Abstract
Self-incompatibility (SI) is one of several mechanisms that have evolved to prevent inbreeding in plants. SI in Brassica is controlled by the polymorphic S locus complex. Two S locus-encoded proteins are coordinately expressed in the stigma epidermis: the cell wall-localized S locus glycoprotein (SLG) and the plasma membrane-anchored S receptor kinase (SRK). These proteins are thought to recognize a pollen factor that leads to the rejection of self-pollen. Evidence has accumulated that indicates that both proteins are necessary for the ability of the stigma to inhibit self-pollen. However, it has not been possible to prove this necessity definitively or to demonstrate that these genes are sufficient for this phenotype, because previous attempts to transfer this phenotype via transformation have not been successful. In this study, two overlapping S locus genomic clones, which cover ≈55 kilobases of DNA and contain the SLG, SRK, and an anther-expressed gene in the region common to the two, were introduced into a self-compatible Brassica napus line. The resulting transgenic plants were shown to carry the female part of the SI phenotype, rejecting pollen in a haplotype-specific manner. However, the pollen SI phenotype was not found in any of the transgenic plants. These data show that the SLG and SRK are sufficient for the female side but not the male side of the SI phenotype in Brassica and that there must be an independent pollen S factor encoded outside the cloned region.
Self-incompatibility (SI) is one of the mechanisms evolved by higher plants to promote outbreeding and is also a good genetic model to study cell-to-cell communication. In Brassica species, SI is controlled by haplotypes of the S locus complex (1, 2). The control is of the sporophytic type in which the pollen parent genotype determines the phenotype of the pollen (1). When a haplotype in the pollen parent matches that of the pistil, pollen germination is arrested at the stigma surface. Thus, sporophytic SI not only prevents self-fertilization but also prevents crossing between genetically related individuals. The S haplotypes are generally codominant with each other, and SI is dominant over self-compatibility (3, 4).
Two genes have been found to be associated with the SI phenotype. The first encodes a secreted S locus glycoprotein (SLG; ref. 5), and the second encodes an S receptor kinase (SRK; ref. 6). The latter protein has a receptor-like domain very similar to SLG, a transmembrane domain, and a kinase domain. Both genes are expressed primarily in stigma papilla cells (5–7). Although some transcripts have been detected in anther tissue, both proteins could be detected only in stigma (8–10). Consistent with this finding, accumulated genetic evidence indicates that expression of both genes in papillar cells is required for the operation of SI (11–17), though recently the necessity of SLG for SI has been questioned (18). Several self-compatible mutant strains of Brassica have been identified in which self-compatibility is associated with spontaneous mutations at the S locus causing disruption of the SRK gene (13, 14). In addition, spontaneous mutations at loci unlinked to the S locus that down-regulate the SLG gene (12) and transgene-induced mutations that down-regulate the SLG (11, 17) and SRK (15, 16) genes are associated with loss of the pistil's ability to reject self-pollen.
A model for the Brassica SI response has been proposed (6, 7) based on the above observations and on what is known about receptor kinase interactions with their specific ligands in mammalian cells (19). The SRK protein would form a receptor complex in the stigma papilla cells that recognizes an allele-specific ligand present on the pollen surface causing a cascade of reactions leading to rejection of self-pollen. The SLG protein either would be involved in bringing the ligand into contact with the SRK protein or would serve as part of the receptor complex. The pollen ligand is thought to be a protein encoded by a separately linked gene and recognized by the receptor complex in a haplotype-specific manner. Although this hypothesis is reasonable, a number of aspects of this model need to be tested experimentally. For example, although the data mentioned above indicate that both SLG and SRK are necessary for SI, it is not clear whether these are sufficient for operation of the SI response in the pistil. Attempts to modify SI specificity by transformation experiments with the SLG and/or SRK genes have thus far been unsuccessful, largely as a result of transgene-induced cosuppression (15–17).
Brassica napus is an amphidiploid plant that is normally self-compatible, whereas its presumptive progenitors, Brassica oleracea and Brassica rapa, are diploid SI species (20). SI haplotypes can be transferred into B. napus from the progenitor species via interspecific crosses, with the resulting individuals being repeatedly backcrossed to the B. napus parent, thus generating SI lines (21, 22). The SI B. napus line W1 was produced by introducing a functional S haplotype called 910 into the self-compatible Westar line (21). Other than the region involved in SI, these lines are virtually isogenic. We have recently isolated and characterized a ≈65-kilobase (kb) fosmid contig spanning the SLG-910 and SRK-910 genes (23). Herein, we present the results of the transgenic analysis of the fosmid clones in the Westar background to determine whether the SLG and SRK genes are sufficient for the stigma side of the SI phenotype. Further, we wished to see which, if any, of the genes encoded in this region would confer the male part of SI.
Materials and Methods
Transgene Constructs, Plant Transformation, and Cross-Pollination Analysis.
The inserts of the Fos20 and Fos22 genomic clones that overlap with S locus linked 1 (SLL1), SLG, and SRK in the overlapping region (23) were released by NotI digestion, recloned as NotI fragments into the plant transformation vector binary bacterial artificial chromosome 2 (BIBAC2; refs. 24 and 25), and introduced into Agrobacterium tumefaciens strain COR338 (24) via triparental mating (26). The self-compatible B. napus Westar line was used as recipient for the two transgene constructs. Plant transformation was performed by using Agrobacterium-mediated methods (27). The NPTII- and SLG-910-specific primers used for testing the presence of the transgenes have been described (21, 24). SI was tested by measuring seed set. Pollination bags were applied 2 days before anthesis for both self- and cross-pollination. Cross-pollinations were performed on fully open flowers 2 days after emasculation.
Genomic DNA and RNA Gel Blot Analysis.
For pulsed field gel electrophoresis analysis, high molecular mass genomic DNA was isolated from nuclei of young leaf tissue and embedded into agarose plugs as described by Zhang et al. (28). For regular genomic DNA blot analysis, DNA was extracted from floral buds as described (29). Approximately 5 μg of DNA was digested, size fractionated through agarose gel, and transferred to nylon membrane in 20× SSC (1× SSC is 150 mM NaCl/15 mM sodium citrate).
Total RNA was extracted from stigmas isolated from 5- to 6-mm-long floral buds by using a hot phenol method (30). Approximately 6 μg of each RNA sample was fractionated through a 1.2% agarose-formaldehyde gel and transferred to nylon membrane in 20× SSC. Equal loading of RNA samples was indicated by rehybridization of the membrane by using the cDNA to 18S rRNA from B. rapa (31) as probe. Prehybridization and hybridization steps were performed as described (29). Membranes were washed for 50 min at a final stringency of 0.1× SSC and 0.1% SDS at 65°C. Gel-purified DNA fragments labeled by random priming (32) were used as probes.
Results
BIBAC–Fosmid Constructs and Transformation of Self-Compatible Brassica.
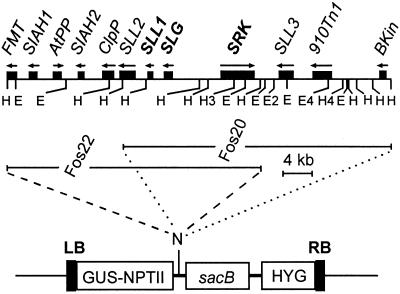
Previously, the entire SLG-SRK region was not cloned on a single genomic fragment (29, 33, 34). Transformation with single genes driven by heterologous promoters, thus potentially generating gain-of-function plants, has been previously unsuccessful (15). We have recently isolated a ≈65-kb fosmid contig covering the SLG-910 and SRK-910 genes from the SI B. napus line W1 (23). The distance between the two S genes is only ≈6 kb of DNA, which is the shortest among the S haplotypes characterized to date (23, 34, 35). A vector for the transformation of large pieces of DNA into plants called the BIBAC vector has been constructed (24, 25), which makes the transformation of the entire fosmid genomic clones (≈40 kb) possible. A method for routine introduction of DNA fragments larger than ≈25 kb into plant nuclear genome had not yet been demonstrated (24). Two fosmid clones, Fos20 and Fos22, were chosen for cloning into the BIBAC vector to transform the self-compatible Westar line. As shown in Fig. 1, Fos22 contains the SRK, SLG, the anther-specifically expressed gene SLL1, and several upstream genes, whereas Fos20 contains SLL1, SLG, SRK, and the gene SLL3, which encodes a putative small secreted protein, as well as a transposon and a nonfunctional kinase gene. The two genomic clones overlap in the region containing the SLL1, SLG, and SRK genes. SLL1 is an anther-specific gene (29) and should not play a role in the stigma. Thus, these clones provide us an opportunity for gain-of-function transgenic studies to address the issue of whether the SLG and SRK genes are sufficient for conferring the SI phenotype on the stigma side and whether any other genes from this region are functionally relevant.
Figure 1.
Schematic representation of the transgene constructs. (Upper) The 910 S haplotype genomic region covered by the Fos20 and Fos22 genomic clones (23). FMT, methionyl-tRNA transformylase; SIAH, Drosophila seven-in-absentia; AtPP, Arabidopsis putative protein; ClpP, Clp protease; 910Tn1, a Spm-like transposon; Bkin, a nonfunctional kinase. (Lower) Part of the BIBAC2 vector (24, 25). GUS, β-glucuronidase; NPTII, neomycin phosphotransferase II; sacB, levansucrase; HYG, hygromycin phosphotransferase. RB and LB denote right and left borders of T-DNA. E, EcoRI; H, HindIII; N, NotI. The number after the letters indicates multiple sites.
The two BIBAC–fosmid constructs were transformed into the self-compatible cultivar Westar line, which carries the nonfunctional A10 S haplotype, likely because of a 1-bp deletion in the SRK-A10 gene (13). Six putative transgenic plants (T0) that regenerated shoots and roots on selective medium were analyzed subsequently by genomic DNA gel blot analysis by using the selectable marker gene NPTII as a probe. Further, the PCR with primers specific to the NPTII and SLG-910 genes was used to ascertain presence of the transgenes (data not shown). Consequently, one of the six was found to be a false positive. Among the five positive transgenic lines, four were from the Fos20 construct, and one was from Fos22. One of the four Fos20 transgenic lines was male sterile (no pollen) and was eliminated from further analysis. The other four lines set self-seeds, suggesting that neither of the two fosmid clones contains the complete functional S haplotype. Self-seeds (T1) were collected for further analysis.
Phenotype of the Transgenic Lines.
T1 progeny were grown from each of the four primary transgenic lines for analysis of pollination phenotypes. Presence of the transgenes was indicated by PCR with primers specific to the SLG-910 and NPTII genes. All progeny plants that inherited the transgenes set roughly the same amount of self-seeds as plants that did not inherit the transgenes (data not shown). To determine the pollination phenotype of these plants in the stigma and pollen, large numbers of reciprocal crosses between transgenic plants and the SI W1 as well as the self-compatible Westar lines were performed. As negative controls, the same crosses were also performed for the progeny plants that did not inherit the transgenes. The pollination results are summarized in Table 1.
Table 1.
Average seed set per pod for the transgenic lines (T1 generation) when reciprocally crossed with parental lines
| Female × Pollen | Transgene* | Seeds per pod† | No. of pods | No. of plants |
|---|---|---|---|---|
| W1 (S910S910) × (Self) | 0.2 | 85 | 4 | |
| W1 (S910S910) × Westar | 18.3 | 48 | 3 | |
| Tg‡ (Fos20) × W1 (S910S910) | + | 1.6 | 446 | 19 |
| − | 18.7 | 196 | 10 | |
| Tg (Fos20) × Westar | + | 20.5 | 279 | 19 |
| − | 20.1 | 145 | 10 | |
| Tg (Fos20) × T2 (SA14SA14) | + | 19.3 | 82 | 7 |
| W1 (S910S910) × Tg (Fos20) | 21.8 | 49 | 3 | |
| Tg (Fos22) × W1 (S910S910) | + | 0.7§ | 269 | 7 |
| − | 22.2 | 175 | 8 | |
| Tg (Fos22) × Westar | + | 12.8§ | 112 | 7 |
| − | 22.2 | 100 | 8 | |
| Tg (Fos22) × T2 (SA14SA14) | + | 12.4§ | 23 | 2 |
| W1 (S910S910) × Tg (Fos22) | 11.4§ | 52 | 3 |
*“+” indicates presence of the transgene; “−” indicates loss of the transgene caused by segregation.
†For the Fos20 transgenic lines, the data are the combination of all three lines (3A, 5B, and 6A).
‡Tg, transgenic lines in the self-compatible Westar background.
§The somewhat lower seed set for some of the plants was caused by the conditions in the growth chamber.
Among the progeny of all three Fos20-transformed lines (3A, 5B, and 6A), all 19 plants that inherited the transgene were SI on the stigma side as indicated by the cross-pollination results of transgenic stigmas with W1 and Westar pollen. When transgenic stigmas were fertilized with Westar pollen, the resulting pods were fully developed, and normal numbers of seeds were set (average of ≈20.5 per pod). These results were similar to those obtained when W1 was fertilized with Westar pollen (average of ≈18.3 per pod). In contrast, those fertilized with W1 pollen set very few seeds (average of ≈1.6 per pod), which is typical of the strong SI response shown by W1 when self-pollinated (average of ≈0.2 per pod). All 10 plants that did not inherit the transgene because of segregation were compatible (average of ≈18.7 and 20.1 per pod when pollinated with pollen from W1 and Westar, respectively), confirming that the change in phenotype was indeed caused by the transgene. The T2 line is another SI B. napus line that contains the functional A14 S haplotype (22). Pollen from this line was used to fertilize the transgenic lines yielding fully developed pods having a normal seed set (averaged ≈19.3 per pod) comparable to that seen for Westar, indicating that the rejection of W1 pollen by the transgenic stigmas is a haplotype-specific phenomenon. The pollen from all of the transgenic plants was able to fertilize the W1 line (average of ≈21.8 per pod), indicating that the male part of the phenotype remains unchanged. This result is consistent with the earlier observations that all of the transgenic lines set self-seeds, even though the female SI phenotype was expressed. We did not notice any significant differences in pollination phenotype among plants that inherited the transgene, suggesting that there was no obvious relationship between stigma SI phenotype and transgene dosage.
Similar pollination results were obtained with progeny of the Fos22-transformed line (389A). Nine progeny plants that inherited the transgene as judged by PCR were analyzed (along with eight plants that did not). Of the plants that inherited the transgene, seven plants showed the strong gain-of-function SI phenotype on the stigma side, similar to that seen for the Fos20-transformed lines (Table 1). Further, the response was again haplotype-specific, and the pollen phenotype was not changed (Table 1). The typical phenotypes of seed pods produced after various cross-pollinations are shown in Fig. 2. Pollination of the transgenic stigmas with W1 pollen yielded very few seeds, whereas pollination of the transgenic stigmas with Westar or T2 pollen produced fully developed pods, very similar to those seen in the self-compatible Westar line. Two progeny plants carrying the transgene (389A-5 and 389A-7) did not show any significant differences when pollinated by W1 or Westar pollens. This result correlated with a lack of expression of the SLG and SRK genes in these plants (see below).
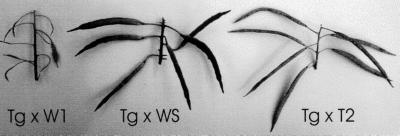
Figure 2.
Comparison of mature B. napus pods produced when transgenic stigmas were pollinated with pollen from W1 (Tg × W1), Westar (Tg × WS), and T2 (Tg × T2).
Genomic DNA Gel Blot Analysis of the Brassica Transgenic Lines.
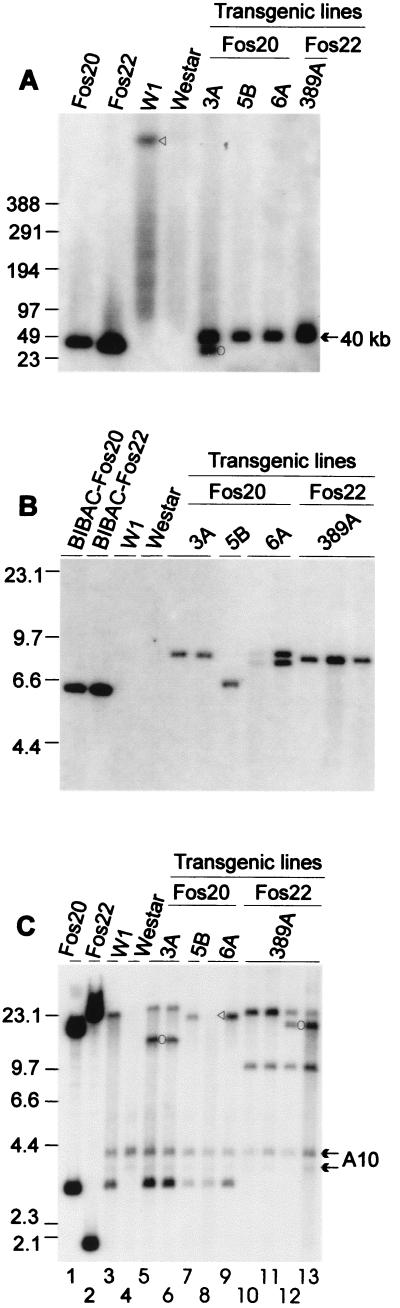
Genomic DNA was isolated from the progeny of each transgenic Brassica line, cleaved with various restriction endonucleases, and analyzed by hybridization with several probes. This analysis was used to determine whether the fosmid genomic inserts were transferred intact into the plant chromosome, to estimate the approximate copy number, and to identify unique band patterns for each of the transgenic lines. First, DNA blots of NotI-digested high molecular mass genomic DNA were probed with cDNAs for the SLG-910 and SRK-910 kinase portion to determine whether the fosmid inserts were transferred intact into the plant chromosomes. As shown in Fig. 3A, progeny of all four lines had a ≈40-kb band, indicating that the transgene fragments were intact. In addition to the ≈40-kb fragment, the 3A line had a shorter and weaker band, which might have been caused by the transfer of a partial T-DNA fragment during transformation.
Figure 3.
Genomic DNA gel blot analysis of the T1 progeny plants. Molecular size markers are indicated at left in kilobases. Sources of DNA are labeled above each lane. Each lane represents DNA from one progeny plant. The transgene constructs, W1, and Westar DNAs were all included as controls. (A) Gel-blot hybridization of NotI-digested high molecular mass DNA with SLG-910 and SRK-910 cDNAs. Restriction fragments were separated by pulsed field gel electrophoresis in a CHEF-DRIII apparatus (Bio-RAD) in 1× Tris-acetate-EDTA at 3 V/cm with a pulse-switching interval ramped from 5 to 120 s over 42 h. The triangle indicates the ≈1,000-kb NotI fragment encompassing both the SLG-910 and SRK-910 genes, which was identified previously by pulsed field gel electrophoresis analysis (29). The circle indicates a truncated copy in 3A line. (B) Gel-blot hybridization of HindIII-digested DNA with NPTII-specific DNA. (C) Gel-blot hybridization of EcoRI-digested DNA with SLG-910 and SRK-910 cDNAs. The two fosmids were cut with NotI and EcoRI. The ≈12-kb band in lanes 5 and 6 and the ≈16-kb band in lanes 12 and 13 (indicated by circles) represent truncated copies of the transgenes. The triangle in lane 8 indicates a band visible after longer exposure. The arrows indicate the bands likely representing the endogenous A10 haplotype present in Westar.
DNA blots of HindIII-digested genomic DNA were then probed with the NPTII probe to estimate the copy number of the transgenes. As shown in Fig. 3B, only the 6A line progeny had two different bands representing two copies at different chromosomal sites, whereas the other three lines had only one copy. The various sizes of the restriction fragments identified by hybridization to the NPTII-specific probe indicated that the T-DNA integrated into a unique site in each of the transformed lines. The 3A line contained two copies that hybridized to the SLG-910 and SRK-910 cDNAs, and only one fragment hybridized to the NPTII-specific left border probe. The shorter copy was likely caused by the partial transfer of the T-DNA, because it is thought that the T-DNA is transferred linearly from the right border to the left border (24).
Finally, DNA blots of EcoRI-digested genomic DNA were probed with cDNAs for SLG-910 and SRK-910 kinase portion (Fig. 3C) to establish the unique band pattern of each of the transgenic lines. EcoRI was chosen, because the 25-kb EcoRI fragment hybridizing to both SLG and SRK probes is a hallmark of the 910 S haplotype (29). In addition, there is a ≈3-kb fragment that hybridizes to the SRK kinase portion probe, because there is an EcoRI site within the SRK gene. All progeny of the Fos20-transformed lines produced the expected ≈3-kb band and various bands larger than ≈17 kb, which represent the ≈17-kb EcoRI–HindIII transgene fragment plus flanking regions from the host genome. The intermediate bands (≈12 kb) very likely resulted from the truncated copy shown on the NotI-digested blot (Fig. 3A). All the 389A line progeny yielded the expected ≈25-kb band as well as a ≈10-kb band, which represents the 1.8-kb EcoRI–HindIII transgene fragment plus the flanking region from the host genome. Surprisingly, some of the 389A progeny yielded an unexpected ≈16-kb band, which might have been created by a postintegration rearrangement event.
Expression of the SLG-910 and SRK-910 Transgenes in the Self-Compatible Westar Line.
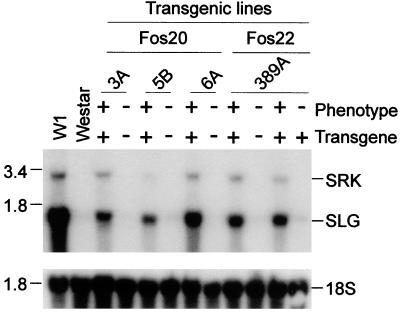
The fact that both Fos20 and Fos22 constructs gave rise to the same phenotype indicates that only the overlapping region between the two is responsible for the SI function. From our recent transcriptional mapping studies of the cloned 910 S haplotype region (23), it is known that only three genes are encoded in this overlapping region: SLL1, SLG, and SRK. SLL1 is specifically expressed in anthers (29) and thus should not function in stigmas. To determine whether the phenotype is indeed conferred by expression of the SLG-910 and SRK-910 genes, RNA gel blot analysis was performed with the stigma RNA to examine expression of the two genes. As shown in Fig. 4, all the transgenic progeny plants that gained the SI phenotype also expressed both genes at significant levels. The only exception was 389A-5, which inherited the transgenes but showed barely detectable expression and did not gain the SI phenotype. As expected, progeny plants lacking the transgenes through segregation had no expression as did the negative control Westar (faint signals could be observed after a longer exposure because of the presence of the nonfunctional A10 S haplotype in Westar that cross-hybridizes with the SLG-910 and SRK-910 probes; data not shown). Thus, the expression studies provide a direct correlation between gain of the stigma SI phenotype and expression of the SLG-910 and SRK-910 genes in the same tissue. S gene expression levels vary among the transgenic lines and are all somewhat lower than the expression level of W1 but were sufficient to confer the phenotype.
Figure 4.
Expression of the SLG and SRK transgenes in the stigmas of the T1 progeny plants. Sources of stigma tissues are indicated above each lane (the last lane is 389A-5). “+” indicates presence of the transgene; “−” indicates loss of the transgene caused by segregation. The blot was probed with SLG-910 and SRK-910 cDNAs. “Phenotype” indicates the gain-of-function SI phenotype in the stigmas. Molecular size markers are indicated at left in kilobases. (Lower) Rehybridization of the same blot to 18S rDNA.
Discussion
The results reported herein, when combined with information on sequencing and transcriptional mapping of the cloned 910 S haplotype region (23), show that SLG and SRK are sufficient to confer the stigma SI phenotype in Brassica. The common region shared by the Fos20 and Fos22 genomic clones does not contain any additional genes or significant ORFs except for SLG, SRK, and the anther-specific SLL1. Nevertheless, our results do not mean that both genes are required to determined haplotype specificity on the pistil side. Further, these results show that the SLG, SRK, SLL1, SLL3, as well as other genes encoded in this region do not function as the pollen determinant of SI or at the very least are not sufficient for determining the pollen phenotype. This finding is consistent with results from previous biochemical (8), genetic (11–14), and transgenic (15–17) studies and supports the current model for the Brassica SI response that proposes SRK as a key signal transmitter and an independent pollen component.
A gain-of-function phenotype by gene transformation provides the most direct proof that the transgene or transgenes are not only necessary but are also sufficient for function, and therefore this phenotype is an important means to study gene function. Unfortunately, previous attempts at introducing S genes into Brassica have been unsuccessful, perhaps because of homology-dependent cosuppression of the S gene family members (15, 16). The experiments reported herein may have been successful in comparison to those previously published, as well as our own previous unsuccessful efforts, for several reasons. First, native promoters may be important to drive the S gene expression to appropriate levels. In the work by Conner et al. (15), the SRK gene was driven by an SLG promoter, which may drive SRK expression to an abnormally high level and thus trigger the cosuppression response. Second, it may be necessary for the S genes to be linked physically to ensure the coordinated spatial and temporal expression as well as their relative level. This physical linkage is a major advantage of large insert transformation via the BIBAC system, because it allows expression of plant genes or gene clusters in their native genomic context and might eliminate site-dependent gene expression. This lack of consistent transgene expression might be an explanation for the previous transformation studies conducted in our laboratory (K.-R. J. Stahl and S.J.R., unpublished data). The two S genes with their native promoters were transformed individually into Westar and then crossed into one line. A significant number of transgenic lines were analyzed, but no phenotypic changes were observed. Our success in obtaining the gain-of-function SI phenotype by introducing the cloned SLG-SRK genomic region into Brassica opens the way for additional functional studies of the Brassica S locus. For example, with the transgenic approach, we can now address the issue whether SRK alone is sufficient and/or whether SLG is indeed required for haplotype-specific rejection of self-pollen.
Our results also suggest that there must be a pollen S gene outside the ≈55-kb genomic region covered by the Fos20 and Fos22 constructs. This gene has been confirmed by recent findings. After years of intensive search conducted by several laboratories employing various approaches (23, 29, 35–37), the male determinant of SI specificity in Brassica has just been identified (38). The recently identified pollen S gene, SCR (for S locus cysteine-rich protein), is predicted to encode a small secreted cysteine-rich protein. Putative alleles of SCR have been identified in our sequenced A14 and 910 S haplotypes (ref. 23 and N.B., Y.C., Y.-M.B., and S.J.R., unpublished data). The putative SCR-910 is located just outside the Fos20 clone, ≈2 kb downstream of the Bkin gene, and is located in Fos16, a clone that was not used in this transformation study (for a map of the 910 S haplotype, see ref. 23). Our data, together with those of Schopfer et al. (38), strongly suggest a direct interaction between SCR and the SLG/SRK receptors during the initial contact/recognition reaction between self-pollen and stigmatic papillar cells. The interaction is thought to activate the SRK kinase and, subsequently, a signal transduction pathway that leads to self-pollen rejection.
In addition, it should be noted with regards to the evolution of SI that genes for pollen and stigma self-recognition are different and that they are separated by at least three intervening genes.
Acknowledgments
We thank Shoba Sivasankar and Nic Bate for critical reading of the manuscript and Carol M. Hamilton for the BIBAC2 vector and the Agrobacterium strain COR338. Y.C. was partly supported by an Ontario Graduate Scholarship, Canada. This work was founded by the Natural Sciences and Engineering Research Council of Canada.
Abbreviations
- SI
self-incompatibility
- SLG
S locus glycoprotein
- SRK
S receptor kinase
- kb
kilobase
- BIBAC
binary bacterial artificial chromosome
- SLL1
S locus linked 1
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050480297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050480297
References
- 1.Bateman A J. Heredity. 1955;9:52–68. [Google Scholar]
- 2.Nasrallah J B, Nasrallah M E. Plant Cell. 1993;5:1325–1335. doi: 10.1105/tpc.5.10.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ockendon D J. Heredity. 1974;33:159–171. [Google Scholar]
- 4.Ockendon D J. Euphytica. 1982;31:325–331. [Google Scholar]
- 5.Nasrallah J B, Kao T H, Goldberg M L, Nasrallah M E. Nature (London) 1985;318:617–618. [Google Scholar]
- 6.Stein J C, Howlett B, Boyes D C, Nasrallah M E, Nasrallah J B. Proc Natl Acad Sci USA. 1991;88:8816–8820. doi: 10.1073/pnas.88.19.8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goring D R, Rothstein S J. Plant Cell. 1992;4:1273–1281. doi: 10.1105/tpc.4.10.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stein J C, Dixit R, Nasrallah M E, Nasrallah J B. Plant Cell. 1996;8:429–445. doi: 10.1105/tpc.8.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kandasamy M K, Paolillo D J, Faraday C D, Nasrallah J B, Nasrallah M E. Dev Biol. 1989;134:462–472. doi: 10.1016/0012-1606(89)90119-x. [DOI] [PubMed] [Google Scholar]
- 10.Delorme V, Giranton J-L, Hatzfeld Y, Friry A, Heizmann P, Ariza M J, Dumas C, Gaude T, Cock J M. Plant J. 1995;7:429–440. doi: 10.1046/j.1365-313x.1995.7030429.x. [DOI] [PubMed] [Google Scholar]
- 11.Toriyama K, Stein J C, Nasrallah M E, Nasrallah J B. Theor Appl Genet. 1991;81:769–776. doi: 10.1007/BF00224988. [DOI] [PubMed] [Google Scholar]
- 12.Nasrallah M E, Kandasamy M J, Nasrallah J B. Plant J. 1992;2:497–506. [Google Scholar]
- 13.Goring D R, Glavin T L, Schafer U, Rothstein S J. Plant Cell. 1993;5:531–539. doi: 10.1105/tpc.5.5.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nasrallah J B, Rundle S J, Nasrallah M E. Plant J. 1994;5:373–384. [Google Scholar]
- 15.Conner J A, Tantikanjana T, Stein J C, Kandasamy M K, Nasrallah J B, Nasrallah M E. Plant J. 1997;11:809–823. [Google Scholar]
- 16.Stahl R J, Arnoldo M, Glavin T L, Goring D R, Rothstein S J. Plant Cell. 1998;10:209–218. doi: 10.1105/tpc.10.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takasaki T, Hatakeyama K, Watanabe M, Toriyama K, Isogai A, Hinata K. Plant Mol Biol. 1999;40:659–668. doi: 10.1023/a:1006274525421. [DOI] [PubMed] [Google Scholar]
- 18.Cabrillac D, Delorme V, Garin J, Ruffio-Chable V, Giranton J, Dumas C, Gaude T, Cock J M. Plant Cell. 1999;11:971–986. doi: 10.1105/tpc.11.5.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ullrich A, Schlessinger J. Cell. 1990;61:203–212. doi: 10.1016/0092-8674(90)90801-k. [DOI] [PubMed] [Google Scholar]
- 20.Downey R K, Rakow G F W. In: Principles of Cultivar Development. Fehr W R, editor. New York: MacMillan; 1987. pp. 437–486. [Google Scholar]
- 21.Goring D R, Banks P, Beversdorf W D, Rothstein S J. Mol Gen Genet. 1992;234:185–192. doi: 10.1007/BF00283838. [DOI] [PubMed] [Google Scholar]
- 22.Goring D R, Banks P, Fallis L, Baszczynski C L, Beversdorf W D, Rothstein S J. Plant J. 1992;2:983–989. [PubMed] [Google Scholar]
- 23.Cui Y, Brugière N, Jackman L, Bi Y-M, Rothstein S J. Plant Cell. 1999;11:2217–2231. doi: 10.1105/tpc.11.11.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamilton C M, Frary A, Lewis C, Tanksley S D. Proc Natl Acad Sci USA. 1996;93:9975–9979. doi: 10.1073/pnas.93.18.9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamilton C M. Gene. 1997;200:107–116. doi: 10.1016/s0378-1119(97)00388-0. [DOI] [PubMed] [Google Scholar]
- 26.Draper J, Scott R, Armitage P, Walden R. Plant Genetic Transformation and Gene Expression: A Laboratory Manual. Oxford: Blackwell; 1988. [Google Scholar]
- 27.Moloney M M, Walker J, Sharma K. Plant Cell Rep. 1989;8:238–242. doi: 10.1007/BF00778542. [DOI] [PubMed] [Google Scholar]
- 28.Zhang H-B, Zhao X, Ding X, Paterson A H, Wing R A. Plant J. 1995;7:175–184. [Google Scholar]
- 29.Yu K, Schafer U, Glavin T L, Goring D R, Rothstein S J. Plant Cell. 1996;8:2369–2380. doi: 10.1105/tpc.8.12.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verwoerd T C, Dekker B N M, Hoekema A. Nucleic Acids Res. 1983;17:2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Da Rocha P S C F, Bertrand H. Eur J Biochem. 1995;229:550–557. doi: 10.1111/j.1432-1033.1995.tb20497.x. [DOI] [PubMed] [Google Scholar]
- 32.Feinberg A P, Vogelstein B. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 33.Boyes D C, Nasrallah M E, Vrebalov J, Nasrallah J B. Plant Cell. 1997;9:237–247. doi: 10.1105/tpc.9.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conner J A, Conner P, Nasrallah M E, Nasrallah J B. Plant Cell. 1998;10:801–812. doi: 10.1105/tpc.10.5.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki G, Kai N, Hirose T, Fukui K, Nishio T, Takayama S, Isogai A, Watanabe M, Hinata K. Genetics. 1999;153:391–400. doi: 10.1093/genetics/153.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyes D C, Nasrallah J B. Plant Cell. 1995;7:1283–1294. doi: 10.1105/tpc.7.8.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephenson A G, Doughty J, Dixon S, Elleman C J, Hiscock S J, Dickinson H G. Plant J. 1997;12:1351–1359. [Google Scholar]
- 38.Schopfer C R, Nasrallah M E, Nasrallah J B. Science. 1999;286:1697–1700. doi: 10.1126/science.286.5445.1697. [DOI] [PubMed] [Google Scholar]