Abstract
The first step in the encounter between a host and a pathogen is attachment to the host epithelium. For uropathogenic Escherichia coli, these interactions are mediated by type 1 and P adhesive pili, which are long (∼1 μm) rods composed of more than 1000 protein subunits arranged in a helical structure. Here we used single-molecule atomic force microscopy to study the mechanical properties of type 1 pili. We found that type 1 pili readily extend under an applied force and that this extensibility is the result of unwinding the pilus rod's helical quaternary structure. The forced unraveling is also reversible, with helical rewinding taking place under considerable forces (∼60 pN). These data are similar to those obtained on P pili using optical tweezers, indicating that these are conserved properties of uropathogenic E. coli pili. We also show that our data can readily be reproduced using Monte Carlo simulation techniques based on a two-state kinetic model. This model provides a simple way to extrapolate the mechanical behavior of pili under a wide range of forces. We propose that type 1 pilus unraveling is an essential mechanism for absorbing physiological shear forces encountered during urinary tract infections and probably essential for adhesion and colonization of the bladder epithelium.
INTRODUCTION
Adhesion of many bacteria to host tissues is the first step in successful colonization and infection (1,2). This adhesive interaction is typically mediated by pili, which are long (∼1 μm) rods composed of more than 1000 protein subunits (immunoglobulin (Ig)-like domains) that form a helical structure that is anchored to the outer bacterial membrane (3) (Fig. 1 A). Pilus assembly in S. typhimurium, B. pertusssis, K. pneumoniae, uropathogenic E. coli (UPEC), and a multitude of other gram-negative pathogens requires a conserved chaperone-usher pathway to produce fibers important in gastroenteritis, whooping cough, pneumonia, urinary tract infections, and a variety of other diseases (4,5). Uropathogenic strains of E. coli use type 1 and P pili to colonize the bladder and kidney, respectively. P and type 1 pili produced by UPEC have been shown to be important for numerous functions including mediating colonization, invasion, and biofilm formation (6–8). These pili, therefore, serve as a lifeline for UPEC. Without strong binding and the ability to withstand fluid forces in the bladder, the bacteria would easily be cleared in the urine. Shear forces in the bladder and kidney caused by fluid and urine flow are a major factor that UPEC must resist to persist in the urinary tract.
FIGURE 1.
(A, left) Negative-stain electron micrograph of uropathogenic E. coli (UTI89) cell expressing type 1 pili (scale bar = 200 nm). (A, right) A cartoon diagram showing the structural subunits of type 1 pili. FimH is the adhesin at the distal end of the tip fibrillum, followed by FimG and FimF. Oligos of FimA form the helical pilus rod structure that is visible in the micrograph. (B) Two-dimensional diagram of donor-strand exchange between FimA Ig-like domains. Each pilin structural subunit donates its N-terminal extension to complete its neighbor's fold and form a protein chain. The N-terminal extension is held in place through noncovalent interactions (shaded region) such as hydrogen bonds and hydrophobic interactions. Each pilin subunit has one disulfide bond (red) between the A and B β-strands, close to where the N-terminal extension ends. (C) Diagram of the single-molecule AFM. Purified pili were adsorbed onto a glass substrate and then stretched using the tip of a cantilever (see Materials and Methods).
Type 1 pili are encoded by the fim gene cluster, fimA-I (9) and mediate binding to mannose receptors expressed on the surface of the bladder epithelium (10). This triggers the invasion of UPEC into the superficial umbrella cells of the bladder, where they multiply and form intracellular bacterial communities (IBCs) (7,8). The formation of IBCs allows UPEC to evade host defenses and persist in the bladder epithelia and urinary tract (8,11). The tips of type 1 pili are short and comprise only three proteins (FimF, G, H, where FimH is the adhesin protein and FimF and FimG are adaptor proteins (Fig. 1 A) (5)). The rigid pilus rod is comprised of repeating monomers of FimA subunits that form a helical quaternary structure 6–7 nm thick with a helical cavity 20 Å wide and 3.1 subunits per turn (2–5,12). The helical rod of the pilus has an interesting architecture in that none of the subunits that comprise them associates with another through covalent interactions (Fig. 1 B). Instead, the entire pilus is held together noncovalently by hydrogen-bonding networks and hydrophobic interactions (13). Pilin subunits have an Ig-like fold, but they are missing their seventh (G) β-strand (Fig. 1 B). The absence of the C-terminal seventh β-strand results in a deep groove on the surface of the pilin that exposes its hydrophobic core. During pilus biogenesis, an N-terminal extension that is present on every structural subunit completes the Ig fold of its neighbor in a process termed donor-strand exchange (13). The importance of this structural framework in pathogenesis is unknown.
Recent mechanical measurements done on P pili using optical tweezers show that P pili readily extend under an applied force (14–16). Type 1 pili are structurally similar to P pili and are very important in IBC formation (7) and shear-dependent binding (17,18), yet the mechanical properties of this ubiquitous type of pilus are not known (type 1 pili are expressed by ∼80% of all E. coli strains). Here we use single-molecule atomic force microscopy (AFM) techniques (19–21) to measure the mechanical properties of UPEC type 1 pili. We found that, like P pili, the rods of type 1 pili are highly extensible. This dramatic extension is a result of unwinding the pilus rod's helical quaternary structure when exposed to mechanical stress. The forced unraveling of type 1 pili is also reversible, with helical rewinding taking place under considerable forces (∼60 pN). These results demonstrate that type 1 pili are dynamic structures with spring-like properties under applied forces. The “spring forces” were also shown to be additive, whereby the simultaneous unwinding of several pili required a much larger force, proportional to the number of pili being extended. To better understand the molecular origin of the elastic properties of the helical rod, we used a simple two-state kinetic model and Monte Carlo simulation techniques. We show that this model closely reproduces the experimental data and provides a simple way to predict the mechanical behavior of pili under a wide range of physiological forces. This model predicts that pili elasticity serves as a mechanism for extending the lifetime of the adhesin-receptor interaction and explains the mechanism by which bacteria remain bound under shear forces. Our results show that reversible unraveling of type 1 pili is essential for absorbing physiological shear forces encountered during urinary tract infections and that this mechanism might be essential for successful colonization and invasion of host tissues.
MATERIALS AND METHODS
Strains and plasmids
The E. coli K-12 strains HB101 and ORN103 were used as host strains for cloning and expression. The plasmid pPap5 carries the entire pap operon with its natural promoter (22), and plasmid pSH2 carries the entire fim operon with its natural promoter (23).
Whole pilus purification
To induce pilus expression, the HB101 strain containing pPap5 was grown on tryptic soy agar (TSA) plates containing the appropriate antibiotic for 36 h. The ORN103 strain containing pSH2 was grown in Luria broth with antibiotics for 48 h at static conditions to induce pilus expression. Bound P pili were heat purified as described by Kuehn et al. (24). Cells expressing type 1 pili were pelleted and purified as described previously (5).
AFM
The mechanical properties of single pili were studied using a home-built single-molecule AFM (19-21) that consists of a detector head (Veeco, Santa Barbara, CA) mounted on top of a single-axis piezoelectric positioner with a strain gauge sensor (P841.10, Physik Instruments, Kartsruhe, Germany). The P841 has a total travel of 15 μm and is attached to two piezoelectric positioners (P280.10A, Physik Instruments) that are used to control the x and y positions. This system has a z axis resolution of a few nanometers and can measure forces in the range of 10–10,000 pN. The monitoring of the force reported by the cantilever and the control of the movement of the piezoelectric positioners are achieved by means of two data acquisition boards (PCI 6052E, PCI 6703, National Instruments, Austin, TX) and controlled by custom-written software (LabView, National Instruments; and Igor, WaveMetrics, Lake Oswego, OR). The spring constant of each individual cantilever (MLCT-AUHW: silicon nitride gold-coated cantilevers; Veeco Metrology Group, Santa Barbara, CA) was calculated using the equipartition theorem (25). Cantilever spring constants varied between 20 and 50 pN/nm and rms force noise (1-kHz bandwidth) was ∼10 pN. Unless noted, the pulling speed of the different force–extension curves was in the range of 1–3 nm/ms.
Pili mechanical measurements
In a typical experiment, a small aliquot of the purified pili (∼1–50 μl, 10 μg/ml) was allowed to adsorb to a clean glass coverslip (for ∼10 min) and then rinsed with PBS, pH 7.4. Before use the glass coverslips were cleaned by sonication in acetone for 20 min followed by boiling for 10 min in 3 N KOH and then 30% H2O2. Between steps, the coverslips were rinsed and sonicated with MilliQ water (>18.2 MΩ × cm). The coverslips were dried in a stream of N2 gas. Segments of a pilus were then picked up randomly by adsorption to the cantilever tip, which was pressed down onto the sample for 1–2 s at forces of several nanonewtons and then stretched for several micrometers. The probability of picking up a pilus was typically kept low (less than one in 50 attempts) by controlling the number of pili used to prepare the coverslips.
Analysis of force-extension curves
The elasticity of the stretched pili was analyzed using the worm-like chain (WLC) model of polymer elasticity (26,27):
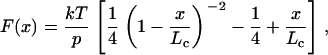 |
(1) |
where F is force, p is the persistence length, x is end-to-end length, and Lc is contour length of the stretched protein. The adjustable parameters are the persistence length, p, and the contour length, Lc.
RESULTS
Stretching E. coli type 1 pili using AFM
To study the mechanical properties of type 1 pili we used the AFM tip to pick up random segments of purified pili. Fig. 2, A–C shows typical force-extension curves obtained after stretching type 1 pili at a pulling speed of 1–3 nm/ms. One feature of these patterns is the presence of several distinct regions (vertical dashed lines ,Fig. 2 A). The first region probably corresponds to nonspecific interactions between the cantilever tip and surface. The second region most likely represents the unwinding of the pilus helical quaternary structure. This extension takes place at a constant force of ∼60 pN (63 ± 17 pN, n = 564, 94 different pili; Fig. 2 D) and is seen as a plateau in the force-extension curve (dotted line). Interestingly, this force is within physiological levels of shear flow (up to ∼90 pN/bacterium; (28)). We found that this plateau region can be very long, reaching up to 4 μm in some cases, indicating that type 1 pili can elongate several times their unstretched length. In order to quantify the extensibility of individual type 1 pili, we used the WLC model (Eq. 1), which predicts the relationship between the extension of a polymer and the entropic restoring force generated (26,27). The solid line in Fig. 2 A shows the prediction of the WLC equation using a contour length Lc = 3.7 μm and a persistence length p = 1.2 nm. The average values are Lc ∼ 2 μm (1.9 ± 0.7 μm, n = 230) and p = 3.3 ± 1.6 nm (n = 36). Once the rod has been completely unraveled, a larger force is then required to stretch the chain. This is seen as an increase in the slope of the force-extension curve (Fig. 2 A, region III). We interpret the rupture force (height of the final peak; Fig. 2, A–C) as the detachment of the pilus from the AFM tip or substrate. The FimA subunits have an Ig-like fold, and several single-molecule force spectroscopy studies have demonstrated that Ig-like domains unfold at forces of 50–300 pN (19,29,30). Because FimA domains are linked head-to-tail via noncovalent interactions (Fig. 1 B), unfolding of a subunit will cause the whole pilus fiber to break. Hence, we cannot exclude the possibility that some of the rupture forces may result from the breakage of the connections between rod subunits. However, our data show that the noncovalent interactions that link subunits together must be very strong because these can survive long extensions (several micrometers) and very high forces (up to 500 pN in some cases).
FIGURE 2.
Force-extension curves obtained after stretching type 1 pili. (A–C) Examples of force-extension curves for type 1 pili. There are three distinct regions of pili stretching, which are marked by the vertical dotted lines (I, II, and III). The first region represents nonspecific interactions between the AFM tip and sample. The second region is a force plateau, Fplateau (dashed line) that corresponds to the unwinding of the pilus rod at a constant force. The third region is the final stretching of the completely unraveled pilus rod. The continuous line shows the prediction of the WLC equation using a persistence length, p, of 1.2 nm. (D and E) Frequency histograms for the plateau force, Fplateau, and contour length, Lc. Gaussian fits gave a mean Fplateau of ∼60 pN (63 ± 17 pN, n = 564, 94 different pili) and Lc ≈ 2 μm (1.9 ± 0.7 μm, n = 230).
Stretching E. coli P pili using AFM
We also measured the mechanical properties of E. coli P pili using AFM techniques. Because P pili have been studied recently using optical tweezers techniques (14), these experiments should also allow a direct comparison between the different techniques. The P and type 1 pilus rods have very similar structures and lengths (2,31,32). FimA makes up the type 1 rod, whereas PapA makes up the P pilus rod. PapA and FimA are 45% homologous over 95% of the protein sequence, so major differences in structural dynamics between the two pilus systems would be surprising. Fig. 3 shows several examples of force-extension patterns obtained after stretching random stretches of P pili. We found that, similar to type 1 pili, P pili are highly extensible with an average contour length, Lc, ∼3 μm (2.9 ± 1.8 μm, n = 130) and a plateau force, Fplateau, of ∼35 pN (34 ± 14 pN, n = 246, 48 different pili). One interesting feature of P pili force-extension curves is the presence of a “hump” in the last elongation region (Fig. 3 A, dashed area). This may correspond to the simultaneous stretching of individual PapA subunits after unwinding of the rod helical structure (14). These force-extension patterns for P pili are remarkably similar to those obtained with optical tweezers (14), which shows that P pili extend at a constant force of ∼30 pN (27 ± 2pN) with a clear “hump” before the detachment force peak.
FIGURE 3.
Force-extension patterns for P pili. (A–C) Examples of force-extension curves for several P pili. (A) Solid line shows the prediction of the WLC equation using a persistence length, p, of 1.6 nm. (D and E) Frequency histograms for the plateau force, Fplateau, and contour length, Lc. The lines correspond to Gaussian fits, which gave mean Fplateau of ∼35 pN (34 ± 14 pN, n = 246, 48 different pili) and Lc ≈ 3 μm (2.9 ± 1.8 μm, n = 130).
Our results show that type 1 and P Pili have similar but not identical mechanical properties. Both types of pili undergo a massive structural transition at high forces in which the rod extends to several times its original length. Our data show that the plateau region for P pili is seen at lower forces than type 1 pili (35 vs. 60 pN), suggesting a weaker interaction between adjacent turns in the helical rod. This is consistent with scanning EM measurements of P and type 1 pili and suggests that the interaction between single turns of P pilus helix is significantly weaker than that in type 1 pili (32). Another difference is the lack of a “hump” in the force-extension patterns of type 1. The structural significance of these differences needs to be further investigated. Our results also suggest that physiological levels of shear flow (up to ∼90 pN/bacterium (28)) are likely to trigger unwinding of the helical region of both types of pili.
The unraveling of the type 1 helical rod structure is fully reversible
We found that type 1 pili could be stretched and relaxed repeatedly provided that we limited the extension so that the pilus did not detach from any of the attachment points (i.e., the AFM tip or the substrate). To measure the unraveling and refolding of the pilus quaternary structure, we used a double-pulse protocol (15,29,33,34). Fig. 4 shows consecutive stretch/relaxation curves obtained on a type 1 pilus. These are a series of two pulling (black, forward arrow) and relaxing (gray, backward arrow) cycles of a pilus (i and ii). The last trace shows the spontaneous detachment of the pilus (Fig. 4 iii). This recording has the typical force-extension pattern for type 1 pili and demonstrates that the previous traces corresponded to the reversible extension of a single pilus. The force-relaxation patterns in Fig. 4, i and ii, follow almost exactly the same trace as those during pulling. The second extension-relaxation cycle, ii, starts at ∼90 nm away from the coverslip to prevent picking up more pili. These data show that type 1 pili are able to refold quickly after being extended.
FIGURE 4.
The forced unraveling of the helical rod structure is fully reversible. Consecutive force-extension and relaxation curves for a single type 1 pilus using a double-pulse stretching protocol (i–iii) in which the pilus was first extended (black traces) and then relaxed (gray traces). The time delay between stretching pulses was 10 s. The second extension-relaxation cycle (ii) starts at ∼90 nm away from the coverslip to prevent picking up more pili. The third trace (iii) corresponds to the spontaneous detachment of the pilus from the AFM tip.
Our data demonstrate that the helical rod of a type 1 pilus is a truly elastic structure that can accommodate large increases in its length and quickly (<1 s) refold to its resting length. Our results also show that the refolding of the helical rod can take place under considerable force. For example, Fig. 4 ii shows that most of the refolding takes place at forces of ∼60 pN (we obtained the zero-force baseline from Fig. 4 iii). At this force, denoted as Fplateau, the helical rod structure is in equilibrium between the unraveled and folded conformations. Hence, very little energy is dissipated during the extension/relaxation cycles where most of the stretching energy is used during the relaxation. These results are similar to force extension/relaxation studies done on P pili (15), indicating that this is a conserved property of UPEC pili. This observation is in contrast to most of the other proteins investigated mechanically so far (such as titin, fibronectin, or tenascin), which show a large hysteresis in the unfolding and refolding curves (19,29,33–35). Hence, E. coli type 1 pili are spring-like organelles designed to extend quickly and relax under force with very little hysteresis.
Multimodal and stepwise unraveling
Bacteria bind host cells in a multivalent fashion, usually being tethered to the host surface by more than one receptor-adhesin interaction. Each E. coli bacterium can express ∼100 type 1 pili on its surface (Fig. 1 A), making it likely that multiple pili attach simultaneously to host receptors. In order to mimic the effect of multiple pili binding, we increased the concentration of type 1 pili adsorbed to glass coverslips. Fig. 5 A shows two typical recordings under these conditions. The characteristic feature is a stair-step pattern on the force-extension curve. Once a pilus detaches or breaks, the force on the cantilever drops, and the pili that remain attached continue to unravel. Hence, the traces in Fig. 5 A correspond to the sequential detachment of many (∼10) pili that connected the AFM tip to the substrate. The stair-step patterns in these force-extension curves display multimodal properties. The force from baseline to each plateau region (before region III) was measured (Fstep) and plotted as a frequency histogram (Fig. 5 B). This histogram shows multiple force peaks, 50, 112, 192, 272, 336, and 400 pN, reflecting the quantized nature of pilus detachment. These data show that unraveling forces are additive and may mimic what occurs in vivo when multiple pili from a single E. coli bind individual host receptors; unraveling forces add up, and the group of pili bound can now withstand much greater forces (up to 600 pN when 10 pili are bound).
FIGURE 5.
Simultaneous stretching of multiple type 1 pili. (A) To promote numerous pili binding to the cantilever, a large concentration of type 1 pili was allowed to adsorb to the coverslips. Force-extension curves show a stair-step pattern, corresponding to the stretching and detachment of multiple pili. The number of pili being stretched was estimated by counting the number of back steps in the force-extension curves. (B) Frequency histogram for the force between steps, Fstep (dotted lines in bottom trace), reveals a multimodal force detachment pattern. The mean peaks are at found at 50, 112, 192, 272, 336, and 400 pN (n = 130 steps) with a mean Fstep of 69 pN.
Monte Carlo simulations of pilus extensibility
To better understand the molecular origin of the elastic properties of the helical rod, we used a simple two-state kinetic model and Monte Carlo simulation techniques (19,33,35) as opposed to a sticky-chain model as described previously (16). We modeled the extensibility of the helical rod with an entropic elasticity as described by the WLC equation and a force-dependent all-or-none unraveling of the individual turns. This model divides the helical rod into small folded segments of contour length lF that can undergo an all-or-none transition into a stretched, unwound state of contour length lU (Fig. 6 A). The increase in contour length on unwinding of a turn is ΔLc = lU − lF = 5 nm, where lF ≈ 3 nm and lU ≈ 8 nm.
FIGURE 6.
Monte Carlo simulation of type 1 pili elasticity. (A) A simple model for the reversible unwinding of the pili helical rod under a stretching force. This model divides the helical rod into small folded segments of contour length lF that can undergo an all-or-none transition into a stretched, unwound state of contour length lU. The increase in contour length on unwinding of a turn is ΔLc = lU − lF, where lF ≈ 3 nm and lU ≈ 8 nm. (B) The unraveling of the helical rod was modeled as a two-state Markovian process in which the rate constants for unwinding, α, and rewinding, β, are force dependent and are given by α = α0exp(FΔxu/kT) and β = βoexp(−FΔxf/kT), where F is the applied force, Δxu and Δxf are the unwinding and rewinding distances, and α0 and β0 are the rate constants at zero force. (C) Experimental force-extension curve for a type 1 pilus. (D) Monte Carlo simulation of a force-extension curve obtained by stretching, at a constant speed (1 nm/ms), 245 subunits. The kinetic parameters are: αo = 5 × 10−2 s−1; βo = 7 × 102 s−1; Δxu= 0.2 nm; and Δxf = 0.5 nm. (E) Superimposition of the experimental trace (orange) and the simulated trace (green). (F) Monte Carlo simulation of force-extension (black trace) and force-relaxation (red trace) curves obtained by stretching/relaxing 320 subunits, using the same kinetic parameters as in panel D.
The unraveling of the helical rod was modeled as a two-state first-order (Markov) process, where the unwinding probability of adjacent turns of the helix was Pu = Nf*α*Δt, where Nf is the number of folded turns and Δt is the polling interval (19,33). The rewinding probability was Pf = Nu*β*Δt where Nu is the number of unwound turns. The rate constants for unwinding, α, and rewinding, β, are force dependent and are given by α(F) = α0*exp(F*Δxu/kT) and β(F) = β0*exp(−F*Δxf/kT), where F is the applied force and Δxu and Δxf are the unwinding and rewinding distances, which in an energy diagram correspond to the distance to the transition state (Fig. 6 B (36)). α0 and β0 are the rate constants in the absence of an applied force, and k and T have their usual meanings.
To simulate the extension of the helical rod, the force experienced by the pili during a stretching pulse at a constant speed is calculated using the WLC equation. We use this force value to compute the probability of unraveling of a turn using the Monte Carlo approach. Fig. 6 D shows a Monte Carlo simulation of a force-extension curve obtained by stretching, at a constant speed (1 nm/ms), 245 subunits. As shown in Fig. 6 E, this simple model (green trace) closely reproduces the experimental force-extension data (orange trace). Furthermore, as shown in Fig. 6 F, this model accurately simulates the consecutive unwinding (black trace) and rewinding (red trace) of a single type 1 pilus (see Fig. 4). Fig. 6 F shows a Monte Carlo simulation of force-extension (black trace) and force-relaxation (red trace) curves obtained by stretching/relaxing 320 subunits.
From these simulations, we find that the parameters that best describe the experimental data are the following: αo = 5 × 10−2 s−1; βo = 7 × 102 s−1; Δxu= 0.2 nm; and Δxf = 0.5 nm. We can use these kinetic parameters to estimate the free energies for the unbinding and rebinding of adjacent pilin subunits. The free energies can be calculated from the rate constants α0 and β0 using Eyring rate theory: ΔG = kT*ln(rate/A). Assuming a preexponential factor of 106 s−1 (37), we estimate a ΔGu = 17 kT and a ΔGf = 7 kT. These values are similar to those estimated for P pili (16) and correspond to the energy of a typical protein-ligand bond (range 5–30 kT (38)).
In summary, a simple two-step Monte Carlo simulation accurately simulates the unwinding and rewinding of type 1 pili and offers an alternative model to the sticky-chain model (16).
Pili extensibility can dramatically affect the lifetime of the bonds between bacteria pili and host receptors
Shear forces in the bladder and kidney caused by fluid and urine flow are a major factor that UPEC must subvert to persist in the urinary tract. Recent work by Thomas et al. (17,18) showed that E. coli expressing type 1 pili bind more tightly to target cells when under increased shear force and attributed this to force-driven conformational changes in the adhesin domain, FimH. However, an alternative scenario is that the receptor-ligand interaction could be modulated by force-driven elongation of the pilus rod, as proposed by Bullitt and Makowski (31). To explore this idea, we quantified the effect of pili elasticity on bond lifetime, using Monte Carlo techniques as described by Oberhauser et al. (33). According to this model, a protein-ligand bond will break under an applied force as described by Bell (36): koff(F) = koff*exp(F*d/kT), where koff is the spontaneous off rate (at zero force) and d is the distance that will destabilize the bond and lead to failure. Although the parameters are not known for the specific binding affinities between FimH and mannose, we simulated the effect of pili elasticity on the lifetime of the receptor bonds under a stretching force using the P selectin-leukocyte rolling interaction as a model because it is a bond exposed to shear forces. For this bond, the estimated rupture distance, d, is 0.04 nm, and the off rate is koff = 0.95 s−1 (39).
We assumed three extreme cases: 1), a bond linked to a rigid rod made of 500 nonextensible turns in which each turn can extend by only ΔLc= 0.05 nm (Fig. 7 A); 2), a bond linked to a semirigid rod made of 400 nonextensible (ΔLc = 0.05 nm) plus 100 extensible (ΔLc= 5 nm) turns (Fig. 7 B); and 3), a bond linked to an extensible rod made of 500 turns in which the unwinding of each turn leads to an increase in contour length, ΔLc= 5 nm (Fig. 7 C). For these simulations, we used the kinetic parameters estimated in the previous simulations (Fig. 6). The pulling speed was 2 μm/s. As the simulations show (Fig. 7, Ai, Bi, and Ci), the elastic properties of the pili can have a dramatic effect on the lifetime of the receptor bond. A bond linked to a rigid rod tends to break at high forces (∼600 pN; Fig. 7 Aii) and to survive relatively short times (∼0.5 s; Fig. 7 Aiii). A bond linked to a semirigid rod tends to break at lower forces (∼450 pN; Fig. 7 Bii) and to survive longer times (∼1 s; Fig. 7 Biii). A bond linked to a fully extensible rod tends to break at very low forces (∼60 pN; Fig. 7 Cii) and to survive much longer times (∼up to 4 s; Fig. 7 Ciii).
FIGURE 7.
Effect of pili elastic properties on bond lifetime. (A–C) Monte Carlo simulations of the effect of pili mechanical properties on the lifetime of the receptor bonds under a stretching force. (A) Simulation of a bond linked to a rigid rod made of 500 nonextensible turns in which each turn can extend by only ΔLc = 0.05 nm. The bond survives for 0.5 ± 0.3 s (iii) and breaks at 611 ± 148 pN, n = 170 (ii). (B) Simulation of a bond linked to a semirigid rod made of 400 nonextensible (ΔLc = 0.05 nm) plus 100 extensible (ΔLc = 5 nm) turns. The bond survives for 0.9 ± 0.3 s (iii) and breaks at 455 ± 231 pN, n = 172 (ii). (C) Simulation of a bond linked to an extensible rod made of 500 turns in which the unwinding of each turn leads to an increase in contour length, ΔLc = 5 nm. The bond survives for 1.9 ± 1.0 s (iii) and breaks at 62 ± 19 pN, n = 176 (ii).
Although speculative, our simulation results clearly show that the elastic properties of the pili can have a dramatic effect on the lifetime of the receptor bond. We hypothesize that the dynamic extensibility of the long helical rod could allow the pili-receptor bond to persist over long extensions. This is important because longer persistence may lead to greater chances of activating signaling events within either the bacteria or a host cell. These events may be the actin rearrangement within the host that allows bacterial invasion or the signaling of genes within the bacteria that are activated during intracellular replication. Maier et al. (40) attribute the force-dependent elongation of type IV pili as the ability to release tension generated during retraction without breaking the pilus. Thus, the same may be true for type 1 pili in controlling pilus-receptor interactions for shear-dependent binding and signaling.
DISCUSSION
The first step in the encounter between a host and a pathogen is the attachment of the pathogen to the host epithelium. This interaction begins a dynamic molecular cross talk that ultimately determines the outcome of the infectious process. Bacterial entry into superficial umbrella cells lining the bladder lumen is a critical event in urinary tract infections. Bacterial entry into umbrella cells activates a complex genetic cascade leading to the formation of intracellular bacterial communities that undergo a defined maturation and differentiation program that is critical in disease. Type 1 and P pili are critical organelles and virulence factors for UPEC.
Our results demonstrate that pili are dynamic structures that function as molecular springs under applied forces. Using single-molecule AFM, we found that the rods of both P pili and type 1 pili are highly extensible (they can be extended about two to four times their resting length). However, the required forces to unravel each of these pili are different (35 vs. 60 pN). This could possibly represent a specific adaptation to the biological niche in which type 1 pili promote bacterial colonization. The structural significance for this is to be further investigated. The forced unraveling of type 1 pili is also reversible, taking place under considerable forces (∼60 pN). Together, these results suggest a conserved structural mechanism that is used in bacterial pathogenesis.
Importantly, type 1 pili elasticity provides a mechanism for extending the lifetime of its adhesin-receptor interaction. It has been shown that, under shear flow, bacteria can transition among unbound, rolling, and stationary states (17,28). Pilus unraveling provides a simple mechanism for explaining these transitional changes. When shear flow is low, drag forces are not enough to induce pilus unraveling, and it acts as a rigid rod, promoting short-lived FimH-mannose interactions. As shear forces increase from moderate to high flow, bacteria transition to rolling and stationary states. At high shear forces, the probability of unraveling also increases, promoting longer FimH-mannose interactions. Therefore, at moderate to high shear stress (drag forces <30 pN), a rolling state would be expected in which only a small population of the pili are unraveling under force and extending their bond lifetime. Additionally, the number of unraveling pili necessary to keep the bacterium stationary is achieved only under high shear stress.
Adhesin-receptor interactions are critical in the pathogenic cascade for binding and invasion; however, pilus dynamics may also play a major role in pathogenesis. Being able to unravel under applied forces, pili increase the lifetime of their adhesin-receptor interaction. Being able to refold under considerable force, pili are capable of acting as molecular springs and shock absorbers. And being able to achieve multimodal properties, pili are able to work in unison and withstand greater shear forces as they arise. The ability of these protein complexes to behave in such a manner is remarkable. These results not only lend a new understanding to how bacteria combat hostile environments within the host but also offer new insight into protein interactions and the functionality of protein complexes.
Acknowledgments
This work was funded by National Institutes of Health/National Institute of Allergy and Infectious Diseases grants AI029549 and AI48689 (S.J.H.), NIH grant DK067443 (A.F.O.), the John Sealy Memorial Endowment Fund for Biomedical Research 2531-03 (A.F.O.), Polycystic Kidney Foundation grant 116a2r (A.F.O), and a training fellowship from the Keck Center for Computational and Structural Biology of the Gulf Coast Consortia (National Library of Medicine grant 5T15LM07093 to T.G.).
References
- 1.Langermann, S., S. Palaszynski, M. Barnhart, G. Auguste, J. S. Pinkner, J. Burlein, P. Barren, S. Koenig, S. Leath, et al. 1997. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science. 276:607–611. [DOI] [PubMed] [Google Scholar]
- 2.Bullitt, E., and L. Makowski. 1998. Bacterial adhesion pili are heterologous assemblies of similar subunits. Biophys. J. 74:623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gong, M., and L. Makowski. 1992. Helical structure of P pili from Escherichia coli. Evidence from x-ray fiber diffraction and scanning transmission electron microscopy. J. Mol. Biol. 228:735–742. [DOI] [PubMed] [Google Scholar]
- 4.Soto, G. E., and S. J. Hultgren. 1999. Bacterial adhesins: common themes and variations in architecture and assembly. J. Bacteriol. 181:1059–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones, C. H., J. S. Pinkner, R. Roth, J. Heuser, A. V. Nicholes, S. N. Abraham, and S. J. Hultgren. 1995. FimH adhesin of type 1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc. Natl. Acad. Sci. USA. 92:2081–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mulvey, M. A., Y. S. Lopez-Boado, C. L. Wilson, R. Roth, W. C. Parks, J. Heuser, and S. J. Hultgren. 1998. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science. 282:1494–1497. [DOI] [PubMed] [Google Scholar]
- 7.Anderson, G. G., J. J. Palermo, J. D. Schilling, R. Roth, J. Heuser, and S. J. Hultgren. 2003. Intracellular bacterial biofilm-like pods in urinary tract infections. Science. 301:105–107. [DOI] [PubMed] [Google Scholar]
- 8.Justice, S. S., C. Hung, J. A. Theriot, D. A. Fletcher, G. G. Anderson, M. J. Footer, and S. J. Hultgren. 2004. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc. Natl. Acad. Sci. USA. 101:1333–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hultgren, S. J., S. Normark, and S. N. Abraham. 1991. Chaperone-assisted assembly and molecular architecture of adhesive pili. Annu. Rev. Microbiol. 45:383–415. [DOI] [PubMed] [Google Scholar]
- 10.Abraham, S. N., D. Sun, J. B. Dale, and E. H. Beachey. 1988. Conservation of the d-mannose-adhesion protein among type 1 fimbriated members of the family Enterobacteriaceae. Nature. 336:682–684. [DOI] [PubMed] [Google Scholar]
- 11.Mulvey, M. A., J. D. Schilling, and S. J. Hultgren. 2001. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect. Immun. 69:4572–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brinton, C. C., Jr. 1965. The structure, function, synthesis and genetic control of bacterial pili and a molecular model for DNA and RNA transport in gram negative bacteria. Trans. N. Y. Acad. Sci. 27:1003–1054. [DOI] [PubMed] [Google Scholar]
- 13.Sauer, F. G., J. S. Pinkner, G. Waksman, and S. J. Hultgren. 2002. Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation. Cell. 111:543–551. [DOI] [PubMed] [Google Scholar]
- 14.Jass, J., S. Schedin, E. Fallman, J. Ohlsson, U. J. Nilsson, B. E. Uhlin, and O. Axner. 2004. Physical properties of Escherichia coli P pili measured by optical tweezers. Biophys. J. 87:4271–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fallman, E., S. Schedin, J. Jass, B. E. Uhlin, and O. Axner. 2005. The unfolding of the P pili quaternary structure by stretching is reversible, not plastic. EMBO Rep. 6:52–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersson, M., E. Fallman, B. E. Uhlin, and O. Axner. 2006. A sticky chain model of the elongation and unfolding of Escherichia coli P pili under stress. Biophys. J. 90:1521–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas, W. E., E. Trintchina, M. Forero, V. Vogel, and E. V. Sokurenko. 2002. Bacterial adhesion to target cells enhanced by shear force. Cell. 109:913–923. [DOI] [PubMed] [Google Scholar]
- 18.Thomas, W., M. Forero, O. Yakovenko, L. Nilsson, P. Vicini, E. Sokurenko, and V. Vogel. 2006. Catch-bond model derived from allostery explains force-activated bacterial adhesion. Biophys. J. 90:753–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rief, M., M. Gautel, F. Oesterhelt, J. M. Fernandez, and H. E. Gaub. 1997. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 276:1109–1112. [DOI] [PubMed] [Google Scholar]
- 20.Fisher, T. E., A. F. Oberhauser, M. Carrion-Vazquez, P. E. Marszalek, and J. M. Fernandez. 1999. The study of protein mechanics with the atomic force microscope. Trends Biochem. Sci. 24:379–384. [DOI] [PubMed] [Google Scholar]
- 21.Fisher, T. E., M. Carrion-Vazquez, A. F. Oberhauser, H. Li, P. E. Marszalek, and J. M. Fernandez. 2000. Single molecular force spectroscopy of modular proteins in the nervous system. Neuron. 27:435–446. [DOI] [PubMed] [Google Scholar]
- 22.Lindberg, F. P., B. Lund, and S. Normark. 1984. Genes of pyelonephritogenic E. coli required for digalactoside-specific agglutination of human cells. EMBO J. 3:1167–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orndorff, P. E., and S. Falkow. 1984. Organization and expression of genes responsible for type 1 piliation in Escherichia coli. J. Bacteriol. 159:736–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuehn, M. J., J. Heuser, S. Normark, and S. J. Hultgren. 1992. P pili in uropathogenic E. coli are composite fibres with distinct fibrillar adhesive tips. Nature. 356:252–255. [DOI] [PubMed] [Google Scholar]
- 25.Florin, E.-L., M. Rief, H. Lehmann, M. Ludwig, C. Dornmair, V. T. Moy, and H. E. Gaub. 1995. Sensing specific molecular interactions with the atomic force microscope. Biosens. Bioelectron. 10:895–901. [Google Scholar]
- 26.Bustamante, C., J. F. Marko, E. D. Siggia, and S. Smith. 1994. Entropic elasticity of lambda-phage DNA. Science. 265:1599–1600. [DOI] [PubMed] [Google Scholar]
- 27.Marko, J. F., and E. D. Siggia. 1995. Stretching DNA. Macromolecules. 28:8759–8770. [Google Scholar]
- 28.Thomas, W. E., L. M. Nilsson, M. Forero, E. V. Sokurenko, and V. Vogel. 2004. Shear-dependent ‘stick-and-roll’ adhesion of type 1 fimbriated Escherichia coli. Mol. Microbiol. 53:1545–1557. [DOI] [PubMed] [Google Scholar]
- 29.Oberhauser, A. F., P. E. Marszalek, M. Carrion-Vazquez, and J. M. Fernandez. 1999. Single protein misfolding events captured by atomic force microscopy. Nat. Struct. Biol. 6:1025–1028. [DOI] [PubMed] [Google Scholar]
- 30.Li, H., W. A. Linke, A. F. Oberhauser, M. Carrion-Vazquez, J. G. Kerkvliet, H. Lu, P. E. Marszalek, and J. M. Fernandez. 2002. Reverse engineering of the giant muscle protein titin. Nature. 418:998–1002. [DOI] [PubMed] [Google Scholar]
- 31.Bullitt, E., and L. Makowski. 1995. Structural polymorphism of bacterial adhesion pili. Nature. 373:164–167. [DOI] [PubMed] [Google Scholar]
- 32.Hahn, E., P. Wild, U. Hermanns, P. Sebbel, R. Glockshuber, M. Haner, N. Taschner, P. Burkhard, U. Aebi, and S. A. Muller. 2002. Exploring the 3D molecular architecture of Escherichia coli type 1 pili. J. Mol. Biol. 323:845–857. [DOI] [PubMed] [Google Scholar]
- 33.Oberhauser, A. F., P. E. Marszalek, H. P. Erickson, and J. M. Fernandez. 1998. The molecular elasticity of the extracellular matrix protein tenascin. Nature. 393:181–185. [DOI] [PubMed] [Google Scholar]
- 34.Oberhauser, A. F., C. Badilla-Fernandez, M. Carrion-Vazquez, and J. M. Fernandez. 2002. The mechanical hierarchies of fibronectin observed with single-molecule AFM. J. Mol. Biol. 319:433–447. [DOI] [PubMed] [Google Scholar]
- 35.Tskhovrebova, L., and J. Trinick. 2001. Flexibility and extensibility in the titin molecule: analysis of electron microscope data. J. Mol. Biol. 310:755–771. [DOI] [PubMed] [Google Scholar]
- 36.Bell, G. I. 1978. Models for the specific adhesion of cells to cells. Science. 200:618–627. [DOI] [PubMed] [Google Scholar]
- 37.Chakrapani, S., and A. Auerbach. 2005. A speed limit for conformational change of an allosteric membrane protein. Proc. Natl. Acad. Sci. USA. 102:87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Protein ligand database. http://www-mitchell.ch.cam.ac.uk/pld. Accessed October, 2006. [Online].
- 39.Alon, R., S. Chen, K. D. Puri, E. B. Finger, and T. A. Springer. 1997. The kinetics of l-selectin tethers and the mechanics of selectin-mediated rolling. J. Cell Biol. 138:1169–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maier, B., M. Koomey, and M. P. Sheetz. 2004. A force-dependent switch reverses type IV pilus retraction. Proc. Natl. Acad. Sci. USA. 101:10961–10966. [DOI] [PMC free article] [PubMed] [Google Scholar]









