Abstract
Four different tandem EGFPs were constructed to elucidate the nuclear microenvironment by quantifying its diffusional properties in both aqueous solution and the nuclei of living cells. Diffusion of tandem EGFP was dependent on the length of the protein as a rod-like molecule or molecular ruler in solution. On the other hand, we found two kinds of mobility, fast diffusional mobility and much slower diffusional mobility depending on cellular compartments in living cells. Diffusion in the cytoplasm and the nucleoplasm was mainly measured as fast diffusional mobility. In contrast, diffusion in the nucleolus was complex and mainly much slower diffusional mobility, although both the fast and the slow diffusional mobilities were dependent on the protein length. Interestingly, we found that diffusion in the nucleolus was clearly changed by energy depletion, even though the diffusion in the cytoplasm and the nucleoplasm was not changed. Our results suggest that the nucleolar microenvironment is sensitive to energy depletion and very different from the nucleoplasm.
INTRODUCTION
The cell nucleus contains many proteins that form a multimolecular complex or a material such as chromatin and a nucleolus. Most of the proteins in the nucleus are concerned with molecular processing such as ribosome biogenesis, mRNA synthesis, transcription and molecular transportation to and from the nucleus. For these processes to be accomplished properly, proteins related to each process are expected to act dynamically and precisely in the nucleus. Consequently, the dynamics of various molecules such as RNAs and nuclear proteins in living cells have become a subject of major interest because mobilities of such molecules in the nucleus could provide important information about the molecular functions of the nucleus (1–3). On the other hand, such mobility of functional protein molecules in the nucleus might be mainly affected by the nuclear architecture and microenvironment (1,4) as well as their function because the chromosomes and the nucleoli occupy a large portion of the nuclear space and changes depending on many factors such as gene expression, cell cycle progression, and other metabolic state of the cell. Therefore, for understanding the relation between functional proteins and nuclear microenvironment, it is helpful to analyze mobility of standard protein molecules with well-defined hydrodynamic properties as well as functional nuclear proteins (1,5,6) or labeled macromolecules (7).
In the last few years, many studies based on fluorescence microscopic techniques such as FRAP, single particle tracking (SPT), and fluorescence correlation spectroscopy (FCS) have been carried out for cell biology (8–15). The studies showed that a variety of small fluorescent probes such as BCECF (9), fluorescein-labeled macromolecules (dextran and Ficoll) from 3 to 1000 kD (13), and monomeric EGFP (14), move rapidly in the cytoplasm, whereas labeled linear dsDNA diffuses very slowly and has a size dependence of the diffusion constant (16). The key point of these studies is that the diffusion of small dextrans and Ficolls in the cytoplasm is only restricted mildly whereas that for large macromolecules can be greatly slowed.
On the other hand, a few studies of protein mobility in the cell nucleus have been carried out (10,13,14) with biologically inert protein, even though many studies have been carried out with nuclear proteins (1,3,6). A study based on FRAP and microinjection with diverse sizes of fluorescein-labeled dextrans (13) showed that diffusion in the nucleus was slowed approximately fourfold compared with their diffusion in water. However, more variability in the measured data for the nucleus was found than for cytoplasm. Monomer GFP molecule showed much more complex diffusion in nucleus than in cytoplasm (14). Recent studies of FRAP (1,17) and FCS combined with FRAP experiment (18) using living cells have shown that various EGFP-fused nuclear proteins diffuse at different rates depending on their localization and function. Nuclear proteins could interact with target molecules or immobile structures such as chromatin, which slowed down the mobility of the proteins (5,6,19). An FCS experiment with monomeric EGFP showed that diffusion of EGFP, which is presumably inert to other proteins, was restricted depending on the position in the nucleus compared to diffusion in the cytoplasm (14). Furthermore, whether intranuclear mobility of many molecules results from passive diffusion or active transport is still controversial (3,20). The nuclear microenvironment, which may be one of the reasons, has not yet been clearly quantified under various physiological conditions.
FCS has been applied as a powerful technique for assessing biomolecular diffusion and interactions both in aqueous conditions and in living cells with single-molecule sensitivity (21–26). FCS detects fluorescence intensity fluctuations caused by Brownian motion of fluorescent probe molecules in a tiny detection volume (∼0.3 fL) generated by confocal illumination. Through time correlation analysis of the fluorescence fluctuations, the diffusion coefficient, the molecular concentration, and the molecular interaction of probe molecules are accessible. Because FCS need only a very small detection volume and has high sensitivity, it will also be useful to measure diffusional mobility of proteins in very small regions of subnuclear microenvironments in living cells. Although FRAP is adequate for measuring the diffusion of fluorescent molecules and possible exchange in target organelles in the living cell (3,7), the measurable minimal fluorescent intensity and diffusional speed range are limited to brighter and slower ranges than those for FCS. Therefore, we can anticipate that FCS will provide complementary information for faster movement at lower expression levels of various functional proteins in the nucleus.
EGFP is a powerful fluorescent bioprobe molecule with a well-known cylindrical structure (27–29). It has recently been used for various cell measurements in fluorescent imaging of cells as well as for analysis of molecular diffusion using FRAP and FCS. To develop a standard and reproducible method for diffusion analysis of proteins, we designed multiple oligomeric EGFPs with different molecular weights, which can be used as molecular rulers (MRs) for quantification of protein mobility in the nucleus. For this purpose, we constructed plasmids with different levels of oligomeric EGFPn (EGFP2–EGFP5, n = 2–5) with molecular weights of 60, 90, 120, and 150 kD, respectively, tandemly linked by a random amino acid linker. Using multiple oligomeric EGFPs and FCS, we determined the diffusion of the proteins in the cytoplasm, nucleoplasm, and nucleoli of living HEK293, HeLa, and COS7 cells. For strict recognition of the two compartments in the nucleus, mRFP-fibrillarin and H2B-mRFP were used as red fluorescent markers for the nucleolus and the nucleoplasm, respectively.
In this study, FCS analysis by a one-component model showed that the diffusional mobility of EGFPn in aqueous solution was dependent on the length of EGFPn and was well consistent with the diffusion model of a rod-like structure. On the other hand, the diffusion of EGFPn in living cells analyzed by a two-component model showed that fast diffusional mobility in the cytoplasm and the nucleoplasm was consistent with the model of a rod-like molecule as shown in aqueous solution. The fast diffusion rates in the cytoplasm and the nucleoplasm were almost the same, and ∼3.5-fold slower than in solution, regardless of the size of tandem EGFPn and cell type. Mobilities of tandem EGFPn found in the nucleoli of HeLa and COS7 cells were fivefold and sevenfold slower than the fast diffusional mobility in the cytoplasm and the nucleoplasm, respectively. Moreover, the much slower diffusional mobility in the nucleolus was also dependent on the length of EGFPn, demonstrating tandem EGFP molecules were well-defined both in solution and in living cells. Interestingly, the slow diffusion in the nucleolus was related to the energy level of the living cell, because the slow diffusion of EGFP5 in the nucleolus, but not in the cytoplasm and the nucleoplasm, was further slowed by ATP depletion.
MATERIALS AND METHODS
Plasmid construction of tandem EGFP
Plasmids expressing each tandem EGFPn were synthesized with the plasmid expressing EGFP-C1 (Clontech, Palo Alto, CA). The EGFP-C1 was excised at the NdeI and the SmaI restriction sites and ligated between the NdeI and Eco47 III restriction sites of another EGFP-C1. The linker between EGFPn containing 25 random amino acid residues (SGLRSRAQASNSAVDGTAGPLPVAT) originated from the remaining bases of the multiple-cloning site. Plasmid constructs of H2B-mRFP and mRFP-fibrillarin were obtained as gifts from Drs. H. Kimura (Kyoto University, Kyoto, Japan) (30,31) and T. Saiwaki (Osaka University, Osaka, Japan) (32), respectively. All plasmid constructs for transfection were purified using a plasmid DNA midiprep kit (QIAGEN, Hilden, Germany).
Cell culture and expression of tandem EGFPn proteins
For transient expression of tandem EGFPn, human embryonic kidney 293 (HEK293), HeLa, and COS7 cells were plated at confluence levels of 10–20% on LAB-TEK chambered coverslips with eight wells (Nalge Nunc International, Rochester, NY) for 12 or 24 h before transfection. Cells were transfected with a EGFPn vector or cotransfected with a vector of EGFPn and H2B-mRFP or mRFP-fibrillarin, and grown in a 5% CO2 humidified atmosphere at 37°C in Dulbecco's modified Eagle's medium (DMEM, Sigma-Aldrich, St. Louis, MO) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 10 mg/ml streptomycin. Transfection was carried out with FuGENE 6 (Roche Molecular Biochemicals, Mannheim, Germany) or Effectene (QIAGEN) as indicated by the manufacturer. The transfected cells were incubated for 24 or 48 h and washed with Opti-MEM to remove phenol red dye in DMEM, and then the medium was replaced by Opti-MEM before LSM and FCS measurements. Energy depletion was performed by addition of 6 mM 2-deoxyglucose (2-DG, Sigma-Aldrich) and 10 mM sodium azide (NaN3, Sigma) to the culture medium (3,20). LSM images were collected for the same cells before and after 2-DG and NaN3 treatment.
Western immunoblotting
The immunoblot analysis was performed according to the standard method. Cells expressing tandem EGFPn were grown on 10-cm culture plates for 48 h after transfection, the BD Living Colors A.v. peptide antibody (BD Biosciences Clontech, Mountain View, CA) was used as the primary antibody. Primary antibody-bound protein bands were detected with an alkaline phosphatase-conjugated secondary antibody (mouse anti-rabbit IgG, Chemicon International, Temecula, CA) by BCIP/NBT dye solution (Sigma-Aldrich).
Cell homogenization
After FCS measurements, the cultured cells on a Lab-Tek (Nalge Nunc International) chambered coverslip were collected by centrifugation at 1500 rpm for 5 min and then the pellets of cells were homogenized in 50 μl of buffer (10 mM Hepes pH 7.9 containing 10 mM NaCl, 3 mM MgCl2, 1 mM DTT, 0.4 mM PMSF, and 0.1 mM sodium orthovanadate). Each EGFPn protein solution was collected from the supernatant after centrifugation at 100,000 rpm for 20 min and measured by FCS again.
Live cell imaging
Fluorescence microscopy was performed using an LSM510 inverted confocal laser scanning microscopy (LSM; Carl Zeiss, Jena, Germany). LSM observations were all performed at 25°C. EGFPn was excited at 488 nm of a CW Ar+ laser through a water immersion objective lens (C-Apochromat, 40×, 1.2 NA; Carl Zeiss) with emission detected above 505 nm for single scanning experiments using cells expressing EGFPn. Monomeric RFP-fibrillarin or H2B-mRFP was imaged using a 543-nm laser light and detection was above 560 nm. The pinhole diameters for confocal imaging were adjusted to 70 μm and 80 μm for EGFP and mRFP, respectively. To avoid bleed-through effects in double-scanning experiments, EGFP and mRFP were scanned independently in a multitracking mode.
FCS measurements and quantitative analysis
FCS measurements were all performed at 25°C on a ConfoCor 2 (Carl Zeiss) as described previously (24,33). Excitation of EGFP was carried out at 488 nm and 6.3 mW by adjusting an acousto-optical tunable filter (AOTF) to 0.1%. Fluorescence autocorrelation functions ((FAF) G (τ)), from which the average residence time (τi) and the absolute number of fluorescent proteins in the detection volume were obtained as follows:
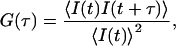 |
(1) |
where I (t + τ) is the fluorescence intensity in single photon counting method obtained from the detection volume at delay time τ. Brackets denote ensemble averages. The curve fitting for the multicomponent model is given by:
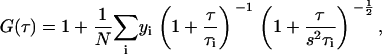 |
(2) |
where yi and τi are the fraction and diffusion time of component i, respectively. N is the number of fluorescent molecules in the detection volume defined by the beam waist w0 and the axial radius z0, s is the structure parameter representing the ratio of w0 and z0. The detection volume made by w0 and z0 was approximated as a cylinder.
All FAFs in aqueous solutions were measured for 30 s five times at 5-s intervals. In the case of intracellular measurement, FAFs were measured for 15 s one or three times, and very low fluorescent cells under concentration of 20 molecules (<0.1 M) per detection volume (0.3 fL) were chosen for FCS measurement. Under these conditions, the effect of photobleaching on FCS analysis was minimized. The measurement position was chosen in the LSM image. Because the optical passes of LSM and FCS are not the same, the real position of FCS measurement was tuned to the position on LSM images with a coverglass coated by dried rhodamine 6G (Rh6G), following the protocol provided by the manufacturer (34). The real position of FCS measurement was also checked with bleaching of H2B-mRFP in living cells. Although there was no significant difference between the position of FCS measurements checked by a coverglass and living cells, misalignment under 1 μm was found. This range of misalignment may not affect analysis of diffusion in the region of nucleoplasm and nucleolus, diameters of which were of the order of 10 and 2 μm, respectively. The detection pinhole for FCS was fixed to a diameter of 70 μm and emission was recorded through a 505–550-nm bandpass filter for measurement of cells expressing EGFPn or through a 505–530-nm bandpass filter for measurement of cells coexpressing EGFPn and mRFP tagged proteins for excluding any cross-talk signal from mRFP. In practice, there was almost no cross-talk signal from mRFP using the three cell types expressing mRFP only. The fluorescence of cells expressing mRFP shows almost the same background fluorescence signal under 15 cps when a 505–550-nm bandpass filter was used. All measured FAFs were fitted by the fit program installed on the ConfoCor 2 system using the model Eq. 2. FAFs in aqueous solutions were fitted by a one-component model (i = 1), and FAFs in cells by a one- or two-component model (i = 1 or 2) to consider free diffusion and restricted diffusion, respectively (see also the text). The pinhole adjustment of the FCS setup, structure parameter, and detection volume were calibrated everyday by FCS measurements of Rh6G solution with a concentration of 10−7 M. Although the structure parameters determined by Rh6G after the pinhole adjustment were changed and ranged from 4 to 8 each day, FCS analysis was carried out with data sets with structure parameters ranging from 5 to 6, which are known to be a stable condition for FCS measurement. An average value of structure parameter was fixed for FCS analysis of all data carried out in a day under the same conditions. Diffusion time of component i, τi, is related to the translational diffusion constant D of component i by
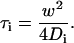 |
(3) |
Diffusion of a spheroidal molecule is related to various physical parameters by the Stokes-Einstein equation as follows
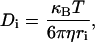 |
(4) |
where T is the absolute temperature, ri is the hydrodynamic radius of the spheroidal molecule, η is the fluid-phase viscosity of the solvent, and kB is the Boltzman constant. Because τi is proportional to viscosity, the relative viscosity (τcell/τsolution) can be easily estimated. When the diffusion time of Rh6G is measured and the molecular weight of the sample molecule is known, the diffusion time of the sample molecule as a spherical shape can be simply calculated by the following equation (23).
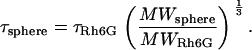 |
(5) |
The diffusion time τ is also related to the frictional coefficient of the diffusing molecules, which depends on the shapes of molecules undergoing diffusion in a solution of defined viscosity. The ratio of the frictional coefficient between spheroidal (f0) and ellipsoidal (f) molecules and the relationship between the diffusion time and frictional coefficient are given by Perrin's equation (35,36)
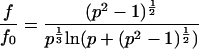 |
(6) |
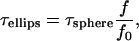 |
(7) |
where f and f0 are frictional coefficients of ellipsoidal and spherical molecules, respectively, p is the axial ratio of the ellipsoidal molecule, τellips and τsphere are the diffusion times of ellipsoidal and spherical molecules, respectively. Based on the known size of the EGFP molecule, 4 nm in length and 3 nm in diameter, and the average length of 25 amino acids, the predicted diffusion time of tandem EGFPn was calculated for spherical and ellipsoidal models using Eqs. 5, 6, and 7. The lengths of amino acid linkers used for the calculation were 3.7 nm for an α-helix structure and 9.1 nm for a simple linear structure of 25 amino acids. The diffusion constants of EGFPns (Dgfpn) in the solution and cells were calculated from the published diffusion constant of Rh6G, DRh6G (280 μm2/s) (37), and measured diffusion times of Rh6G (τRh6G) and EGFPns (τgfpn) as follows:
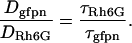 |
(8) |
RESULTS
LSM observation
Expressed oligomeric EGFPn localized in the nucleus
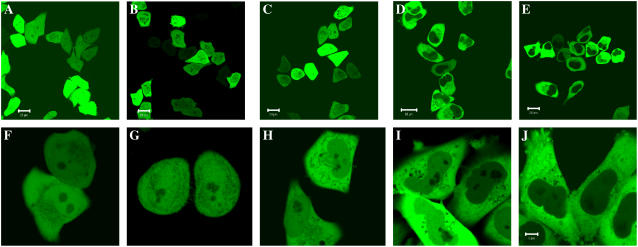
To observe the distribution and localization of monomer EGFP and oligomeric EGFPn in HEK293, COS7, and HeLa cells, the cells were transiently transfected with DNA plasmids encoding EGFPn or cotransfected with plasmids encoding each EGFPn and H2B-mRFP. Cells expressing each oligomeric EGFP were observed at 24–48 h after transfection. Typical LSM images of HeLa cells expressing each EGFPn taken at 24 h after transfection are shown in Fig. 1. Monomer EGFP and EGFP2 were uniformly distributed through the cytoplasm and nucleus in each cell except in the nucleolus (Fig. 1 A (F) and B (G)). In contrast, EGFP3, EGFP4, and EGFP5 showed different distribution patterns in the cytoplasm and the nucleus. In the case of EGFP3, the fluorescent intensity of proteins in the cytoplasm was higher than that in the nucleus, although the difference was not significant (Fig. 1, C and H). For EGFP4 and EGFP5, the fluorescent intensity in the nucleus was much weaker than that in the cytoplasm (Fig. 1 D (I) and E (J)). However, the fluorescence intensity of EGFP4 and EGFP5 in the nucleus was sufficient to be detected by LSM measurement (Fig. 1, I and J). The fluorescent intensities in the nucleus for EGFP2, EGFP3, EGFP4, and EGFP5 at 48 h were increased compared with these at 24 h. For all oligomeric EGFPn, there was no speckled or aggregated distribution in the cytoplasm and the nucleoplasm and the fluorescence in the nucleoplasm except in the nucleolus had a uniform pattern (Fig. 1, F–J). This uniform pattern of fluorescence in the nucleoplasm was confirmed by comparing the fluorescence of tandem EGFP with that of H2B-mRFP on HeLa or COS7 cells coexpressing EGFP3 and H2B-mRFP or EGFP5 and H2B-mRFP, respectively (Fig. S1 in Supplementary Material), because it is known that H2B-GFP show heterogeneous fluorescent pattern in the nucleus depending on the density of chromatin (4,31,38). In the case of HEK293 and COS7 cells transfected with the tandem EGFPn, the difference of fluorescence intensity between the cytoplasm and nucleus was clearly discriminated from EGFP3 regardless of the expression level, and the fluorescent intensity in the nucleus was decreased with the increase in size of tandem GFP (C. Pack and M. Kinjo, unpublished data).
FIGURE 1.
Tandem EGFP located in the nucleus. LSM images of HeLa cells expressing (A) monomer EGFP, (B) EGFP2, (C) EGFP3, (D) EGFP4, and (E) EGFP5, are shown. The images were taken at 24 h and 48 h after transfection with EGFP and tandem EGFPn, respectively. Bars, 20 μm. Panels F and J show enlarged images of cells expressing EGFPn from EGFP1 to EGFP5, respectively, showing their location on the nucleus. Bars, 10 μm. The fluorescent intensity in the nucleus decreased with the increase in size of the oligomers. Tandem EGFPs, regardless of their size, were equally distributed in the nucleoplasm except the nucleolus (see also supplementary Fig. S1).
LSM observations of HeLa cell indicated that tandem EGFP bigger than EGFP3 (>90 kD) had difficulty localizing in the nucleus. The localization of EGFPn in the nucleus was dependent on the size of the EGFPn molecule. Although all types of tandem GFPn could be localized in the nucleus, there was less tandem EGFP4 and EGFP5 in the nucleus than monomeric EGFP, EGFP2, and EGFP3. The small number of EGFP molecules in the nucleus (from 50 to 5 molecules in the detection volume of 0.3 fL) might be sufficient to be detected by FCS measurement even in very weak fluorescent cells. For the weak fluorescent intensity in the nucleus for EGFP3, EGFP4, and EGFP5, we did not need to select weakly expressing cells as explained in Materials and Methods, and could easily perform FCS measurement in the nucleus.
FCS measurement in aqueous solution
Tandem EGFPn diffuse in solution like a rod-like molecule
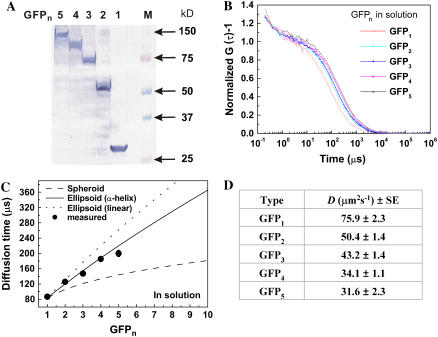
For analysis of the diffusion properties of monomer EGFP and oligomeric EGFPn in aqueous solution, cells transfected with the EGFPn were homogenized and the proteins from the cell lysate were extracted and measured in aqueous solution. There was no drastic change or burst of average fluorescent intensity during FCS measurement resulting from aggregated EGFP molecules or contaminants from the homogenized cell extracts during the measurement time of 60 s. The FAF of each tandem EGFPn was analyzed by a one-component model (Eq. 2, i = 1) and was well fitted. Fig. 2 A shows typical FAFs of EGFPn obtained from aqueous solution. For comparison of the extents of diffusion speeds, the amplitude of G (τ) (G (0) − 1) was normalized to unity. The autocorrelation functions of EGFPn shifted gradually to the right depending on the molecular weight of tandem EGFPn (Fig. 2 B). Diffusion times corresponding to the FAFs of EGFP1∼5 were 86.8 ± 3.9 μs, 125.8 ± 2.7 μs, 147.4 ± 4.8 μs, 185.4 ± 5.4 μs, and 200 ± 8.5 μs, respectively (Fig. 2 C). These results indicated that the diffusional mobility of EGFPn decreased with increasing molecular weight. That there was no degradation of monomeric and tandem EGFPn was also confirmed by the Western blotting results (Fig. 2 B), which were well consistent with the expected molecular weight of each oligomeric EGFPn. Diffusion constants of monomeric and oligomeric EGFPn in solution are summarized in Fig. 2 D. The diffusion constant (76 μm2s−1) of monomeric EGFP (263 amino acids, 30 kD) was similar to those (87 μm2s−1) of previous studies (24,39,40) with recombinant GFP (238 amino acids, 27 kD) synthesized by bacterial expression.
FIGURE 2.
FCS measurement of tandem EGFPn in aqueous solution. (A) Immunoblots of EGFP and tandem EGFPn and (B) normalized FAFs of the proteins in aqueous solution are shown. Lysates from HEK293 cells expressing monomer EGFP and oligomer EGFPn were blotted. The amplitude of FAF, G (0) − 1, was normalized to unity for comparison of the extent of diffusional speed. Diffusion times obtained by fitting the functions (B) with a one-component model (Eq. 2, i = 1) are plotted in panel C. Error bar shows mean ± SD of three measurements. Dashed line shows diffusion times calculated by Eq. 5 using the molecular weight and the measured diffusion time of Rh6G assuming the oligomeric EGFPn is spherical. Solid and dotted lines show the calculated diffusion times assuming that the linker between EGFP forms a rigid α-helix and a linear shape, respectively. (D) Diffusion constants for EGFP and tandem EGFPn in aqueous solution. Each diffusion constant was calculated from Eq. 8 using the known diffusion constant (280 μm2s−1) and the measured diffusion time (21 ± 2 μs) of Rh6G. Data are averaged from five independent experiments. (Mean ± SE of five independent experiments.)
Oligomeric EGFPn contains a linker of 25 random amino acids connecting monomer EGFP molecules. Consequently, oligomeric EGFPn can have different molecular shapes from spherical to linear. Because the linker can change p, the axial ratio of the protein molecule (Eq. 6), the diffusional mobilities of tandem types of EGFPn from EGFP2 to EGFP5 may reflect the diffusional mobility of an ellipsoidal or rod-like molecule. For this case, diffusion times of oligomeric EGFPn from EGFP2 to EGFP5 could be much slower than those of the proteins in spherical shape. Fig. 2 C shows a plot of the measured diffusion time (solid circles) of each EGFP and three plots of predicted diffusion times calculated by diffusion models for the spherical shape and two rod-like shapes with different p-values (Eqs. 5–7). Enhanced EGFP has a well-known cylindrical structure with a diameter of ∼3 nm and height of ∼4 nm (27). For simplification, monomer EGFP was assumed to be a spherical molecule and then the diffusion time of oligomeric EGFPn was calculated as a spherical molecule or rod-like molecule by Eq. 5. The measured diffusion time of monomer EGFP (30 kD) agreed well with the calculated value obtained from Eq. 5 using the empirical diffusion time (21 ± 2 μs) and the known molecular weight (0.479 kD) of Rh6G. The dashed line in Fig. 2 C plots the calculated diffusion time of oligomeric EGFPn with a spherical shape. The other two lines plot the predicted diffusion times of rod-like oligomeric EGFPn assuming that the amino acid linkers have an α-helix (solid line) or a linear structure (dotted line) with lengths of ∼3.7 nm and ∼9.1 nm, respectively. With this simple assumption, EGFP1∼5 have longitudinal lengths of 4, 12, 20, 28, and 36 nm, respectively, for an α-helix linker and 4, 17, 30, 43, and 56 nm, respectively, for a linear linker. As shown in Fig. 2 C, the measured diffusion times of oligomeric EGFPn (solid circles) are much longer than the calculated diffusion times of the EGFPn as a spherical molecule and well agreed with the rod-like model for the α-helix linker, even though the diffusion time of EGFP5 was slightly shorter than the calculated value. This indicated that diffusion of monomer and oligomeric EGFPn from EGFP2 to EGFP5 in solution reflected free diffusion of rod-like molecules and depended on the putative length of the oligomeric EGFP. Consequently, we concluded that monomeric and oligomeric EGFPn could be used as molecular rulers that change the diffusion time according to their own longitudinal length. This property of tandem EGFPn will be useful to analyze mobility of proteins in organelles, particularly in the subnuclear microenvironment.
LSM and FCS measurement in cells
FCS measurements of oligomeric EGFPn in vivo were performed using three cell lines, HEK293, COS7, and HeLa. Cells expressing a comparatively low concentration of EGFPn under ∼20 molecules (<0.1 μM) per detection volume (0.3 fL) were chosen because a dilute concentration of fluorescent molecules is adequate for FCS measurement. Even with this condition, there might be photobleaching effect on FCS measurements. Recently, a method combining FCS with photobleaching analysis was reported for studying intracellular binding and diffusion of functional proteins (41). This study suggested that the method is applicable to analyze mobility of monomer EGFP even in highly fluorescent cells. Nevertheless, it is noted that our study focused on the mobility of freely moving tandem EGFPn in the microenvironment containing the detection volume, but not that of immobile tandem EGFPn, which gives rise to a photobleaching and make FCS analysis more complex. For excluding a possible photobleaching effect, we carefully selected cells with weak fluorescence or without photobleaching during FCS measurement.
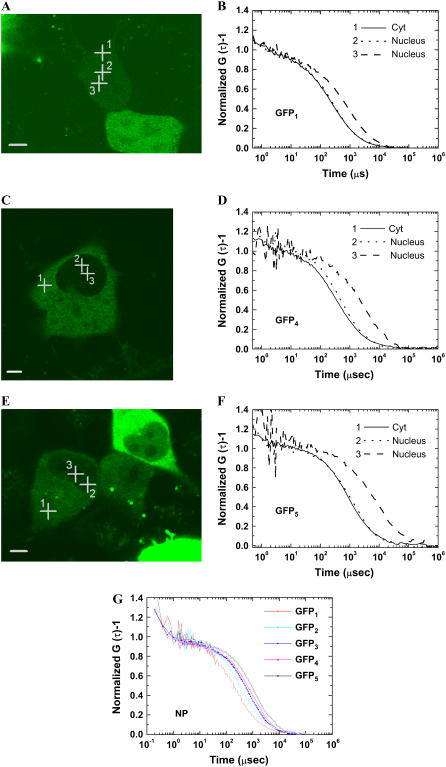
All FCS measurements were performed after taking LSM images and multiple positions for FCS measurements in the cytoplasm excepting endoplasmic reticulum and plasma membrane, and multiple positions in the nucleus were chosen in the LSM image of a cell. After FCS measurements, an LSM image was taken again to check whether measured positions of FCS were deviated from the LSM images. In weakly fluorescent cells, it was not easy to discriminate the nucleolus from the nucleoplasm, particularly, in cells expressing EGFP3, EGFP4, and EGFP5, in which most of the proteins were located in the cytoplasm and only a few EGFP molecules were located in the nucleus. Fig. 3 shows typical examples of LSM and FCS measurements for the three cell lines. LSM images for FCS measurement of a HEK cell expressed by EGFP1, a COS7 cell by EGFP4, and a HeLa cell by EGFP5 are shown in Fig. 3, A, C, and E, respectively. On the weakly fluorescent HEK cell expressing EGFP (Fig. 3 A), the boundary between the cytoplasm and nucleus was not clear. On the other hand, the cells expressing EGFP4, and EGFP5 (Fig. 3, C and E) show a clear contrast of the boundary resulting from the difference of fluorescence intensity between the cytoplasm and nucleus. The boundary between the cytoplasm and nucleus was not clear for weakly fluorescent cells expressing EGFP1 and EGFP2, regardless of the cell type. However, the boundary was clearly visible with cells expressing EGFP3, EGFP4, and EGFP5, depending on the size of tandem EGFPn, even though the fluorescence signals of the cells were weak. The clear boundary between the cytoplasm and nucleus for EGFP3, EGFP4, and EGFP5 made it easy to discriminate the two.
FIGURE 3.
Two kinds of diffusional mobility in the nucleus. For FCS analysis, very weakly fluorescent and nonphotobleaching cells less than the molecular number of N = 20 in the detection volume of FCS, which corresponds to a concentration under 10−7 M, were selected at 24 h after transfection. After recording LSM images of selected (A) HEK, (C) COS7, and (E) HeLa cells, FCS measurements were performed on multiple places in the cytoplasm and in the nucleus. Bars, 5 μm. For clarification, only three typical and normalized FAFs of EGFP1, EGFP4, and EGFP5 in the cytoplasm (curve 1) and the nucleus (curves 2 and 3) in HEK, COS7, and HeLa cells are shown in panels B, D, and F, respectively. The rightward shift of the FAF curve indicates the slow-diffusion-mobility. The nucleus has both fast diffusion (curve 2) and much slower diffusion (curve 3). The normalized FAFs of EGFP and tandem EGFPn in the nucleus of a HeLa cell with the fast-diffusion-mobility (curve 2) are summarized in panel G. The normalized FAFs of the fast-diffusion-mobility were gradually shifted to the right according their molecular size. (Cyt, cytoplasm).
FCS analysis in living cells
For all cells expressing EGFP1 or tandem EGFPn, diffusive fluorescent regions in the cytoplasm and multiple positions in the nucleus were measured by FCS. Examples of FAFs of EGFP in HEK, EGFP4 in COS7, and EGFP5 in HeLa cells are shown in Fig. 3, B, D, and F, respectively. Cross-hairs in the LSM images correspond to the FCS measurement points. In Fig. 3 B, position 1 of FCS measurement point was chosen for measuring cytoplasm, and positions 2 and 3 were presumed to be in the nucleus. In Fig. 3, D and F, position 1 of the cross-hair corresponds to a point in the cytoplasm and positions 2 and 3 to random points in the nucleus. The amplitudes of all FAF (G (0) − 1) were normalized to unity for comparison of the shift of the curve. One of two FAFs obtained from the nucleus showed no or a small difference from that in the cytoplasm (curve 2 in Fig. 3, B, D, and F). Interestingly, other FAFs obtained from the nucleus largely shifted to the right, indicating much slower diffusional mobility (curve 3 in Fig. 3, B, D, and F). This slower diffusion was occasionally found in nuclei of all cells expressing monomeric and oligomeric EGFPn regardless of the cell type. This indicated that there were two types of diffusional mobility in the three compartments: the fast-diffusion-mobility (FAF curves 1 and 2 in Fig. 3) in the cytoplasm and the nucleus, and the slow-diffusion-mobility (FAF curve 3 in Fig. 3) in the nucleus (summarized in Table 1). Fluorescent intensity at the point of slower diffusion was weak compared to that at other places in the nucleus. However, we could not specify the precise position of the slow-diffusion-mobility in the cell nucleus because of the very weak fluorescence in the nucleus. Fig. 3 G shows a plot superimposing normalized FAFs of EGFPn measured in the nucleus of HeLa cell excluding the slow-diffusion-mobility. The FAF of each oligomeric EGFP in the cell nucleus shifted to the right with the size of tandem EGFPn. This shift was well consistent with the result in aqueous solution (Fig. 2 A). This consistency suggests that the fast-diffusion-mobility of oligomeric EGFPn in the cell nucleus might follow the diffusion model of a rod-like molecule.
TABLE 1.
Diffusion constants of monomeric EGFP and tandem EGFPn in the cytoplasm, the nucleoplasm, and the nucleolus in three different living cells (23)
| Fast (fraction) | Slow (fraction) | Very slow (fraction) | |
|---|---|---|---|
| *Fast-diffusion-mobility (Cyt and NP) | (>90%) | – | (<10%) |
| Slow-diffusion-mobility (NL) | (<80%) | (20%∼100%) | – |
| Cyt (fast/first components)
|
NP (fast/first components)
|
||||
|---|---|---|---|---|---|
| †Cell line | Type of GFP | D (μm2s−1) | Fraction (%) | D (μm2s−1) | Fraction (%) |
| HeLa | GFP1 | 23.4 ± 2.5 | 95 | 22.7 ± 2.3 | 96 |
| GFP2 | 16.4 ± 0.8 | 92 | 14.9 ± 0.8 | 93 | |
| GFP3 | 13.1 ± 1.4 | 91 | 11.6 ± 1.7 | 91 | |
| GFP4 | 9.0 ± 0.8 | 93 | 8.6 ± 0.6 | 90 | |
| GFP5 | 8.3 ± 1.1 | 90 | 7.8 ± 1.2 | 90 | |
| COS7 | GFP1 | 27.1 ± 3.0 | 90 | 22.3 ± 1.3 | 95 |
| GFP2 | 15.8 ± 1.0 | 93 | 14.9 ± 0.3 | 92 | |
| GFP3 | 14.7 ± 0.6 | 91 | 11.1 ± 0.8 | 94 | |
| GFP4 | 10.8 ± 1.0 | 92 | 9.2 ± 0.8 | 91 | |
| GFP5 | 9.5 ± 1.0 | 90 | 7.9 ± 0.4 | 90 | |
| HEK | GFP1 | 21.0 ± 1.6 | 95 | 24.9 ± 2.1 | 93 |
| GFP2 | 17.4 ± 3.5 | 92 | 16.4 ± 2.2 | 95 | |
| GFP3 | 14.1 ± 1.4 | 91 | 13.2 ± 0.9 | 91 | |
| GFP4 | 10.3 ± 1.1 | 90 | 10.5 ± 1.8 | 90 | |
| GFP5 | 10.4 ± 0.5 | 92 | 9.6 ± 0.4 | 92 | |
| NL (fast/first components)
|
NL (slow/second components)
|
||||
| Cell line | Type of GFP | D‡ (μm2s−1) | Fraction (%) | D‡ (μm2s−1) | Fraction (%) |
| HeLa | GFP1 | 24.2 ± 1.3 | – | 5.8 ± 1.6 | – |
| GFP2 | 17.6 ± 1.2 | – | 3.8 ± 0.5 | – | |
| GFP3 | 13.0 ± 1.0 | – | 2.1 ± 0.3 | – | |
| GFP4 | 11.0 ± 0.7 | – | 1.9 ± 0.1 | – | |
| GFP5 | 8.9 ± 0.4 | – | 1.6 ± 0.4 | – | |
| COS7 | GFP1 | 22.8 ± 1.7 | – | 5.7 ± 1.6 | – |
| GFP2 | 16.1 ± 2.9 | – | 3.7 ± 0.8 | – | |
| GFP3 | 13.3 ± 1.5 | – | 1.7 ± 0.2 | – | |
| GFP4 | 9.0 ± 0.3 | – | 1.3 ± 0.2 | – | |
| GFP5 | 9.1 ± 0.8 | – | 1.1 ± 0.2 | – | |
Two kinds of mobility, fast-diffusion-mobility and slow-diffusion-mobility, were differentiated by two-component analysis of G (τ). The fast-diffusion-mobility has “fast” and “very slow” components. In contrast, the slow-diffusion-mobility has “fast” and “slow” components. These three components have different ranges for the diffusion time and fraction.
The diffusion constants and fractions in cytoplasm (Cyt) and nucleoplasm (NP) correspond to the average diffusion constants and fractions for the first component of the fast-diffusion-mobility (“fast” in top part of table). The diffusion constant and fractions of the second component (“very slow” in top part of table) is not shown. The diffusion constant in the nucleolus (NL) corresponds to those for the first component (“fast” in top part of table) and the second component (“slow” in top part of table) of the slow-diffusion-mobility. The fraction in the NL was not shown because the values were very variable (see also the text). The values of D in NL indicated those having a fraction >50%. Data were averaged over 15–20 cells for HeLa, COS7, and HEK293 (Mean ± SE of three independent experiments).
The average values of D in the nucleolus were obtained from cells only expressing monomer EGFP and tandem EGFPn without mRFP-fibrillarin. These values were consistent with the result of Fig.5 E obtained by coexpressed cells.
Analysis of FAF in cells was performed with a two-component model ((Eq. 2), i = 1 and 2), a fast diffusing component (first component) and a slower diffusing component (second component), because FAF of each tandem EGFPn cannot be fitted by a one-component model, but best fitted by the two-component model. However, some FAFs were best fitted by a one-component model. In this case, we adopted the result of one-component analysis (supplementary Fig. S2). The first component was considered to be a freely diffusing component and the second component was assumed to be a slowly diffusing component (14,24,42). High density of the cellular solutes and some restricted mobility in a cellular microstructure may slow down free diffusion. With conditions of cells having a concentration under 20 EGFPn molecules (<0.1 μM) and a comparatively short measurement time under 30 s, the influence of photobleaching on diffusion time, which gives rise to a very long diffusion time and an increase of the fraction (yi value in Eq. 2), could be minimized. Photobleaching effects were checked from the time trace of fluorescent intensity for all FCS data (supplementary Fig. S3). Increasing the incubation time after transfection for a few days made the effect of photobleach on FCS measurement much stronger, because the promoter for protein expression is strong and not a controlled one. In practice, photobleaching effect was very small for weakly fluorescent cells at an early stage after expression of tandem GFP (supplementary Fig. S4). Background fluorescent signals under 2 × 103 cps and 10 × 103 cps were detected in medium and nontransfected HeLa, HEK, and COS7 cells (14). No significant correlation amplitudes were detected in the culture medium. In contrast, very weak correlations with very long diffusion times above 105 μs were sometimes detected in each cell type when FCS measurement was carried out over longer duration over 60 s. This was derived from very slow and large fluctuation of fluorescence but not from photobleaching. To solve the background with very slow fluctuation, we adapted a shorter measurement time as described above. Considering each tandem EGFP has much larger brightness per molecule than that of monomer EGFP (C. Pack and M. Kinjo, unpublished data) and the diffusion time of the proteins was an order of millisecond ranges, the short measurement time of FCS might be enough to obtain a reliable autocorrelation function.
Two diffusional mobility in the nucleus
The fast-diffusion-mobility in the cytoplasm and the nucleus
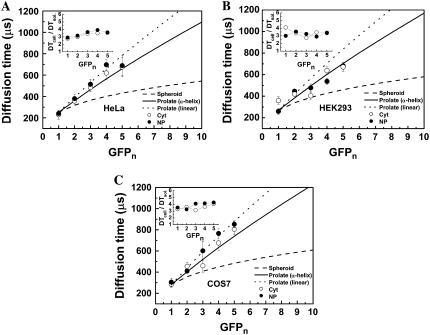
Fig. 4 A shows a plot of the diffusion time of first component obtained from FAFs representing the fast-diffusion-mobility in the cytoplasm and in the nucleus of HeLa cells (curves 1 and 2 in Fig. 3, B, D, and F). For these FAFs of the fast-diffusion-mobility, >90% of the fraction (y1 in Eq. 2) was defined as the first component, which represents free diffusion. These results were highly reproducible. As shown in Fig. 4 A (solid circles), the diffusion times of the fast-diffusion-mobility in the nucleus were gradually increased with the increase in the molecular size of tandem EGFPn. In the cytoplasm (Fig. 4 A, open circles), the diffusion time of the first component for EGFPn also increased with size. No significant difference between the first components in the cytoplasm and nucleus was found. Average diffusion times of monomeric and oligomeric EGFP in HEK and COS7 cells also increased with increasing the size both in the cytoplasm and in the nucleus (Fig. 4, B and C). The ratio of the diffusion time of the first component in the cytoplasm and the nucleus of each cell type to that in aqueous solution (DTcell/DTsol.), which indicates the ratio of viscosity (Eqs. 3 and 4), is shown in inserts in Fig. 4, A, B and C, respectively. Regardless of the cell type, the average ratios of viscosities in the cytoplasm and the nucleus were not significantly different and 3.5-fold higher than that in solution. Moreover, there was no dependency of the viscosity ratio on the size of oligomeric EGFPn. These results agreed with previous results obtained from microinjected fluorescent macromoleucules and monomeric EGFP (7,13,42).
FIGURE 4.
Fast diffusional mobility of tandem EGFPn is dependent on the molecular length. FAFs were fitted with a two-component model using Eq. 2, i = 2 for analyzing the diffusion times and the fractions of components 1 and 2. For the FAFs of the fast-diffusion-mobility in Fig. 3 G, the fraction of the first component was >90% for EGFP and tandem EGFPn in the cytoplasm and the nucleus. The diffusion times of the first components in HeLa cells (A), HEK cells (B), and COS7 cells (C) are plotted. The plots of measured diffusion times shown in panels A, B, and C show the average values of five cells for each protein. The error bars represent mean ± SD. The diffusion times of the first components were increased according to the molecular weight. As shown in Fig. 2, the calculated diffusion times for a spherical and a rod-like molecule were plotted for comparison. Dashed line shows diffusion times calculated by Eq. 5 using the molecular weight and the measured diffusion time of Rh6G assuming the oligomeric EGFPn are spherical. Solid and dotted lines show the calculated diffusion times assuming that the linker between EGFP forms a rigid α-helix and a linear shape, respectively. Inserts show the ratio of diffusion times of the first component in the cytoplasm and the nucleus to that in aqueous solution.
Using the result that the average viscosity in the cytoplasm and nucleus was 3.5-fold higher than that in solution, the expected diffusion times of tandem EGFPn in the cell were calculated. As shown in Fig. 2, the measured diffusion times of first components in living cells were also compared with three calculated diffusion times (Fig. 4, A, B, and C) assuming the shape of oligomeric EGFP to be spherical (dashed lines) or rod-like with the expected linker lengths of 4 nm (solid lines) and 9 nm (dotted lines). Dependency of the diffusion times on the size of oligomeric EGFP both in the cytoplasm and in the nucleus was consistent with that of a rod-like molecule rather than a spherical one (dashed lines). Based on the result that the diffusion properties of rod-like molecules of oligomeric EGFP in the cytoplasm and the nucleus are equivalent and consistent with the result in aqueous solution, the oligomeric EGFPn located in the nucleus was not truncated or degraded. Consequently, our results suggested that the diffusion of oligomeric EGFPn as a rod-like molecule was well conserved in the cellular circumstance in all of three cell lines.
Other diffusion times (second component) of the FAFs for the fast-diffusion-mobility (curves 1 and 2 in Fig. 3, B, D, and F) were very slow, and ranged from 104 to 105 μs in the nucleus as well as in the cytoplasm. The range of these long diffusion times was very broad and so it is not clear that the diffusion time of the second component was also dependent on the size of tandem EGFPn. The fraction of the second component (y2 in Eq. 2) was very small (<10%), regardless of the size of tandem EGFPn. The slow drift of fluorescence could come from cell mobility or very large organelles such as vesicles in cytoplasm (24,42) and such as a compact structure of chromatin in nucleus (4) during FCS measurement. Otherwise very weak photobleaching might be not completely excluded, even though data of photobleached samples were checked and excluded. However, a possibility of trapped diffusion in complex chromatin structures cannot be completely excluded. To analyze an effect of chromatin structures on the very slow diffusion time, we treated cells coexpressing tandem EGFP3 and H2B-mRFP or EGFP5 and H2B-mRFP, respectively, with Trichostatin A (TSA) (supplementary Fig. S5). It was previously reported that TSA inhibits histone deacetylation and so increases chromatin accessibility of relatively larger dextrans (4,43). In LSM observation, no significant changes of fluorescent pattern for tandem EGFP3 and EGFP5 were found, although that of H2B-mRFP was significantly changed after TSA treatment (supplementary Figs. S1 and S5). This result suggests that tandem EGFPn can freely and equally access to all regions of euchromatin and heterochromatin and so no effect of TSA treatment occurred. Moreover, there were no significant changes of diffusion time and fraction for the very slow component in the nucleoplasm after TSA treatment when FCS measurements on euchromatin (dilute H2B-mRFP fluorescent region) and heterochromatin (dense H2B-mRFP fluorescent region) were carried out) (C. Pack and M. Kinjo, unpublished data). Because the fraction of the very slow component was very small (<10%) and the diffusion times were very broad with large standard deviation even before TSA treatment, it is likely that the effect of TSA treatment on mobility of tandem EGFPn in the nucleus cannot be detectable in our experimental system. Nevertheless, the result of LSM observation was consistent with the result of FCS measurement. Details and discussion of such very slow diffusion can be omitted in this article because the fraction is small and we focus on well-defined diffusion property of tandem EGFPn as molecular ruler.
The slow-diffusion-mobility in the nucleus
On the other hand, the right-shifted FAFs for tandem EGFPn, which represent the slow diffusion-mobility, found in the nucleus (curves 3, dashed lines in Fig. 3, B, D, and F) showed a different range of diffusion times and a different fraction for the second component compared to those for the fast-diffusion-mobility (curves 1 and 2, solid and dotted lines in Fig. 3, B, D, and F). Obviously, although the diffusion times of the first component for the slow-diffusion-mobility in the nucleus were consistent with those for the fast-diffusion-mobility in the cytoplasm and in the nucleus (Table 1), the diffusion times of the second component for the slow-diffusion-mobility ranged from 800 to 5000 μs, increasing with the size of tandem EGFP (e.g., curve 3 in Fig. 3, B, D, and F). Moreover, the fraction of the second component for the slow-diffusion-mobility varied from 20 to 100% depending on the cells, and even the measured position in the same nucleus. This observation was very reproducible, and was consistent among the three cell types. Obviously, our results indicated that the protein mobility in the nuclear microenvironment might be separated into two kinds of diffusing species (i.e., the first component of fast-diffusion-mobility and the second component of slow-diffusion-mobility). These two kinds of diffusing species had different ranges of diffusion time (or apparent viscosity) depending on the position inside the nucleus.
The slow-diffusion-mobility of tandem EGFPn in the nucleolus
Fluorescent intensity at the position of the slow-diffusion-mobility (i.e., the right-shifted FAFs) in the nucleus (position 3 of Fig. 3, A, C, and E) was weak compared to other places inside the nucleus. In addition, the slow-diffusion-mobility was often found in the nucleolus in the cells expressing EGFP4 and EGFP5 with large and clear nucleoli. The density, the number, and the morphology of the nucleolus changed according to the cell cycle as well as cell type and other cell conditions. Recently, the nucleolus has been detected by fluorescence microscopy in cell lines expressing fluorescent protein-tagged nucleolar proteins such as fibrillarin and B23 (32,44). Fibrillarin is related to various steps of pre-rRNA processing and ribosome assembly and located in the dense fibrillar component (DFC) of the nucleolus during interphase (45). Using a nucleolar protein tagged with different fluorescent proteins will help in discriminating the nucleolar structures from nucleoplasm and tracing the changes of the nuclear structure during the cell cycle or depending on physiological cell conditions.
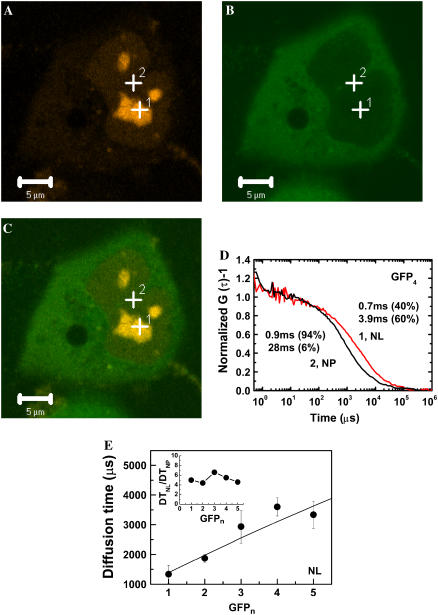
Fig. 5 shows an LSM image and FCS measurement of a HeLa cell coexpressing EGFP4 and mRFP-fibrillarin. The strong red fluorescence in the nucleus (Fig. 5 A) indicates the nucleolus. A weak green fluorescence signal was also detected in the nucleoplasm (Fig. 5 B). This LSM observation for fibrillarin agreed with the previous results (12,32). The shape, the size, and the number of nucleoli were different from cell to cell. Using cotransfected HeLa cells, FCS measurement was carried out for positions of green fluorescent nucleoplasm and the red fluorescent nucleolus with a diameter of over 2 μm in the x-y plane of the LSM image. FAF inside the nucleolus (Fig. 5 D, red line) shifted to the right compared to that in the nucleoplasm (Fig. 5 D, black line), which meant that the diffusion in the nucleolus was much slower than that in the nucleoplasm. The FAFs obtained from nucleolus fit well with the two-component model. Occasionally, some FAFs fit well even in the one-component model. The slow-diffusion-mobility in the nucleolus (Fig. 5 D; curve 1) consisted of the first component of 700 μs (40%) and the second component of 3900 μs (60%). In contrast, the fast-diffusion-mobility in the nucleoplasm (Fig. 5 D; curve 2) consisted of the first component of 900 μs (94%) and the second component of 28,000 μs (6%). Diffusion times (1 × 103–4 × 103 μs) of second components for FAFs measured in the nucleolus (Fig. 5 E) were much shorter than those of second components measured in the nucleoplasm and the cytoplasm (ranging from 104 to 105 μs). The time range (Fig. 5 E) was consistent with those obtained from Fig. 3, B, D, and F. In contrast, almost no such diffusional component in the range of 1 × 103–4 × 103 μs was found in places other than the nucleolus. On the other hand, the diffusion time of the first component in the nucleolus was the same as those in the cytoplasm and the nucleoplasm. The fraction of the first component was decreased with the increased fraction of the second component. The diffusion times of second components in the nucleoli increased with the size of EGFP1, EGFP2, EGFP3, and EGFP4, even though there was little difference between EGFP4 and EGFP5 (solid circles in Fig. 5 E). The inset in Fig. 5 E shows the average ratio of the diffusion time of the second component in the nucleolus to the diffusion time of the first component in the nucleoplasm (DTNL/DTNP). There was no dependency of the ratio on the size of EGFPn and average value of the ratio for all tandem EGFPn was ∼5.2. The solid line in Fig. 5 E shows the calculated diffusion times of tandem EGFPn as a rod-like molecule with an α-helix linker when the relative viscosity in the nucleolus is fixed by the average ratio of diffusion time (Fig. 5 E, inset). The measured diffusion times of tandem EGFPn were consistent with the calculated values. Our results indicated that the slow-diffusion-mobility in the nucleolus also reflected the diffusion of a rod-like molecule rather than a spherical molecule.
FIGURE 5.
Slow diffusional mobility of oligomeric EGFP found in the nucleolus is much slower than that in the nucleoplasm. For clear discrimination of nucleoplasm from nucleolus in cells weakly expressing EGFP and tandem EGFPn, a nucleolar protein of mRFP-fibrillarin was coexpressed. (A) LSM images of HeLa cells coexpressing mRFP-fibrillarin and (B) tetrameric EGFP, and (C) a merged image are shown. Bars, 5 μm. FCS measurement was carried at 24h after cotransfection of genes encoding mRFP-fibrillarin and EGFPn. Nucleoli with a diameter of over 2 μm were selected for FCS measurement. FCS measurements were performed on multiple places in the nucleoplasm and the nucleolus. For clarification, (D) two normalized FAFs (G (0) − 1 = 1) of EGFP4 measured on two points in the nucleoplasm (black curve) and the nucleolus (solid curve) of a single cell are shown. The normalized FAF indicated that the diffusion in the two positions was clearly different. The error bars represent mean ± SD for three measurements of a single cell. (E) The diffusion times of the second components (solid circles) for EGFP and tandem EGFPn in the nucleoli in HeLa cells are plotted. The insert shows the ratio of diffusion time in the nucleolus to that in the nucleoplasm (DTNL/DTNP). Solid line shows the calculated diffusion time using the rod-like model (with an α-helix linker) assuming that the apparent viscosity of the nucleolus is 5.2-fold higher than the nuceloplasm (insert). The plots of measured diffusion times shown in panel E represent the average values of six cells for each protein. The error bars represent mean ± SE. (NP, nucleoplasm; NL, nucleolus.)
Table 1 summarizes the diffusion constants of the fast-diffusion-mobility (the first component with a fraction >90%) found in the cytoplasm and the nucleoplasm, and the diffusion constants of the slow-diffusion-mobility (first and second components) found in the nucleolus. The average values were obtained from living cells only expressing monomer EGFP and tandem EGFPn without mRFP-fibrillarin. Diffusion constants of the fast-diffusion-mobility both in the nucleoplasm and in the cytoplasm decreased with the length of tandem EGFPn in HeLa, COS7, and HEK cells, even though the diffusion constants of EGFP4 and EGFP5 in the cytoplasm of HEK cells did not change. Diffusion constants of the first and the second components in the nucleoli of HeLa and COS7 cells also decreased with the length of EGFPn. There was little difference between diffusion constants in the cytoplasm and the nucleoplasm of HEK293 and HeLa cells. In contrast, diffusion constants in the cytoplasm of COS7 cells were slightly larger than those in the nucleoplasm. Based on these results, it was concluded that the diffusional motion of tandem EGFPn in the nucleous as well as in the cytoplasm and the nucleoplasm was well consistent with free diffusion of rod-like molecules, regardless of the cell type. It is emphasized that the microenvironment of the nucleolus as well as the nucleoplasm and the cytoplasm could be quantitatively understood by diffusion analysis of the oligomeric EGFPn as molecular rulers (MR). Moreover, our results indicated that the microenvironment and apparent viscosity of the cytoplasm and the nucleoplasm were almost same, even though the constituents of the two compartments were very different.
Compared with those of the first component in the nucleoplasm, the fractions of the second components in the nucleoli were significantly changed from 20 to 100% depending on the nucleolus, even in the same cell (C. Pack and M. Kinjo, unpublished data). Because the length of the z axis (optical axis) of detection volume (<2 μm) was six times longer than the diameter in the x-y plane (<0.2 μm), FCS measurement of a nucleolus with a length in the z axis shorter than 2 μm might contain both the nucleoplasm and the nucleolus. This might affect the variability of the fraction. However, it is also presumed that the diffusion of oligomeric EGFPn in the nucleolus has more variability than that found in the cytoplasm and the nucleoplasm, indicating the dynamic change of the nucleolar microenvironment or the complexity of subnucleolar structures such as DFC, fibrillar centers, and the granular region (45,46). More detailed study using two-color 3D imaging combined with FCS measurement is in progress for elucidating the large diffusion changes in the nucleolus according to a long-time scale or the cell cycle of a single cell. Nevertheless, our results showed that the mobility of MR in the nucleolus was dependent onto length of them, but was much slower than those in the cytoplasm and the nucleoplasm. Consequently, it was concluded that the diffusion of protein in the nucleus must be separated into two significant diffusing components, fast-diffusion-mobility in the nucleoplasm and slow-diffusion-mobility in the nucleolus.
Nucleolar microenvironment is sensitive to energy depletion
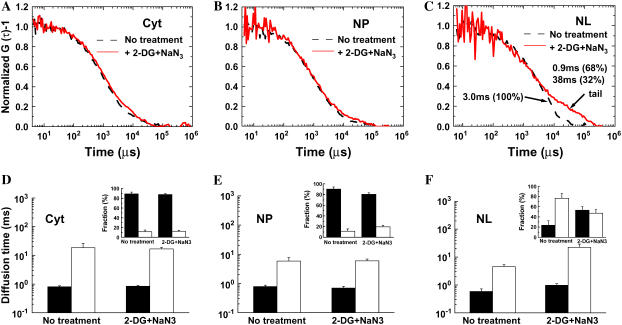
To examine effect of energy depletion on the mobility of oligomeric EGFP in the nuclear microenvironment, the culture medium containing HeLa cells expressing EGFP4 or EGFP5 was perfused with 2-DG and NaN3 solution (3,20) at 25 or 37°C. LSM and FCS measurements were carried out with HeLa cells expressing EGFP5 or coexpressing EGFP5 and mRFP-fibrillarin (or H2B-mRFP). For FCS measurement of cells transfected with EGFP5, HeLa cells with clear and large nucleoli (>4 μm in diameter) were chosen despite the fluorescence signals of the cytoplasm being a little strong (for example, right upper cell in Fig. 3 E). FCS measurement was carried on the same position of single cells before and after the energy depletion. We confirmed the redistribution of H2B-mRFP and nuclear shrinkage through LSM images of cells coexpressing H2B-mRFP and EGFP5 after ATP depletion at room temperature for 30 min (C. Pack and M. Kinjo, unpublished data). This result was consistent with a previous study (20). With cells expressing EGFP5, Fig. 6, A–C, show FAFs of EGFP5 at the same positions in the cytoplasm, the nucleoplasm, and the nucleolus of a single HeLa cell, respectively, before (dashed black lines) and after the energy depletion (solid red lines). FAFs of EGFP5 both in the cytoplasm and in the nucleoplasm were slightly shifted to the right by the energy depletion (Fig. 6, A and B). In contrast, the FAF of EGFP5 in the nucleolus was significantly changed in the longer time range as shown in Fig. 6 C. The energy depletion induced a big tail on the FAF, which indicates that a fraction with much slower mobility was newly produced. The diffusion time corresponding to the tail found in the nucleolus was 13-fold slower than that of the second component found before energy depletion, and the fraction of the new slower component was increased from 0% up to ∼32% (Fig. 6 C).
FIGURE 6.
MR mobility in the nucleolus is changed by the energy depletion. HeLa cells were transiently transfected with EGFP5, or cotransfected with EGFP5 and H2B-mRFP. The FAF of EGFP5 in single HeLa cells is shown before (dashed black lines) and 30 min after addition (solid red lines) of 2-deoxyglucose (6 mM) and sodium-azide (10 mM) to culture medium at room temperature. The redistribution of H2B-mRFP on the cells coexpressed with EGFP5 was confirmed by LSM before and after the treatment. The changes of the normalized FAF on (A) cytoplasm, (B) nucleoplasm, and (C) nucleolus of a single cell induced by energy depletion are shown. The amplitude of FAF, G (0) − 1, was normalized to unity. The average diffusion times and the average fractions (insert) of the first (solid bars) and the second (open bars) components for EGFP5 in the cytoplasm, the nucleoplasm, and the nucleolus before and after ATP depletion are shown in panels D, E, and F, respectively. The error bars are the measured mean ± SE (n = 8 cells).
Fig. 6, D–F, shows the average change of the diffusion time and the fraction in each cellular compartment of five HeLa cells expressing EGFP5. Averaged diffusion times in the cytoplasm (0.8 ± 0.04 ms and 0.85 ± 0.04 ms) and the nucleoplasm (0.79 ± 0.06 ms and 0.7 ± 0.08 ms) before and after ATP depletion, respectively, were not changed (solid bars in Fig. 6, D and E). Instead, the fractions of first components were slightly decreased in the cytoplasm (∼1%) and the nucleoplasm (∼9%) (Fig. 6, D and E, solid bars in inset). On the other hand, diffusion times of the second component in the nucleolus were increased from 4.6 ± 0.8ms to 22.4 ± 6.7 ms (Fig. 6 F, open bars), even though the fraction of the second component was decreased from 77 to 47% by the energy depletion (Fig. 6 F, open bars in inset). In addition to the change of the second component in the nucleolus, the diffusion time of the first component in the nucleolus was also increased from 0.58 ± 0.1 ms to 1.0 ± 0.1 ms (Fig. 6 F, solid bars). This indicated that the microenvronment inside the nucleolus, which was reflected by diffusion of EGFP5 molecules, was more sensitive to energy depletion than those of the nucleoplasm and the cytoplasm.
DISCUSSION
Tandemly linked EGFPn proteins were constructed for modeling rod-like molecules. The diffusion properties of the proteins were quantitatively dependent on their length. These series of standard proteins allowed us to analyze protein mobility in living cells. LSM observation of HeLa cells expressing monomer EGFP and four different kinds of tandem EGFPn showed that the proteins could be distributed to the cell nucleus regardless of their molecular weights. Monomeric EGFP, EGFP2, and EGFP3 were easily distributed in the nucleus. In contrast, the fluorescent intensities in the nuclei of cells expressing EGFP4 and EGFP5 were lower than in the cytoplasm, even though they were also located in the cell nuclei. Although the tendency of fluorescence intensity was very much different for EGFPn in the nucleus, all tandem proteins were detected by LSM and could also be detected by FCS. Many studies have shown that the transport of inert molecules to the nucleus depends inversely on molecular size with an exclusion limit at ∼5–10 nm in diameter or 40–60 kD in molecular weight (47,48). These studies discussed only the exclusion limits of spherical molecules. Our results for tandem EGFPn with molecular weights of 60, 90, 120, and 150 kD showed that rod-like proteins could localize to the cell nucleus within 24h after transfection depending on size, even though the mechanism for their transport to the nucleus was not clear.
Western blots of tandem proteins from cell lysates showed that the molecular weights of proteins synthesized in cells were well consistent with those expected from their numbers of amino acids. FCS measurement of monomer and tandem EGFPn in aqueous solution showed that their diffusion times also increased with molecular weight. Comparison of the measured diffusion time with the calculated diffusion time according to Perrin's equation (35,36) indicated that the tandemly linked EGFPn behaved like rod-like molecules. The fact that diffusion times of tandem series of EGFP are proportional to their lengths in aqueous solution indicates that the proteins could be employed as molecular rulers (MR) in living cells.
Combining a well-defined MR with the high sensitivity of FCS measurement make possible analysis of protein mobility in living cells, in particular in the nucleus. In contrast to the cytoplasm, our results showed that there were two kinds of diffusional mobility in the nucleus, both of which also depended on the length of MR as shown in solution and cytoplasm. One was the fast-diffusion-mobility of tandem EGFPn found in the nucleoplasm as well as in the cytoplasm, in which the first component had a fraction above 90%, reflecting the free diffusion of the MR (represented by D of the first components in NP; Table 1). The other was the slow-diffusion-mobility (represented by D of the second components in NL; Table 1) observed in the nucleolus. The second component of the fast-diffusion-mobility in nucleoplasm showed a very slow diffusion (104–105 μs) with fractions under 10%, indicating no length dependency and no significant change by TSA treatment. The first component of the slow-diffusion-mobility (represented by D of the first components in NL, Table 1) in nucleolus was almost equivalent to the first component of the fast-diffusion-mobility in nucleoplasm (represented by D of the first components in NP, Table 1). There was no significant change in the diffusion time and the fraction for the first component of the fast-diffusion-mobility and for the second component of the slow-diffusion-mobility by TSA treatment (C. Pack and M. Kinjo, unpublished data). The result of FCS before and after TSA treatment was well consistent with LSM observation using two-color imaging (supplementary Figs. S1 and S5). Previous studies (4,38,43) using LSM observation of labeled dextran with various sizes showed that a globular protein with molecular weight of 1 MD (an apparent pore size of 14 nm) might be no limitation in access to chromatin. Because molecular weights of tandem EGFPn are much smaller than 1 MD, tandem EGFPn might freely access the two types of chromatin.
A study of FCS and monomer EGFP using both a two-component model and an anomalous subdiffusion model analysis (14) has shown that the diffusion of EGFP in the nucleus was much more complex than in the cytosol. The study described averaged diffusional mobility of EGFP in the entire nucleus but not in each compartments in the nucleus such as the nucleolus, and suggested that the ratio of diffusion mobilities in cells and in solution was not dependent on the two models used. The fast-diffusion-mobility of tandem GFPn in the cytoplasm and the nucleoplasm was dependent on length. The ratio of diffusion time in each compartment to that in solution showed that the apparent viscosities of the cytoplasm and nucleoplasm were identical. In addition, the apparent viscosity in the three cell lines (HeLa, COS7, and HEK293) was found to be ∼3.5-fold higher than in aqueous solution. The viscosities in the cytoplasm and the nucleoplasm were well consistent with previous studies using FRAP (9,13) and using FCS (14).
We investigated the protein mobility in the nucleolar microenvironment of living cells in detail. The size and shape of the nucleolus during each phase of the cell cycle are not constant. Moreover, it was not easy to discriminate between the nucleoplasm and nucleolus in the cells weakly expressing the monomer and tandem EGFPn. We marked the nucleolus with mRFP-tagged fibrillarin to distinguish it from the nucleoplasm. Our observations in the nucleolus (Fig. 5 E and Table 1) indicated that mobility of the inert EGFP and tandem EGFPn in the nucleolus was also dependent on the length of the protein, but that the mobility was ∼17-fold slower for HeLa and 24-fold slower for COS7 than in aqueous solution. Nevertheless, assuming a random walk model, the result suggested that it would take the tandem proteins just a few seconds to travel a distance of 4 μm, roughly the diameter of a nucleolus. Rapid association or exchange of GFP-fibrillarin (0.046 μm2s−1) (12) and GFP-B23 (0.08 μm2s−1) (32) in the nucleolus was observed by FRAP. These results suggested that the nucleolus is not a static protein mass such as aggregates, and that proteins were dynamically exchanged between the nucleoplasm and the nucleolus. EGFP tagged fibrillarin was shown to have diffusion constants of 0.53 μm2s−1 even in the nucleoplasm (12). On the other hand, diffusion of the MR in the nucleolus was much faster than for the nucleolar proteins (Table 1). For instance, the diffusion constants of tandem EGFP2 were 14.9 ± 0.8 and 3.8 ± 0.5 μm2s−1 in the nuceloplasm and the nucleolus of the HeLa cell, respectively, although the molecular weight and shape of EGFP2 might be similar to EGFP-tagged fibrillarin (60 kD). Our observations indicated that the architecture of nucleolus was not very tight and some proteins, at least GFPn, could be almost freely accessible inside of the compartment, because the mobility of GFPn was only slowed down about one-fifth compared with the nucleoplasm and the cytoplasm. Consequently, our study of MR mobility in the nucleoplasm and the nucleolus might be very helpful to understand the variability of mobility of microinjected labeled macromolecules in the nucleus (3,13) or the restricted mobility of monomeric EGFP (14) and various nuclear proteins (12,20,32,41,49). In those studies, the complex microenvironment inside of the nucleolus was not considered in detail, even though the mobilities of the nuclear proteins were measured in the nucleolus and the interactions with nucleoli were analyzed.
Recent LSM observation of human U2OS cells expressing yellow fluorescent protein tagged H2B and electron microscopic observation of ATP-depleted cells have shown that the chromatin structure changes with nuclear shrinkage under energy depletion, and suggest that movement of mRNA-protein complexes (mRNPs) is constrained by the structural changes in the nucleus (20). It would be interesting to know whether the redistribution of the chromatin structure by energy depletion also affects other small proteins, and whether the nucleolar microenvironment is also changed by ATP depletion. To determine whether the diffusion of the longest EGFP5 in the nucleolar microenvironment was affected by the cellular metabolism, we treated HeLa cells expressing EGFP5 with metabolic inhibitors 2-DG and NaN3. Interestingly, our results showed that the diffusion of EGFP5 in the nucleolus was slowed down by ATP depletion, but that in the cytoplasm and the nucleoplasm it was only slightly changed. The small change of EGFP5 mobility (Fig. 6 B) in the nucleoplasm suggested that the microenvironment of nucleoplasm was not so changed. This result indicated that the mobility of proteins smaller than mRNP complex was not sensitive to the structural change in the nucleoplasm (20). Otherwise, the energy depletion would change large nuclear matrix structures (50–52), which affect the much larger molecular size of mRNP (r ∼ 133 nm as a circular mRNP with 2.8 kb) rather than that of EGFP5 (longitudinal length, ∼28 nm). Recent reports have indicated that nuclear diffusion can be limited by a mesoscale viscosity for particles that are larger than 100 nm in diameter (53). In contrast, the change of EGFP5 mobility in the nucleolus induced by ATP depletion suggested that the effect of energy depletion on the microenvironment of the nucleolus was bigger than that of the nucleoplasm and cytoplasm, even though the origin of the significant mobility change in the nucleolus was not clear. Nevertheless, our results clearly indicate that the microenvironment of the nucleolus is physiologically very different from that of the nucleoplasm. It is interesting to note that the two motor proteins, nuclear actin and myosin I are related to rDNA and are required for RNA polymerase I transcription (54). Such ATP-binding motor proteins can modify the nucleolar microenvironment.
In this study, we have demonstrated that combination of FCS and oligomeric EGFPn with different lengths is a novel method to elucidate the nuclear microenvironment of living cells. The microenvironment of the two compartments in the nucleus can now be differentiated and analyzed by using tandem MR, two-color imaging, and FCS. We found that MR EGFPn, which is presumably inert, could rapidly diffuse inside of the cell nucleolus as well as the nucleoplasm depending only on the length of the protein. Our experimental system can be applied to understanding the mobility of other functional proteins in the nucleolus as well as in the cytoplasm and the nucleoplasm. More importantly, it is also suggested that the microenvironment of the nucleolus is very sensitive to pharmacological energy depletion compared to that of the cytoplasm and the nucleoplasm. Consequently, it is concluded that the physiological state of the nucleolar microenvironment can be understood through mobility analysis of tandem MR in living cells. Combining this method with other fluorescence microscopic methods such as time-lapse microscopy will allow complementary analysis of the nucleolar microenvironment of various cell types and single cells while varying the cell cycle or other physiological conditions such as cell stresses. Effects of GTP depletion or specific inhibitors such as actinomycin D, which primarily affects ribosome biogenesis in the nucleolus through the inhibition of RNA polymerase transcription, will also be important to understand the relations between the nucleolar microenvironment and physiological conditions in detail.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Acknowledgments
The authors thank Professor Hiroshi Kimura (Kyoto University, Japan) and Dr. Takuya Saiwaki (Osaka University, Japan) for providing H2B-mRFP and mRFP-fibrillarin clones, respectively. The authors also thank Dr. Isse Nagao for technical advice for the Western blot experiment.
This research was partly supported by KAKENHI, Grand-in-Aid for Scientific Research (B) by Japan Society for the Promotion of Science (15370062) and Grand-in-Aid for Scientific Research on Priority Areas “Nuclear Dynamics” by MEXT (17050001). C.P. gratefully acknowledges support from 21st Century COE Program for “Advance Life Science on the Base of Bioscience and Nanotechnology” in Hokkaido University, and for “Topological Science and Technology” in Hokkaido University. C.P. is a Postdoctoral Fellow of the JSPS (1705456) in Japan.
Abbreviations used: cps, count per second; Cyt, cytoplasm; 2-DG, 2-deoxyglucose; DT, diffusion time; EGFPn, tandemly linked oligomeric EGFP; FAF, fluorescence autocorrelation function; MR, molecular ruler; FCS, fluorescence correlation spectroscopy; FRAP, fluorescence recovery after photobleach; mRFP, monomeric red fluorescent protein; NP, nucleoplasm; NL, nucleolus; SPT, single particle tracking; TSA, Trichostatin A.
References
- 1.Misteli, T. 2001. Protein dynamics: implications for nuclear architecture and gene expression. Science. 291:843–847. [DOI] [PubMed] [Google Scholar]
- 2.Pederson, T. 2000. Diffusional protein transport within the nucleus: a message in the medium. Nat. Cell Biol. 2:E73–E74. [DOI] [PubMed] [Google Scholar]
- 3.Carmo-Fonseca, M., M. Platani, and J. R. Swedlow. 2002. Macromolecular mobility inside the cell nucleus. Trends in Cell Biol. 12: 491–5. [DOI] [PubMed] [Google Scholar]
- 4.Görisch, S. M., P. Lichter, and K. Rippe. 2005. Mobility of multi-subunit complexes in the nucleus: accessibility and dynamics of chromatin subcompartments. Histochem. Cell Biol. 123:217–228. [DOI] [PubMed] [Google Scholar]
- 5.Houtsmuller, A. B., S. Rademakers, A. L. Nigg, D. Hoogstraten, J. H. J. Hoeijmakers, and W. Vermeulen. 1999. Action of DNA repair endonuclease ERCC1/XPF in living cells. Science. 284:958–961. [DOI] [PubMed] [Google Scholar]
- 6.Phair, R. D., P. Scaffidi, C. Elbi, J. Vecerová, A. Dey, K. Ozato, D. T. Brown, G. Hager, M. Bustin, and T. Misteli. 2004. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 24:6393–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verkman, A. S. 2002. Solute and macromolecule diffusion in cellular aqueous compartments. Trends Biochem. Sci. 27:27–33. [DOI] [PubMed] [Google Scholar]
- 8.Brock, R., M. A. Hink, and T. M. Jovin. 1998. Fluorescence correlation microscopy of cells in the presence of autofluorescence. Biophys. J. 75:2547–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kao, H. P., J. R. Abney, and A. S. Verkman. 1993. Determinants of the translational mobility of a small solute in cell cytoplasm. J. Cell Biol. 120:175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kues, T. R. Peters, and U. Kubitscheck 2001. Visualization and tracking of single protein molecules in the cell nucleus. Biophys. J. 80: 2954–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luby-Phelps, K. P.E. Castle, D.L. Taylor, and F. Lanni 1987. Hindered diffusion of inert tracer particles in the cytoplasm of mouse 3T3 cells. Proc. Natl. Acad. Sci. USA. 84: 4910–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature. 404:604–609. [DOI] [PubMed] [Google Scholar]
- 13.Seksek, O., J. Biwersi, and A. S. Verkman. 1997. Translational diffusion of macromolecule-sized solutes in cytoplasm and nucleus. J. Cell Biol. 138:131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wachsmuth, M., W. Waldeck, and J. Langowski. 2000. Anomalous diffusion of fluorescent probes inside living cell nuclei investigated by spatially resolved fluorescence correlation spectroscopy. J. Mol. Biol. 298:677–689. [DOI] [PubMed] [Google Scholar]
- 15.Weiss, M. M. Elsner, F. Kartberg, and T. Nilsson. 2004. Anomalous subdiffusion is a measure for cytoplasmic crowding in living cells. Biophys. J. 87: 3518–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lukacs, G. L., P. Haggie, O. Seksek, D. Lechardeur, N. Freedman, and A. S. Verkman. 2000. Size-dependent DNA mobility in cytoplasm and nucleus. J. Biol. Chem. 275:1625–1629. [DOI] [PubMed] [Google Scholar]
- 17.Sprague, B. L., R. L. Pego, D. A. Stavreva, and J. G. McNally. 2004. Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys. J. 86:3473–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmiedeberg, L., K. Weisshart, S. Diekmann, G. M. Hoerste, and P. Hemmerich. 2004. High- and low-mobility populations of HP1 in heterochromatin of mammalian cells. Mol. Biol. Cell. 15:2819–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNally, J. G., W. G. Müller, D. Walker, R. Wolford, and G. L. Hager. 2000. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 287:1262–1265. [DOI] [PubMed] [Google Scholar]
- 20.Shav-Tal, Y., X. Darzacq, S. M. Shenoy, D. Fusco, S. M. Janicki, D. L. Spector, and R. H. Singer. 2004. Dynamics of single mRNPs in nuclei of living cells. Science. 304:1797–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinjo, M., and R. Rigler. 1995. Ultrasensitive hybridization analysis using fluorescence correlation spectroscopy. Nucleic Acids Res. 23:1795–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nomura, Y., H. Tanaka, L. Poellinger, F. Higashino, and M. Kinjo. 2001. Monitoring of in vitro and in vivo translation of green fluorescent protein and its fusion proteins by fluorescence correlation spectroscopy. Cytometry. 44:1–6. [PubMed] [Google Scholar]
- 23.Pack, C., K. Aoki, H. Taguchi, M. Yoshida, M. Kinjo, and M. Tamura. 2000. Effect of electrostatic interactions on the binding of charged substrate to GroEL studied by highly sensitive fluorescence correlation spectroscopy. Biochem. Biophys. Res. Commun. 267:300–304. [DOI] [PubMed] [Google Scholar]
- 24.Saito, K., E. Ito, Y. Takakuwa, M. Tamura, and M. Kinjo. 2003. In situ observation of mobility and anchoring of PKCβI in plasma membrane. FEBS Lett. 541:126–131. [DOI] [PubMed] [Google Scholar]
- 25.Terada, S., M. Kinjo, and N. Hirokawa. 2000. Oligomeric tubulin in large transporting complex is transported via kinesin in squid giant axons. Cell. 103:141–55. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida, N., M. Kinjo, and M. Tamura. 2001. Microenvironment of endosomal aqueous phase investigated by the mobility of microparticles using fluorescence correlation spectroscopy. Biochem. Biophys. Res. Commun. 280:312–318. [DOI] [PubMed] [Google Scholar]
- 27.Ormö, M., A. B. Cubitt, K. Kallio, L. A. Gross, R. Y. Tsien, and S. J. Remington. 1996. Crystal structure of the Aequorea Victoria green fluorescent protein. Science 273:1392–5. [DOI] [PubMed] [Google Scholar]
- 28.Tsien, R. Y. 1998. The green fluorescent protein. Annu. Rev. Biochem. 57:509–544. [DOI] [PubMed] [Google Scholar]
- 29.Yang, F., L. G. Moss, and G. N. Phillips. 1996. The molecular structure of green fluorescent protein. Nat. Biotechnol. 14:1246–1251. [DOI] [PubMed] [Google Scholar]
- 30.Kimura, H., and P. R. Cook. 2001. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 153:1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanda, T., K. F. Sullivan, and G. M. Wahl. 1998. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 8:377–385. [DOI] [PubMed] [Google Scholar]
- 32.Chen, D., and S. Huang. 2001. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. Cell Biol. 153:169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saito, K., M. Tamura, and M. Kinjo. 2004. Direct detection of caspase-3 activation in single live cells by cross-correlation analysis. Biochem. Biophys. Res. Commun. 324:849–854. [DOI] [PubMed] [Google Scholar]
- 34.Weisshart, K., V. Jungel, and S. J. Briddon. 2004. The LSM 510 META-ConfoCor 2 system: an integrated imaging and spectroscopic platform for single-molecule detection. Cur. Pharm. Biotech. 5:135–154. [DOI] [PubMed] [Google Scholar]
- 35.Cantor, C. R., and P. R. Schimmel. 1980. In Biophysical Chemistry Part II: Chapter 10. Techniques for the Study of Biological Structure and Function. W. H. Freeman, New York. 560–567.
- 36.Björling, S., M. Kinjo, Z. Földes-Papp, E. Hagman, P. Thyberg, and R. Rigler. 1998. Fluorescence correlation spectroscopy of enzymatic DNA polymerization. Biochemistry. 37:12971–12978. [DOI] [PubMed] [Google Scholar]
- 37.Rigler, R., Ü. Mets, J. Widengren, and P. Kask. 1993. Fluorescence correlation spectroscopy with high count rate and low background: analysis of translational diffusion. Eur. Biophys. J. 22:169–175. [Google Scholar]
- 38.Görisch, S. M., K. Richter, M. O. Scheuermann, H. Herrmann, and P. Lichter. 2003. Diffusion-limited compartmentalization of mammalian cell nuclei assessed by microinjected macromolecules. Exp. Cell Res. 289:282–294. [DOI] [PubMed] [Google Scholar]
- 39.Swaminathan, R., C. P. Hoang, and A. S. Verkman. 1997. Photobleaching recovery and anisotropy decay of green fluorescent protein EGFP-S65T in solution and cells: cytoplasmic viscosity probed by green fluorescent protein translational and rotational diffusion. Biophys. J. 72:1900–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terry, B. R., E. K. Matthews, and J. Haseloff. 1995. Molecular characterization of recombinant green fluorescent protein by fluorescence correlation microscopy. Biochem. Biophys. Res. Commun. 217: 21–7. [DOI] [PubMed] [Google Scholar]
- 41.Wachsmuth M., T. Weidemann, G. Müller, U. W. Hoffmann-Rohrer, T. A. Knoch, W. Waldeck, and J. Langowski. 2003. Analyzing intracellular binding and diffusion with continuous fluorescence photobleaching. Biophys. J. 84:3353–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larson, D. R., Y. M. Ma, V. M. Vogt, and W. W. Webb. 2003. Direct measurement of Gag-Gag interaction during retrovirus assembly with FRET and fluorescence correlation spectroscopy. J. Cell Biol. 162:1233–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Görisch, S. M., M. Wachsmuth, K. Fejes Tóth, P. Lichter, and K. Rippe. 2005. Histone acetylation increases chromatin accessibility. J. Cell Sci. 118:5825–5834. [DOI] [PubMed] [Google Scholar]
- 44.Leung, A. K., D. Gerlich, G. Miller, C. Lyon, Y. W. Lam, D. Lleres, N. Daigle, J. Zomerdijk, J. Ellenberg, and A. I. Lamond. 2004. Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J. Cell Biol. 166:787–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olson, M. O. J., and M. Dundr. 2005. The moving parts of the nucleolus. Histochem. Cell Biol. 123:203–216. [DOI] [PubMed] [Google Scholar]
- 46.Olson, M. O. J., M. Dundr, and A. Szebeni. 2000. The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 10:189–196. [DOI] [PubMed] [Google Scholar]
- 47.Peters, R. 1983. Nuclear envelope permeability measured by fluorescence microphotolysis of single liver cell nuclei. J. Biol. Chem. 258:11427–11429. [PubMed] [Google Scholar]
- 48.Davis, L. I. 1995. The nuclear pore complex. Annu. Rev. Biochem. 64:865–896. [DOI] [PubMed] [Google Scholar]
- 49.Weidemann T., M. Wachsmuth, T. A. Knoch, G. Müller, W. Waldeck, and J. Langowski 2003. Counting nucleosomes in living cells with a combination of fluorescence correlation spectroscopy and confocal imaging. J. Mol. Biol. 334:229–40. [DOI] [PubMed] [Google Scholar]
- 50.Nickerson, J. A. 2001. Experimental observations of a nuclear matrix. J. Cell Sci. 114:463–474. [DOI] [PubMed] [Google Scholar]
- 51.Rando, O. J., K. Zhao, and G. R. Crabtree. 2000. Searching for a function for nuclear actin. Trends Cell Biol. 10:92–97. [DOI] [PubMed] [Google Scholar]
- 52.Pederson, T., and U. Aebi. 2003. Actin in the nucleus: what form and what for? J. Struct. Biol. 140:3–9. [DOI] [PubMed] [Google Scholar]
- 53.Tseng, Y., J. S. Lee, T. P. Kole, I. Jiang, and D. Wirtz. 2004. Micro-organization and visco-elasticity of the interphase nucleus revealed by particle nanotracking. J. Cell Sci. 117:2159–2167. [DOI] [PubMed] [Google Scholar]
- 54.Philimonenko, V. V., J. Zhao, S. Iben, H. Dingova, K. Kysela, M. Kahle, H. Zentgraf, W. A. Hofmann, P. Lanerolle, P. Hozak, and I. Grummt. 2004. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat. Cell Biol. 6:1165–1172. [DOI] [PubMed] [Google Scholar]