths reveal a functional microscopic ordering of the sample, consistent with a robust sample preparation and energetically favorable conformation of the MSP1 protein in Nanodiscs.
TABLE 1.
Resonance positions and line widths of 15 randomly selected crosspeaks in 2D 13C-13C chemical shift correlation spectrum with 200 ms DARR mixing time
| ω1-13C (ppm) | ω2-13C (ppm) | ω1 FWHM* (Hz) | ω2 FWHM* (Hz) |
|---|---|---|---|
| 18.3 | 25.9 | 98 | 89 |
| 39.1 | 22.9 | 107 | 80 |
| 48.5 | 26.2 | 88 | 60 |
| 43.6 | 23.2 | 92 | 78 |
| 36.8 | 30.7 | 89 | 82 |
| 63.8 | 31.9 | 90 | 74 |
| 62.8 | 32.6 | 84 | 79 |
| 183.4 | 180.4 | 86 | 78 |
| 63.0 | 28.3 | 90 | 82 |
| 50.0 | 179.1 | 82 | 66 |
| 31.3 | 61.1 | 92 | 77 |
| 180.6 | 55.8 | 82 | 63 |
| 176.2 | 56.7 | 77 | 77 |
| 175.6 | 26.7 | 89 | 61 |
| 31.7 | 40.8 | 97 | 77 |
Data were processed with 30 Hz net line broadening (Lorentzian-to-Gaussian apodization). The full width at half-maximum height (FWHM) was determined using Sparky (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco).
Proline conformations support the belt model
In all of the proposed models of apo A-I in HDL particles (25,26,28,61,62), proline residues create a break between helices. These breaks take the form of β-turns in the picket fence model or in the belt model, provide kinks which help to orient the hydrophobic side of the amphiphatic helices properly toward the lipid interface for protein-lipid interaction (63). Proline has sterically constrained φ- and ψ-angles and can be found predominantly in one of the two regions in Ramachandran space, with a common allowed φ-angle of ∼−60° and two ranges of ψ-angles with middle values of ∼ −45° and ∼135° (64). As shown in Fig. 4, A and B, proline spin systems can be easily identified because of their characteristic chemical shifts. To relate the experimental chemical shifts to the conformation of prolines, the chemical shifts and the structural information of several model proteins including SH3 (38), ubiquitin (39,40), and Crh (42) were used, as both high-resolution x-ray structure and SSNMR chemical shifts are available for these proteins. Table 2 shows φ- and ψ -angles of prolines in the crystal structures of these proteins and their SSNMR 13C chemical shifts. All of the backbone torsion angles fall into one of the two energetically favorable regions and an obvious pattern emerges where high positive ψ-values (within one allowed region) result in Cα chemical shifts that are shifted upfield to 61–62 ppm, whereas negative ψ-angle values (prolines in the other allowed region) result in downfield shifted Cα values of ∼65–66 ppm. This is consistent with the general trend of Cα chemical shifts as a function of secondary structure, whereas Cβ, Cγ, and Cδ chemical shifts do not appear to be sensitive to differences in the backbone conformation of proline, instead depending more upon ring conformation. Thus the Cα chemical shift analysis is most informative with respect to the backbone conformation. The chemical shifts of individual proline peaks, as well as the chemical shift range of the more congested proline regions (Cα between ∼65.3 and 66.8 ppm), are all consistent with prolines with ψ-angles close to zero.
TABLE 2.
Torsion angles and solid-state 13C chemical shifts of prolines in several model proteins
| Protein | PDB ID | Residue | φ (°) | ψ (°) | Cα (ppm) | Cβ (ppm) | Cγ (ppm) | Cδ (ppm) |
|---|---|---|---|---|---|---|---|---|
| Ubiquitin | 1UBQ | Pro-19 | −54.9 | −24.5 | 64.8 | 32.0 | 27.8 | 50.2 |
| Pro-37 | −57.0 | 137.0 | 61.5 | 31.8 | 28.4 | 50.8 | ||
| Pro-38 | −57.2 | −32.2 | 66.0 | 33.2 | 27.7 | 51.2 | ||
| Crh | 1MU4 | Pro-18 | −60.2 | −35.5 | 65.1 | 31.6 | 28.2 | 49.2 |
| SH3 | 1SHG | Pro-20 | −55.0 | −36.6 | 65.0 | 32.1 | 27.3 | 50.1 |
| Pro-54 | −63.5 | 129.1 | 61.6 | 30.1 | 28.3 | 50.0 |
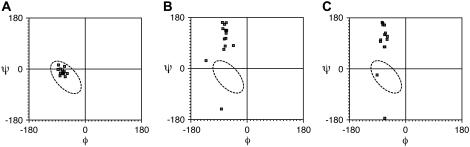
We compared the proline conformations in different models including the belt model (27) and the picket fence model with two different alignments (25). Fig. 5 shows the Ramachandran plots of prolines in each model. One of the two energetically favorable and also statistically populated regions (64) that is consistent with our experimental data is circled in each plot. The most striking observations are the constantly large positive values for ψ-angles in the two picket fence models where prolines are found in β-turns between helices. The dihedral angle values (ψ ∼ 0) indicative of observed chemical shifts (65.5–66.5 ppm) of proline in MSP1 in a Nanodisc are consistent with those found in the double belt model and do not agree at all with those that would be expected for prolines in the picket fence models. Since the proline residues are the key conformational difference between the picket fence and the belt model, this result strongly supports the belt model and contradicts any model in which prolines are in β-turn conformations. It is possible, but unlikely, that a single hairpin could be formed without a proline in a β-turn conformation; therefore it is not possible to rule out all possible hairpin models.
FIGURE 5.
Ramachandran plots for proline torsion angles in MSP1 dimer in (A) belt model, (B) head-to-head picket fence model, and (C) head-to-tail picket fence model. The circled region represents schematically one of the two energetically favorable regions that are consistent with the experimental chemical shifts.
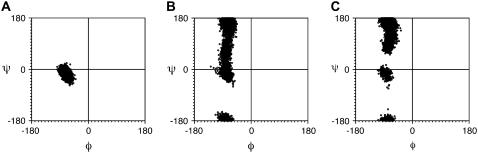
To examine whether there is strong correlation between the proline conformation and the overall arrangements of the helices, molecular dynamics simulations were performed according to a previously published protocol (27). In these simulations, the previously reported computational models (25,27) were directly used as starting structures without further energy minimization. Fig. 6 shows the torsion angles of prolines extracted from the trajectory of 1 ns dynamics simulations of three different models. For the belt model, the ψ-angles of all prolines remain close to zero during the entire trajectory, which demonstrates the extremely high-energy barrier for prolines to undergo a conformational change within the Nanodisc. Similarly, the transition of individual proline from one favorable conformation to the other has been observed for neither the head-to-head nor the head-to-tail picket fence models, indicating the stability of both models. Most prolines remain to be in the conformation with large positive ψ-angles in the entire trajectory. Therefore, the relative orientation of helices appears to dictate the conformation of prolines in each model. The fact that only prolines with one favorable conformation (ψ close to zero) are observed in our experimental spectra strongly supports the belt model as the correct model of MSP1 in Nanodiscs.
FIGURE 6.
The Ramachandran plots of proline torsion angles extracted from the trajectories of 1 ns molecular dynamics simulations of (A) belt model, (B) head-to-head picket fence model, and (C) head-to-tail picket fence model.
Complete comparison of experimental and predicted chemical shifts
It has been well established that Cα and Cβ chemical shifts depend on both the amino acid type and on the backbone conformation (58,65–67). To take the chemical shift analysis procedure further, we simulated the 13C-13C correlation spectra of MSP1 in various models, employing the program ShiftX (68), which utilizes a semiempirical approach to calculate protein chemical shifts from their structures. Although the program is based on solution NMR chemical shift data, it now has been thoroughly demonstrated in small globular proteins that solution and solid-state chemical shifts are strongly correlated (39–41). Among all the chemical shifts predicted by this program, the correlations between predicted and experimental values are best for Cα and Cβ shifts (0.98 and 0.996 correlation coefficients, respectively) (68). Therefore, we compared the predicted Cα and Cβ shifts with the experimental 2D 13C-13C spectra. A similar method was utilized by Baldus and co-workers in a different context (69); they calculated correlation factors for Cα and Cβ (0.9606 and 0.9947, respectively) SSNMR chemical shifts in three model proteins: SH3, ubiquitin, and Crh.
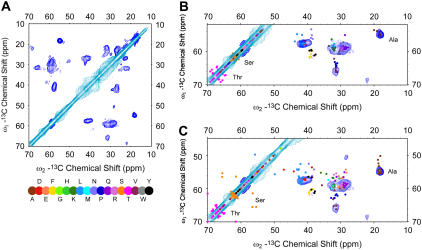
To compare the simulated and experimental spectra, a 2D 13C-13C chemical shift correlation spectrum with polarization transferred by SPC-5 recoupling scheme (70) was acquired (Fig. 7 A). In this spectrum, the mixing time was chosen to ensure that crosspeaks result only from one-bond correlations, enabling Cα-Cβ crosspeaks to be unambiguously identified. Fig. 7, B and C, shows two examples of the overlaid experimental and simulated spectra for the belt model (Fig. 7 B) and one of the picket fence models with head-to-head alignment (Fig. 7 C). Besides the aforementioned differences in Pro signals, clear variations are also observed in the Ser, Thr, and Ala regions. For the picket fence model shown, several Ser and Thr Cα-Cβ crosspeaks deviate far from the diagonal in the simulated spectra, clearly indicating nonhelical conformations; these features are not observed in the experimental spectra. In the Ala Cα-Cβ crosspeak region, multiple predicted crosspeaks from the head-to-head picket fence model fail to overlap with the experimental data. Likewise in the Leu, Asp, and Asn Cα-Cβ crosspeak regions—although for both models shown there are signals in the simulated spectra that do not agree perfectly with experimental data—the deviations are substantially more significant for the picket fence model.
FIGURE 7.
(A) 2D 13C-13C chemical shift correlation spectrum of MSP1 acquired at 500 MHz 1H frequency using SPC-5 recoupling scheme. The contours were cut at 5 σ noise level with positive contours in cyan and negative contours in blue. The data were acquired at −10°C and 11.111 kHz spinning frequency for 31 h. The other acquisition parameters are 0.8 ms 1H-13C CP contact time, 78 kHz 1H TPPM decoupling (6.5 μs, 15°), 7.68 ms t1 acquisition time (768 × 10 μs) with TPPI detection, 20.48 ms t2 acquisition time (2048 × 10 μs), 1.5 s pulse delay. The data were processed with 50 Hz net line broadening (Lorentzian-to-Gaussian apodization), ∼63° phase shifted sine bell functions, and zero filled to 8192 (ω2) × 2048 (ω1) complex points before Fourier transformation. Overlay of the simulated 2D 13C-13C spectra of (B) belt model and (C) head-to-head picket fence model and the experimental spectra shown in A. The chemical shifts used to generate the simulated spectra are predicted by the program ShiftX (68).
To quantify the results of this overall comparison, the simulated crosspeaks that overlap with any contours greater than the 5 σ noise floor were counted for each model. The uncertainty of ±0.5 ppm was used for both Cα and Cβ predicted chemical shifts. Because helical Ser and Thr crosspeaks are close to the diagonal and generally obscured by the strong diagonal peaks, they were excluded from the quantitative comparison. The Thr and Ser Cα and Cβ chemical shifts can instead be observed in 2D double quantum/zero quantum 13C-13C correlation spectra in which diagonal peaks do not exist (shown in the Supplementary Material). Gly was also excluded because of the lack of a Cβ atom. The results are summarized in Table 3. In all molecular models, at least 70% of the MSP1 residues are helical and therefore should have very similar chemical shifts. A smaller number of residues differ among the models; these differences correspond to those key residues that have multiple energetically allowed secondary structures, which in turn are the important determinants of the relative orientation of adjacent helices. We find that overall agreement between the experimental data and the belt model is best, consistent with the detailed analysis of proline residues as presented above.
TABLE 3.
Overall comparison of simulated and experimental 2D 13C-13C correlation spectrum
| Model | Total number of residues | Number of residues excluding G, S, T | Number of overlapping Cα-Cβ crosspeaks | Percentage of overlapping Cα-Cβ crosspeaks |
|---|---|---|---|---|
| Belt | 195 | 166 | 159 | 95.8% |
| Head-to-head picket fence | 191 | 162 | 147 | 90.7% |
| Head-to-tail picket fence | 191 | 162 | 145 | 89.5% |
*The uncertainty of ±0.5 ppm was used for simulated crosspeak positions and 5 σ noise cutoff was used for experimental spectra.
Although our study does not provide every detail about the structure of discoidal HDL at atomic resolution, it illustrates an approach that can be applied to other lipid-bound states of apo A-I. Considering the fact that the x-ray structure of lipid-free full length apo A-I has recently been solved (20), more structural information about the lipid-bound states will likely help in understanding the pathway by which lipid-free/poor apo A-I is converted to discoidal HDL and finally to mature HDL. A detailed understanding of the structural transformation of apo A-I will lend structural insight into the interactions of apo A-I with other essential components of the reverse cholesterol transport pathway, such as LCAT and SR-BI. The results therefore contribute both fundamental knowledge as well as an approach that may eventually lead to therapeutic strategies for reverse cholesterol transfer.
CONCLUSIONS
To define the structure of discoidal lipoprotein particles, we have presented an SSNMR study of uniformly 13C, 15N-labeled MSP1 encircling the phospholipid bilayer Nanodisc. We demonstrate that precipitated Nanodiscs have excellent microscopic order for high-resolution multi-dimensional SSNMR studies, as indicated by the narrow inhomogeneous line widths. Our results further show that lipid-protein complexes possess the conformational homogeneity required for SSNMR studies and yield line widths comparable to small globular proteins. Our experiments reveal tensor properties, including the CSA of carbonyl sites and the dipolar couplings among protonated sites, which are consistent with a sample that is microscopically rigid on the NMR timescale. These favorable properties are essential for detailed structural study by SSNMR.
The NMR data verify that MSP1 maintains its predominantly helical conformation in PEG-precipitated Nanodiscs. In particular, the proline Cα chemical shifts provide useful structural constraints. The experimentally determined proline backbone conformations are in complete agreement with those in the belt model but disagree with the majority of those in the picket fence model. This conclusion is further supported by empirical comparisons of the chemical shifts among the majority of Cα and Cβ sites in MSP1 and is also consistent with the results from molecular dynamics simulations, which show that the arrangement of helices in the picket fence model does not energetically favor the proline conformations observed from experimental spectra. The results therefore support the belt model for Nanodisc structure.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Acknowledgments
The authors thank Dr. Paul Molitor (VOICE NMR Facility, School of Chemical Sciences, University of Illinois) for technical assistance, Dr. Donghua H. Zhou for advice on performing 750 MHz magic-angle spinning NMR, Amy Y. Shih and Prof. Klaus Schulten for providing the MD simulation trajectories, Dr. Ilia G. Denisov for a careful reading of the manuscript, and Yelena V. Grinkova for sample preparation advice.
This research was supported by the University of Illinois startup funds to C.M.R. and the National Institutes of Health GM33775 grant to S.G.S.
References
- 1.Gordon, T., W. P. Castelli, M. C. Hjortland, W. B. Kannel, and T. R. Dawber. 1977. High density lipoprotein as a protective factor against coronary heart disease. The Framingham Study. Am. J. Med. 62:707–714. [DOI] [PubMed] [Google Scholar]
- 2.Asztalos, B. F., L. A. Cupples, S. Demissie, K. V. Horvath, C. E. Cox, M. C. Batista, and E. J. Schaefer. 2004. High-density lipoprotein subpopulation profile and coronary heart disease prevalence in male participants of the Framingham Offspring Study. Arterioscler. Thromb. Vasc. Biol. 24:2181–2187. [DOI] [PubMed] [Google Scholar]
- 3.Glomset, J. A. 1968. The plasma lecithins:cholesterol acyltransferase reaction. J. Lipid Res. 9:155–167. [PubMed] [Google Scholar]
- 4.Fielding, C. J., and P. E. Fielding. 1995. Molecular physiology of reverse cholesterol transport. J. Lipid Res. 36:211–228. [PubMed] [Google Scholar]
- 5.Barter, P. J., and K. A. Rye. 1996. Molecular mechanisms of reverse cholesterol transport. Curr. Opin. Lipidol. 7:82–87. [DOI] [PubMed] [Google Scholar]
- 6.Colvin, P. L., and J. S. Parks. 1999. Metabolism of high density lipoprotein subfractions. Curr. Opin. Lipidol. 10:309–314. [DOI] [PubMed] [Google Scholar]
- 7.Jonas, A. 2000. Lecithin cholesterol acyltransferase. Biochim. Biophys. Acta. 1529:245–256. [DOI] [PubMed] [Google Scholar]
- 8.Krieger, M. 1999. Charting the fate of the “good cholesterol”: identification and characterization of the high-density lipoprotein receptor SR-BI. Annu. Rev. Biochem. 68:523–558. [DOI] [PubMed] [Google Scholar]
- 9.Segrest, J. P., S. C. Harvey, and V. Zannis. 2000. Detailed molecular model of apolipoprotein A-I on the surface of high-density lipoproteins and its functional implications. Trends Cardiovasc. Med. 10:246–252. [DOI] [PubMed] [Google Scholar]
- 10.Bayburt, T. H., Y. V. Grinkova, and S. G. Sligar. 2002. Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett. 2:853–856. [Google Scholar]
- 11.Denisov, I. G., Y. V. Grinkova, A. A. Lazarides, and S. G. Sligar. 2004. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J. Am. Chem. Soc. 126:3477–3487. [DOI] [PubMed] [Google Scholar]
- 12.Baas, B. J., I. G. Denisov, and S. G. Sligar. 2004. Homotropic cooperativity of monomeric cytochrome P450 3A4 in a nanoscale native bilayer environment. Arch. Biochem. Biophys. 430:218–228. [DOI] [PubMed] [Google Scholar]
- 13.Bayburt, T. H., and S. G. Sligar. 2002. Single-molecule height measurements on microsomal cytochrome P450 in nanometer-scale phospholipid bilayer disks. Proc. Natl. Acad. Sci. USA. 99:6725–6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Civjan, N. R., T. H. Bayburt, M. A. Schuler, and S. G. Sligar. 2003. Direct solubilization of heterologously expressed membrane proteins by incorporation into nanoscale lipid bilayers. Biotechniques. 35:556–560, 562–553. [DOI] [PubMed] [Google Scholar]
- 15.Duan, H., N. R. Civjan, S. G. Sligar, and M. A. Schuler. 2004. Co-incorporation of heterologously expressed Arabidopsis cytochrome P450 and P450 reductase into soluble nanoscale lipid bilayers. Arch. Biochem. Biophys. 424:141–153. [DOI] [PubMed] [Google Scholar]
- 16.Bayburt, T. H., Y. V. Grinkova, and S. G. Sligar. 2006. Assembly of single bacteriorhodopsin trimers in bilayer Nanodiscs. Arch. Biochem. Biophys. 450:215–222. [DOI] [PubMed] [Google Scholar]
- 17.Bayburt, T. H., and S. G. Sligar. 2003. Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Sci. 12:2476–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leitz, A. J., T. H. Bayburt, A. N. Barnakov, B. A. Springer, and S. G. Sligar. 2006. Functional reconstitution of beta2-adrenergic receptors utilizing self-assembling Nanodisc technology. Biotechniques. 40:601–612. [DOI] [PubMed] [Google Scholar]
- 19.Frank, P. G., and Y. L. Marcel. 2000. Apolipoprotein A-I: structure-function relationships. J. Lipid Res. 41:853–872. [PubMed] [Google Scholar]
- 20.Ajees, A. A., G. M. Anantharamaiah, V. K. Mishra, M. M. Hussain, and H. M. Murthy. 2006. Crystal structure of human apolipoprotein A-I: insights into its protective effect against cardiovascular diseases. Proc. Natl. Acad. Sci. USA. 103:2126–2131. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Borhani, D. W., D. P. Rogers, J. A. Engler, and C. G. Brouillette. 1997. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc. Natl. Acad. Sci. USA. 94:12291–12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koppaka, V. 2001. Structural studies of discoidal lipoprotein A-I. Cell. Mol. Life Sci. 58:885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brouillette, C. G., G. M. Anantharamaiah, J. A. Engler, and D. W. Borhani. 2001. Structural models of human apolipoprotein A-I: a critical analysis and review. Biochim. Biophys. Acta. 1531:4–46. [DOI] [PubMed] [Google Scholar]
- 24.Nissen, S. E., T. Tsunoda, E. M. Tuzcu, P. Schoenhagen, C. J. Cooper, M. Yasin, G. M. Eaton, M. A. Lauer, W. S. Sheldon, C. L. Grines, S. Halpern, T. Crowe, J. C. Blankenship, and R. Kerensky. 2003. Effect of recombinant ApoA-I Milano on coronary atherosclerosis in patients with acute coronary syndromes: a randomized controlled trial. JAMA. 290:2292–2300. [DOI] [PubMed] [Google Scholar]
- 25.Phillips, J. C., W. Wriggers, Z. Li, A. Jonas, and K. Schulten. 1997. Predicting the structure of apolipoprotein A-I in reconstituted high-density lipoprotein disks. Biophys. J. 73:2337–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segrest, J. P., M. K. Jones, A. E. Klon, C. J. Sheldahl, M. Hellinger, H. De Loof, and S. C. Harvey. 1999. A detailed molecular belt model for apolipoprotein A-I in discoidal high density lipoprotein. J. Biol. Chem. 274:31755–31758. [DOI] [PubMed] [Google Scholar]
- 27.Shih, A. Y., I. G. Denisov, J. C. Phillips, S. G. Sligar, and K. Schulten. 2005. Molecular dynamics simulations of discoidal bilayers assembled from truncated human lipoproteins. Biophys. J. 88:548–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogers, D. P., L. M. Roberts, J. Lebowitz, G. Datta, G. M. Anantharamaiah, J. A. Engler, and C. G. Brouillette. 1998. The lipid-free structure of apolipoprotein A-I: effects of amino-terminal deletions. Biochemistry. 37:11714–11725. [DOI] [PubMed] [Google Scholar]
- 29.Maiorano, J. N., R. J. Jandacek, E. M. Horace, and W. S. Davidson. 2004. Identification and structural ramifications of a hinge domain in apolipoprotein A-I discoidal high-density lipoproteins of different size. Biochemistry. 43:11717–11726. [DOI] [PubMed] [Google Scholar]
- 30.Koppaka, V., L. Silvestro, J. A. Engler, C. G. Brouillette, and P. H. Axelsen. 1999. The structure of human lipoprotein A-I—evidence for the “belt” model. J. Biol. Chem. 274:14541–14544. [DOI] [PubMed] [Google Scholar]
- 31.Panagotopulos, S. E., E. M. Horace, J. N. Maiorano, and W. S. Davidson. 2001. Apolipoprotein A-I adopts a belt-like orientation in reconstituted high density lipoproteins. J. Biol. Chem. 276:42965–42970. [DOI] [PubMed] [Google Scholar]
- 32.Silva, R. A., G. M. Hilliard, L. Li, J. P. Segrest, and W. S. Davidson. 2005. A mass spectrometric determination of the conformation of dimeric apolipoprotein A-I in discoidal high density lipoproteins. Biochemistry. 44:8600–8607. [DOI] [PubMed] [Google Scholar]
- 33.Lins, L., S. Piron, K. Conrath, B. Vanloo, R. Brasseur, M. Rosseneu, J. Baert, and J. M. Ruysschaert. 1993. Enzymatic hydrolysis of reconstituted dimyristoylphosphatidylcholine-apo A-I complexes. Biochim. Biophys. Acta. 1151:137–142. [DOI] [PubMed] [Google Scholar]
- 34.Culot, C., F. Durant, S. Lazarescu, P. A. Thiry, B. Vanloo, M. Y. Rosseneu, L. Lins, and R. Brasseur. 2004. Structural investigation of reconstituted high density lipoproteins by scanning tunnelling microscopy. Appl. Surf. Sci. 230:151–157. [Google Scholar]
- 35.Mishra, V. K., G. M. Anantharamaiah, J. P. Segrest, M. N. Palgunachari, M. Chaddha, S. W. Sham, and N. R. Krishna. 2006. Association of a model class A (apolipoprotein) amphipathic alpha helical peptide with lipid: high resolution NMR studies of peptide.lipid discoidal complexes. J. Biol. Chem. 281:6511–6519. [DOI] [PubMed] [Google Scholar]
- 36.Opella, S. J., and F. M. Marassi. 2004. Structure determination of membrane proteins by NMR spectroscopy. Chem. Rev. 104:3587–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griffin, R. G. 1998. Dipolar recoupling in MAS spectra of biological solids. Nat. Struct. Biol. 5:508–512. [DOI] [PubMed] [Google Scholar]
- 38.Pauli, J., M. Baldus, B. van Rossum, H. de Groot, and H. Oschkinat. 2001. Backbone and side-chain 13C and 15N resonance assignments of the alpha-spectrin SH3 domain by magic angle spinning solid state NMR at 17.6 Tesla. Chembiochem. 2:272–281. [DOI] [PubMed] [Google Scholar]
- 39.Igumenova, T. I., A. E. McDermott, K. W. Zilm, R. W. Martin, E. K. Paulson, and A. J. Wand. 2004. Assignments of carbon NMR resonances for microcrystalline ubiquitin. J. Am. Chem. Soc. 126:6720–6727. [DOI] [PubMed] [Google Scholar]
- 40.Igumenova, T. I., A. J. Wand, and A. E. McDermott. 2004. Assignment of the backbone resonances for microcrystalline ubiquitin. J. Am. Chem. Soc. 126:5323–5331. [DOI] [PubMed] [Google Scholar]
- 41.Franks, W. T., D. H. Zhou, B. J. Wylie, B. G. Money, D. T. Graesser, H. L. Frericks, G. Sahota, and C. M. Rienstra. 2005. Magic-angle spinning solid-state NMR spectroscopy of the beta1 immunoglobulin binding domain of protein G (GB1): 15N and 13C chemical shift assignments and conformational analysis. J. Am. Chem. Soc. 127:12291–12305. [DOI] [PubMed] [Google Scholar]
- 42.Bockmann, A., A. Lange, A. Galinier, S. Luca, N. Giraud, M. Juy, H. Heise, R. Montserret, F. Penin, and M. Baldus. 2003. Solid state NMR sequential resonance assignments and conformational analysis of the 2 × 10.4 kDa dimeric form of the Bacillus subtilis protein Crh. J. Biomol. NMR. 27:323–339. [DOI] [PubMed] [Google Scholar]
- 43.Egorova-Zachernyuk, T. A., J. Hollander, N. Fraser, P. Gast, A. J. Hoff, R. Cogdell, H. J. M. de Groot, and M. Baldus. 2001. Heteronuclear 2D-correlations in a uniformly [C-13, N-15] labeled membrane-protein complex at ultra-high magnetic fields. J. Biomol. NMR. 19:243–253. [DOI] [PubMed] [Google Scholar]
- 44.Gammeren, A. J., F. B. Hulsbergen, J. G. Hollander, and H. J. Groot. 2005. Residual backbone and side-chain 13C and 15N resonance assignments of the intrinsic transmembrane light-harvesting 2 protein complex by solid-state magic angle spinning NMR spectroscopy. J. Biomol. NMR. 31:279–293. [DOI] [PubMed] [Google Scholar]
- 45.Holdeman, T. C., and K. H. Gardner. 2001. Letter to the Editor: H-1, C-13 and N-15 chemical shift assignments of the N-terminal PAS domain of mNPAS2. J. Biomol. NMR. 21:383–384. [DOI] [PubMed] [Google Scholar]
- 46.Stringer, J. A., C. E. Bronnimann, C. G. Mullen, D. H. Zhou, S. A. Stellfox, Y. Li, E. H. Williams, and C. M. Rienstra. 2005. Reduction of RF-induced sample heating with a scroll coil resonator structure for solid-state NMR probes. J. Magn. Reson. 173:40–48. [DOI] [PubMed] [Google Scholar]
- 47.Hediger, S., B. H. Meier, N. D. Kurur, G. Bodenhausen, and R. R. Ernst. 1994. NMR cross-polarization by adiabatic passage through the Hartmann-Hahn condition (APHH). Chem. Phys. Lett. 223:283–288. [Google Scholar]
- 48.Bennett, A. E., C. M. Rienstra, M. Auger, K. V. Lakshmi, and R. G. Griffin. 1995. Heteronuclear decoupling in rotating solids. J. Chem. Phys. 103:6951–6958. [Google Scholar]
- 49.Delaglio, F., S. Grzesiek, G. W. Vuister, G. Zhu, J. Pfeifer, and A. Bax. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 6:277–293. [DOI] [PubMed] [Google Scholar]
- 50.Morcombe, C. R., and K. W. Zilm. 2003. Chemical shift referencing in MAS solid state NMR. J. Magn. Reson. 162:479–486. [DOI] [PubMed] [Google Scholar]
- 51.Martin, R. W., and K. W. Zilm. 2003. Preparation of protein nanocrystals and their characterization by solid state NMR. J. Magn. Reson. 165:162–174. [DOI] [PubMed] [Google Scholar]
- 52.Baldus, M., A. T. Petkova, J. Herzfeld, and R. G. Griffin. 1998. Cross polarization in the tilted frame: assignment and spectral simplification in heteronuclear spin systems. Mol. Phys. 95:1197–1207. [Google Scholar]
- 53.BioMagResBank. http://www.bmrb.wisc.edu.
- 54.Takegoshi, K., S. Nakamura, and T. Terao. 2001. C-13-H-1 dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 344:631–637. [DOI] [PubMed] [Google Scholar]
- 55.Wishart, D. S., and B. D. Sykes. 1994. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR. 4:171–180. [DOI] [PubMed] [Google Scholar]
- 56.Cornilescu, G., F. Delaglio, and A. Bax. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 13:289–302. [DOI] [PubMed] [Google Scholar]
- 57.Luca, S., D. V. Filippov, J. H. van Boom, H. Oschkinat, H. J. de Groot, and M. Baldus. 2001. Secondary chemical shifts in immobilized peptides and proteins: a qualitative basis for structure refinement under magic angle spinning. J. Biomol. NMR. 20:325–331. [DOI] [PubMed] [Google Scholar]
- 58.Iwadate, M., T. Asakura, and M. P. Williamson. 1999. C alpha and C beta carbon-13 chemical shifts in proteins from an empirical database. J. Biomol. NMR. 13:199–211. [DOI] [PubMed] [Google Scholar]
- 59.Li, L., J. Chen, V. K. Mishra, J. A. Kurtz, D. Cao, A. E. Klon, S. C. Harvey, G. M. Anantharamaiah, and J. P. Segrest. 2004. Double belt structure of discoidal high density lipoproteins: molecular basis for size heterogeneity. J. Mol. Biol. 343:1293–1311. [DOI] [PubMed] [Google Scholar]
- 60.Lorch, M., S. Fahem, C. Kaiser, I. Weber, A. J. Mason, J. U. Bowie, and C. Glaubitz. 2005. How to prepare membrane proteins for solid-state NMR: a case study on the alpha-helical integral membrane protein diacylglycerol kinase from E. coli. ChemBioChem. 6:1693–1700. [DOI] [PubMed] [Google Scholar]
- 61.Rogers, D. P., L. M. Roberts, J. Lebowitz, J. A. Engler, and C. G. Brouillette. 1998. Structural analysis of apolipoprotein A-I: effects of amino- and carboxy-terminal deletions on the lipid-free structure. Biochemistry. 37:945–955. [DOI] [PubMed] [Google Scholar]
- 62.Brouillette, C. G., and G. M. Anantharamaiah. 1995. Structural models of human apolipoprotein A-I. Biochim. Biophys. Acta. 1256:103–129. [DOI] [PubMed] [Google Scholar]
- 63.Klon, A. E., J. P. Segrest, and S. C. Harvey. 2002. Comparative models for human apolipoprotein A-I bound to lipid in discoidal high-density lipoprotein particles. Biochemistry. 41:10895–10905. [DOI] [PubMed] [Google Scholar]
- 64.Lovell, S. C., I. W. Davis, W. B. Arendall 3rd, P. I. de Bakker, J. M. Word, M. G. Prisant, J. S. Richardson, and D. C. Richardson. 2003. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins. 50:437–450. [DOI] [PubMed] [Google Scholar]
- 65.Spera, S., and A. Bax. 1991. Empirical correlation between protein backbone conformation and C-alpha and C-beta C-13 nuclear-magnetic-resonance chemical-shifts. J. Am. Chem. Soc. 113:5490–5492. [Google Scholar]
- 66.Kuszewski, J., J. Qin, A. M. Gronenborn, and G. M. Clore. 1995. The impact of direct refinement against 13C alpha and 13C beta chemical shifts on protein structure determination by NMR. J. Magn. Reson. B. 106:92–96. [DOI] [PubMed] [Google Scholar]
- 67.Beger, R. D., and P. H. Bolton. 1997. Protein phi and psi dihedral restraints determined from multidimensional hypersurface correlations of backbone chemical shifts and their use in the determination of protein tertiary structures. J. Biomol. NMR. 10:129–142. [DOI] [PubMed] [Google Scholar]
- 68.Neal, S., A. M. Nip, H. Zhang, and D. S. Wishart. 2003. Rapid and accurate calculation of protein 1H, 13C and 15N chemical shifts. J. Biomol. NMR. 26:215–240. [DOI] [PubMed] [Google Scholar]
- 69.Heise, H., S. Luca, B. L. de Groot, H. Grubmuller, and M. Baldus. 2005. Probing conformational disorder in neurotensin by two-dimensional solid-state NMR and comparison to molecular dynamics simulations. Biophys. J. 89:2113–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hohwy, M., C. M. Rienstra, C. P. Jaroniec, and R. G. Griffin. 1999. Fivefold symmetric homonuclear dipolar recoupling in rotating solids: application to double quantum spectroscopy. J. Chem. Phys. 110:7983–7992. [Google Scholar]