Abstract
The expression patterns of five genes (PsGA20ox1, PsGA20ox2, PsGA3ox1, PsGA2ox1, and PsGA2ox2) encoding five regulatory gibberellin (GA) biosynthesis enzymes (two GA 20-oxidases, a GA 3β-hydroxylase, and two GA 2β-hydroxylases) were examined to gain insight into how these genes coordinate GA biosynthesis during germination and early postgermination stages of the large-seeded dicotyledonous plant pea (Pisum sativum). At the time the developing embryo fills the seed coat, high mRNA levels of PsGA20ox2 (primarily responsible for conversion of C20-GAs to GA20), PsGA2ox1 (primarily responsible for conversion of GA20 to GA29), and PsGA2ox2 (primarily responsible for conversion of GA1 to GA8) were detected in the seeds, along with high GA20 and GA29 levels, the enzymatic products of these genes. Embryo maturation was accompanied by a large reduction in PsGA20ox2 and PsGA2ox1 mRNA and lower GA20 and GA29 levels. However, PsGA2ox2 transcripts remained high. Following seed imbibition, GA20 levels in the cotyledons decreased, while PsGA3ox1 mRNA and GA1 levels increased, implying that GA20 was being used for de novo synthesis of GA1. The presence of the embryo axis was required for stimulation of cotyledonary GA1 synthesis at the mRNA and enzyme activity levels. As the embryo axis doubled in size, PsGA20ox1 and PsGA3ox1 transcripts increased, both GA1 and GA8 were detectable, PsGA2ox2 transcripts decreased, and PsGA2ox1 transcripts remained low. Cotyledonary-, root-, and shoot-specific expression of these GA biosynthesis genes and the resultant endogenous GA profiles support a key role for de novo GA biosynthesis in each organ during germination and early seedling growth of pea.
GAs are known to play important roles in germination of a wide range of dicotyledonous plant species (Koornneef and van der Veen, 1980; Groot and Karssen, 1987; Hilhorst and Karssen, 1988; Nambara et al., 1991; for review, see Bewley and Black, 1994; Sponsel and Hedden, 2004). In addition to weakening the endosperm and testa, which surrounds the radicle in certain seeds at maturity (Groot and Karssen, 1987; Debeaujon and Koornneef, 2000), GAs appear to control the growth of the embryo axis (Groot and Karssen, 1987), as well as growth of the rapidly developing shoot and root tissues (Reid et al., 2004).
These GA-mediated events in postimbibition germination and seedling growth are thought to be regulated, at least in part, by the modulation of tissue- and cell-specific GA levels and also by altering the ability of the cells to respond to GA (Richards et al., 2001). Monitoring the expression of genes encoding enzymes involved in GA biosynthesis and catabolism and quantitating the endogenous GA profile is a valuable approach for gaining insight into how higher plants regulate levels of the growth-active GAs, especially during germination and early seedling growth. For example, analysis of whole seed extracts of germinating Arabidopsis (Arabidopsis thaliana) seeds has revealed up-regulation of several GA biosynthesis genes (GA20ox and GA3ox) that encode enzymes responsible for synthesis of the growth-active GAs (Yamaguchi et al., 1998; Ogawa et al., 2003). This gene expression pattern led to a parallel detectable increase in the growth-active GA4 (Ogawa et al., 2003). The up-regulation of GA20ox and GA3ox occurs while there is minimal transcription of GA2ox genes, which encode 2β-hydroxylases that can reduce the pool of bioactive GA1 or GA4, or their immediate precursors, GA20 and GA9, respectively, during seed germination (Ogawa et al., 2003). Even so, the tissue-specific regulation and coordination of the expression of these gene families and the developmental changes of endogenous GAs within the embryo tissues during germination and early seedling growth are not well understood in any plant species.
Our approach in this study was to define the roles of GAs and the regulation of GA biosynthesis during germination and early seedling growth of the large-seeded dicot pea (Pisum sativum). Pea has been used as a model system for explaining the role of GAs in plant growth and development of large-seeded dicots (Reid et al., 2004; Sponsel and Hedden, 2004). Comparative developmental studies across a range of higher plant species are crucial since a number of physiological and morphological differences exist during germination and early seedling growth between large-seeded species (such as pea) and small-seeded species, including Arabidopsis and tomato (Lycopersicon esculentum). For example, the cotyledons of pea contain large storage reserves and pea also exhibits hypogeal germination (cotyledons of germinating seeds remain under the soil), compared to epigeal germination in many other model plants (including Arabidopsis and tomato, where the cotyledons emerge from the soil, expand, and carry out photosynthesis). In addition, the pea seed is nonendospermic at maturity and its seed coat does not act as a mechanical barrier for radical protrusion, like tomato and Arabidopsis seeds (Petruzzelli et al., 1995).
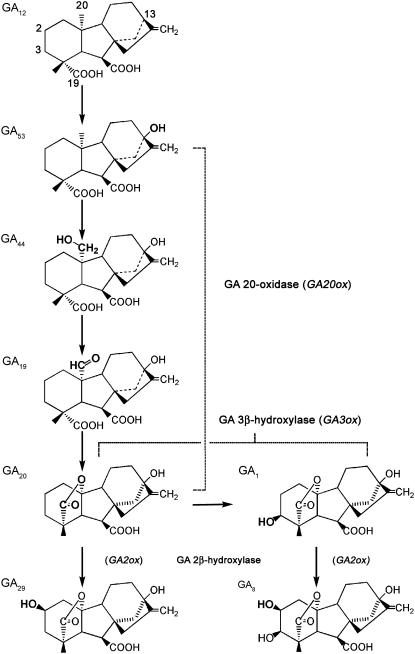
In the pea seedling, GA biosynthesis occurs via the early 13-hydroxylation pathway (Sponsel, 1995): GA12→GA53→GA44→GA19→GA20→GA1 (Fig. 1). Thus, GA 20-oxidases catalyze the conversion of GA53 via GA44 and GA19 to GA20, GA 3β-hydroxylase converts GA20 to bioactive GA1, and GA 2β-hydroxylases convert GA20 to biologically inactive GA29 and GA1 to biologically inactive GA8 (Hedden and Phillips, 2000).
Figure 1.
GAs in pea shoots and roots are synthesized via the early 13-hydroxylation pathway. GA12 is 13-hydroxylated to GA53; C20 is sequentially oxidized by a GA 20-oxidase from GA53 to GA44, to GA19, and finally to GA20. GA20 is then oxidized by a 3β-hydroxylase to GA1 (a growth-active GA). GA20 and GA1 can each be oxidized by a 2β-hydroxylase to GA29 and GA8, respectively, the latter conversion inactivating GA1.
GA biosynthesis inhibitor studies using compounds that inhibit several early steps in GA biosynthesis (the cyclization of geranylgeranyl diphosphate to ent-copalyl diphosphate: Sponsel, 1983; oxidation of ent-kaurene to ent-kaurenoic acid: Graebe, 1986) as well as later steps (3β-hydroxylation of GA20 to GA1; Ross et al., 1993) suggested that de novo GA biosynthesis was not essential for pea seed germination per se, but was, however, essential for the maintenance of normal seedling growth soon after germination (i.e. 4 d after imbibition [DAI]). Furthermore, the na mutation of the pea GA biosynthesis gene NA (PsKAO1, encodes ent-kaurenoic acid oxidase and is mainly expressed in vegetative tissues; Davidson et al., 2003) provides genetic evidence for the role of GAs in early pea vegetative growth. Thus, pea seeds with the na mutation germinate readily (the homolog PsKAO2 is highly expressed in the developing seeds, and this results in normal GA levels in na seeds; Potts and Reid, 1983; Davidson et al., 2003). Subsequently, though, expanding shoot tissues of na show dramatically decreased levels of the growth-active GA1 (Proebsting et al., 1992), and this results in severely dwarfed plants with extremely short internodes (Potts and Reid, 1983) and a markedly reduced taproot length (Yaxley et al., 2001). Additionally, Ross et al. (1993) have shown that GA biosynthesis occurs in shoots of rapidly growing young pea seedlings. The above data, taken together, strongly suggest that, following germination, de novo GA biosynthesis is required for normal pea seedling shoot and root growth.
Although these studies support the importance of GA biosynthesis in pea, the exact timing and tissue-specific nature of de novo GA biosynthesis during these early events in seedling shoot and root growth still are not known (in pea or in other plant species). Since bioactive GA1 in young seedling shoots of pea is low (Ross et al., 1992) and GAs can also be readily transported between tissues (Proebsting et al., 1992), profiling the expression patterns of the genes that code for the key regulatory GA biosynthesis enzymes, along with quantitation of the endogenous GAs in each organ of pea seeds and seedlings, would greatly increase our understanding of the spatial and temporal regulation of GA biosynthesis during germination and early seedling growth. Further, previous studies on the relationship of the embryo axis and the cotyledons in germinating seeds and seedlings of pea have indicated that the presence of the embryo axis is necessary for complete subcellular organization in the cotyledons (Bain and Mercer, 1966b). However, how the embryo axis presence affects GA biosynthesis in the cotyledons of germinating seeds is not known.
The following late GA biosynthesis genes have been identified and reported to be expressed in both vegetative and developing seeds of pea (with one exception): two GA20ox genes, PsGA20ox1 (Martin et al., 1996; Garcia-Martinez et al., 1997; van Huizen et al., 1997) and PsGA20ox2 (only reported in developing seeds; Ait-Ali et al., 1997); one GA3ox gene, PsGA3ox1 (Lester et al., 1997; Martin et al., 1997; Ozga et al., 2003); and two GA2ox genes, PsGA2ox1 and PsGA2ox2. PsGA2ox1 plays a major role in GA20 deactivation, while PsGA2ox2 is most likely important for GA1 deactivation in pea vegetative and reproductive tissues (Lester et al., 1999; Martin et al., 1999).
This study characterizes the expression pattern of these five key GA biosynthesis genes in developing seeds and mature embryos and in shoots, roots, and cotyledons of pea from 0 to 6 DAI. It also examines the coordination of gene expression among these GA gene family members during the critical stages for germination and seedling establishment in two distinctly different cultivars of pea (‘Alaska,’ a model cultivar for tall [LE] vining pea, and ‘Carneval,’ a model cultivar for semidwarf [le], semileafless field pea). Further, it tests if the embryo axis regulates GA biosynthesis in the cotyledons of germinating seeds. Real-time reverse transcription (RT)-PCR, the current method of choice for obtaining sensitive, specific, and reproducible quantification of mRNA (Bustin, 2000), was used to quantify gene expression. To coordinate gene expression patterns with product levels, endogenous GAs that are products of enzymes coded for by these GA biosynthesis genes were also quantified by gas chromatography-mass spectrometry-selected ion monitoring (GC-MS-SIM) using the stable isotope dilution method. Additionally, enzyme activities of GA 3-oxidase and GA 2-oxidases were monitored in the cotyledons by profiling the metabolism of exogenous [14C]GA20.
RESULTS AND DISCUSSION
GA Biosynthesis in Developing Seeds and Mature Embryos
High levels of mRNA were detected for PsGA20ox2, PsGA2ox1, and PsGA2ox2 in developing seeds 20 d after anthesis (DAA; Table I). These are the genes that encode the enzymes that convert GA53 to GA20 (GA 20-oxidase), and GA20 to GA29 and/or GA1 to GA8 (GA 2β-hydroxylases), respectively (Fig. 1). Additionally, high levels of GA20 and GA29 were also present (Table II). In contrast, the transcript abundance of PsGA20ox1 was low in the developing seeds (approximately 3,000-fold lower than PsGA20ox2). This suggests that PsGA20ox2 codes for the majority of the GA 20-oxidase that is responsible for the biosynthesis of GA20 in the developing seeds (see also Ait-Ali et al., 1997). PsGA3ox1 mRNA levels were also low, and growth-active GA1 as well as its immediate biologically inactive catabolite, GA8, were not detected in the 20-DAA developing seeds (Tables I and II). These data are consistent with previous gene expression studies (Ait-Ali et al., 1997; Lester et al., 1999), with feeding experiments that indicate that the developing pea cotyledon is a site for 2β-hydroxylation of GA20 into GA29 (Sponsel, 1983) and with an emerging hypothesis that levels of growth-active GA are minimized in the developing embryo to allow for seed maturation processes to proceed (Gazzarrini et al., 2004). Curaba et al. (2004) found that the embryonic transcription regulators LEC2 and FUS3, involved in multiple aspects of Arabidopsis seed development (including repression of leaf traits and premature germination, and activation of seed storage protein genes), down-regulate AtGA3ox2 gene expression, thereby resulting in lower levels of growth-active GA4 and GA1 in the maturing embryos (as determined using the lec2 and fus3 Arabidopsis mutants).
Table I.
Relative transcript levels of PsGA20ox1, PsGA20ox2, PsGA3ox1, PsGA2ox1, and PsGA2ox2 in developing seeds, mature embryos, and 0.5- and 1-DAI embryo axes and cotyledonsa
| Gene | Developing Seeds 20 DAA | Mature Embryos 0 DAI | Cotyledons
|
Embryo Axis
|
||
|---|---|---|---|---|---|---|
| 0.5 DAI | 1 DAI | 0.5 DAI | 1 DAI | |||
| ‘Alaska’ | ||||||
| PsGA20ox1 | 9.5 ± 4.8b | 1.1 ± 0.3 | 2.1 ± 1.1 | 28.6 ± 17.3 | 66.8 ± 24.7 | 870.7 ± 106.3 |
| PsGA20ox2 | 28,848.3 ± 2,623.8 | 43.3 ± 4.9 | 91.6 ± 29.3 | 145.4 ± 74.4 | 5.9 ± 1.0 | 25.3 ± 1.7 |
| PsGA3ox1 | 301.1 ± 126 | 7.2 ± 0.3 | 8.0 ± 4.4 | 183.8 ± 76.1 | 354.8 ± 79.4 | 5,361.8 ± 973.1 |
| PsGA2ox1 | 15,234.9 ± 2,332.8 | 401.9 ± 7.1 | 533.2 ± 153.6 | 111.1 ± 14.9 | 14.0 ± 2.3 | 15.1 ± 7.0 |
| PsGA2ox2 | 4,973.6 ± 755.7 | 3,313.2 ± 701.9 | 5,518.2 ± 867.2 | 2,193.9 ± 940.8 | 8,235.6 ± 971.4 | 2,250.7 ± 1,145.5 |
| ‘Carneval’ | ||||||
| PsGA20ox1 | 1.0 ± 0.0 | 0.8 ± 0.7 | 14.9 ± 14.3 | 46.3 ± 22.5 | 768.0 ± 79.2 | |
| PsGA20ox2 | 338.3 ± 217.3 | 180.9 ± 143.6 | 1,098.9 ± 472.5 | 4.9 ± 0.7 | 7.6 ± 0.7 | |
| PsGA3ox1 | 4.6 ± 0.6 | 48.9 ± 40.1 | 44.8 ± 32.1 | 689.2 ± 308.3 | 4,312.4 ± 606.9 | |
| PsGA2ox1 | 2,259.6 ± 433.4 | 549.6 ± 193.5 | 1,441.6 ± 51.3 | 31.3 ± 14.4 | 31.8 ± 1.9 | |
| PsGA2ox2 | 19,668.1 ± 5,228.3 | 4,038.0 ± 1,849.6 | 2,436.1 ± 1,020.2 | 6,896.0 ± 1,672.5 | 1,687.6 ± 309.1 | |
Transcript levels were compared across genes, genotypes, developmental stages, and tissues using the average of mature embryo PsGA20ox1 samples of ‘Carneval’ as a reference for normalization.
Data are means ± se, n = 2 to 3.
Table II.
Endogenous GAs in developing seeds, mature embryos, embryonic axes, and cotyledons of 1-DAI seeds, and in the cotyledons, shoots, and roots of young seedlings of ‘Alaska’
| Tissue | DAI | GA19 | GA20 | GA29 | GA1 | GA8 |
|---|---|---|---|---|---|---|
| ng gfw−1 | ||||||
| 20-DAA seed | 21.8 ± 3.9a | 445.9 ± 96.3 | 188.7 ± 34.0 | ndb | nd | |
| Mature embryo | 0 | 1.8c | 22.0 ± 4.0 | 84.0 ± 1.2 | <0.3d | 0.5 ± 0.2 |
| Cotyledon | 1 | 1.1 ± 0.1 | 9.8 ± 0.7 | 23.3 ± 1.4 | 1.5 ± 0.1 | 0.7 ± 0.2 |
| Cotyledon | 2 | <0.3 | 3.2 ± 0.3 | 39.2 ± 18.5 | nd | 0.6 ± 0.0 |
| Cotyledon | 4 | <0.4 | <0.4 | 20.6 ± 0.7 | nd | 1.0 ± 0.2 |
| Embryo axis | 1 | 3.7 ± 0.2 | 11.6 ± 2.2 | 148.6 ± 27.0 | 1.2 ± 0.1 | 10.1 ± 0.9 |
| Shoot | 2 | 2.5 ± 0.1 | 2.3 ± 0.3 | 145.8 ± 10.0 | 1.0 ± 0.1 | 14.8 ± 0.2 |
| Shoot | 4 | 2.0 ± 0.0 | 0.6 ± 0.0 | 93.2 ± 20.5 | 1.0 ± 0.0 | 14.3 ± 2.2 |
| Root | 2 | 2.7 ± 0.2 | 2.7 ± 0.2 | 137.7 ± 9.7 | 0.7 ± 0.0 | 10.8 ± 0.1 |
| Root | 4 | 1.3 ± 0.2 | 0.9 ± 0.0 | 67.5 ± 4.0 | 0.7 ± 0.1 | 6.2 ± 0.1 |
| pg organ−1 | ||||||
| 20-DAA seed | 9,570 ± 1,700 | 196,330 ± 42,410 | 83,080 ± 14,960 | nd | nd | |
| Mature embryo | 0 | 420 | 5,190 ± 933 | 19,850 ± 285 | <70 | 110 ± 52 |
| Cotyledon pair | 1 | 420 ± 36 | 3,610 ± 267 | 8,550 ± 520 | 540 ± 22 | 260 ± 56 |
| Cotyledon pair | 2 | <110 | 1,310 ± 126 | 15,870 ± 7,500 | nd | 240 ± 9 |
| Cotyledon pair | 4 | <180 | <170 | 9,270 ± 295 | nd | 430 ± 102 |
| Embryo axis | 1 | 40 ± 2 | 140 ± 26 | 1,740 ± 316 | 10 ± 1 | 120 ± 10 |
| Shoot | 2 | 70 ± 2 | 60 ± 8 | 3,940 ± 271 | 30 ± 2 | 400 ± 4 |
| Shoot | 4 | 150 ± 3 | 50 ± 1 | 6,670 ± 1,465 | 70 ± 2 | 1,020 ± 156 |
| Root | 2 | 140 ± 10 | 130 ± 9 | 6,940 ± 489 | 40 ± 0 | 540 ± 3 |
| Root | 4 | 120 ± 14 | 80 ± 4 | 6,390 ± 378 | 70 ± 5 | 590 ± 8 |
Data are means ± se, n = 2.
No endogenous GA was detected in either replicate tissue sample, although the stable isotope-labeled GA internal standard was found.
One tissue sample only.
No endogenous GA was detected in one of the two samples.
Further maturation of the pea embryo resulted in a large reduction in PsGA20ox2 (665-fold) and PsGA2ox1 (38-fold), but there was no significant change in PsGA2ox2 mRNA levels. This finding is consistent with the decreased levels of GA20 (20-fold) and GA29 (2.3-fold) that we observed in the embryo at maturity (Tables I and II). We would suggest that the high levels of PsGA2ox2 mRNA in 20 DAA and mature embryos, as well as in the 0.5-DAI embryo axis (Table I), and the apparent preference of PsGA2ox2 for GA1 as a substrate (Lester et al., 1999) indicate a key role for GA 2-oxidases in the later phase of seed development, thereby assuring minimal levels of growth-active GA1. This would allow the seed to complete normal maturation and possibly aid in prevention of precocious germination. Synthesis of GA1 and GA8 in the embryo during the latter stage of pea seed development was very low or not detectable (Table II; Sponsel, 1983), and these GAs do not accumulate in the mature embryo (Table II; Ross et al., 1993).
GA Biosynthesis in the Cotyledonary Tissue during Germination and Early Seedling Growth
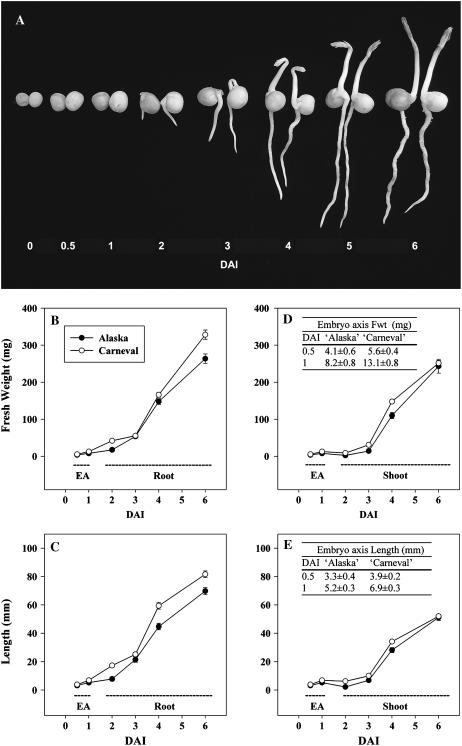
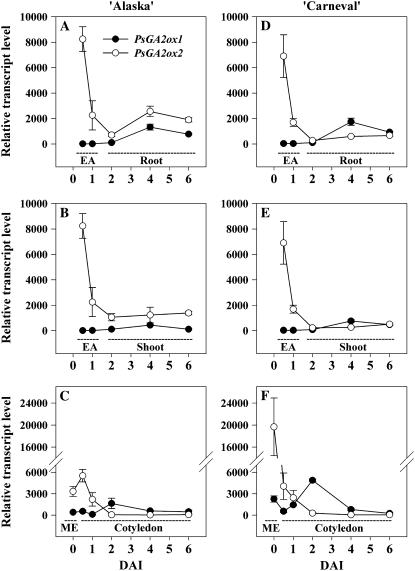
During the first 0.5 DAI, mature air-dried seeds of ‘Carneval’ absorbed more water (1.7-fold) than ‘Alaska’ (45.7 and 26.6% relative water content [RWC] at 0.5 DAI, respectively; Fig. 2). At 1 DAI, the RWC of cotyledons from both cultivars was similar (50% in ‘Alaska’ and 55% in ‘Carneval’), and the RWC gradually increased from 1 to 6 DAI, reaching 67% to 68% by 6 DAI in both cultivars. The transcript levels of GA20ox, GA3ox, and GA2ox genes in the cotyledons remained relatively constant in ‘Alaska’ (Table I; Figs. 3C, 4C, and 5C) during the first 0.5 DAI. ‘Carneval’ cotyledons exhibited a 4- to 5-fold decrease in PsGA2ox1 and PsGA2ox2 mRNA levels during their first half-day of imbibition. The much higher levels of these transcripts in the mature seeds of ‘Carneval’ relative to those in ‘Alaska’ are a likely reason for the large decline in their abundance observed during the first half-day of ‘Carneval’ seed imbibition (Table I; Fig. 5F). By 1 DAI, the RWC of ‘Alaska’ cotyledons had increased to 50%, the transcript abundance of PsGA2ox1 had decreased (3.6-fold; Table I), and, coincidentally, the level of GA29 decreased (3.6-fold; Table II).
Figure 2.
Germinating pea seeds and actively growing seedlings of ‘Alaska’ (left in each pair) and ‘Carneval’ (right in each pair) from mature dry seed (0 DAI) to 6 DAI (A). Embryo axis (EA) FW and length from 0.5 to 1 DAI are given in the tables in D and E, respectively. Root FW (B) and length (C) and shoot FW (D) and length (E) are shown for 2 to 6 DAI. Data are means ± se, n = 15 to 28.
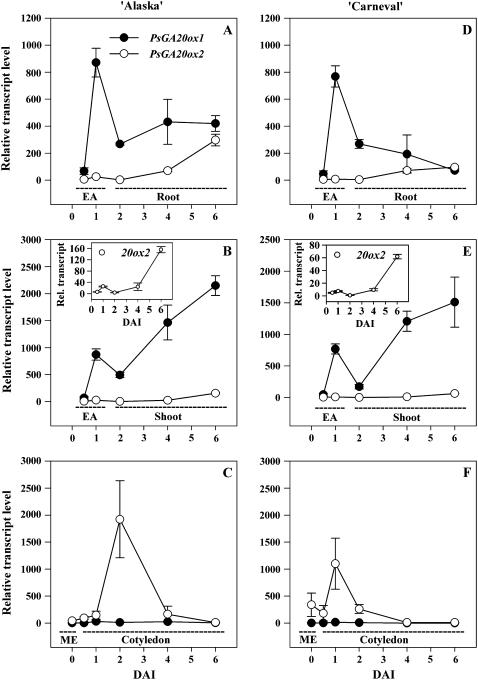
Figure 3.
Relative transcript levels of two GA 20-oxidase genes, PsGA20ox1 and PsGA20ox2, during seed germination and early seedling growth of ‘Alaska’ (A–C) and ‘Carneval’ (D–F). Relative transcript levels were determined in mature embryos (ME; C and F), embryo axes (EA; 0.5 and 1 DAI; A, B, D, and E), roots (2–6 DAI; A and D), shoots (2–6 DAI; B and E), and cotyledons (0.5–6 DAI; C and F) of the imbibed pea seed and growing seedling. Inset graphs in B and E are the relative transcript levels of PsGA20ox2 using a smaller y axis scale. Transcript levels were compared across all genes, genotypes, developmental stages, and tissues using the average Ct value of PsGA20ox1 from the mature embryo of ‘Carneval’ (Ct = 34.2 ± 0.1) as the reference sample. Data are means ± se, n = 2 to 3.
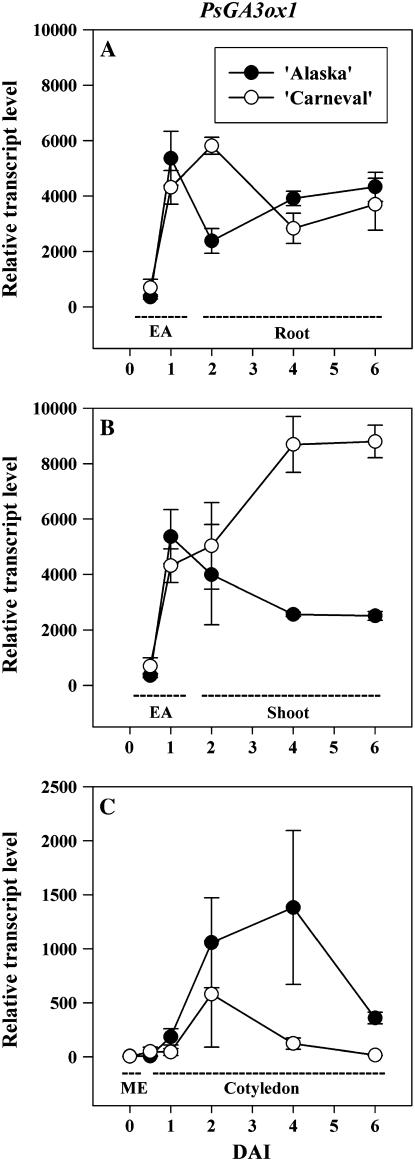
Figure 4.
Relative transcript levels of the GA 3-oxidase gene PsGA3ox1 during seed germination and early seedling growth of ‘Alaska’ and ‘Carneval.’ Relative mRNA transcript levels were determined in mature embryos (ME; C), embryo axes (EA; 0.5 and 1 DAI; A and B), roots (2–6 DAI; A), shoots (2–6 DAI; B), and cotyledons (0.5–6 DAI; C) of pea. Transcript levels were compared as described for Figure 3. Data are means ± se, n = 2 to 3.
Figure 5.
Relative mRNA transcript levels of two GA 2-oxidase genes, PsGA2ox1 and PsGA2ox2, during seed germination and early seedling growth of ‘Alaska’ (A–C) and ‘Carneval’ (D–F). Relative mRNA transcript levels were determined in the mature embryos (ME; C and F), embryo axes (EA; 0.5 and 1 DAI; A, B, D, and E), roots (2–6 DAI; A and D), shoots (2–6 DAI; B and E), and cotyledons (0.5–6 DAI; C and F) of pea. Transcript levels were compared as described for Figure 3. Data are means ± se, n = 2 to 3.
The decrease in 2β-hydroxylation of GA20 to GA29 in cotyledons at 1 DAI, together with the increase in transcription of PsGA3ox1 (25-fold; Table I) and in the production of GA1 (5-fold; Table II), suggests that the cotyledonary GA20 serves as substrate for in situ 3β-hydroxylation into bioactive GA1 (note that GA20 levels also decreased 2.3-fold by 1 DAI; Table II). Although bioactive GAs have a well-defined role in coordinating mobilization of the reserve materials in cereals (Jacobsen et al., 1995), their possible role in mobilization of cotyledonary reserves in dicots remains unclear (Bewley and Black, 1994). The growth-active GA1 in pea cotyledons would not be involved in subsequent growth or photosynthetic processes as the hypogeal cotyledons do not develop into green leaf-like structures that photosynthesize and their cells have a short but normal life span of 2 to 3 weeks (Smith and Flinn, 1967). However, since cell wall loosening enzymes have been shown to be GA inducible during germination of other species (tomato and Arabidopsis; Chen et al., 2001; Ogawa et al., 2003), the bioactive GA1 present in these cotyledons could be necessary for promoting the formation of the large reticulum of intercellular spaces in this tissue during germination (Smith and Flinn, 1967). The increase in cotyledonary intercellular spaces could increase the oxygen diffusion rates through the tissue to support the high respiratory activity of the cotyledonary storage parenchyma cells during storage mobilization (Bain and Mercer, 1966a). Transport of GA20 from the cotyledon to the embryo axis to support synthesis of growth-active GA1 for embryo axis expansion also can occur (Ross et al., 1993).
Radicle protrusion occurred between 1 and 2 DAI (Fig. 2A) and it was accompanied by an increase in the expression of cotyledonary PsGA20ox2 (13-fold in ‘Alaska’; Fig. 3C) and PsGA3ox1 (6-fold in ‘Alaska’; Fig. 4C), together with a decrease in cotyledonary GA20 and GA1 levels (Table II). The cotyledonary PsGA20ox2 mRNA levels that we observed (Fig. 3C) were not consistent with GA feedback regulation, i.e. PsGA20ox2 transcripts increased sharply from 1 to 2 DAI then decreased by 4 DAI, while during this same period GA20 levels decreased from 1 to 4 DAI (Table II). Cotyledonary PsGA3ox1 mRNA levels increased during the first DAI, at the same time endogenous GA1 levels increased (‘Alaska’; Tables I and II). Subsequently, though, from 1 to 2 DAI, PsGA3ox1 transcript abundance in both cultivars continued to increase, while cotyledon GA1 content decreased to undetectable levels (Table II; Fig. 4C). Although feedback up-regulation of PsGA3ox1 transcription by low levels of bioactive GA could be occurring in the cotyledons after 1 DAI (‘Alaska’), the increase in cotyledonary PsGA20ox2 and PsGA3ox1 transcript levels observed during or soon after radicle protrusion in both cultivars (1–2 DAI; Fig. 4C) suggested that a signal from the embryo axis may induce expression of GA biosynthesis genes in the cotyledon. Indeed, Bain and Mercer (1966b) found that the presence of the axis is required for 2 d following imbibition if complete subcellular organization of the cotyledon is to occur.
Effect of the Embryo Axis on GA Biosynthesis in the Cotyledon
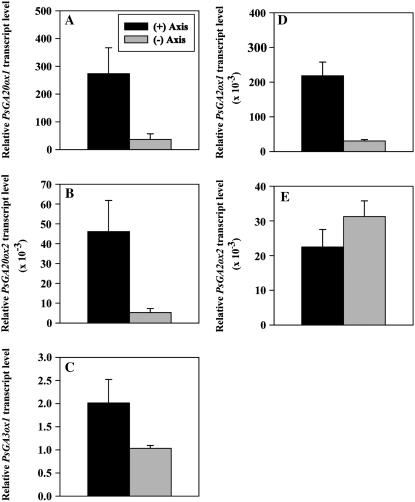
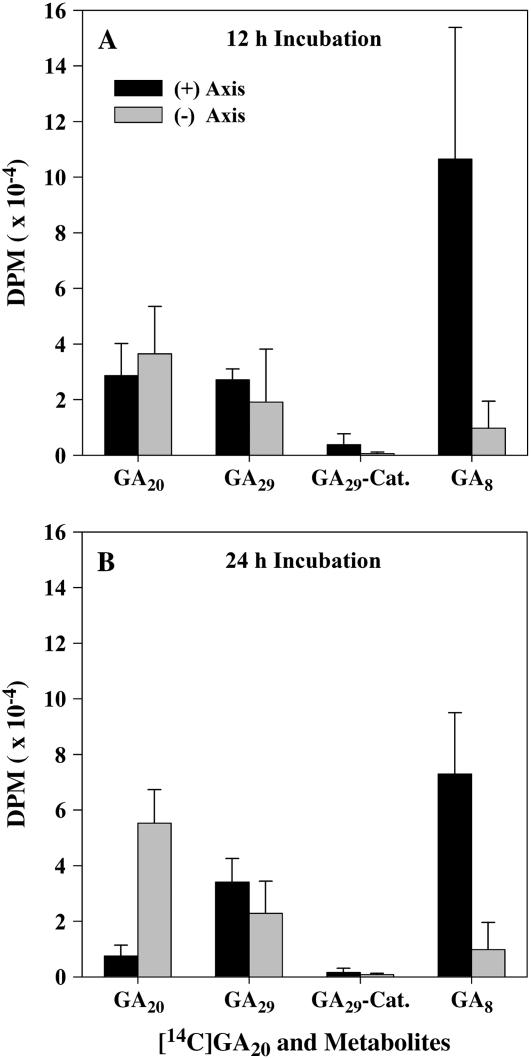
To examine whether the presence of the embryo axis is required to induce the expression of GA biosynthesis genes and, in turn, the metabolism of GAs in the cotyledons of germinating pea seeds, expression of PsGA20ox1, PsGA20ox2, PsGA3ox1, PsGA2ox1, and PsGA2ox2 and metabolism of [14C]GA20 were compared in the cotyledons of ‘Alaska’ imbibed for 2 d with or without the presence of the embryo axis (axis excised within 2 h after imbibition). Removal of the embryo axis from the cotyledons 2 h after imbibition reduced the transcript abundance of cotyledonary PsGA20ox1 (3-fold), PsGA20ox2 (9-fold), PsGA3ox1 (2-fold), and PsGA2ox1 (7-fold), but had no effect on PsGA2ox2 transcript levels after 2 d of imbibition (Fig. 6). Embryo axis removal also resulted in more than a 7-fold reduction in the conversion of [14C]GA20 to [14C]GA8 in the 2-DAI cotyledons (Fig. 7). Since PsGA2ox2 transcript levels were not affected by embryo axis removal, the reduction in conversion of [14C]GA20 to [14C]GA8 is likely due to the lower transcript abundance of cotyledonary PsGA3ox1 (Fig. 6) leading to reduced GA 3β-hydroxylase activity, and then less [14C]GA1 substrate for conversion to [14C]GA8.
Figure 6.
Relative transcript levels of PsGA20ox1 (A) and PsGA20ox2 (B), PsGA3ox1 (C), and PsGA2ox1 (D) and PsGA2ox2 (E) in ‘Alaska’ cotyledons imbibed for 2 d with or without the embryo axis. Transcript levels were compared across genes and tissues using the average Ct value of PsGA3ox1 from the cotyledon with no axis sample (Ct = 37.5 ± 0.1) as a reference for normalization. Data are means ± se, n = 3.
Figure 7.
Amount (dpm) of [14C]GA20 and its metabolites, [14C]GA29, [14C]GA29-catabolite, and [14C]GA8, detected over a 12-h (A) or 24-h (B) period in ‘Alaska’ cotyledons imbibed for 2 d with or without the embryo axis. Data are means ± se, n = 2 to 3. [14C]GA29 and [14C]GA8 identities were confirmed by GC-MS-SIM analysis. The characteristic MS fragment ions at the correct retention times for both [14C]GA29 and [14C]GA8 samples were identified: [14C]GA29 [M+506, 100; 508, 61; 491, 16; 493, 10; KRI, 2720]; GA29 standard [M+506, 100; 508, 15; 491, 12; 493, 2; KRI, 2718]; [14C]GA8 [M+594, 86; 596, 100; 579, 10; 581, 11; KRI, 2844]; and GA8 standard [M+594, 100; 596, 34; 579, 11; 581, 2.5; KRI, 2845]. The intensity of the M+2 ions for both [14C]GA29 and [14C]GA8 samples was significantly above the natural abundance of the M+2 ion in their respective protio-GA standards (46% greater for [14C]GA29 and 86% greater for [14C]GA8 samples as calculated using the M+ and M+2 ions), identifying the presence of 14C-GA29 and 14C-GA8 in these samples. [14C]GA29-catabolite identity is putative as it is based on HPLC retention times of both its free and methyl-ester forms.
GA Biosynthesis in the Embryo Axis
Consistent with a faster initial rate of water imbibition, initiation of radicle protrusion was observed in 53% of the ‘Carneval’ seeds by 1 DAI, and the percentage of germinated seeds increased to 77% by 1.5 DAI. In contrast, only 27% of ‘Alaska’ seeds exhibited radicle emergence at 1.5 DAI. Germination percentages then increased to 90% in ‘Carneval’ and 65% in ‘Alaska’ by 2 DAI.
From 0.5 to 1 DAI, embryonic-axis fresh weight (FW) doubled and axis length increased 1.6-fold for both cultivars (Fig. 2, D and E, see inset tables). During the same period, transcript abundance increased markedly for embryo-axis-derived PsGA20ox1 (13-fold in ‘Alaska’) and also for PsGA3ox1 (15-fold in ‘Alaska’; Table I; Figs. 3 and 4). These large increases in gene expression of the GA20ox and GA3ox biosynthesis genes in the embryo axis occur concomitantly with a substantial decrease in PsGA2ox2 transcript abundance (approximately 4-fold) and maintenance of low levels of PsGA2ox1 transcript (Table I; Fig. 5). These data suggest an increased capacity to synthesize and maintain growth-active GA1 in the embryo axis for the rapid expansion that occurs soon after imbibition. The presence of GA20, GA1, and GA8 in the 1-DAI embryo axis is supportive of this hypothesis, especially since GA1 levels were much lower in the mature embryo (0 DAI) than in the 1-DAI embryo axis (Table II).
Increases in GA20ox (AtGA20ox1 and AtGA20ox3), GA3ox (AtGA3ox1 and AtGA3ox2) transcript abundance, and also in the levels of the growth-active GA4 were observed after imbibition of wild-type Arabidopsis seeds (Ogawa et al., 2003). However, as whole seed extracts were analyzed, their data did not address the possibility of tissue specificity for GA biosynthesis gene expression (and, thus, GA production) during the seed germination process or during early seedling development.
GA Biosynthesis in Young Seedling Roots and Shoots
Following seed germination (which occurred approximately 2 DAI; Fig. 2A), the seedling plumules of both cultivars began to appear (4 DAI), and by 5 DAI the shoots of 73% of ‘Alaska’ and 88% of ‘Carneval’ seedlings had emerged. Complete emergence of all germinated seedlings had taken place by 6 DAI for both cultivars. Root FW and length generally exceeded that of the shoot from 2 to 6 DAI for both cultivars (Fig. 2). Finally, both cultivars had similar patterns of shoot and root growth, with a slower growth rate from 2 to 3 DAI followed by a higher rate of growth from 3 to 6 DAI (Fig. 2).
Transcript levels in the shoot for PsGA20ox1 were more abundant (141-fold in ‘Alaska’) than for PsGA20ox2 at 2 DAI, and they increased markedly from 2 to 6 DAI as the shoot elongated rapidly (Figs. 2, D and E, and 3, B and E). Concomitant with these increases in transcript level of PsGA20ox1, endogenous levels of GA20 decreased 5-fold in the embryo axis from 1 to 2 DAI, and GA20 levels continued to decrease from 2 to 4 DAI in the shoot (3.5-fold; Table II). Since the abundance of PsGA20ox1 mRNA was shown to be regulated by the levels of growth-active GA in the shoots of ‘Alaska’ seedlings (application of GA3 reduced and application of GA biosynthesis inhibitor Prohexadione increased PsGA20ox1 mRNA levels; Ayele et al., 2006), these data suggest that the large increase in shoot-derived PsGA20ox1 mRNA is likely due to a feedback regulation mechanism, i.e. as a result of low growth-active GA1 at this stage of rapid shoot growth.
Since the expression of shoot-derived PsGA2ox1 remained low (Fig. 5B) and the GA29 concentration (ng g−1 FW) actually decreased in the shoot from 2 to 4 DAI (Table II), we conclude that the reduction in shoot GA20 levels is not a result of increased 2β-hydroxylation of GA20 to GA29. Rather, the ability of the shoot to maintain a stable but relatively low level of PsGA3ox1 transcripts and a low level of GA1 (ng g−1 FW; see Fig. 4B and Table II) in the pea shoot from 2 to 4 DAI in ‘Alaska’ (Fig. 4B; Table II), while GA8 levels remain stable and relatively high (Table II), suggests that a significant portion of the pool of GA20 in the young shoot is being used as a substrate for synthesis of GA1, but the GA1 is then rapidly converted to GA8 in situ. It is also possible that some shoot-derived GA20 was transported to the root (Proebsting et al., 1992) for subsequent synthesis of GA1 in the root tissue. Shoot-derived PsGA2ox2 transcript abundance was also maintained at the same level from 2 to 6 DAI in both cultivars (Fig. 5, B and E), and that is also in agreement with the maintenance of relatively high and constant levels of GA8 in the ‘Alaska’ shoots from 2 to 4 DAI (Table II). Root growth was rapid by 2 DAI and transcript levels of PsGA20ox1 were 60- to 120-fold higher than for PsGA20ox2 (Fig. 3, A and D). Also, PsGA3ox1 (Fig. 4A) transcripts were abundant in the rapidly growing roots. As was the case for shoot tissues, GA20 levels in the roots decreased during this rapid growth phase (Table II). Feedback regulation of mRNA levels in the roots by an applied growth-active GA has been observed for PsGA20ox1 (although not for PsGA20ox2) during the 4- to 6-DAI period (Ayele et al., 2006). However, in the present experiment, as the roots continued to grow and mature (4–6 DAI), PsGA20ox1 transcript levels either remained the same (‘Alaska’) or declined (‘Carneval’), and transcripts of PsGA20ox2 increased to levels similar to that of PsGA20ox1 by 6 DAI (Fig. 3, A and D). These data suggest that roots, which are more sensitive to applied GAs (Tanimoto, 1990) and contain lower levels of GA1 and GA8 than shoots (ng g−1 FW; Table II), may limit endogenous GA1 production by maintaining lower GA20ox transcript levels than are observed in the shoot (Figs. 3, A, B, D, and E). We should also note that our trends in mRNA abundance of GA biosynthesis genes were also similar to those of Lange et al. (2005), where CmGA20ox3 expression in young pumpkin (Cucurbita pepo) seedlings was higher in the shoot tip and hypocotyl than in the root and root tip.
PsGA3ox1 (LE) transcript levels in the roots were maintained at moderately high levels as the roots of both cultivars grew from 2 to 6 DAI (Fig. 4A; Martin et al., 1997), and for our pea seedlings GA1 levels remained constant and relatively low from 2 to 4 DAI (Table II). Yaxley et al. (2001) found that roots from 12-d-old pea plants isogenic for LE and le-1 had similar levels of endogenous GA1 and similar root lengths as did a second pair of isogenic lines for the null mutation le-2 (truncated le-1; Martin et al., 1997) and le-1 (Yaxley et al., 2001). As a result, Yaxley et al. (2001) proposed that another GA3ox gene exists in pea roots that can substitute for the loss of LE. Our data are consistent with the hypothesis that LE does contribute to the mRNA pool for GA3ox in the root, but it does not rule out the possibility that another GA3ox gene may also be expressed in pea roots, especially when PsGA3ox1 mRNA levels are reduced.
To further localize expression of GA biosynthesis genes in the root, we analyzed the expression pattern of the GA biosynthesis genes in root tips (4-mm apex, which constitutes about 2.4%–3.3% of the total root FW) and also in the remaining part of the root from 6-DAI seedlings. PsGA20ox2 transcript abundance was similar to that of PsGA20ox1 in the more mature root tissue, but was markedly lower (12- to 24-fold lower) in root tips for both cultivars (Table III). In ‘Alaska’ pea roots, the first 2 to 3 mm of the root apex consists of the root cap and the root meristem, which contains mostly dividing cells. Behind this zone of cell division is a zone consisting of mainly elongating and differentiating cells (from 2–3 to 11 mm or greater; Rost and Baum, 1988; Rost et al., 1988). The root-specific expression pattern of the GA20ox genes suggests that PsGA20ox1 is the major gene for GA20 synthesis in the root tip's dividing cells, while PsGA20ox1 and PsGA20ox2 transcripts both contribute to the GA20ox transcript pool in more mature root tissues. A similar expression pattern for PsGA3ox1 was evident between the root and root tip at 6 DAI (Table III), which is consistent with the pattern seen in developing pumpkin seedlings (7 d old), where expression of CmGA3ox3 was similar between the root and root tip tissues (Lange et al., 2005).
Table III.
Relative transcript levels of PsGA20ox1, PsGA20ox2, PsGA3ox1, PsGA2ox1, and PsGA2ox2 in 6-DAI roots and root tips of ‘Alaska’ and ‘Carneval’
| Tissue | PsGA20ox1 | PsGA20ox2 | PsGA3ox1 | PsGA2ox1 | PsGA2ox2 |
|---|---|---|---|---|---|
| ‘Alaska’ | |||||
| Roota | 419.4b ± 58.4c | 296.6 ± 43.1 | 4,332.6 ± 523.0 | 771.0 ± 54.4 | 1,896.8 ± 174.8 |
| Root tipd | 1,031.4 ± 5.6 | 11.0 ± 5.4 | 4,668.5 ± 1,088.3 | 587.6 ± 34.6 | 3,032.1 ± 1,091.5 |
| ‘Carneval’ | |||||
| Root | 71.8 ± 5.7 | 96.3 ± 6.8 | 3,700.2 ± 936.3 | 935.3 ± 52.6 | 665.7 ± 50.7 |
| Root tip | 82.3 ± 11.8 | 6.5 ± 2.3 | 2,807.1 ± 132.2 | 1,044.5 ± 5.9 | 1,522.5 ± 97.4 |
Remaining portion of the 6-DAI root after 4 mm of its tip is removed.
Transcript levels were compared across genes, genotypes, developmental stages, and tissues using the average of mature embryo PsGA20ox1 samples of ‘Carneval’ as a reference for normalization.
Data are means ± se, n = 2 to 3.
Four-millimeter root tip.
Cultivar-Specific GA Gene Expression
The relatively similar GA gene expression patterns between the two different genotypes indicate the general nature (and likely importance) of the spatial and temporal regulation of these GA biosynthesis genes in facilitating the establishment of the pea seedling in the first few days after germination. Specific expression pattern differences between the genotypes for the GA3ox and GA2ox genes do occur, and these seem likely to be related to the LE (PsGA3ox1) gene. ‘Carneval’ carries the le-1 mutation (Ayele, 2006), and the GA 3β-hydroxylase enzyme coded by the le-1 gene appears to be substantially less efficient than the wild-type enzyme (Lester et al., 1997; Martin et al., 1997). In ‘Carneval’ shoots, the PsGA3ox1 mRNA levels were approximately 3-fold greater than those in ‘Alaska’ (LE) shoots from 4 to 6 DAI (shoots were in the dark from 2–4 DAI since emergence from the potting medium did not begin until 4 DAI; Fig. 4B). The greater abundance of PsGA3ox1 transcripts in the shoots of ‘Carneval’ may be a result of negative feedback control of GA3ox expression, as has been observed in a number of other studies (Hedden and Phillips, 2000). Overall, our data are consistent with the view that pea shoots are dependent on PsGA3ox1 transcripts (LE gene) for conversion of GA20 to growth-active GA1, the latter GA being causal for shoot elongation in pea (Ingram et al., 1984; Ross et al., 1992).
Additionally, we found that from 2 to 6 DAI, PsGA2ox2 transcript abundance is lower in the actively growing shoots and roots of ‘Carneval’ (le-1) than in ‘Alaska’ (LE; Fig. 5, A, B, D, and E). This suggests that one mechanism to compensate for the reduction in enzyme efficiency of the GA 3β-hydroxylase coded for by le-1 (PsGA3ox1 results in lower levels of GA1 in shoots of le-1 compared to wild-type LE; Ross et al., 1993) is the reduction in levels of PsGA2ox2 transcripts. PsGA2ox2 has been implicated as playing a major role in GA1 deactivation in the pea shoot (Lester et al., 1999).
In summary, although embryo development is a complex process and the expression patterns of these GA biosynthesis genes and GA1 content are not necessarily the causal factors for specific developmental events, our findings support the emerging hypothesis that endogenous growth-active GA (GA1 in pea) is minimized in the developing embryo to allow for seed maturation processes to proceed. However, GA gene expression and endogenous GA profiles are consistent with previous findings that GA20 (the immediate precursor to the growth-active GA1) is sequestered in the developing pea embryo and significant amounts of GA20 exist in the embryo at maturity. Additionally, high PsGA2ox2 message levels in the mature quiescent embryo and in the embryo axis at 0.5 DAI likely reflect a mechanism where growth-active GA1 is maintained at minimal levels to prevent embryo axis expansion during the later phases of seed maturation and/or under nonoptimal germination conditions.
During seed imbibition, the expression pattern of this suite of GA biosynthesis genes and the concomitant levels of endogenous GAs suggest that pea cotyledons are serving as a reservoir of GA20 (both preexisting and newly synthesized), which is then used as a substrate for GA 3β-hydroxylase in situ or in the embryo axis after transport from cotyledons. The growing embryo axis regulates GA biosynthesis in the cotyledons by increasing the transcript abundance of cotyledonary GA biosynthesis genes PsGA20ox1, PsGA20ox2, and PsGA3ox1 (but not PsGA2ox2), leading to increased conversion of GA20 to GA8 via GA1.
As the embryo axis initiates growth (by 1 DAI), a dramatic change in the expression patterns of these regulatory GA biosynthesis genes occurs in the axis tissue, providing the embryo axis with a very much increased capacity to produce growth-active GA1 for axis expansion. In the rapidly growing young seedling (2–6 DAI), both shoots and roots display unique expression patterns, which likely provide for coordination of GA biosynthesis within and between these organs. Overall, our results show that coordination of these key GA biosynthesis genes during germination and early seedling growth is highly regulated, and they also suggest that each organ modulates the levels of GA biosynthesis gene transcripts to maintain specific pools of both precursors and growth-active GA1 during seed maturation, germination, and active growth phases (early seedling growth) of the plant.
MATERIALS AND METHODS
Plant Material
The pea (Pisum sativum) cv ‘Alaska’ (I3) was chosen as a model vining-type pea plant. ‘Alaska’ has normal leaflet morphology (AF), wild-type internode length (LE), white flowers and green cotyledons at maturity, and it begins to flower at approximately the 10th node under long- or short-day conditions. ‘Carneval’ was chosen as a model for semidwarf (semileafless; af) field pea, which is used extensively in crop agriculture. ‘Carneval’ has white flowers and yellow cotyledons at maturity, begins to flower at about the 15th to 17th node under long-day conditions, and was found to contain Mendel's dwarfing gene, le-1 (data not shown). Both cultivars readily germinate upon imbibition with water at 15°C to 25°C.
Growth Conditions and Tissue Harvesting
Mature air-dry seeds of ‘Alaska’ (5.4% RWC) and ‘Carneval’ (5.8% RWC) were planted at a depth of approximately 2.5 cm into moist sterilized sand in 3-L plastic pots (10 seeds per pot), the pots were placed in a growth chamber (Conviron) at 22°C/20°C (day/night) in a 16/8-h photoperiod with cool-white fluorescent and incandescent lights (205 μE m−2 s−1) until harvest. For germination and growth measurements, seeds of each cultivar were harvested at 0.5, 1, 2, 3, 4, and 6 DAI from the sand medium, and separated by dissection into cotyledons and embryo axes (0.5 and 1 DAI) or into cotyledons, roots, and shoots (2–6 DAI; 15–30 seeds or seedlings per time point). Seeds were scored as being germinated when protrusion of the radicle (2–5 mm) through the seed coat was visible. The RWC of the cotyledons was determined by comparing the sample weights before and after drying for 72 h at 60°C, and are expressed on a FW basis. For RNA extraction, seedlings at 0.5, 1, 2, 4, and 6 DAI were separated either into cotyledons and embryo axes (0.5 and 1 DAI), or cotyledons, shoots, and roots (2 and 4 DAI), or cotyledons, shoots, root tips (approximately 4 mm), and remainder of roots (6 DAI), and immediately frozen in liquid N2 and stored at −80°C until extraction. From 2 DAI, only seeds classed as germinated were used. To examine the mRNA levels in the mature embryos (0 DAI), seeds of the two cultivars were immersed in ice:water (1:1, w/v) for 4 h to facilitate seed coat removal, and the embryos (cotyledon plus embryo axis) were immediately frozen in liquid nitrogen and stored at −80°C until RNA extraction.
To study whether the presence of the embryo axis was required to induce the expression of GA biosynthesis genes and, in turn, the metabolism of GAs in the cotyledons of germinating pea seeds, mature seeds of ‘Alaska’ were surface sterilized in 1.2% sodium hypochlorite solution for 25 min and rinsed five times with sterile deionized water. Seeds for embryo axis removal were kept immersed in sterile water for 2 h after surface sterilization. After the 2-h imbibition period, the testa and embryo axis were removed without damage to the cotyledon using a scalpel. The cotyledons without an embryo axis or the intact seeds (for cotyledon with embryo axis treatment) were then placed in a 9-cm sterile petri plate (20 seeds per plate) on a sterile Whatman #1 filter paper wetted with 10 mL of sterile deionized water and imbibed in darkness at 22°C/20°C 16/8 h for 2 d. After the 2 d of incubation, a portion of the cotyledons from each treatment (with and without embryo axis) was harvested into liquid nitrogen and stored at −80°C for RNA extraction. The remaining cotyledons or intact seedlings were transferred to petri plates for the [14C]GA20 metabolism study.
RNA Isolation
Tissues were finely ground in liquid N2, and 200 to 550 mg FW (20-DAA seeds, mature embryos, embryo axes, shoots, roots, or root tips) or 100 to 250 mg FW (cotyledons) subsamples were used for total RNA isolation using a modified TRIzol (Invitrogen) protocol (cotyledon subsamples were taken from two ground cotyledons). After initial extraction with the TRIzol reagent and centrifugation, the supernatant was cleaned by chloroform partitioning (0.2 mL mL−1 TRIzol). The resulting supernatant fraction was then precipitated by first using an isopropanol solution (0.25 mL mL−1 TRIzol) followed by a high salt solution (1.2 m sodium citrate and 0.8 m NaCl) to remove polysaccharides and proteoglycans. The RNA sample was further precipitated with 4 m LiCl and finally followed by a mixture of 3 m sodium acetate (pH 5.2):100% ethanol (1:20, v/v). The precipitate was dissolved in diethylpyrocarbonate-treated water. The integrity of the RNA was verified both electrophoretically and by the average absorption ratio 260 to 280 nm. The total RNA samples of all tissues were then digested with DNase (DNA-free kit; Ambion), and the cotyledonary total RNA samples were further purified with RNeasy columns (Qiagen). Sample RNA concentration was determined in duplicate by A260 measurement, and then the samples were stored at −80°C until quantitation by real-time RT-PCR.
Gene Expression Analysis
Primers and Probes
Primers and probes for the target gene quantifying amplicons GA3ox1-87 (used for PsGA3ox1 quantification) and for the reference gene amplicon 18S-62 (used for pea 18S rRNA quantification) were designed by Ozga et al. (2003; Table IV). The target gene quantifying amplicons GA20ox1-104 (used for PsGA20ox1 quantification), GA20ox2-88 (used for PsGA20ox2 quantification), GA2ox1-73 (used for PsGA2ox1 quantification), and GA2ox2-83 (used for PsGA2ox2 quantification) were designed by Ayele et al. (2006; Table IV).
Table IV.
Primer and probe sequences used in the quantification of relative mRNA levels
| Gene | Type | Quantifying (5′ to 3′) |
|---|---|---|
| Amplicon: GA20ox1-104 | ||
| PsGA20ox1 | FPa | GCATTCCATTAGGCCAAATTTC |
| RPb | CCACTGCCCTATGTAAACAACTCTT | |
| Probe | CCTTCATGGCTCTTTC | |
| Amplicon: GA20ox2-88 | ||
| PsGA20ox2 | FP | AATACATCTTCTCTACCGTTGCAAAT |
| RP | TTGGCGGTGTTAAACAAGGTT | |
| Probe | ACATACCCTCAGAGTTC | |
| Amplicon: GA3ox1-87 | ||
| PsGA3ox1 | FP | TTCGAGAACTCTGGCCTCAAG |
| RP | ATGTTCCTGCTAACTTTTTCATGGTT | |
| Probe | ACAATATCACAGAATCTGGT | |
| Amplicon: GA2ox1-73 | ||
| PsGA2ox1 | FP | TTCCTCCTGATCATAGCTCCTTCT |
| RP | TTGAACCTCCCATTAGTCATAACCT | |
| Probe | GAGAATCACCAACATT | |
| Amplicon: GA2ox2-83 | ||
| PsGA2ox2 | FP | AACACAACAAAGCCTAGAATGTCAA |
| RP | ACCATCTTCGATAACGGGCTTAT | |
| Probe | TGTATTTTGCAGCACCACC | |
| Amplicon: 18S-62 | ||
| Ps18S rRNA | FP | ACGTCCCTGCCCTTTGTACA |
| RP | CACTTCACCGGACCATTCAAT | |
| Probe | ACCGCCCGTCGCTCCTACCG |
FP, Forward primer.
RP, Reverse primer
All probes were TaqMan MGB and were labeled at the 5′ end with fluorescent reporter dye 6-carboxyfluorescein (target gene probes) or VIC (18S-62 reference gene probe), and at the 3′ end with nonfluorescent quencher dye (Applied Biosystems). To confirm the PCR product produced by the quantifying primers, RT-PCR amplification products were separated and identified using 1% agarose gel electrophoresis and ethidium bromide staining.
Real-Time RT-PCR Assay
Real-time RT-PCR assays were performed on a model 7700 sequence detector (Applied Biosystems) using a TaqMan One-Step RT-PCR Master Mix Reagent kit (Applied Biosystems). Total reaction volume was either 25 or 50 μL, and the reaction components were adjusted accordingly to maintain the same concentration. For each 50-μL reaction, 5 μL of sample RNA (200 ng of total RNA for PsGA20ox1, PsGA20ox2, PsGA3ox1, PsGA2ox1, and PsGA2ox2 or 10 pg of total RNA for 18S rRNA quantitation) was mixed with 25 μL of 2× Master Mix (containing AmpliTaq Gold DNA polymerase), 1.25 μL of 40× MultiScribe (reverse transcriptase and RNase inhibitor mix), 3 μL of forward primer (5 μm; final concentration 300 nm), 3 μL of reverse primer (5 μm; final concentration 300 nm), 1 μL of probe (5 μm; final concentration 100 nm), and 11.75 μL of diethylpyrocarbonate-treated water. Samples were subjected to thermal cycling conditions of RT at 48°C for 30 min, DNA polymerase activation at 95°C for 10 min, and 40 cycles of denaturation at 95°C for 15 s followed by anneal extension at 60°C for 1 min. PCR amplification of each sample was carried out in duplicate in 96-well optical reaction plates covered with optical caps (Applied Biosystems), and the average of the two subsamples was used to calculate the sample transcript abundance. Total RNA extracts from each tissue were pooled across all time points per cultivar, and this pooled sample was run on each plate and used as a control to correct for plate-to-plate amplification differences. A pooled sample from one real-time RT-PCR run was taken arbitrarily as the standard for normalizing the Ct values of samples in other runs as follows:
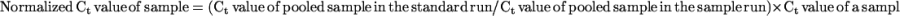 |
The relative transcript abundance of the target genes in the individual plant samples was determined by the 2−ΔCt method (Livak and Schmittgen, 2001), where ΔCt was the difference between the target sample Ct and average Ct of the reference sample. Transcript levels were compared across all genes, genotypes, developmental stages, and tissues using the average Ct value of PsGA20ox1 from the mature embryo of ‘Carneval’ (Ct = 34.2 ± 0.1) as the reference sample for data in Figures 3, 4, and 5. For data in Figure 6, transcript levels were compared across genes and tissues using the average Ct value of PsGA3ox1 from the cotyledon with no axis sample (Ct = 37.5 ± 0.1) as a reference sample. At least two and often three replicate plant samples were assayed.
GA Metabolism
To study GA metabolism in the cotyledons with the axis attached or the axis removed, [14C]GA20 (specific activity of 34 μCi/μmol) was injected into one cotyledon of 2-DAI ‘Alaska’ seedlings (with axis treatment) or one 2-DAI cotyledon (without axis treatment) at two spots (a total of 2.5 μL of 50% aqueous ethanol; a total of approximately 82,000 dpm). The [14C]GA20-labeled cotyledons were incubated for 12 or 24 h on filter paper moistened with 10 mL of sterile water in 15-cm petri plates (five cotyledons or seedlings per plate) placed at 22°C/20°C (day/night) in a 16/8-h photoperiod as described previously. The incubation period started 4 h into the photoperiod for both treatments. After incubation with the [14C]GA20 substrate, the [14C]GA20-treated cotyledons were harvested onto dry ice and stored at −80°C until extraction.
The [14C]GA20-treated cotyledons (five per sample) were homogenized in cold 80% methanol (10 mL per sample) in 30-mL Corex tubes using a polytron homogenizer. 17-[14C]GA7 (approximately 11,000 dpm) was added at homogenization to each sample extract as an external standard for recovery determination of radioactive metabolites at the HPLC step. The extracts were mixed overnight on a shaker (150 rpm) at 4°C in darkness, and then centrifuged for 30 min at 10,000g. The methanolic supernatant was removed, and the residue was resuspended in 5 mL of homogenization solvent and shaken for at least 4 h. The residue extracts were centrifuged for 30 min at 10,000g, and the pooled methanolic extracts were evaporated to the aqueous phase using a SpeedVac concentrator. After adjusting the pH of the aqueous extract to 8.0 with 0.1 n NH4OH, the extract was partitioned four times against n-hexane (5 mL) in 20-mL glass scintillation vials. The aqueous phase was then adjusted to pH 3 with 0.1 n HCl and partitioned five times against ethyl acetate (5 mL). The combined ethyl acetate extract was reduced in volume using the SpeedVac concentrator, and partitioned four times against 5% (w/v) aqueous NaHCO3 (2 mL). The combined NaHCO3 extract was transferred into a 30-mL Pyrex tube placed on ice, pH was adjusted to 3 with 6 n HCl, and then it was partitioned four times against ethyl acetate (5 mL). The ethyl acetate extracts were pooled and evaporated to complete dryness using a SpeedVac concentrator prior to HPLC purification.
For HPLC analysis, the ethyl acetate extracts were dissolved in 400 μL of 20% MeOH, filtered through a 0.45-μm nylon filter (Whatman International), and injected onto a 4.6- × 250-mm C18 column (5 μm; Beckman Instruments). The samples were eluted at 1 mL min−1 flow rate using the following linear gradient of methanol (solvent A) and aqueous 0.01% TFA (solvent B): 20% solvent A for 1 min, gradient to 100% solvent A in 45 min, and isocratic 100% solvent A for 5 min. Radioactivity in the sample effluent was monitored using a flow-through radiochemical detector. Fractions eluting at the retention times of GA8 (8.6 min), GA29 (10.7 min), GA1 (16.1 min), GA29-catabolite (16.4 min), GA20 (24.2 min), and GA7 (27.6 min) were collected and reduced to dryness. The putative GA8, GA29, GA1, and GA29-catabolite fractions were pooled across treatments, methylated using diazomethane, and rechromatographed as their methyl-esters by C18 HPLC, using the same solvent system and radiochemical detection. The [14C]GA methyl-ester fractions that chromatographed at the retention time of the GA methyl-ester standards confirmed the presence of the respective [14C]GA. As the [14C]GA29 and [14C]GA8 methyl-ester fractions were of sufficient quantity, they were converted to their trimethylsilyl ether derivatives (Gaskin and MacMillan, 1991), and GA identity was confirmed by GC-MS-SIM as described by van Huizen et al. (1995).
Analysis of Endogenous GA Levels
Developing seeds (20 DAA), mature embryos (cotyledon plus embryo axis), embryo axes, cotyledons, shoots, and roots were freeze-dried and subsequently ground to a fine powder in a mortar and pestle (0.6–11 g dry weight) with liquid N2 and washed sea sand (Fisher Scientific). The tissue powder was homogenized with 80% (v/v) aqueous methanol, and 20 ng of [17,17-2H2]GA1, [17,17-2H2]GA19, [17,17-2H2]GA20, and [17,17-2H2]GA29 and 33 ng of [17,17- 2H2]GA8 (obtained from Prof. L.N. Mander, Research School of Chemistry, Australian National University, Canberra, ACT, Australia) was added to each extract as internal standards for recovery determination at the GC-MS-SIM step. The methanolic extracts were filtered through a 55-mm filter paper (Whatman #2; Whatman International), eluted through a C18 preparative column (3 g of C18 preparative reversed-phase material [Waters] preconditioned with 100% methanol followed by 80% (v/v) aqueous methanol; Koshioka et al., 1983), and then dried in vacuo at 35°C.
The extract residue was dissolved in 1 mL of 10% aqueous methanol with 1% acetic acid and injected onto a C18 (μ-Bondapak) Radial-PAK (8-mm × 10-cm) column connected to a Waters HPLC system. The samples were eluted at 2 mL min−1 using the following linear gradient of 10% methanol in 1% acetic acid [water:MeOH:acetic acid, 89:10:1 (v/v); solvent A] and 100% MeOH (solvent B): 0 to 10 min 100% solvent A, 10 to 50 min gradient to 30% solvent A, and 50 to 80 min gradient to 100% solvent B. Fractions eluting at the corresponding retention times of the GAs of interest were collected and dried in vacuo at 35°C.
Fractions containing the putative GAs were dissolved in methanol, methylated using diazomethane, then taken to dryness and trimethylsilylated using BSTFA with 1% TMCS (Gaskin and MacMillan, 1991). Identification and quantification of GAs were carried out using an Agilent 6890 gas chromatograph connected to an Agilent 5973 mass spectrometer (GC-MS). The derivatized samples were injected onto a DB-1701 capillary column (30-m × 0.25-mm × 0.25-μm film thickness; J&W Scientific) with an initial column temperature at 60°C for 1 min followed by temperature programming at 25°C min−1 to 240°C and then 5°C min−1 to 280°C. Helium was used as the carrier gas at a flow rate of 1 mL min−1.
The endogenous GAs were identified by SIM of three prominent ions (including the molecular ion M+, except for GA19: i.e. GA8, 596/594, 581/579, 450/448; GA1, 508/506, 493/491, 450/448; GA29, 508/506, 493/491, 449/447; GA20, 420/418, 377/375, 405/403; GA19, 436/434, 376/374, 404/402) characteristic to the corresponding GAs, as well as comparison of the GC retention times of the endogenous GAs with their respective [2H2]GA standards. Endogenous GA concentrations were calculated by reference to the stable isotope-labeled internal standard using equations for isotope dilutions analysis adapted from Gaskin and MacMillan (1991) as described by Jacobsen et al. (2002).
Acknowledgments
We thank Ashley Durec, Bridget McLeod, and Dr. David W. Pearce for technical assistance.
This work was supported by the Alberta Agricultural Research Institute and the Natural Sciences and Engineering Research Council of Canada (to J.A.O.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jocelyn A. Ozga (jocelyn.ozga@ualberta.ca).
References
- Ait-Ali T, Swain SM, Reid JB, Sun T, Kamiya Y (1997) The LS locus of pea encodes the gibberellin biosynthesis enzyme ent-kaurene synthase A. Plant J 11: 443–454 [DOI] [PubMed] [Google Scholar]
- Ayele BT (2006) Gibberellin biosynthesis during germination and young seedling growth of pea. PhD thesis. University of Alberta, Edmonton, Alberta, Canada [DOI] [PMC free article] [PubMed]
- Ayele BT, Ozga JA, Reinecke DM (2006) Regulation of GA biosynthesis genes during germination and young seedling growth of pea (Pisum sativum L.). J Plant Growth Regul doi/10.1007/s00344-006-0007-8 [DOI] [PMC free article] [PubMed]
- Bain JM, Mercer FV (1966. a) Subcellular organization of the cotyledons in germinating seeds and seedlings of Pisum sativum L. Aust J Biol Sci 19: 69–84 [Google Scholar]
- Bain JM, Mercer FV (1966. b) The relationship of the axis and the cotyledons in germinating seeds and seedlings of Pisum sativum L. Aust J Biol Sci 19: 85–96 [Google Scholar]
- Bewley JD, Black M (1994) Seeds: Physiology of Development and Germination, Ed 2. Plenum Press, New York
- Bustin SA (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25: 169–193 [DOI] [PubMed] [Google Scholar]
- Chen F, Dahal P, Bradford KJ (2001) Two tomato expansin genes show divergent expression and localization in embryos during seed development and germination. Plant Physiol 127: 928–936 [PMC free article] [PubMed] [Google Scholar]
- Curaba J, Moritz T, Blervaque R, Parcy F, Raz V, Herzog M, Vachon G (2004) AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol 136: 3660–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SE, Elliott RC, Helliwell CA, Poole AT, Reid JB (2003) The pea gene NA encodes ent-kaurenoic acid oxidase. Plant Physiol 131: 335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Koornneef M (2000) Gibberellin requirement for Arabidopsis seed germination is determined both by the testa characteristics and embryonic abscisic acid. Plant Physiol 122: 415–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martinez JL, Lopez-Diaz I, Sanchez-Beltran MJ, Phillips AL, Ward DA, Gaskin P, Hedden P (1997) Isolation and transcript analysis of gibberellin 20-oxidase genes in pea and bean in relation to fruit development. Plant Mol Biol 33: 1073–1084 [DOI] [PubMed] [Google Scholar]
- Gaskin P, MacMillan J (1991) GC-MS of the Gibberellins and Related Compounds: Methodology and a Library of Spectra. University of Bristol (Cantock's Enterprises), Bristol, UK
- Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P (2004) The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev Cell 7: 373–385 [DOI] [PubMed] [Google Scholar]
- Graebe JE (1986) Gibberellin biosynthesis from gibberellin A12-aldehyde. In M Bopp, ed, Plant Growth Substances 1985. Springer-Verlag, New York, pp 74–82
- Groot SPC, Karssen CM (1987) Gibberellins regulate seed germination in tomato by endosperm weakening: a study with gibberellin mutants. Planta 171: 525–531 [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL (2000) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5: 523–530 [DOI] [PubMed] [Google Scholar]
- Hilhorst HWM, Karssen CM (1988) Dual effect of light on the gibberellin- and nitrate-stimulated seed germination of Sisymbrium officinale and Arabidopsis thaliana. Plant Physiol 86: 591–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram TJ, Reid JB, Murfet IC, Gaskin P, Willis CL, MacMillan J (1984) Internode length in Pisum: the Le gene controls the 3β-hydroxylation of gibberellin A20 to gibberellin A1. Planta 160: 455–463 [DOI] [PubMed] [Google Scholar]
- Jacobsen JV, Gubler F, Chandler PM (1995) Gibberellin action in germinated cereal grains. In PJ Davies, ed, Plant Hormones: Physiology, Biochemistry and Molecular Biology, Ed 2. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 246–271
- Jacobsen JV, Pearce DW, Poole AT, Pharis RP, Mander LN (2002) Abscisic acid, phaseic acid and gibberellin contents associated with dormancy and germination in barley. Physiol Plant 115: 428–441 [DOI] [PubMed] [Google Scholar]
- Koornneef M, van der Veen JH (1980) Induction and analysis of gibberellin-sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor Appl Genet 58: 257–263 [DOI] [PubMed] [Google Scholar]
- Koshioka M, Takeno K, Beall FD, Pharis RP (1983) Purification and separation of plant gibberellins from their precursors and glucosyl conjugates. Plant Physiol 73: 398–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange T, Kappler J, Fischer A, Frisse A, Padeffke T, Schmidtke S, Lange MJP (2005) Gibberellin biosynthesis in developing pumpkin seedlings. Plant Physiol 139: 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Davies PJ, Reid JB (1997) Mendel's stem length gene (Le) encodes a gibberellin 3β-hydroxylase. Plant Cell 9: 1435–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Smith JJ, Elliott RC, Reid JB (1999) Gibberellin 2-oxidation and the SLN gene of Pisum sativum. Plant J 19: 65–73 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Martin DN, Proebsting WM, Hedden P (1997) Mendel's dwarfing gene: cDNAs from the Le alleles and function of the expressed proteins. Proc Natl Acad Sci USA 94: 8907–8911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DN, Proebsting WM, Hedden P (1999) The SLENDER gene of pea encodes a gibberellin 2-oxidase. Plant Physiol 121: 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DN, Proebsting WM, Parks TD, Dougherty WG, Lange T, Lewis MJ, Gaskin P, Hedden P (1996) Feed-back regulation of gibberellin biosynthesis and gene expression in Pisum sativum L. Planta 200: 159–166 [DOI] [PubMed] [Google Scholar]
- Nambara E, Akazawa T, McCourt P (1991) Effects of the gibberellin biosynthetic inhibitor uniconazol on mutants of Arabidopsis. Plant Physiol 97: 736–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15: 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozga JA, Yu J, Reinecke DM (2003) Pollination-, development-, and auxin-specific regulation of gibberellin 3β-hydroxylase gene expression in pea fruit and seeds. Plant Physiol 131: 1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruzzelli L, Harren F, Perrone C, Reuss J (1995) On the role of ethylene in seed germination and early growth of Pisum sativum. J Plant Physiol 145: 83–86 [Google Scholar]
- Potts WC, Reid JB (1983) Internode length in Pisum. III. The effect and interaction of the Na/na and Le/le gene differences on endogenous gibberellin-like substances. Physiol Plant 57: 448–454 [Google Scholar]
- Proebsting WM, Hedden P, Lewis MJ, Croker SJ, Proebsting LN (1992) Gibberellin concentration and transport in genetic lines of pea: effects of grafting. Plant Physiol 100: 1354–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards DE, King KE, Ait Ali T, Harberd NP (2001) How gibberellin regulates plant growth and development: a molecular genetic analysis of gibberellin signaling. Annu Rev Plant Physiol Plant Mol Biol 52: 67–88 [DOI] [PubMed] [Google Scholar]
- Reid JB, Symons GM, Ross JJ (2004) Regulation of gibberellin and brassinosteroid biosynthesis by genetic, environmental and hormonal factors. In PJ Davies, ed, Plant Hormones: Biosynthesis, Signal Transduction, Action. Ed 3. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 179–203
- Ross JJ, Reid JB, Dungey HS (1992) Ontogenetic variation in levels of gibberellin A1 in Pisum: implications for the control of stem elongation. Planta 186: 166–171 [DOI] [PubMed] [Google Scholar]
- Ross JJ, Reid JB, Swain SM (1993) Control of stem elongation by gibberellin A1: evidence from genetic studies including the slender mutant sln. Aust J Plant Physiol 20: 585–599 [Google Scholar]
- Rost TL, Baum S (1988) On the correlation of primary root length, meristem size and protoxylem tracheary element position in pea seedlings. Am J Bot 75: 414–424 [Google Scholar]
- Rost TL, Jones TJ, Falk RH (1988) Distribution and relationship of cell division and maturation events in Pisum sativum (Fabaceae) seedling roots. Am J Bot 75: 1571–1583 [Google Scholar]
- Smith DL, Flinn AM (1967) Histology and histochemistry of the cotyledons of Pisum arvense L. during germination. Planta 74: 72–85 [DOI] [PubMed] [Google Scholar]
- Sponsel VM (1983) The localization, metabolism and biological activity of gibberellins in maturing and germinating seeds of Pisum sativum cv. Progress No. 9. Planta 159: 454–468 [DOI] [PubMed] [Google Scholar]
- Sponsel VM (1995) The biosynthesis and metabolism of gibberellins in higher plants. In PJ Davies, ed, Plant Hormones: Physiology, Biochemistry and Molecular Biology, Ed 2. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 66–97
- Sponsel VM, Hedden P (2004) Gibberellin biosynthesis and inactivation. In PJ Davies, ed, Plant Hormones: Biosynthesis, Signal Transduction, Action. Ed 3. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 63–94
- Tanimoto E (1990) Gibberellin requirement for the normal growth of roots. In N Takahashi, B Phinney, J MacMillan, eds, Gibberellins. Springer-Verlag, New York, pp 229–240
- van Huizen R, Ozga JA, Reinecke DM (1997) Seed and hormonal regulation of gibberellin 20-oxidase expression in pea pericarp. Plant Physiol 115: 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Huizen R, Ozga JA, Reinecke DM, Twitchin B, Mander LN (1995) Seed and 4-chloroindole-3-acetic acid regulation of gibberellin metabolism in pea pericarp. Plant Physiol 109: 1213–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Smith MW, Brown RGS, Kamiya Y, Sun TP (1998) Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10: 2115–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaxley JR, Ross JJ, Sherriff LJ, Reid JB (2001) Gibberellin biosynthesis mutations and root development in pea. Plant Physiol 125: 627–633 [DOI] [PMC free article] [PubMed] [Google Scholar]