Abstract
The nuclear receptor superfamily comprises ligand-regulated transcription factors that control various developmental and physiological pathways. These receptors share a common modular structure and regulate gene expression through the recruitment of a large set of coregulatory proteins. These transcription cofactors regulate, either positively or negatively, chromatin structure and transcription initiation. One of the first proteins to be identified as a hormone-recruited cofactor was RIP140. Despite its recruitment by agonist-liganded receptors, RIP140 exhibits a strong transcriptional repressive activity which involves several inhibitory domains and different effectors. Interestingly, the RIP140 gene, located on chromosome 21 in humans, is finely regulated at the transcriptional level by various nuclear receptors. In addition, the protein undergoes several post-translational modifications which control its repressive activity. Finally, experiments performed in mice devoid of the RIP140 gene indicate that this transcriptional cofactor is essential for female fertility and energy homeostasis. RIP140 therefore appears to be an important modulator of nuclear receptor activity which could play major roles in physiological processes and hormone-dependent diseases.
History
In the early 90’s, one of the main goals for several laboratories working on nuclear receptor (NR) signaling was to identify associated proteins which could act as transcriptional coregulators. The efforts were initially focused on partners of the ligand binding domain (LBD) encompassing the ligand-dependent activating function (AF-2) because it was the most convenient (due to the existence of inactivating mutations and to the use of antagonist ligands).
RIP140 (Receptor Interacting Protein of 140 kDa) was one of the first NR transcriptional cofactors to be isolated. It was first identified by far-western blotting in human cancer cell extracts using a chimeric radiolabeled probe containing the ligand binding domain (LBD) of the mouse ERα fused to the glutathione-S-transferase [Cavailles et al., 1995]. In the presence of estradiol, this probe detected several bands corresponding to RIP140 and to the p160 family of coactivators. Using the same strategy, the RIP140 cDNA was then isolated from a cDNA expression library established from ZR75-1 breast cancer cells [Cavailles et al., 1995].
The mouse RIP140 cDNA was isolated 3 years later from a mouse embryonic library using a yeast two hybrid strategy using the LBD of the orphan TR2 receptor as a bait [Lee et al., 1998]. Currently, the cDNA sequences from several species including rat, dog, chicken, xenopus and zebra fish have been deposited in databases. The RIP140 gene is also known as NRIP1 (Nuclear Receptor-Interacting Protein 1) which is the official symbol provided by the HUGO gene nomenclature committee.
Protein domain structure
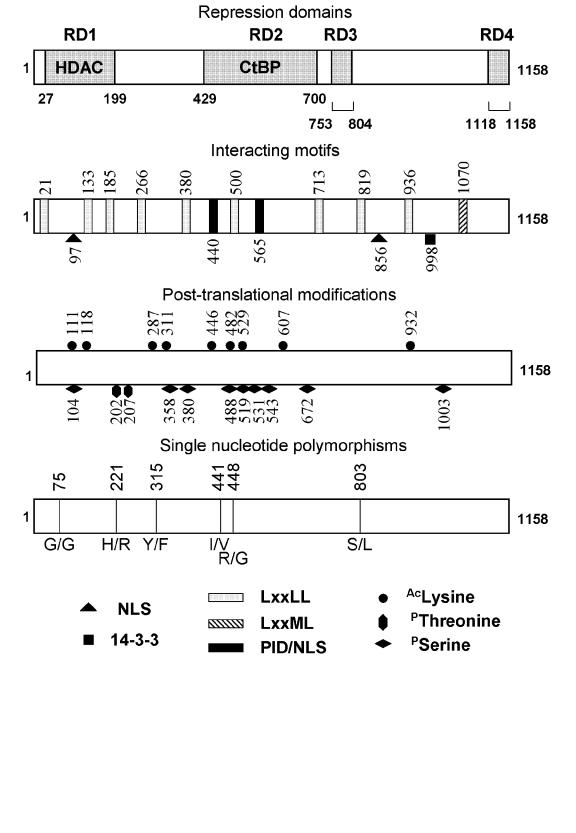
The human RIP140 protein comprises 1158 amino acids with an overall important identity between species (83% of amino acid identity between human and mouse sequences). When the RIP140 cDNA was isolated, no known conserved motifs were found in the deduced primary sequence by comparison to databases. However, several important functional domains allowing interaction with different partners (i.e., nuclear receptors and downstream effectors) have now been identified (see the sections on Interactions with nuclear receptors, Role in Transcription Control, and Post-translational modifications and Figure 1). In addition, and as expected for a transcriptional regulator, the RIP140 protein contains two putative nuclear localization signals (NLS) located at position 97 (KRKR monopartite NLS) and 856 (KKRKx10KKMK bipartite motif).
Figure 1. Schematic representation of RIP140 structure.
The boxes represent the RIP140 molecule showing respectively the different functional domains together with post-translational modifications and single nucleotide polymorphisms. The numbers correspond to the amino acid position on the molecule. A) Repression domains (RD). The four domains (RD1 to 4) map respectively to the regions located between residues 27 and 199 429 and 700, 753 to 804 and 1118 to 1158 (Castet et al. 2004; Christian et al. 2004; Vo et al. 2001; Wei et al. 2000; Wei et al. 2001). B) Interacting motifs. The ten LxxLL NR binding sites (Heery et al. 1997) and the atypical LxxML motif (Chen et al. 2002) are shown together with the two putative nuclear localization signals at position 97 and 856, the 14-3-3 binding motif (RTFSYP) (Zilliacus et al. 2001) and the CtBP binding sites PIDLS and PINLS at position 440 and 565 (Castet et al. 2004; Christian et al. 2004; Vo et al. 2001). C) Post-translational modifications. The figure shows phosphorylation (S104, S358, S380, S488, S519, S531, S543, S672, S1003, T202 and T207) (Huq et al. 2005a; Gupta et al. 2005) and acetylation sites (K111, K118, K287, K211, K446, K482, K529, K607, K932) (Huq and Wei 2005b; Vo et al. 2001). D) Single nucleotide polymorphism (SNPs). The six SNPs causing amino acid sequence changes in the human RIP140 protein correspond to G75G, H221R, Y315F, I441V, R448G, S803L (Caballero et al. 2005).
Interactions with nuclear receptors
Identification of interacting motifs
Since its isolation as an ERα partner, RIP140 was shown to interact with many other nuclear receptors (NRs) such as RARα/β, RXRα/β, TRα/β [L'Horset et al., 1996; Treuter et al., 1998], GR [Subramaniam et al., 1999; Windahl et al., 1999], AR [Bevan et al., 1999; Carascossa et al., 2006; Ikonen et al., 1997], VDR [Masuyama et al., 1997], PPARα/γ/δ [Lim et al., 2004; Treuter et al., 1998; Windahl et al., 1999], LXRα [Miyata et al., 1998], PXR [Masuyama et al., 2001], LXRβ, HNF4α and RORβ [Albers et al., 2005], ERRα/β/γ [Castet et al., 2006; Sanyal et al., 2004], SF1 and DAX-1 [Sugawara et al., 2001], TAK1/TR4 [Yan et al., 1998] and TR2 [Lee et al., 1998].
As shown in Figure 1, the RIP140 protein contains nine NR interacting boxes (LxxLL motifs) spread throughout the molecule [Heery et al., 1997]. Interestingly, the binding of RIP140 to RAR and RXR requires a slightly divergent sequence corresponding to an LxxML motif and located between amino acids 1070 and 1074 [Chen et al., 2002]. The binding of RIP140 to NRs primarily requires an active AF-2 domain since it is disrupted by mutations in the helix 12 [Cavailles et al., 1994]. The RIP140 binding site overlaps that of coactivators such as members of the SRC family (p160 proteins) [Eng et al., 1998], thus explaining why the two types of molecules compete for interaction with NRs [Eng et al., 1998; Treuter et al., 1998]. The various LxxLL motifs present in the RIP140 sequence display distinct preferences for nuclear receptors. In vitro interaction assays showed that liganded TRβ has a clear preference for RIP140 LxxLL motifs 3, 5 and 8 [Moore et al., 2004]. Moreover, when evaluated in yeast [Heery et al., 2001] or mammalian [Hu and Funder, 2006] two-hybrid assays, the nine LxxLL motifs exhibited different relative affinities for steroid or retinoid receptors, the central LxxLL motif 6 (residues 500 to 504) being the most efficient in interacting with GR, ERα and RARα.
RIP140 also interacts with other transcription factors like c-jun [Teyssier et al., 2003] or the aryl hydrocarbon receptor (AhR) [Kumar et al., 1999]. The interaction with AhR occurs independently of NR boxes through a domain that was mapped between amino acid residues 154 and 350 [Kumar et al., 1999].
Recruitment of RIP140 on target genes
During the past five years, chromatin immunoprecipitation (ChIP) assays have been used to confirm the interaction of transcription factors on their target genes and to describe the cyclical recruitment of the transcription machinery [Metivier et al., 2006]. However, in the case of RIP140, very few kinetics studies using ChIP assays were reported. In RA-treated P19 cells, RIP140 was detected in a cyclic fashion on RAR-targeted genes such as the RARβ2 and TR2 promoters [Chen et al., 2004; Hu et al., 2004]. Interestingly, these studies presented kinetic evidence for competition of RIP140 with P/CAF for ligand-dependent interactions with RAR/RXR on targeted promoters. Later on, other studies demonstrated the ligand-dependent recruitment of RIP140 on target genes. This was described on the RARα promoter in the presence of E2 [Laganiere et al., 2005] and on the PSA (prostate specific antigen) enhancer and promoter upon treatment of LNCaP cells with R1881 [Carascossa et al., 2006].
Role in transcription control
Repression of transcription
RIP140 appears to be an unconventional NR coregulator. Indeed, despite its recruitment by agonist-liganded receptors, which initially suggested that it might act as a transcriptional coactivator, most of the published data indicates that RIP140 inhibits target gene transcription. It was initially proposed that this transcriptional repression occurred by competition with coactivators [Treuter et al., 1998]. Indeed, evidence was provided that the in vitro binding of RIP140 and SRC-1 to nuclear receptors was competitive and might account for the negative effect of RIP140 on hormone-dependent transcription.
More recently, it was demonstrated that RIP140 displayed active repression. Experiments using the full-length RIP140 sequence tethered to the GAL4-DBD revealed a transrepressive activity on the reporter gene [Lee et al., 1998]. When global analysis of the transcriptome was compared in wild-type and RIP140-null cells, a significant proportion of genes were found to be increased in cells which did not express RIP140 [Christian et al., 2005], thus confirming a direct or indirect RIP140-associated repression of transcription.
Four repressive domains (RD) have been identified in the RIP140 molecule ([Castet et al., 2004; Christian et al., 2004] and Figure 1). The RD1 encompasses a region located between residues 27 and 199 which acts by recruiting class I and II histone deacetylases (HDACs) [Castet et al., 2004; Wei et al., 2001; Wei et al., 2000]. The RD2 maps to the region located between amino acids 429 and 739 and interacts with carboxyl-terminal-binding proteins (CtBP1 and CtBP2) through two conserved motifs (sequences PIDLS and PINLS) [Castet et al., 2004; Vo et al., 2001]. The RD3 and RD4 are located in the carboxyl-terminal region of the molecule and correspond respectively to amino acid residues 753-804 and 1118-1158, but no downstream effectors have yet been identified.
Very interestingly, several post-translational modifications seem to play an important role in controlling the repressive activity of RIP140 (see the section on Post-translational modifications below and Figure 1).
Activation of transcription
Although RIP140 mainly exerts a negative effect on transcription, several reports have described positive effects on gene expression. The most convincing situation concerns the effect in yeast where transactivation was reproducibly observed on different nuclear receptors such as ERα [Nephew et al., 1998; Sheeler et al., 2000], RARα [Joyeux et al., 1997] or GR [Windahl et al., 1999].
In mammalian cells, activation of transcription has also been linked to RIP140 overexpression. In transient transfection experiments, low concentrations of RIP140 expression plasmid led to a slight but significant increase in ERα [Cavailles et al., 1995; Henttu et al., 1997] or AhR activity [Kumar et al., 1999]. More recently, RIP140 was reported to strongly stimulate transactivation by ERRα and ERRγ when they regulated transcription of target genes through Sp1 sites [Castet et al., 2006]. This regulation could be due to an indirect effect of RIP140 overexpression which might titrate away from the promoter (a) transcriptional repressor(s) of Sp1 activity. Based on their ability to interact with both Sp1 and RIP140, HDACs appear to be good candidates to mediate such an indirect regulation.
In addition, RIP140 overexpression is also associated with an increase in transcription when activated nuclear receptors repress gene expression. For instance, RIP140 relieved the negative effect of glucocorticoids on nGRE or NFk-B sites [Subramaniam et al., 1999] or the repression by estrogens on TNFα promoter [An et al., 1999].
Post-translational modifications of the RIP140 protein
Phosphorylation
Phosphorylation of RIP140 was first suggested by experiments showing that its interaction with 14-3-3 was decreased upon treatment with alkaline phosphatase [Zilliacus et al., 2001]. 14-3-3 proteins form a family of highly conserved ubiquitous factors which act as regulators of a wide range of biological processes [Darling et al., 2005]. They interact directly with a large number of target proteins and alter their activity, localization or protein-protein interactions (see below). Recently, the mapping of phosphorylation sites was achieved by liquid chromatography-tandem mass spectroscopy [Huq et al., 2005]. Phosphorylation, which occurs on nine serines and two threonine residues (Figure 1), participates in the regulation of RIP140 biological activity. Experiments using either mutagenesis of the phosphorylated residues or kinase activators or inhibitors suggest that MAPK-phosphorylation of Thr202 and 207 increases the transrepressive activity of RIP140 in part by regulating its interaction with HDAC3 [Gupta et al., 2005].
Acetylation
RIP140 is also modified by acetylation as shown by initial studies demonstrating the acetylation of human RIP140 by CBP [Vo et al., 2001]. In this study, the major (but not the sole acetylation site) was found to be lysine 446. Acetylation of RIP140 inhibits its interaction with CtBP1 and is associated with a lower transrepressive activity. More recently, a proteomic analysis of the acetylation pattern of RIP140 revealed eight acetylated lysines in the amino-terminal and central region of the protein expressed in insect cells (Figure 1) [Huq and Wei, 2005]. Paradoxically, in this study, the use of an HDAC inhibitor increased the repressive activity of RIP140, as previously reported by others [Castet et al., 2004]. This apparent discrepancy concerning the contribution of HDAC enzymatic activity to the transcriptional repression exerted by RIP140 might reflect cell specific effects with, for instance, a different requirement for HDACs and/or CtBPs to inhibit transcription according to the cell type.
Subcellular localization
By indirect fluorescence using a rabbit antiserum raised against a synthetic peptide, the RIP140 protein was localized in nuclear dots and appeared excluded from nucleoli [Cavailles et al., 1995]. This was supported by data obtained using a fluorescent tagged version of the mouse [Lee et al., 1998] or human [Carascossa et al., 2006; Zilliacus et al., 2001] protein showing an intranuclear distribution in foci which appeared to be different from PML bodies [Tazawa et al., 2003]. By mutagenesis, the domain of human RIP140 responsible for its targeting to small nuclear dots was mapped to amino acids 431 to 472 [Tazawa et al., 2003].
The subcellular localization seems to be regulated by different mechanisms. In hyperacetylation conditions (treatment with an HDAC inhibitor) the mouse RIP140 protein, expressed as a GFP fusion protein, appears slightly delocalized into the cytoplasm [Huq and Wei, 2005]. A similar partial relocalization to the cytoplasmic compartment was induced by the 14-3-3 protein which directly interacts with RIP140 through a consensus 14-3-3 binding site (RTFSYP) [Zilliacus et al., 2001]. In addition, upon overexpression of 14-3-3 protein, the nuclear signal was also relocalized from punctuate to a diffuse pattern. Interestingly, the same effect was reported upon activation of glucocorticoid [Tazawa et al., 2003] or androgen [Carascossa et al., 2006] receptors. The exact role of this intranuclear redistribution of RIP140 remains to be defined. One hypothesis could be that the foci represent a compartment dedicated to the storage of a repressive machinery including RIP140.
Structure and expression of the gene
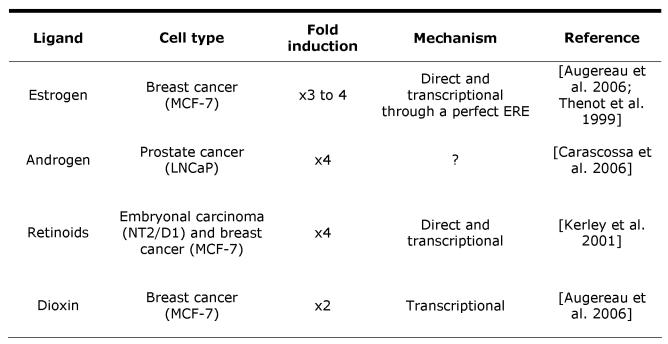
The human RIP140 gene was initially mapped to chromosome 21 in the region q11.2 by fluorescence in situ hybridization [Cavailles et al., 1995]. This was confirmed using artificial chromosomes, showing that the gene maps to a gene-poor region of the genome [Katsanis et al., 1998]. Interestingly, all the RIP140 coding sequence is comprised in a single large exon and it was believed that the gene was intron-less. However, several short non-coding exons (which undergo alternative splicing) have been recently identified in the 5’ region of the gene and the promoter mapped about 100kb upstream of the ATG [Augereau et al., 2006]. RIP140 is a ubiquitously expressed gene. The mRNA was detected in a very large number of human cell lines [Cavailles et al., 1995]. In the mouse, the mRNA was detected in all the tested tissues with a strong expression in the testis and in the brain [Lee et al., 1998]. The accumulation of the RIP140 mRNA is increased by different stimuli (Table 1). In human breast cancer cells, RIP140 mRNA levels are up-regulated by estrogens, retinoids and dioxin [Augereau et al., 2006; Cavailles et al., 1994; Kerley et al., 2001]. In prostate cancer cells, the mRNA steady-state levels are increased by androgens [Carascossa et al., 2006]. More recently, RIP140 expression was shown to be stimulated by ERRα during mouse adipogenesis [Nichol et al., 2006].
Table 1. Regulation of RIP140 mRNA accumulation.
The table shows the nature of the ligand involved in the regulation, the type of cell in which it was observed together with the magnitude of the regulation (fold induction). When indicated, the underlying mechanism (direct or indirect effect at the transcriptional level) is also mentioned.
The regulation by estrogens is direct (i.e., it does not require synthesis of an intermediary protein as judged by the absence of effect of cycloheximide) [Thenot et al., 1999] and operates at the transcriptional level [Augereau et al., 2006]. Transcriptional profilings of ER-regulated genes using stably transfected U2OS cells expressing either ERα or ERβ have also identified RIP140 as an E2-regulated gene although the relative induction by the two isoforms of ER varied according to the study [Monroe et al., 2003; Stossi et al., 2004].
A consensus ERE (which binds the ERα in gel shift and ChIP experiments) has been mapped in the 5’ proximal region of the gene [Augereau et al., 2006; Bourdeau et al., 2004; Lin et al., 2004]. However, it has been proposed that FoxA1 sites might function as a distal enhancer facilitating the recruitment of ERα on the RIP140 promoter [Carroll et al., 2005]. Interestingly, this ERE overlaps with the response element for AhR and transcriptional interference occurs between the two activities [Augereau et al., 2006]. Altogether, these findings highlight the complex regulation of RIP140 expression at the transcriptional level, which involves RIP140 in several feed-back regulatory loops.
Biological roles of RIP140
The physiological importance of RIP140 was evaluated using mice devoid of the RIP140 gene (RIPKO). These RIP140-null mice were generated by replacing almost the entire coding region by a β-galactosidase expressing cassette [White et al., 2000]. RIPKO mice are viable but exhibited several interesting phenotypes. First, the female RIPKO mice are infertile because of defective ovulation (mature follicles fail to release oocytes although granulosa cells undergo luteinization and oocytes could be fertilized in vitro). Embryo transfer experiments indicated that RIP140 was not essential for the preparation of the uterus for implantation. Moreover, ovarian transplantation experiments showed that ovaries are the essential site of RIP140 action in female fertility [Leonardsson et al., 2002].
The second interesting phenotype is at the level of the white adipose tissue. RIPKO mice have a reduced body weight and body fat content. They exhibit a resistance to high-fat diet-induced obesity and increased oxygen consumption [Leonardsson et al., 2004]. Data indicate that RIP140 controls the balance between energy storage and expenditure through the regulation of specific genes involved in energy metabolism [Christian et al., 2005]. Adipocytes isolated from RIPKO mice showed elevated expression of genes such as the uncoupling protein 1 (ucp1) or the carnitine palmitoyltransferase 1b (CPT1b) which are normally directly repressed by RIP140 at the transcriptional level [Christian et al., 2005]. Along with these data, the RIPKO adipocytes exhibited higher levels of total fatty acid oxidation than those of wild-type cells.
More recently, it has been shown that RIPKO animals also exhibit a higher glucose tolerance and insulin responsiveness upon high-fat feeding [Powelka et al., 2006]. Indeed, RIP140 appears to negatively regulate cellular respiration, citric acid cycling, glycolysis and hexose uptake through silencing of genes involved in these metabolic pathways. RIP140 appears therefore to act as a broad negative regulator of multiple metabolic pathways in adipocytes and might therefore be considered as a putative therapeutic target for metabolic syndromes.
At the cellular level, the role of RIP140 has also been evaluated by a knock-down approach. Using siRNA to silence RIP140 expression, it has been suggested that the effects of estrogen on cell proliferation were repressed by RIP140 [White et al., 2005]. Moreover, RIP140 also appeared important for the antiestrogenic effect of retinoic acid on breast cancer cell proliferation.
Alteration of RIP140 expression in human pathology
Single nucleotide polymorphism (SNP)
Several DNA variants have been identified within the coding sequence of RIP140, most of them introducing amino acid changes (Figure 1) [Caballero et al., 2005]. Specific combinations of amino acid variations were found with a higher incidence in patients presenting endometriosis, as compared with the control population. Moreover, the R448G SNP appeared associated with endometriosis in a case-control study comprising 200 samples [Caballero et al., 2005]. The same laboratory reported multilocus analyses of SNP within five genes including RIP140 in two pathologies dealing with estrogen signaling (i.e., osteoporosis [Moron et al., 2006] and male infertility [Galan et al., 2005]). Interestingly, a digenic association involving SNPs in the RIP140 and ERβ genes appeared related to the osteoporosis status. These data need to be confirmed by other studies, but suggest a potential role of RIP140 in bone pathogenesis.
Deregulated expression
Surprisingly, very few studies have analyzed the expression of RIP140 expression in human diseases. Two studies have quantified the accumulation of the mRNA in breast cancers, but on a very limited number of samples [Chan et al., 1999; Rey et al., 2000]. No significant differences in the level of RIP140 mRNA were observed in tamoxifen-resistant breast tumors samples, as compared to tamoxifen-treated or untreated tumors [Chan et al., 1999].
As expected due to its location on chromosome 21, the expression of the RIP140 protein appeared increased in the hippocampus of a patient with Down’s syndrome [Gardiner, 2006]. The gene was also found to be significantly up-regulated in acute myeloid leukemia with complex karyotypes and abnormal chromosome 21 [Baldus et al., 2004].
Conclusions
RIP140 is an unconventional transcriptional cofactor which acts mainly as a negative regulator of hormone-dependent nuclear receptor activity, thus counterbalancing the effect of coactivators. Several inhibitory modules are involved in the transrepressive function of RIP140 which appears to be finely tuned by post-translational modifications. The expression of the gene is under complex regulation and implicates several regulatory loops and cross-talk interactions.
However, although a significant amount of work has been performed on RIP140 to decipher its mechanism of action and to define its biological relevance, several questions still remain unanswered. Does RIP140 act as a repressor for transcription factors other than nuclear receptors? What is the physiological importance of the transcriptional and post-transcriptional regulation of RIP140 expression? What is the exact interplay between the different post-translational modifications and their roles in controlling the biological activity of RIP140? What is the relevance of this coregulator in hormone-dependent carcinogenesis and other pathologies?
Acknowledgments
We are grateful to S Bonnet, M. Fuentès and A. Lucas for technical help. Our work on RIP140 is supported by the "Institut National de la Santé et de la Recherche Médicale", the University of Montpellier I, the "Association pour la Recherche sur le Cancer", the "Association pour la Recherche sur les Tumeurs de la Prostate", the "Ligue Régionale contre le Cancer", the "Fondation Jérôme Lejeune" and the "Agence Nationale pour la Recherche".
Abbreviations
- AF
activating function
- AhR
aryl hydrocarbon receptor
- ChIP
chromatin immunoprecipitation
- CtBP
carboxyl-terminal-binding protein
- E2
17-β-estradiol
- ER
estrogen receptor
- ERR
estrogen receptor-related receptor
- GFP
green fluorescent protein.
- HDAC
histone deacetylase
- LBD
ligand binding domain
- NLS
nuclear localization signals
- NR
nuclear receptor
- NRIP1
nuclear receptor interacting protein 1
- RD
repressive domain
- RIP140
receptor interacting protein of 140 kDa
- SNP
single nucleotide polymorphism
References
- Albers M., Kranz H., Kober I., Kaiser C., Klink M., Suckow J., Kern R., Koegl M. Automated yeast two-hybrid screening for nuclear receptor-interacting proteins. Mol Cell Proteomics. 2005;4:205–13. doi: 10.1074/mcp.M400169-MCP200. [DOI] [PubMed] [Google Scholar]
- An J., Ribeiro R. C., Webb P., Gustafsson J. A., Kushner P. J., Baxter J. D., Leitman D. C. Estradiol repression of tumor necrosis factor-α transcription requires estrogen receptor activation function-2 and is enhanced by coactivators. Proc Natl Acad Sci U S A. 1999;96:15161–6. doi: 10.1073/pnas.96.26.15161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augereau P., Badia E., Fuentes M., Rabenoelina F., Corniou M., Derocq D., Balaguer P., Cavailles V. Transcriptional regulation of the human NRIP1/RIP140 gene by estrogen is modulated by dioxin signalling. Mol Pharmacol. 2006;69:1338–46. doi: 10.1124/mol.105.017376. [DOI] [PubMed] [Google Scholar]
- Baldus C. D., Liyanarachchi S., Mrozek K., Auer H., Tanner S. M., Guimond M., Ruppert A. S., Mohamed N., Davuluri R. V., Caligiuri M. A., Bloomfield C. D., de la Chapelle A. Acute myeloid leukemia with complex karyotypes and abnormal chromosome 21: Amplification discloses overexpression of APP, ETS2, and ERG genes. Proc Natl Acad Sci U S A. 2004;101:3915–20. doi: 10.1073/pnas.0400272101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan C. L., Hoare S., Claessens F., Heery D. M., Parker M. G. The AF1 and AF2 domains of the androgen receptor interact with distinct regions of SRC1. Mol Cell Biol. 1999;19:8383–92. doi: 10.1128/mcb.19.12.8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdeau V., Deschenes J., Metivier R., Nagai Y., Nguyen D., Bretschneider N., Gannon F., White J. H., Mader S. Genome-wide identification of high-affinity estrogen response elements in human and mouse. Mol Endocrinol. 2004;18:1411–27. doi: 10.1210/me.2003-0441. [DOI] [PubMed] [Google Scholar]
- Caballero V., Ruiz R., Sainz J. A., Cruz M., Lopez-Nevot M. A., Galan J. J., Real L. M., de Castro F., Lopez-Villaverde V., Ruiz A. Preliminary molecular genetic analysis of the Receptor Interacting Protein 140 (RIP140) in women affected by endometriosis. J Exp Clin Assist Reprod. 2005;2 doi: 10.1186/1743-1050-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carascossa S., Gobinet J., Georget V., Lucas A., Badia E., Castet A., White R., Nicolas J. C., Cavailles V., Jalaguier S. Receptor-interacting protein 140 is a repressor of the androgen receptor activity. Mol Endocrinol. 2006;20:1506–18. doi: 10.1210/me.2005-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll J. S., Liu X. S., Brodsky A. S., Li W., Meyer C. A., Szary A. J., Eeckhoute J., Shao W., Hestermann E. V., Geistlinger T. R., Fox E. A., Silver P. A., Brown M. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Castet A., Boulahtouf A., Versini G., Bonnet S., Augereau P., Vignon F., Khochbin S., Jalaguier S., Cavailles V. Multiple domains of the Receptor-Interacting Protein 140 contribute to transcription inhibition. Nucleic Acids Res. 2004;32:1957–66. doi: 10.1093/nar/gkh524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castet A., Herledan A., Bonnet S., Jalaguier S., Vanacker J. M., Cavailles V. Receptor-interacting protein 140 differentially regulates estrogen receptor-related receptor transactivation depending on target genes. Mol Endocrinol. 2006;20:1035–47. doi: 10.1210/me.2005-0227. [DOI] [PubMed] [Google Scholar]
- Cavailles V., Dauvois S., Danielian P. S., Parker M. G. Interaction of proteins with transcriptionally active estrogen receptors. Proc Natl Acad Sci U S A. 1994;91:10009–13. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavailles V., Dauvois S., L'Horset F., Lopez G., Hoare S., Kushner P. J., Parker M. G. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. Embo J. 1995;14:3741–51. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. M., Lykkesfeldt A. E., Parker M. G., Dowsett M. Expression of nuclear receptor interacting proteins TIF-1, SUG-1, receptor interacting protein 140, and corepressor SMRT in tamoxifen-resistant breast cancer. Clin Cancer Res. 1999;5:3460–7. [PubMed] [Google Scholar]
- Chen Y., Hu X., Wei L. N. Molecular interaction of retinoic acid receptors with coregulators PCAF and RIP140. Mol Cell Endocrinol. 2004;226:43–50. doi: 10.1016/j.mce.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Chen Y., Kerimo A., Khan S., Wei L. N. Real-time analysis of molecular interaction of retinoid receptors and receptor-interacting protein 140 (RIP140) Mol Endocrinol. 2002;16:2528–37. doi: 10.1210/me.2002-0124. [DOI] [PubMed] [Google Scholar]
- Christian M., Tullet J. M., Parker M. G. Characterization of four autonomous repression domains in the corepressor receptor interacting protein 140. J Biol Chem. 2004;279:15645–51. doi: 10.1074/jbc.M313906200. [DOI] [PubMed] [Google Scholar]
- Christian M., Kiskinis E., Debevec D., Leonardsson G., White R., Parker M. G. RIP140-targeted repression of gene expression in adipocytes. Mol Cell Biol. 2005;25:9383–91. doi: 10.1128/MCB.25.21.9383-9391.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling D. L., Yingling J., Wynshaw-Boris A. Role of 14-3-3 proteins in eukaryotic signaling and development. Curr Top Dev Biol. 2005;68:281–315. doi: 10.1016/S0070-2153(05)68010-6. [DOI] [PubMed] [Google Scholar]
- Eng F. C., Barsalou A., Akutsu N., Mercier I., Zechel C., Mader S., White J. H. Different classes of coactivators recognize distinct but overlapping binding sites on the estrogen receptor ligand binding domain. J Biol Chem. 1998;273:28371–7. doi: 10.1074/jbc.273.43.28371. [DOI] [PubMed] [Google Scholar]
- Galan J. J., Buch B., Cruz N., Segura A., Moron F. J., Bassas L., Martinez-Pineiro L., Real L. M., Ruiz A. Multilocus analyses of estrogen-related genes reveal involvement of the ESR1 gene in male infertility and the polygenic nature of the pathology. Fertil Steril. 2005;84:910–8. doi: 10.1016/j.fertnstert.2005.03.070. [DOI] [PubMed] [Google Scholar]
- Gardiner K. Transcriptional dysregulation in Down syndrome: predictions for altered protein complex stoichiometries and post-translational modifications, and consequences for learning/behavior genes ELK, CREB, and the estrogen and glucocorticoid receptors. Behav Genet. 2006;36:439–53. doi: 10.1007/s10519-006-9051-1. [DOI] [PubMed] [Google Scholar]
- Gupta P., Huq M. D., Khan S. A., Tsai N. P., Wei L. N. Regulation of co-repressive activity of and HDAC recruitment to RIP140 by site-specific phosphorylation. Mol Cell Proteomics. 2005;4:1776–84. doi: 10.1074/mcp.M500236-MCP200. [DOI] [PubMed] [Google Scholar]
- Heery D. M., Kalkhoven E., Hoare S., Parker M. G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–6. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Heery D. M., Hoare S., Hussain S., Parker M. G., Sheppard H. Core LXXLL motif sequences in CREB-binding protein, SRC1, and RIP140 define affinity and selectivity for steroid and retinoid receptors. J Biol Chem. 2001;276:6695–702. doi: 10.1074/jbc.M009404200. [DOI] [PubMed] [Google Scholar]
- Henttu P. M., Kalkhoven E., Parker M. G. AF-2 activity and recruitment of steroid receptor coactivator 1 to the estrogen receptor depend on a lysine residue conserved in nuclear receptors. Mol Cell Biol. 1997;17:1832–9. doi: 10.1128/mcb.17.4.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq M. D., Khan S. A., Park S. W., Wei L. N. Mapping of phosphorylation sites of nuclear corepressor receptor interacting protein 140 by liquid chromatography-tandem mass spectroscopy. Proteomics. 2005a;5:2157–66. doi: 10.1002/pmic.200401090. [DOI] [PubMed] [Google Scholar]
- Huq M. D., Wei L. N. Post-translational modification of nuclear co-repressor receptor-interacting protein 140 by acetylation. Mol Cell Proteomics. 2005b;4:975–83. doi: 10.1074/mcp.M500015-MCP200. [DOI] [PubMed] [Google Scholar]
- Hu X., Chen Y., Farooqui M., Thomas M. C., Chiang C. M., Wei L. N. Suppressive effect of receptor-interacting protein 140 on coregulator binding to retinoic acid receptor complexes, histone-modifying enzyme activity, and gene activation. J Biol Chem. 2004;279:319–25. doi: 10.1074/jbc.M307621200. [DOI] [PubMed] [Google Scholar]
- Hu X., Funder J. W. The evolution of mineralocorticoid receptors. Mol Endocrinol. 2006;20:1471–8. doi: 10.1210/me.2005-0247. [DOI] [PubMed] [Google Scholar]
- Ikonen T., Palvimo J. J., Janne O. A. Interaction between the amino- and carboxyl-terminal regions of the rat androgen receptor modulates transcriptional activity and is influenced by nuclear receptor coactivators. J Biol Chem. 1997;272:29821–8. doi: 10.1074/jbc.272.47.29821. [DOI] [PubMed] [Google Scholar]
- Joyeux A., Cavailles V., Balaguer P., Nicolas J. C. RIP 140 enhances nuclear receptor-dependent transcription in vivo in yeast. Mol Endocrinol. 1997;11:193–202. doi: 10.1210/mend.11.2.9884. [DOI] [PubMed] [Google Scholar]
- Katsanis N., Ives J. H., Groet J., Nizetic D., Fisher E. M. Localisation of receptor interacting protein 140 (RIP140) within 100 kb of D21S13 on 21q11, a gene-poor region of the human genome. Hum Genet. 1998;102:221–3. doi: 10.1007/s004390050682. [DOI] [PubMed] [Google Scholar]
- Kerley J. S., Olsen S. L., Freemantle S. J., Spinella M. J. Transcriptional activation of the nuclear receptor corepressor RIP140 by retinoic acid: a potential negative-feedback regulatory mechanism. Biochem Biophys Res Commun. 2001;285:969–75. doi: 10.1006/bbrc.2001.5274. [DOI] [PubMed] [Google Scholar]
- Kumar M. B., Tarpey R. W., Perdew G. H. Differential recruitment of coactivator RIP140 by Ah and estrogen receptors. Absence of a role for LXXLL motifs. J Biol Chem. 1999;274:22155–64. doi: 10.1074/jbc.274.32.22155. [DOI] [PubMed] [Google Scholar]
- Laganiere J., Deblois G., Giguere V. Functional genomics identifies a mechanism for estrogen activation of the retinoic acid receptor alpha1 gene in breast cancer cells. Mol Endocrinol. 2005;19:1584–92. doi: 10.1210/me.2005-0040. [DOI] [PubMed] [Google Scholar]
- Lee C. H., Chinpaisal C., Wei L. N. Cloning and characterization of mouse RIP140, a corepressor for nuclear orphan receptor TR2. Mol Cell Biol. 1998;18:6745–55. doi: 10.1128/mcb.18.11.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardsson G., Jacobs M. A., White R., Jeffery R., Poulsom R., Milligan S., Parker M. Embryo transfer experiments and ovarian transplantation identify the ovary as the only site in which nuclear receptor interacting protein 1/RIP140 action is crucial for female fertility. Endocrinology. 2002;143:700–7. doi: 10.1210/endo.143.2.8656. [DOI] [PubMed] [Google Scholar]
- Leonardsson G., Steel J. H., Christian M., Pocock V., Milligan S., Bell J., So P. W., Medina-Gomez G., Vidal-Puig A., White R., Parker M. G. Nuclear receptor corepressor RIP140 regulates fat accumulation. Proc Natl Acad Sci U S A. 2004;101:8437–42. doi: 10.1073/pnas.0401013101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Horset F., Dauvois S., Heery D. M., Cavailles V., Parker M. G. RIP-140 interacts with multiple nuclear receptors by means of two distinct sites. Mol Cell Biol. 1996;16:6029–36. doi: 10.1128/mcb.16.11.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H. J., Moon I., Han K. Transcriptional cofactors exhibit differential preference toward peroxisome proliferator-activated receptors α and δ in uterine cells. Endocrinology. 2004;145:2886–95. doi: 10.1210/en.2004-0011. [DOI] [PubMed] [Google Scholar]
- Lin C. Y., Strom A., Vega V. B., Kong S. L., Yeo A. L., Thomsen J. S., Chan W. C., Doray B., Bangarusamy D. K., Ramasamy A., Vergara L. A., Tang S., Chong A., Bajic V. B., Miller L. D., Gustafsson J. A., Liu E. T. Discovery of estrogen receptor α target genes and response elements in breast tumor cells. Genome Biol. 2004;5 doi: 10.1186/gb-2004-5-9-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuyama H., Brownfield C. M., St-Arnaud R., MacDonald P. N. Evidence for ligand-dependent intramolecular folding of the AF-2 domain in vitamin D receptor-activated transcription and coactivator interaction. Mol Endocrinol. 1997;11:1507–17. doi: 10.1210/mend.11.10.9990. [DOI] [PubMed] [Google Scholar]
- Masuyama H., Hiramatsu Y., Mizutani Y., Inoshita H., Kudo T. The expression of pregnane X receptor and its target gene, cytochrome P450 3A1, in perinatal mouse. Mol Cell Endocrinol. 2001;172:47–56. doi: 10.1016/s0303-7207(00)00395-6. [DOI] [PubMed] [Google Scholar]
- Metivier R., Reid G., Gannon F. Transcription in four dimensions: nuclear receptor-directed initiation of gene expression. EMBO Rep. 2006;7:161–7. doi: 10.1038/sj.embor.7400626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata K. S., McCaw S. E., Meertens L. M., Patel H. V., Rachubinski R. A., Capone J. P. Receptor-interacting protein 140 interacts with and inhibits transactivation by, peroxisome proliferator-activated receptor α and liver-X-receptor α. Mol Cell Endocrinol. 1998;146:69–76. doi: 10.1016/s0303-7207(98)00196-8. [DOI] [PubMed] [Google Scholar]
- Monroe D. G., Getz B. J., Johnsen S. A., Riggs B. L., Khosla S., Spelsberg T. C. Estrogen receptor isoform-specific regulation of endogenous gene expression in human osteoblastic cell lines expressing either ERalpha or ERbeta. J Cell Biochem. 2003;90:315–26. doi: 10.1002/jcb.10633. [DOI] [PubMed] [Google Scholar]
- Moore J. M., Galicia S. J., McReynolds A. C., Nguyen N. H., Scanlan T. S., Guy R. K. Quantitative proteomics of the thyroid hormone receptor-coregulator interactions. J Biol Chem. 2004;279:27584–90. doi: 10.1074/jbc.M403453200. [DOI] [PubMed] [Google Scholar]
- Moron F. J., Mendoza N., Vazquez F., Molero E., Quereda F., Salinas A., Fontes J., Martinez-Astorquiza T., Sanchez-Borrego R., Ruiz A. Multilocus analysis of estrogen-related genes in Spanish postmenopausal women suggests an interactive role of ESR1, ESR2 and NRIP1 genes in the pathogenesis of osteoporosis. Bone. 2006;39:213–21. doi: 10.1016/j.bone.2005.12.079. [DOI] [PubMed] [Google Scholar]
- Nephew K. P., Sheeler C. Q., Dudley M. D., Gordon S., Nayfield S. G., Khan S. A. Studies of dehydroepiandrosterone (DHEA) with the human estrogen receptor in yeast. Mol Cell Endocrinol. 1998;143:133–42. doi: 10.1016/s0303-7207(98)00128-2. [DOI] [PubMed] [Google Scholar]
- Nichol D., Christian M., Steel J. H., White R., Parker M. G. RIP140 expression is stimulated by ERRalpha during adipogenesis. J Biol Chem. 2006 doi: 10.1074/jbc.M604803200. [DOI] [PubMed] [Google Scholar]
- Powelka A. M., Seth A., Virbasius J. V., Kiskinis E., Nicoloro S. M., Guilherme A., Tang X., Straubhaar J., Cherniack A. D., Parker M. G., Czech M. P. Suppression of oxidative metabolism and mitochondrial biogenesis by the transcriptional corepressor RIP140 in mouse adipocytes. J Clin Invest. 2006;116:125–36. doi: 10.1172/JCI26040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey J. M., Pujol P., Callier P., Cavailles V., Freiss G., Maudelonde T., Brouillet J. P. Semiquantitative reverse transcription-polymerase chain reaction to evaluate the expression patterns of genes involved in the oestrogen pathway. J Mol Endocrinol. 2000;24:433–40. doi: 10.1677/jme.0.0240433. [DOI] [PubMed] [Google Scholar]
- Sanyal S., Matthews J., Bouton D., Kim H. J., Choi H. S., Treuter E., Gustafsson J. A. Deoxyribonucleic acid response element-dependent regulation of transcription by orphan nuclear receptor estrogen receptor-related receptor γ. Mol Endocrinol. 2004;18:312–25. doi: 10.1210/me.2003-0165. [DOI] [PubMed] [Google Scholar]
- Sheeler C. Q., Dudley M. W., Khan S. A. Environmental estrogens induce transcriptionally active estrogen receptor dimers in yeast: activity potentiated by the coactivator RIP140. Environ Health Perspect. 2000;108:97–103. doi: 10.1289/ehp.0010897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stossi F., Barnett D. H., Frasor J., Komm B., Lyttle C. R., Katzenellenbogen B. S. Transcriptional profiling of estrogen-regulated gene expression via estrogen receptor (ER) α or ERbeta in human osteosarcoma cells: distinct and common target genes for these receptors. Endocrinology. 2004;145:3473–86. doi: 10.1210/en.2003-1682. [DOI] [PubMed] [Google Scholar]
- Subramaniam N., Treuter E., Okret S. Receptor interacting protein RIP140 inhibits both positive and negative gene regulation by glucocorticoids. J Biol Chem. 1999;274:18121–7. doi: 10.1074/jbc.274.25.18121. [DOI] [PubMed] [Google Scholar]
- Sugawara T., Abe S., Sakuragi N., Fujimoto Y., Nomura E., Fujieda K., Saito M., Fujimoto S. RIP 140 modulates transcription of the steroidogenic acute regulatory protein gene through interactions with both SF-1 and DAX-1. Endocrinology. 2001;142:3570–7. doi: 10.1210/endo.142.8.8309. [DOI] [PubMed] [Google Scholar]
- Tazawa H., Osman W., Shoji Y., Treuter E., Gustafsson J. A., Zilliacus J. Regulation of subnuclear localization is associated with a mechanism for nuclear receptor corepression by RIP140. Mol Cell Biol. 2003;23:4187–98. doi: 10.1128/MCB.23.12.4187-4198.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teyssier C., Belguise K., Galtier F., Cavailles V., Chalbos D. Receptor-interacting protein 140 binds c-Jun and inhibits estradiol-induced activator protein-1 activity by reversing glucocorticoid receptor-interacting protein 1 effect. Mol Endocrinol. 2003;17:287–99. doi: 10.1210/me.2002-0324. [DOI] [PubMed] [Google Scholar]
- Thenot S., Charpin M., Bonnet S., Cavailles V. Estrogen receptor cofactors expression in breast and endometrial human cancer cells. Mol Cell Endocrinol. 1999;156:85–93. doi: 10.1016/s0303-7207(99)00139-2. [DOI] [PubMed] [Google Scholar]
- Treuter E., Albrektsen T., Johansson L., Leers J., Gustafsson J. A. A regulatory role for RIP140 in nuclear receptor activation. Mol Endocrinol. 1998;12:864–81. doi: 10.1210/mend.12.6.0123. [DOI] [PubMed] [Google Scholar]
- Vo N., Fjeld C., Goodman R. H. Acetylation of nuclear hormone receptor-interacting protein RIP140 regulates binding of the transcriptional corepressor CtBP. Mol Cell Biol. 2001;21:6181–8. doi: 10.1128/MCB.21.18.6181-6188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L. N., Farooqui M., Hu X. Ligand-dependent formation of retinoid receptors, receptor-interacting protein 140 (RIP140), and histone deacetylase complex is mediated by a novel receptor-interacting motif of RIP140. J Biol Chem. 2001;276:16107–12. doi: 10.1074/jbc.M010185200. [DOI] [PubMed] [Google Scholar]
- Wei L. N., Hu X., Chandra D., Seto E., Farooqui M. Receptor-interacting protein 140 directly recruits histone deacetylases for gene silencing. J Biol Chem. 2000;275:40782–7. doi: 10.1074/jbc.M004821200. [DOI] [PubMed] [Google Scholar]
- White K. A., Yore M. M., Deng D., Spinella M. J. Limiting effects of RIP140 in estrogen signaling: potential mediation of anti-estrogenic effects of retinoic acid. J Biol Chem. 2005;280:7829–35. doi: 10.1074/jbc.M412707200. [DOI] [PubMed] [Google Scholar]
- White R., Leonardsson G., Rosewell I., Ann Jacobs M., Milligan S., Parker M. The nuclear receptor co-repressor nrip1 (RIP140) is essential for female fertility. Nat Med. 2000;6:1368–74. doi: 10.1038/82183. [DOI] [PubMed] [Google Scholar]
- Windahl S. H., Treuter E., Ford J., Zilliacus J., Gustafsson J. A., McEwan I. J. The nuclear-receptor interacting protein (RIP) 140 binds to the human glucocorticoid receptor and modulates hormone-dependent transactivation. J Steroid Biochem Mol Biol. 1999;71:93–102. doi: 10.1016/s0960-0760(99)00128-4. [DOI] [PubMed] [Google Scholar]
- Yan Z. H., Karam W. G., Staudinger J. L., Medvedev A., Ghanayem B. I., Jetten A. M. Regulation of peroxisome proliferator-activated receptor α-induced transactivation by the nuclear orphan receptor TAK1/TR4. J Biol Chem. 1998;273:10948–57. doi: 10.1074/jbc.273.18.10948. [DOI] [PubMed] [Google Scholar]
- Zilliacus J., Holter E., Wakui H., Tazawa H., Treuter E., Gustafsson J. A. Regulation of glucocorticoid receptor activity by 14--3-3-dependent intracellular relocalization of the corepressor RIP140. Mol Endocrinol. 2001;15:501–11. doi: 10.1210/mend.15.4.0624. [DOI] [PubMed] [Google Scholar]