Abstract
Improvement of nutritive value of crop plants, in particular the amino acid composition, has been a major long-term goal of plant breeding programs. Toward this end, we reported earlier the cloning of the seed albumin gene AmA1 from Amaranthus hypochondriacus. The AmA1 protein is nonallergenic in nature and is rich in all essential amino acids, and the composition corresponds well with the World Health Organization standards for optimal human nutrition. In an attempt to improve the nutritional value of potato, the AmA1 coding sequence was successfully introduced and expressed in tuber-specific and constitutive manner. There was a striking increase in the growth and production of tubers in transgenic populations and also of the total protein content with an increase in most essential amino acids. The expressed protein was localized in the cytoplasm as well as in the vacuole of transgenic tubers. Thus we have been able to use a seed albumin gene with a well-balanced amino acid composition as a donor protein to develop a transgenic crop plant. The results document, in addition to successful nutritional improvement of potato tubers, the feasibility of genetically modifying other crop plants with novel seed protein composition.
One of the goals of plant genetic engineering has been to create crops that are tailored to provide better nutrition for humans and their domestic animals. A major target has been the improvement of the amino acid composition of seed proteins, because animals, including humans, are incapable of synthesizing 10 of the 20 amino acids needed for protein synthesis, and these “essential” amino acids must therefore be obtained from the diet. Advances in plant tissue culture techniques and gene transfer technology have opened up possibilities for modifying the amino acid contents of plants. One approach has been to manipulate the regulation of amino acid biosynthesis to increase the abundance of a particular amino acid. Mutant selection and engineering genes encoding key enzymes of amino acid biosynthetic pathways have been used to increase amino acids in crop plants (1). However, an increase in the free essential amino acids does not lead to an increase in the fixed content, and the amino acids could be leached out from the plant tissue and lost during boiling and other processing (2, 3). An alternative approach is the insertion and the expression of gene(s) encoding essential amino acid-rich proteins in transgenic plants.
Potato is the most important noncereal food crop and ranks fourth in terms of total global food production, besides being used as animal feed and as raw material for manufacture of starch, alcohol, and other food products. The essential amino acids that limit the nutritive value of potato protein are lysine, tyrosine, and the sulfur-containing amino acids methionine and cysteine (4). In quality improvement programs of potato, the priorities have been to improve disease and pest resistance (5–8), to increase yields (9), and to increase adaptability to biotic and abiotic conditions (10–12), whereas nutritional status has remained secondary. In an attempt to develop economically important crop plants with improved nutritional quality, our laboratory earlier reported the cloning of a gene that encodes a seed-specific protein, amaranth seed albumin (AmA1) from Amaranthus hypochondriacus (13). This gene has been patented (14). The AmA1 protein has great potential as a donor protein for the following reasons: (i) unlike most seed proteins, it is a well-balanced protein in terms of amino acid composition and even better than the values recommended by the World Health Organization for a nutritionally rich protein; (ii) it is a nonallergenic protein in its purified form; and (iii) unlike many seed storage proteins, AmA1 is encoded by a single gene and thus would facilitate gene transfer into target plants with less difficulty. In this paper, we report the tuber-specific as well as constitutive expression of AmA1 in potato by using granule-bound starch synthase (GBSS) and cauliflower mosaic virus (CaMV) 35S promoters, respectively. The expression of AmA1 in transgenic tubers resulted in a significant increase in most essential amino acids. Unexpectedly, the transgenic plants also contained more total protein in tubers compared to control plants. These findings demonstrate the feasibility of using the AmA1 gene in genetic engineering to improve the nutritive value of other nonseed and grain crops.
Materials and Methods
Construction of AmA1 Expression Plasmids.
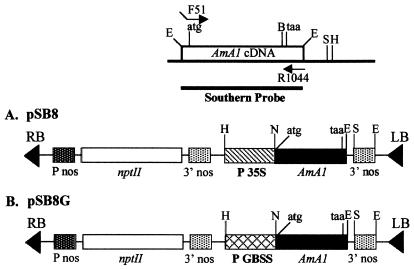
The AmA1 coding region was amplified by PCR using F51 (5′-CACCATGGCGGGATTACCAGTG-3′) and R1044 (5′-CAAGGAAGAACCCTCTTGTTTCC-3′) as forward and reverse primers, respectively. The F51 primer was designed such that the 5′ end had a four-base mismatch with AmA1 gene to create an NcoI site just flanking the start codon ATG for better translational efficiency. The PCR-amplified fragment was digested with BamHI, and the 0.9-kb fragment containing the AmA1 coding region was translationally fused with β-GUS gene at the BamHI site of pBI221 and named as pSB2. The chimeric plasmid pSB6 was constructed by a three-stage cloning wherein the AmA1 C-terminal part was taken as a 0.652-kb NdeI–SacI fragment from pAmA1.3 (13) and a 1.294-kb HindIII–NdeI fragment containing CaMV 35S promoter and AmA1 N-terminal part from pSB2. These two gene pieces were cloned back into the 2.96-kb HindIII–SacI backbone of pBI221. In the next step, pSB8 was made by replacing the HindIII–SacI fragment of pBI121 with the same fragment of pSB6 for use in Agrobacterium-mediated transformation (Fig. 1A).
Figure 1.
Schematic representation of AmA1 expression plasmids containing AmA1 coding sequence. (A) pSB8 plasmid with full-length AmA1 cDNA starting with ATG as translation start and ending with stop signal TAA along with 222 bp of 3′ untranslated region cloned between CaMV 35S promoter and nos terminator in pBI221. (B) pSB8G plasmid, same as pSB8 except that 35S promoter is replaced by GBSS promoter. The forward and reverse primers used in PCR amplification of AmA1 cDNA along with restriction map of pAmA1.3 are shown above the constructs. H, HindIII; B, BamHI; N, NcoI; S, SacI; E, EcoRI; nptII, neomycin phosphotransferase; P35S, CaMV 35S promoter; Pnos, nos promoter; 3′ nos, nos terminator; PGBSS, GBSS promoter.
A GBSS gene promoter was used to obtain tuber-specific high expression of AmA1 in potato. The 0.8-kb CaMV 35S promoter from pSB8 was deleted by HindIII–XbaI digestion and replaced by a 0.8-kb HindIII–XbaI fragment of the GBSS promoter from pPGB-1 (15), and the resulting plasmid was named pSB8G (Fig. 1B). The plasmids pSB8 and pSB8G were mobilized into Agrobacterium strain LBA4404 (16) by using the helper strain HB101∷pRK2013 by a triparental mating technique (17).
Plant Transformation and Selection of Transgenic Lines.
Potato (Solanum tuberosum L. var. A16) shoot cultures were grown in vitro on MS basal medium (18) containing 2.5% sucrose, 0.1 mg/liter indoleacetic acid, and 0.6% agar at 23 ± 2°C in a 16-h photoperiod. The internodal stem segments (3–5 mm) were incubated for 30 min in a saturated culture of Agrobacterium tumefaciens containing the AmA1 expression plasmids, blotted dry on sterile Whatman paper, and transferred onto PR (potato regeneration) medium for 24 h at 23°C under a 16-h photoperiod. The PR medium is a typical MS basal medium containing 2.5% sucrose, 1.5 mg/liter benzylaminopurine, 0.1 mg/liter naphthaleneacetic acid, and 0.1 mg/liter gibberellic acid (pH 5.8), solidified with 0.8% agar. The cocultivation continued for 2 days, and the calli were initiated by transferring the explants onto PR medium containing 250 mg/liter cefotaxime and incubated for 1 wk. To obtain a rapid growth of calli, two to three transfers were made onto fresh PR medium. Shoot-buds were initiated on selective and regenerative medium, PRS (PR medium containing 250 mg/liter cefotaxime and 100 mg/liter kanamycin). Kanamycin-resistant shoot-buds (5–6 mm) were transferred onto rooting medium (MS basal medium containing 2.5% sucrose, 0.2 mg/liter indoleacetic acid, solidified with 0.6% agar) supplemented with 100 mg/liter kanamycin. For each transformation, 40 independent transgenic plants were regenerated.
Nucleic Acid Isolation and Analysis.
DNA from transgenic plantlets was isolated (19) and the approximate copy number of the AmA1 sequence and the site of integration were detected by Southern blot analysis. The intactness of the AmA1 gene in transgenic plants was determined by PCR using F51 and R1044 primers. Total RNA was extracted from leaves, stems, and tubers of pSB8G plants and tubers of pSB8 plants by the method of Powlowski et al. (20). The reverse transcription (RT) was done by using GeneAmp EZ rTth RNA PCR kit (Perkin–Elmer). The RT-PCR was carried out with 20 pmol each of AmA1 gene-specific primers F51 and R1044, 300 μM each dNTP, 2.5 mM Mn(OAc)2, 2.5 units of rTth DNA polymerase, and 1 μg of template RNA in a 25-μl reaction volume. The cDNA was synthesized by initial denaturation at 94°C for 2 min and set to RT at 60°C for 30 min. In the next step, the mixture was again denatured at 94°C for 2 min followed by 40 cycles each of denaturation at 94°C for 1 min, annealing and extension at 60°C for 1 min each, and final extension at 60°C for 7 min.
Protein Extraction and Immunoblot Analysis.
Soluble protein was extracted from 250 mg of tissue of transgenic tubers in 0.5 ml of extraction buffer (25 mM Tris⋅acetate, pH 8.5/0.5 M NaCl/5 mM PMSF). The homogenate was centrifuged at 12,000 × g for 10 min. Protein concentration in the supernatant was measured by using the Bradford protein assay kit (Bio-Rad), and aliquots of 50 and 100 μg of protein from pSB8G and pSB8 tubers, respectively, were precipitated with trichloroacetic acid and denatured by boiling for 5 min in sample loading buffer (21). The proteins resolved in SDS/PAGE were immobilized onto Hybond-C membrane as described earlier (22), and AmA1 was detected with a rabbit polyclonal AmA1 antibody in combination with alkaline phosphatase-conjugated goat anti-rabbit IgG. Coomassie blue-stained gels were loaded with 100 μg of soluble tuber protein for both pSB8 and pSB8G transgenic lines.
Immunoelectron Microscopy.
Tuber sections were fixed in 4% paraformaldehyde and 1% glutaraldehyde in 0.1 M PBS (pH 7.2), dehydrated over grades of ethanol at 4°C, and embedded in LR-white resin (Hard). Thin sections (60–90 nm) lifted onto 400 mesh nickel grids were blocked with 3% BSA in Tris-buffered saline (TBS) for 1 h at room temperature. Immunolabeling was done with polyclonal rabbit anti-AmA1 antibody diluted 1:100 in TBS containing 1% BSA and 0.02% sodium azide overnight at 4°C. Untransformed tuber sections were taken as controls. The grids were incubated with AuroProbeEM GAR G15 (Amersham Pharmacia) at a dilution of 1:50 in TBS containing 0.5% Tween 20 for 1 h at room temperature. Grids were postfixed in 2.5% glutaraldehyde in 0.1 M PBS (pH 7.2) for 10 min and stained with 2% aqueous uranyl acetate for 20 min at room temperature. Electron micrographs were taken with a Philips CM10 transmission electron microscope.
Amino Acid Analysis.
The amino acid composition of pSB8 and pSB8G tubers along with A16 tubers was determined by using a C18 HPLC column equipped with an online Pico Tag amino acid analyzer (Waters). Total soluble protein from wild-type and transgenic tubers was precipitated with 10% trichloroacetic acid on ice for 45 min, washed with ethanol/ether (1:1, vol/vol), and lyophilized. Acid hydrolysis and derivatization of lyophilized protein with phenyl isothiocyanate (PITC) was done as per the Pico Tag manual. The PITC derivative of each amino acid was detected by absorbance at 254 nm.
Results
Construction of Expression Plasmids and Stable Integration of Transgene.
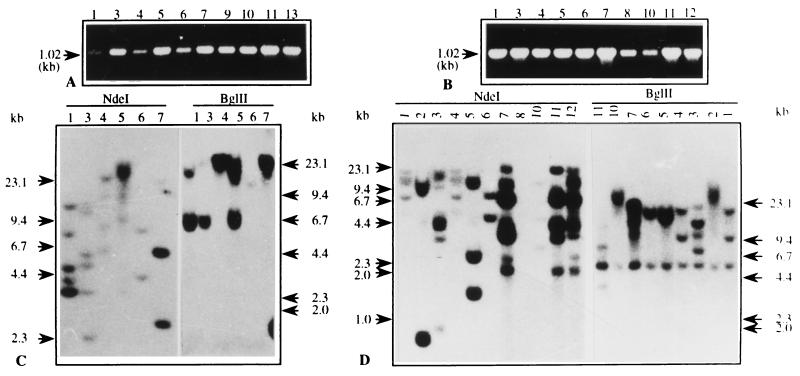
The expression plasmid pSB8 was constructed by using AmA1 coding sequence along with 102 bp of 3′ AmA1 untranslated region under the control of CaMV 35S promoter in pBI121 and pSB8G, wherein 35S promoter was replaced by GBSS promoter (Fig. 1 A and B). The presence of AmA1 gene in transgenic plants was detected by PCR amplification of AmA1 inserts using gene-specific primers. An amplified product of 1.02-kb fragment was detected in both pSB8 and pSB8G plants (Fig. 2 A and B). The approximate copy number and site of integration was detected by Southern hybridization. As the AmA1 full-length coding region was used as a probe and NdeI has a single restriction site in it, the transgenic plants with a single copy insertion should give two bands, whereas plants with two copy insertions should produce four bands and so on. Similarly, BglII having no site in the AmA1 coding region should yield a single band for a single-copy insertion and two bands for two-copy insertions. Southern analysis showed one to three copy insertions in different transgenic lines of pSB8 and pSB8G plants (Fig. 2 C and D). Each transgenic plant showed a characteristic restriction pattern, indicating that it is an independent transformant. The transgenics containing one to three copies of transgene were subsequently transferred to the field and allowed to set tubers for detailed analysis.
Figure 2.
PCR amplification of genomic DNA from pSB8 (A) and pSB8G (B) plantlets. The amplification products and their size are indicated by arrow. The primers and their positions are shown in Fig. 1. In Southern blot analysis, 15 μg of DNA from each transgenic lines of pSB8 (C) and pSB8G (D) was digested with BglII and NdeI and resolved on a 0.7% agarose gel. Digested products were transferred onto GeneScreenPlus membrane and hybridized with 32P-labeled full-length AmA1 cDNA as shown in Fig. 1. Numbers on the lanes are representatives of independent transgenic lines.
Constitutive and Tissue-Specific Accumulation of Transcripts.
Total RNA was isolated from the leaves, stems, and tubers of pSB8G plants and only from the tubers of pSB8 plants. Although there was no RT product in the case of untransformed plants, a 1.02-kb RT product was detected in all transgenic lines, which confirmed the accumulation of mRNAs in transgenic lines. As expected, the transcript level in pSB8G tubers was 5- to 10-fold higher than in pSB8 tubers (data not shown). Based on the band intensity on ethidium bromide-stained gels, it appeared that the transcripts were most abundant in tubers, followed by stem and leaf tissues in pSB8G plants (data not shown). This can be explained by the fact that GBSS promoter is more active in stolons and tubers than in leaves (15).
Immunodetection of AmA1 Protein in Transgenic Tubers.
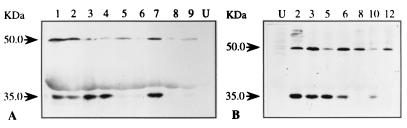
The expression of AmA1 resulted in the accumulation of recombinant proteins at various degrees (10- to 20-fold) among transgenic lines, probably because of position effects (23). This result is similar to that in an earlier report, where it was shown that the expression level in individual transformants varied even at identical gene copy number (24). The 35-kDa AmA1 protein was detected in both pSB8 and pSB8G immunoblots (Fig. 3 A and B); however, as expected, the expression in pSB8G-tubers was much higher as compared with pSB8 tubers. The immunoblots of both pSB8 and pSB8G tubers revealed another distinct band of approximately 50-kDa size, which crossreacted with AmA1 antibody. This higher molecular mass protein band could be the result of glycosylation of AmA1 protein, as glycosylation is a common phenomenon in tuber tissues. Moreover, AmA1 protein has three putative N-linked glycosylation sites (13), and, therefore, it was imperative to check the deglycosylation pattern in tuber proteins. However, there was no deglycosylation in 50-kDa polypeptide on EndoH treatment, although other major tuber proteins such as patatin were deglycosylated (data not shown). Moreover, no major change in tuber soluble protein profiles on Coomassie blue-stained polyacrylamide gels was observed (data not shown).
Figure 3.
Expression of AmA1 protein in pSB8 (A) and pSB8G (B) tubers of transgenic lines by immunoblot analysis. Aliquots of 50 and 100 μg of soluble protein from pSB8G and pSB8 tubers, respectively, were separated by SDS/12.5% PAGE and electroblotted onto Hybond-C membrane. AmA1 protein was detected with a polyclonal anti-AmA1 antibody and alkaline phosphatase-conjugated anti-rabbit IgG antibody. The alkaline phosphatase color development reaction for pSB8G blot (B) was carried out for 5 min, whereas the same reaction for pSB8 blot (A) was prolonged for 20 min. The lane numbers are representatives of the individual transgenic lines, whereas U represents untransformed tuber.
Subcellular Localization of AmA1 Protein in Transgenic Tubers.
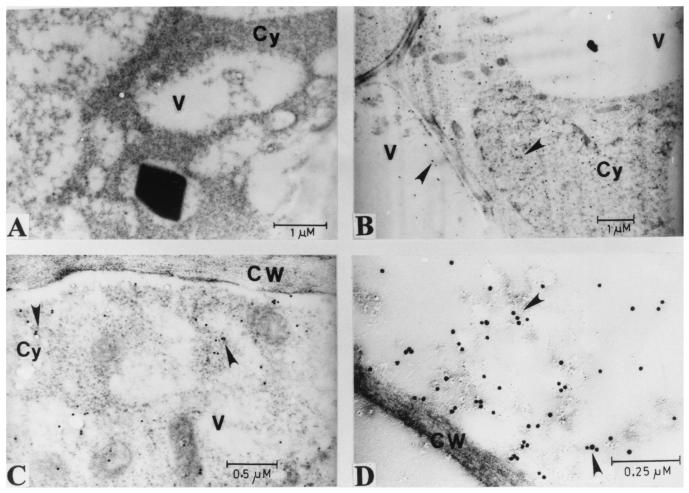
To understand the basis for the stability of AmA1 protein in tuber cells of transgenic plants, the subcellular localization of AmA1 protein in both pSB8 and pSB8G tubers was studied by immunocytochemistry along with transmission electron microscopy. No signal was detected in sections visualized from wild-type tuber (Fig. 4A). The AmA1 was found to be present mainly in the cytoplasm and to a lesser extent in the vacuoles (Fig. 4 B–D), not accumulating into characteristic protein bodies.
Figure 4.
Transmission electron micrographs showing immunolocalization of AmA1 protein in wild-type tuber (A) as well as in pSB8 (B) and pSB8G (C and D) plants. The labeling was done with polyclonal anti-AmA1 antibody followed by 15-nm gold-conjugated goat anti-rabbit IgG antibody. CW, cell wall; Cy, cytoplasm; V, vacuole.
Increased Amino Acid Profile in Transgenic Tubers.
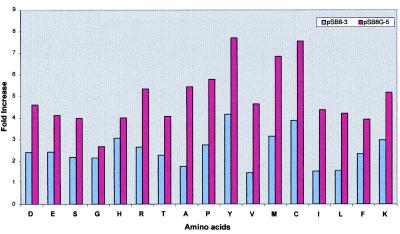
Based on AmA1 expression level, five pSB8G and six pSB8 highly expressed transgenic lines were selected for amino acid analysis. The amino acid content, determined in triplicate for each line, showed an increase in all essential amino acids in both categories of transgenics (Table 1) with respect to wild-type A16 genotype. When compared among pSB8 populations, the highly expressing tubers (pSB8-3) showed a significant 2.5- to 4-fold increase in lysine, methionine, cysteine, and tyrosine contents, which are otherwise limited in potato. As expected, their amounts in the highly expressing pSB8G tubers (pSB8G-5) appeared to be 4- to 8-fold (Fig. 5). These findings are consistent with the immunoblot data (Fig. 3 A and B), wherein the expression of AmA1 was found to be 5- to 10-fold higher in pSB8G tubers than that in pSB8 tubers. The amino acid composition of different proteins separated on the basis of molecular weight and transferred on a poly(vinylidene difluoride) membrane from the same gel did not show any appreciable change (data not shown), thereby suggesting the increased level of essential amino acids was not at the expense of other endogenous proteins of the A16 genotype.
Table 1.
Amino acid profile of tubers of wild-type and transgenic lines
| Amino acid | Micromoles of amino
acid per g of soluble protein
|
||||||
|---|---|---|---|---|---|---|---|
| A16 | pSB8
plants
|
pSB8G plants
|
|||||
| 2 | 3 | 7 | 2 | 3 | 5 | ||
| D | 305 ± 11.6 | 525 ± 15.5 | 734 ± 18.1 | 468 ± 18.0 | 1063 ± 34.0 | 766 ± 27.0 | 1401 ± 56.5 |
| E | 281 ± 9.8 | 440 ± 17.0 | 680 ± 19.8 | 519 ± 20.2 | 900 ± 26.0 | 612 ± 24.7 | 1157 ± 44.8 |
| S | 188 ± 6.5 | 239 ± 15.5 | 410 ± 16.0 | 292 ± 11.0 | 586 ± 12.3 | 492 ± 30.4 | 749 ± 19.2 |
| G | 297 ± 12.5 | 470 ± 17.9 | 639 ± 18.4 | 523 ± 19.5 | 740 ± 18.0 | 640 ± 18.0 | 790 ± 30.0 |
| H | 59 ± 3.5 | 115 ± 10.6 | 180 ± 6.0 | 143 ± 11.5 | 185 ± 8.5 | 117 ± 10.0 | 236 ± 10.5 |
| R | 98 ± 9.0 | 162 ± 8.4 | 259 ± 11.0 | 180 ± 7.0 | 470 ± 14.3 | 308 ± 10.0 | 523 ± 26.0 |
| T | 182 ± 7.0 | 272 ± 10.0 | 413 ± 17.2 | 280 ± 8.7 | 545 ± 24.0 | 301 ± 5.2 | 742 ± 27.5 |
| A | 222 ± 13.2 | 230 ± 13.5 | 389 ± 13.4 | 246 ± 10.5 | 736 ± 15.5 | 414 ± 12.7 | 1206 ± 27.5 |
| P | 248 ± 12.0 | 468 ± 21.0 | 680 ± 15.1 | 485 ± 19.2 | 1042 ± 50.4 | 648 ± 18.0 | 1435 ± 35.2 |
| Y | 9.6 ± 2.0 | 27 ± 1.5 | 40 ± 2.5 | 27 ± 1.0 | 42 ± 2.5 | 34 ± 2.0 | 74 ± 4.5 |
| V | 246 ± 8.6 | 305 ± 12.5 | 356 ± 8.5 | 338 ± 10.5 | 815 ± 18.0 | 516 ± 17.6 | 1142 ± 31.4 |
| M | 10.5 ± 2.0 | 21 ± 1.5 | 33 ± 2.0 | 32 ± 3.0 | 52 ± 4.0 | 31 ± 1.5 | 72 ± 3.5 |
| C | 4.5 ± 0.57 | 15 ± 1.0 | 17.5 ± 1.5 | 10 ± 1.5 | 23 ± 1.0 | 21 ± 2.0 | 34 ± 2.6 |
| I | 206 ± 10.8 | 247 ± 9.2 | 314 ± 10.0 | 264 ± 12.6 | 680 ± 19.8 | 531 ± 12.2 | 898 ± 21.5 |
| L | 303 ± 8.1 | 408 ± 18.6 | 474 ± 19.6 | 392 ± 14.5 | 967 ± 36.0 | 634 ± 21.0 | 1273 ± 29.0 |
| F | 143 ± 9.5 | 227 ± 10.3 | 334 ± 10.0 | 272 ± 11.5 | 478 ± 21.3 | 295 ± 11.6 | 562 ± 21.0 |
| K | 174 ± 11.0 | 435 ± 13.5 | 517 ± 21.1 | 363 ± 15.0 | 584 ± 8.7 | 414 ± 18.0 | 900 ± 25.8 |
Wild-type and transgenic potato plants in restricted experimental plots were grown to maturity in the winter season and tubers were harvested. Protein was extracted from the tubers and used for amino acid analysis. Values are presented as the mean ± SE for three each of wild-type and transgenic plants.
Figure 5.
Amino acid composition of pSB8G-5 and pSB8–3 tubers in comparison with wild-type tubers. The histogram shows fold increase of each amino acid in highly expressed transgenic lines, one each from pSB8G and pSB8 plants, against amino acid level from respective wild-type plants. The quantitative values of each amino acid for transgenic and wild-type tubers are shown in Table 1.
Analysis of Wild-Type and Transgenic Potato Lines in a Restricted Experimental Plot.
The potato tubers were characterized from four plants of A16, and each independent transgenic line was selected on the basis of AmA1 expression. A total of 16 independent pSB8 and eight independent pSB8G plants were tested for two consecutive years, and the results were consistent. A significant change in the forage parts of the transgenic lines as well as in the growth and production of tubers was observed. The most striking change was observed in respect to total protein content (35–45% increase) in transgenic tubers, which was increased in broad correlation with the increase of most essential amino acids (Table 2). There was a more than 2-fold increase in tuber number in both pSB8G and pSB8 lines with higher levels of transgene expression. Besides increase in tuber number, there was a 3.0- to 3.5-fold increase in tuber yield in terms of fresh weight, and a similar trend was observed with most transgenic lines tested.
Table 2.
Characteristics of tubers of wild-type and transgenic lines
| Parameter | A16 | pSB8
plants
|
pSB8G
plants
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 7 | 9 | 2 | 3 | 4 | 5 | 6 | 10 | ||
| Tuber no. per plant | 10.25 ± 2.06 | 19.66 ± 2.08 | 17.75 ± 3.09 | 23.20 ± 2.88 | 17.25 ± 2.21 | 19.75 ± 2.50 | 18.00 ± 2.16 | 19.00 ± 1.82 | 18.00 ± 3.00 | 19.00 ± 2.64 | 20.30 ± 2.08 | 18.0 ± 2.94 | 20.00 ± 3.60 |
| Tuber fresh wt per plant, g | 26.32 ± 3.00 | 136.20 ± 7.71 | 142.30 ± 9.55 | 169.65 ± 5.00 | 117.32 ± 9.29 | 147.7 ± 10.83 | 139.40 ± 7.92 | 132.25 ± 10.08 | 146.56 ± 7.15 | 159.60 ± 7.92 | 175.92 ± 7.0 | 142.80 ± 9.43 | 125.93 ± 10.9 |
| Mean fresh wt per tuber, g | 2.56 | 6.92 | 8.00 | 7.31 | 6.80 | 7.47 | 7.74 | 6.96 | 8.14 | 8.40 | 8.66 | 7.93 | 6.29 |
| Protein content, mg per g tuber | 11.15 ± 0.83 | 15.0 ± 1.05 | 14.57 ± 1.03 | 15.37 ± 1.45 | 14.82 ± 1.38 | 16.12 ± 1.33 | 14.10 ± 0.93 | 15.52 ± 1.07 | 15.25 ± 1.53 | 16.00 ± 0.67 | 16.58 ± 0.20 | 15.45 ± 1.29 | 14.69 ± 0.37 |
Potato plants in restricted experimental plots were grown to maturity in the winter season and tubers were harvested. The tuber number, fresh weight, and protein content were determined. Total tuber protein was extracted with 20% trichloroacetic acid and estimated spectrophotometrically. Values are presented as the mean ± SE for four wild-type and three or four transgenic plants.
Nonallergenic Nature of AmA1 Protein.
In many cases, animal feeding experiments have been suggested for allergenicity tests. However, these tests are plagued with problems such as high levels of variation in results between experimental groups. It is also difficult to feed relevant doses of purified protein or genetically modified foods to laboratory animals for sufficient periods to allow reliable extrapolation of the results to humans. Therefore, to determine whether AmA1 protein is allergenic or not, the following animal experiment was carried out. In this experiment, 6-wk-old BALB/c mice were injected with 30 μg of purified AmA1 protein intranasally as well as intraperitoneally with two successive boosters given at 1-wk intervals. Serum was collected and an ELISA was done to measure IgG and IgE levels. Although the IgG level was high enough, IgE could not be detected.
Discussion
To introduce the amaranth albumin gene into potato, a simple transformation and regeneration protocol was developed. The regeneration protocol involves a single medium composition throughout, and transformants were obtained within 6–8 wk. The diploid potato genotype, A16, that is resistant to Phytophthora infestans, the most devastating pathogen, was used in this study.
Efforts have been made to express seed proteins with unusually high methionine/cysteine content at a high level to increase the nutritive value of legume seeds. Although the 2S albumins from Brazil nut and sunflower contain 18% methionine/8% cysteine and 16% methionine/8% cysteine, respectively, they have lower levels of other essential amino acids. Eventually, when introduced in target plants, they resulted in dramatic increase only in methionine, along with a significant decrease in cysteine content (25, 26). This result reduces the usefulness of such proteins as a means of improving nutritional quality of recipient plants if they are deficient in any other essential amino acids. In contrast, AmA1 protein is well balanced and rich in all essential amino acids, including lysine, tryptophan, tyrosine, and sulfur-containing amino acids (13). More importantly, plans for commercial development of genetically engineered crops to boost their amino acid content by using Brazil nut 2S albumin were abandoned because it is highly allergenic in its purified form and in extracts of transgenic seeds (27). Most allergenic proteins tend to have characteristic sequence stretches. However, we did not find any such sequence homology in AmA1 protein when compared with the existing Protein Data Bank. Literature search has not revealed any allergenicity associated with amaranth grain or amaranth forage. Indeed, amaranth meal or flour has been used in the production of unleavened bread in Mexico and Central and Latin America. Grain amaranth is also used in many other foods throughout the world. The fact that amaranth forage has been an important component of human diet, perhaps the most widely eaten vegetable in the humid tropics for centuries, also suggests its nonallergenic nature. Moreover, animal feeding trials with a rat population eating seed grain of A. hypochondriacus have shown that the grains are suitable as animal feed (28). In addition, the hypersensitivity test in an animal model in this study also showed that AmA1 protein does not evoke any IgE response, which negates the possibility that the protein is allergenic.
The AmA1 protein was found mostly in the cytoplasm and to a lesser extent in the vacuoles of tuber parenchyma of transgenic plants. However, AmA1 protein in its native form in amaranth seeds is present in protein bodies (unpublished data). It is very likely that a vacuolar form of AmA1 also exists in transgenic plants. Solubility of a protein is a pH-dependent phenomenon, and thus solubilization of any protein near its isoelectric point is minimized. Since the isoelectric point of AmA1 is 7.4 (13), it is quite possible that the protein remains soluble in an acidic environment within the tuber vacuole, hence does not aggregate into protein bodies.
At the biochemical level, expression of AmA1 in both categories of transgenics leads to a high increase in all essential amino acids, particularly lysine, tyrosine, and the sulfur amino acids with corresponding increase in total protein content. Although the biological function of AmA1 is not known, it is conceivable that induction of protein synthesis or mitogenic activity might be the result of overexpression of AmA1 or its turned-over products as signal molecule. Because potato constitutes an important part of the diet of many people in developed as well as developing countries, the target of this work is to improve human nutrition. We are not aware of any other report where expressing a foreign protein with a well balanced amino acid composition led to an increase in the nutritional value of a nonseed food crop. We are currently conducting multicentric field trials to assess the agronomic performance of the promising transgenic lines besides assessing their nutritive value in detailed animal feeding trials.
Acknowledgments
We thank Dr. Evert Jacobsen for providing A16 potato genotype and the plasmid pPGB1, and the Department of Biotechnology, Ministry of Science and Technology, Government of India for financial support.
Abbreviations
- AmA1
amaranth seed albumin
- CaMV
cauliflower mosaic virus
- GBSS
granule-bound starch synthase
- PR
potato regeneration
- RT
reverse transcription
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050012697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050012697
References
- 1.Matthews B F, Hughes C A. Amino Acids (Vienna) 1993;4:21–34. doi: 10.1007/BF00805798. [DOI] [PubMed] [Google Scholar]
- 2.Shaul O, Galili G. Plant J. 1992;2:203–209. [Google Scholar]
- 3.Falco S C, Guida T, Locke M, Mauvais J, Sandres C, Ward R T, Webber P. Biotechnology. 1995;13:577–582. doi: 10.1038/nbt0695-577. [DOI] [PubMed] [Google Scholar]
- 4.Jaynes J M, Yang M S, Espinoza N, Dodds J H. Trends Biotechnol. 1986;4:314–320. [Google Scholar]
- 5.During K, Porsch P, Fladung M, Lorz H. Plant J. 1993;3:587–598. [Google Scholar]
- 6.Wu G, Shortt B J, Lawrence E B, Fitzsimmons K C, Shah D M. Plant Cell. 1995;7:1357–1368. doi: 10.1105/tpc.7.9.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moon Y H, Song S K, Choi K W, Lee J S. Mol Cell. 1997;7:807–815. [PubMed] [Google Scholar]
- 8.Perlak F J, Stone T B, Muskopf Y M, Peterson L J, Parker G B, McPherson S A, Wyman J, Love S, Reed G, Biever D, Fischhoff D A. Plant Mol Biol. 1993;22:313–321. doi: 10.1007/BF00014938. [DOI] [PubMed] [Google Scholar]
- 9.Sonnewald U, Hajirezaei M-R, Kossmann J, Heyer A, Trethewey R N, Willmitzer L. Nat Biotechnol. 1997;15:794–797. doi: 10.1038/nbt0897-794. [DOI] [PubMed] [Google Scholar]
- 10.Zhu B, Chen T H, Li P H. Planta. 1996;198:70–77. doi: 10.1007/BF00197588. [DOI] [PubMed] [Google Scholar]
- 11.Bussis D, Heineke D, Sonnewald U, Wilmitzer L, Raschke K, Heldt H W. Planta. 1997;202:126–136. doi: 10.1007/s004250050111. [DOI] [PubMed] [Google Scholar]
- 12.Wallis J G, Wang H, Guerra D J. Plant Mol Biol. 1997;35:323–330. doi: 10.1023/a:1005886210159. [DOI] [PubMed] [Google Scholar]
- 13.Raina A, Datta A. Proc Natl Acad Sci USA. 1992;89:11774–11778. doi: 10.1073/pnas.89.24.11774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Datta, A., Raina, A. & Biswas, S. (1997) U.S. Patent 5,670,635.
- 15.Visser R G F, Stolte A, Jacobsen E. Plant Mol Biol. 1991;17:691–699. doi: 10.1007/BF00037054. [DOI] [PubMed] [Google Scholar]
- 16.Hoekema A, Hirsch P R, Hooykaas P J J, Schilperoort R A. Nature (London) 1983;303:179–180. [Google Scholar]
- 17.Van Haute E, Joos H, Maes S, Warren G, Van Montagu M, Schell J. EMBO J. 1983;2:411–418. doi: 10.1002/j.1460-2075.1983.tb01438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murashige T, Skoog F. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 19.Murray M G, Thompson W F. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pawlowski K, Kunze R, de Vries S, Bisseling T. In: Plant Molecular Biology Mannual. 2nd Ed. Gelvin S B, Schilperoort R A, editors. Boston: Kluwer; 1994. pp. 1–13. [Google Scholar]
- 21.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones J D G, Dunsmuir P, Bedbrook J. EMBO J. 1985;4:2411–2418. doi: 10.1002/j.1460-2075.1985.tb03949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawton M A, Tierney M A, Nakamura I, Anderson E, Komeda Y, Dube P, Hoffman N, Fraley R T, Beachy R N. Plant Mol Biol. 1987;9:315–324. doi: 10.1007/BF00014906. [DOI] [PubMed] [Google Scholar]
- 25.Altenbach S B, Pearson K W, Meecker G, Staraci L C, Sun S S M. Plant Mol Biol. 1989;13:513–522. doi: 10.1007/BF00027311. [DOI] [PubMed] [Google Scholar]
- 26.Molvig L, Tabe L M, Eggum B O, Moore A E, Craig S, Spencer D, Higgins T J V. Proc Natl Acad Sci USA. 1997;94:8393–8398. doi: 10.1073/pnas.94.16.8393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nordlee J A, Taylor S L, Townsend J A, Thomas L A, Bush R K. N Engl J Med. 1996;334:688–692. doi: 10.1056/NEJM199603143341103. [DOI] [PubMed] [Google Scholar]
- 28.Cheeke P R, Bronson J. Proceedings of the Second Amaranth Conference. Emmaus, PA: Rodale; 1980. pp. 5–11. [Google Scholar]