Abstract
To understand the molecular mechanism regulating meristem development in the monocot rice (Oryza sativa), we describe here the isolation and characterization of three floral organ number4 (fon4) alleles and the cloning of the FON4 gene. The fon4 mutants showed abnormal enlargement of the embryonic and vegetative shoot apical meristems (SAMs) and the inflorescence and floral meristems. Likely due to enlarged SAMs, fon4 mutants produced thick culms (stems) and increased numbers of both primary rachis branches and floral organs. We identified FON4 using a map-based cloning approach and found it encodes a small putatively secreted protein, which is the putative ortholog of the Arabidopsis (Arabidopsis thaliana) CLAVATA3 (CLV3) gene. FON4 transcripts mainly accumulated in the small group of cells at the apex of the SAMs, whereas the rice ortholog of CLV1 (FON1) is expressed throughout the SAMs, suggesting that the putative FON4 ligand might be sequestered as a possible mechanism for rice meristem regulation. Exogenous application of the peptides FON4p and CLV3p corresponding to the CLV3/ESR-related (CLE) motifs of FON4 and CLV3, respectively, resulted in termination of SAMs in rice, and treatment with CLV3p caused consumption of both rice and Arabidopsis root meristems, suggesting that the CLV pathway in limiting meristem size is conserved in both rice and Arabidopsis. However, exogenous FON4p did not have an obvious effect on limiting both rice and Arabidopsis root meristems, suggesting that the CLE motifs of Arabidopsis CLV3 and FON4 are potentially functionally divergent.
Plants have the unique ability to generate organs throughout their life cycle because of the continuous activity of meristems. The balance between maintenance of stem cells and the transition of these undifferentiated cells to differentiated cells is critical to normal organ initiation and formation. Stem cells within a small central zone of the shoot apical meristem (SAM) have the ability to grow and divide to replace cells of the SAM flanks, which then drive the formation of lateral organs. Signaling pathways for precise coordination are thought to occur via cell-to-cell communication between and within the stem cells and differentiated cells of the SAM (Clark, 2001; for review, see Bowman and Eshed, 2000).
One of the best-characterized signaling pathways in Arabidopsis (Arabidopsis thaliana) is called the CLAVATA (CLV) pathway because it involves three CLV genes, CLV1 to CLV3. CLV1 is likely an extracellular Leu-rich repeat (LRR) receptor kinase and CLV2 is a LRR protein without a kinase domain (Clark et al., 1997; Jeong et al., 1999). CLV3 is a putative secreted peptide ligand, which likely interacts with the CLV1/CLV2 heterodimeric receptor to limit the size of the stem cell pool in the SAM (Clark et al., 1997; Fletcher et al., 1999; Jeong et al., 1999). Plants defective in any one of the three CLV genes form enlarged SAMs, as well as inflorescence and floral meristems, resulting in increased numbers of flowers and floral organs (Clark et al., 1993, 1995; Kayes and Clark, 1998). In contrast to the CLV genes, WUSCHEL (WUS) and SHOOT MERISTEMLESS genes encoding homeodomain transcription factors have the ability to promote the SAM and reproductive meristem sizes (Barton and Poethig, 1993; Clark et al., 1996; Endrizzi et al., 1996; Laux et al., 1996; Kayes and Clark, 1998; Mayer et al., 1998). WUS is specifically expressed within the organizing center of the SAM and is down-regulated by the activation of CLV3. On the other hand, CLV3 expression is positively regulated by WUS, forming a positive-negative feedback loop to maintain the size of meristems throughout Arabidopsis development (Brand et al., 2000; Schoof et al., 2000; Muller et al., 2006).
CLV3 is a member of the CLV3/ESR-related (CLE) gene family (Cock and McCormick, 2001; Fiers et al., 2005). These genes are not only present in many plant species, but also found in some plant parasites (Wang et al., 2005). CLE genes encode small proteins with a predicted signal peptide at their N termini and a conserved 14-amino acid motif (CLE motif) at or near their C termini (Cock and McCormick, 2001). The importance of the CLE motif is supported by the observations that two independent clv3 alleles (clv3-1 and clv3-5) resulted in the same single amino acid substitution in the CLE motif (encoding clvG75A; Fletcher et al., 1999). In addition, exogenous application of synthetic 14-amino acid peptides, CLV3p (for CLV3), CLE19p (for CLV3/ESR19), and CLE40p (for CLE40), to Arabidopsis roots caused termination of the root apical meristem (Fiers et al., 2005). Recently, the CLE motif of CLV3 was shown to be functional in limiting Arabidopsis SAM size either expressed from a transgene or applied exogenously (Fiers et al., 2006; Ni and Clark, 2006). These results also indicate that CLE domains confer all or most of the function of the CLE proteins.
A growing body of evidence indicates that the CLV pathway for regulating meristem size is functionally conserved in monocots as well as in eudicots. The maize (Zea mays) fasciated ear2 (fea2) locus is the first well-characterized monocot gene involved in the CLV pathway. The fea2 gene encodes a LRR receptor-like protein that is most closely related to CLV2. Loss of function of fea2 causes severe overproliferation of the ear inflorescence meristem and has a more modest effect on floral meristem size and floral organ number (Taguchi-Shiobara et al., 2001). Recently, another maize gene, thick tassel dwarf1 (td1), was reported to function in the inflorescence and flower to limit meristem sizes and encodes a CLV1-like protein (Bommert et al., 2005).
Rice (Oryza sativa) inflorescence architecture is quite different from that of other major cereal crops and is an important trait for agriculture. Instead of two or more florets in one spikelet, as seen in other cereal crops, such as maize and wheat (Triticum aestivum), one rice spikelet has only one floret surrounded by a pair of empty glumes. In addition, rice florets have five types of floral organs with characteristic numbers, one lemma, one palea, two lodicules, six stamens, and one pistil. Recently, a rice mutant called floral organ number1 (fon1) has been described. The fon1 mutant exhibits an enlargement of the floral meristem and an increase in the number of all floral organs (Suzaki et al., 2004). FON1 encodes a protein that is highly similar to td1 and CLV1. Another rice gene, OsLRK1, is also similar to CLV1; silencing of the OsLRK1 gene using RNA interference resulted in plants with an increased floral organ number (Kim et al., 2000). Additionally, other mutants with enlarged floral meristems and increased floral organ numbers have been reported, such as fon2-1, fon2-2, and fon3 (Nagasawa et al., 1996; Jiang et al., 2005). Therefore, members of the grass family also have the components of the CLV pathway. However, to date, a CLV3-like gene has not yet been described in monocots.
Here, we describe three mutant alleles of the rice FON4 gene encoding a CLV3-like protein. The observation that fon4 mutants have abnormal expansion of SAMs and defects in both vegetative and reproductive development further supports conservation of the CLV-signaling pathway in limiting meristem sizes in monocot species. At the same time, we report significant differences of the CLE motif effects between FON4 and CLV3, indicating a need to study the molecular mechanism regulating meristem sizes in crops such as rice.
RESULTS
fon4 Mutants Have Increased Numbers of Floral Organs and Primary Rachis Branches
During rice flower development, the floral meristem first produces a lemma and a palea in opposite positions on the flank of the meristem. Then two lodicules, thought to be homologous to the petal in dicots, are initiated interior to the lemma and near the two lemma margins. Six stamens then emerge in a whorl between the sterile organs (lemma, palea, and lodicules) and the meristem center, and finally a carpel is formed at the center of the flower (Figs. 1A and 2, A–C). To be consistent with the description of the organization of floral organs in Arabidopsis, the regions where lemma/palea, lodicules, stamens, and pistil develop in rice are referred to in this article as whorl 1, whorl 2, whorl 3, and whorl 4, respectively. We have isolated three mutant alleles of the FON4 gene (see “Materials and Methods”). The fon4-1 mutation caused increased numbers of all floral organ types; similarly, the fon4-2 and fon4-3 mutants also showed increased floral organ number (Table I). We observed that the organ number in the inner whorls was more severely affected than those of the outer whorls. Specifically, almost all fon4-1 and fon4-2 flowers, and approximately 78% of fon4-3 flowers, have more than one carpel, ranging from two to 10. In agreement with the increase in the carpel number, some fon4-1, fon4-2, and fon4-3 grains had two seeds with normal embryos (Fig. 1, M and N). We also observed that the stamen number was increased greatly in the three mutants, ranging from six to 10 (Fig. 1, B–D), indicating similar effects of the three mutations.
Figure 1.
Phenotype of the fon4 mutants. A to D, Flower phenotype; arrows indicate pistils. A, Wild-type flower. B, fon4-1 flower with seven stamens and three pistils. C, fon4-2 flower with seven stamens and two pistils. D, fon4-3 flower with seven stamens and two pistils. E, One of the empty glumes was transformed into a lemma and palea-like structure in the fon4-1 flower (arrow). F, Transformation of carpels into lemma and palea-like organs in the fon4-1 flower (arrow). G to I, Pistil morphology of fon4 mutants. G, Pistil transformed from stamen, anther replaced with stigmas and without ovule. H, Pistil with two stigmas in the fon4-1 flower. I, Abnormal pistil with four stigmas in the fon4-1 flower. J to L, Undifferentiated cell mass was formed in fon4-1 (J), fon4-2 (K), and fon4-3 (L) flower (arrows). M and N, Seed morphology of wild type (M) and fon4-1 (N). O and P, Panicle morphology of wild type (O) and fon4-1 (P). Arrows indicate the nodes. Bars = 2 mm in A to N; 20 mm in O and P.
Figure 2.
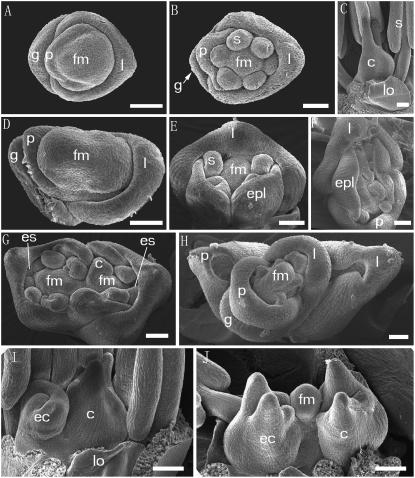
SEM images of the wild type and the fon4-1 mutant. A to C, SEM images of wild type. A, Wild-type flower at initiation of palea primordium. B, Wild-type flower at initiation of stamen primordia. C, Wild-type flower and pistil with two stigmas were formed. D to J, SEM images of fon4-1. D, fon4-1 flower. The floral meristem is overproliferated compare to the wild type. E, fon4-1 flower forms two palea-like organs on the palea side. F, fon4-1 flower forms ectopic palea/lemma-like organs on the lateral of meristem in an additional whorl. G, fon4-1 flower forms ectopic stamens in an additional whorl or in the origin whorl. H, fon4-1 flower in which a secondary flower is developed after a palea and a lemma formed. I, In the fon4-1 flower, an ectopic carpel primordium is produced at the lemma side after the carpel formed. J, Undifferentiated floral meristem-like structure remains in the central region in the fon4-1 flower even after two carpels were formed. fm, Floral meristem; g, glume; p, palea; l, lemma; s, stamen; lo, lodicules; c, carpel; epl, ectopic palea/lemma-like organ; es, ectopic stamen; ec, ectopic carpel. Bars = 50 μm.
Table I.
Number of floral organs in wild-type and fon4 flowers
The mean number of floral organs of 100 flowers of each genotype was counted. The mean numbers of indicated organs are shown with the se.
| Genotype | Lemma/Palea | Lodicule | Stamen | Pistil |
|---|---|---|---|---|
| Wild type | 2.0 ± 0.0 | 2.0 ± 0.0 | 6.0 ± 0.0 | 1.0 ± 0.0 |
| fon4-1 | 2.3 ± 0.6 | 2.2 ± 0.4 | 6.6 ± 0.9 | 3.1 ± 1.1 |
| fon4-2 | 2.0 ± 0.0 | 2.1 ± 0.3 | 6.5 ± 0.8 | 2.9 ± 0.9 |
| fon4-3 | 2.2 ± 0.4 | 2.0 ± 0.0 | 6.2 ± 0.4 | 3.1 ± 1.8 |
In addition to the increase in floral organ number, fon4 mutants also occasionally exhibited homeotic conversion of organ identity; for example, an empty glume was transformed into a lemma-like structure (Fig. 1E); sometimes organs at the positions of lodicules developed into palea/lemma-like organs (Fig. 1F) and also some ectopic stamen-like organs were formed that had stigmas instead of anthers (Fig. 1G). Most of the mutant carpels had normal morphology, with two stigmatic branches (Fig. 1H); however, some pistils possessed stigmas with three to eight stigma branches (Fig. 1I), probably resulting from the fusion of two or more carpels. In all three mutants, undifferentiated cell mass was frequently observed in nearly mature flowers (Fig. 1, J–L).
Besides the abnormal morphology of floral organs, fon4 mutations also affected the inflorescence phyllotaxis. Normally, one primary rachis branch was produced from one node in the wild-type inflorescence axis; however, fon4-1 inflorescences developed more than one primary rachis branch on one axis node (Fig. 1, O and P). We observed that the total number of primary rachis branches of a wild-type inflorescence was 11.4 ± 1.1 (n = 19), whereas a fon4-1 inflorescence had 18.9 ± 2.9 (n = 24) primary rachis branches.
fon4 Flowers Had Abnormal Early Floral Organ Development
To observe the developmental defects of fon4 mutant flowers in more detail, fon4-1 mutant flowers at early developmental stages were fixed and examined by scanning electron microscopy (SEM). Compared with the wild type, the floral meristem of fon4-1 was enlarged (Fig. 2, A and D). In wild-type plants, the lemma and palea were generated from the flank of the floral meristem, the lemma was close to the inflorescence axis, and the palea was opposite the lemma (Fig. 2A). However, ectopic palea/lemma-like organs were observed in the first whorl or in an extra whorl in some fon4-1 flowers (Fig. 2, E and F). In the case of an additional whorl with ectopic palea/lemma-like organs, a secondary flower could be observed at the axil of the lemma and palea after the initiation of these organs (Fig. 2H). This secondary flower produced normal palea, lemma, stamens, and pistil. The formation of a secondary flower might result from the division of the enlarged meristem by the sterile organs. Unlike the wild-type flower that produces six stamens in whorl 3 (Fig. 2B), many fon4-1 flowers formed extra stamens either in the normal whorl or an additional whorl (Fig. 2G).
The effect of fon4 mutations on carpel development is even more dramatic than that on the outer whorls. In the wild type, the carpel primordium is initiated from the floral meristem slightly closer to the lemma than the palea; the carpel primordium then extends as a ridge on the flanks of the meristem toward the opposite side, enclosing the maintaining meristem (Fig. 2C), which then develops into the ovule (Yamaguchi et al., 2004). Thus, the rice flower meristem terminates with the formation of the carpel primordium and the ovule. In contrast, the fon4-1 floral meristem was enlarged and had an elongated dome-like shape (Fig. 2D). The floral meristem was not completely surrounded by the carpel and was able to produce an ectopic carpel (Fig. 2I). Moreover, the region between the two separate carpel primordia remained meristem like in the fon4-1 flower (Fig. 2J) and this meristem was able to produce new organs. As a result, extra carpel primordia were initiated in the fon4-1 flower, resulting in the generation of additional carpels.
To obtain molecular evidence for abnormal organ development, the expression patterns of two rice flower developmental genes, OSH1 and DROOPING LEAF (DL), were analyzed. OSH1 is a rice marker gene for indeterminate meristems (Sato et al., 1996; Yamaguchi et al., 2004). In the wild type, OSH1 expression is relatively strong in the early floral meristem, decreases following carpel primordium initiation, and is undetected after carpel formation (Fig. 3C). However, even after an ectopic carpel was produced in the fon 4-1 mutant, OSH1 expression could be observed in the floral central region (Fig. 3D). DL is a carpel primordial marker and specifies carpel identity (Yamaguchi et al., 2004). It is expressed in the single carpel primordium in the wild-type flower (Fig. 3E). However, DL transcripts in the fon4-1 mutant flower accumulated in the multiple carpel primordia (Fig. 3F). These results indicate that FON4 is required for regulating normal early flower development, including floral organ initiation and floral meristem determinacy.
Figure 3.
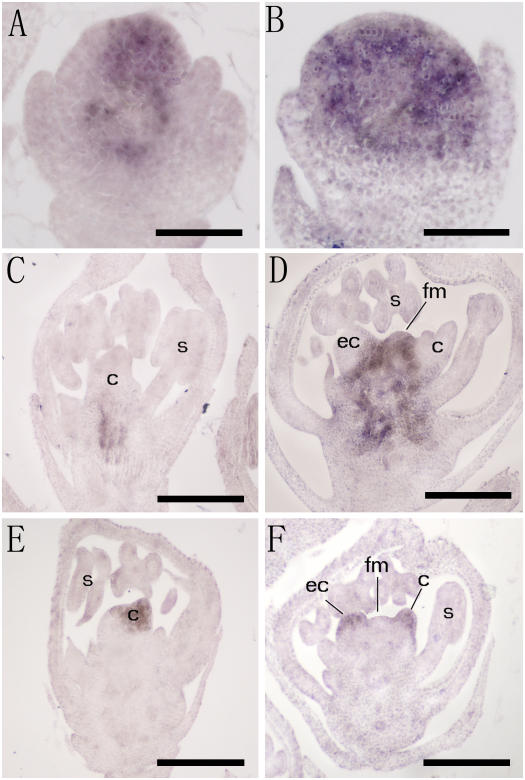
In situ analysis of OSH1 and DL. A to D, Spatial expression of OSH1. OSH1 expression is expanded in the fon4-1 (B) floral meristem compare to the wild type (A). Expression of OSH1 disappears after carpel developed in the wild-type flower (C), whereas its expression is maintained in the center of the flower in the fon4-1 mutant (D). E and F, Spatial expression of DL. Two sets of carpels are developed in the fon4-1 flower (F) compared with the wild type (E). s, Stamen; c, carpel; fm, floral meristem; ec, ectopic carpel. Bars = 50 μm in A and B; 100 μm in C to F.
fon4 Affects Apical Meristem Size
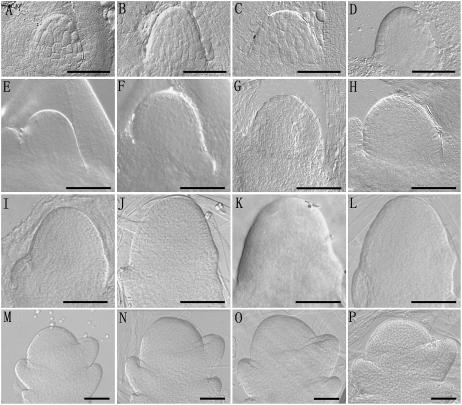
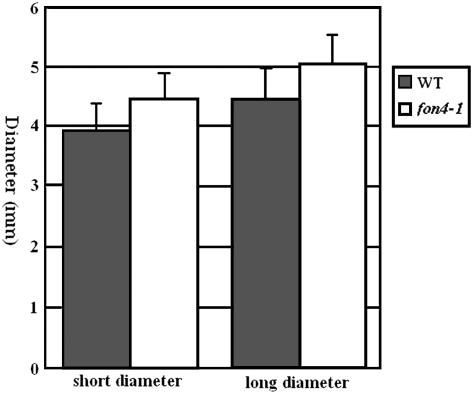
In both Arabidopsis clv mutants and rice fon1 mutants (Clark et al., 1993, 1995; Kayes and Clark, 1998; Suzaki et al., 2004), the fasciated inflorescence and the increase of floral organ number are the consequence of enlarged inflorescence and floral meristems, respectively. To determine whether fon4 mutants have similar defects, we examined fon4-1, fon4-2, and fon4-3 meristems and found that fon4 mutations affected SAM sizes during both vegetative and reproductive development (Fig. 4; Table II). During the embryo development and seedling stages, the sizes of the fon4-1 SAM were significantly larger than those of wild-type plants (Table II). In agreement with these observations, the thickness of fon4-1 mutant culm was slightly larger than that of the wild type (Fig. 5), whereas fon4-1 mutants showed no other abnormalities during vegetative development. The inflorescence meristem of fon4-1 was also larger than that of the wild type and produced more primary rachis branches (Figs. 4 and 1, O and P). Also, enlargement of fon4-1 floral meristems was observed. The width and height of the wild-type floral meristem were about 86.0 ± 5.3 μm (n = 8) and 37.3 ± 6.4 μm (n = 8), respectively; in comparison, the width and height of the fon4-1 floral meristem were 106.8 ± 7.6 μm (n = 8) and 62.7 ± 6.8 μm (n = 8), respectively. The enlarged SAM is supported by the observed OSH1 expression pattern in the fon4-1 mutant (Fig. 3, A and B). These observations suggest strongly that FON4 is important for control of meristem size during vegetative and reproductive development.
Figure 4.
Morphology of the apical meristems of the wild type and the fon4 mutants observed by differential interference contrast optics. A to D, SAMs of the embryos of wild type (A), fon4-1 (B), fon4-2 (C), and fon4-3 (D), respectively. E to H, Vegetative SAMs of wild type (A), fon4-1 (B), fon4-2 (C), and fon4-3 (D), respectively. I to L, Inflorescence meristems of wild type (A), fon4-1 (B), fon4-2 (C), and fon4-3 (D), respectively. M to P, Floral meristems of wild type (A), fon4-1 (B), fon4-2 (C), and fon4-3 (D), respectively. Bars = 50 μm.
Table II.
Size of apical meristems
The mean numbers of indicated size of apical meristems are shown with the se.
| Genotype | Embryo Meristems
|
Vegetative Meristems
|
Floral Meristems
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Width | Height | n | Width | Height | n | Width | Height | n | |
| μm | μm | μm | |||||||
| Wild type | 49.9 ± 3.4 | 42.0 ± 5.1 | 8 | 99.9 ± 7.1 | 61.6 ± 9.3 | 11 | 86.0 ± 5.3 | 37.3 ± 6.4 | 8 |
| fon4-1 | 55.8 ± 6.7 | 53.3 ± 3.7 | 8 | 112.5 ± 9.9 | 94.5 ± 9.6 | 11 | 106.8 ± 7.6 | 62.7 ± 6.8 | 8 |
| t test | 0.02 < P < 0.05 | P < 0.001 | P < 0.01 | P < 0.001 | P < 0.001 | P < 0.001 | |||
Figure 5.
Diameter of the transverse section of the culm at the second internode in rice. The transverse section of the culm was ellipse like; thus, we measured the short diameter and long diameter, respectively.
Isolation of the FON4 Gene
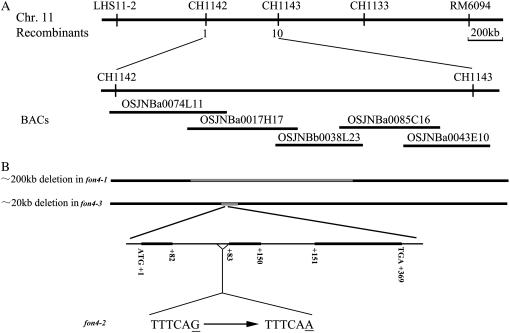
To allow further molecular studies of FON4 functions, we cloned the FON4 gene by using a map-based strategy. The fon4-1 mutant was crossed with Guang-lu-ai4 (cv indica) and 2,100 fon4 mutant plants were selected from F2 progeny. The FON4 locus was then localized within the region of about 400 kb between two polymorphic insertion/deletion markers, CH1142 and CH1143, which are detectable by PCR.
Because the fon4-1 mutant phenotype is similar to Arabidopsis clv mutants, we hypothesized that FON4 is likely to be similar to one of the CLV genes. Using a tBLASTn search in the rice genomic sequence database, one putative gene with high sequence similarity to the CLV3 CLE motif was found in the approximately 400-kb region defined by the CH1142 and CH1143 markers. Because CLE family members are small genes with little sequence identity outside the CLE motif, these genes are often overlooked by automated annotation programs (Ride et al., 1999; Vanoosthuyse et al., 2001). Indeed, this CLV3-like candidate FON4 gene had not yet been annotated in the rice genome sequence. We analyzed the genomic sequence near this putative CLE gene using the GenMARK program (Lukashin and Borodovsky, 1998; Lomsadze et al., 2005) to obtain a putative gene structure. To verify the gene structure, we determined exon-intron junctions by sequencing a cDNA fragment amplified by reverse transcription-PCR by using the FON4-specific primers FP1 and RP1. Comparison with the genomic sequence revealed that the FON4 gene consisted of three exons and two introns and encoded a small peptide of 122 amino acids. This protein also contained a 25-amino acid putative signal sequence at the N terminus according to the SignalP 3.0 program (Bendtsen et al., 2004).
To verify that the CLV3-like gene is indeed FON4, we analyzed the fon4-1, fon4-2, and fon4-3 genomic DNAs using PCR and sequencing. Our results indicated that the fon4-1 and fon4-3 alleles had approximately 200- and 20-kb deletions, respectively; in addition, fon4-2 had a G-to-A base change at the 3′ end of the first intron (Fig. 6B). Within the approximately 200-kb deletion region of fon4-1, there were 45 annotated genes, in addition to the increase of floral organ numbers and enlargement of SAMs; the leaf color of fon4-1 is slightly yellow compared with the wild type. We propose that this phenotype might result from loss of function of other genes in this approximately 200-kb region. This is also supported by the observation that the complementation test using the FON4 genomic sequence cannot rescue the altered leaf color phenotype (data not shown; see below for a description of the complementation experiment). The approximately 20-kb region deleted in fon4-3 has six annotated genes and apparently does not contain the genes for leaf color because the fon4-3 mutant had normal leaf color. Even though fon4-2 has a point mutation, its phenotype is slightly stronger that that of fon4-3; it is possible that this might be due to different subspecies genetic backgrounds of fon4-2 and fon4-3 (see “Materials and Methods”).
Figure 6.
Map-based cloning of FON4. A, Fine mapping of the FON4 gene on chromosome 11. FON4 was finally positioned on chromosome 11 within a 450-kb region flanked by the marker CH1142 and CH1143. Numbers represent recombinant events. B, Schematic representation of FON4 and mutation positions of three fon4 alleles. Black boxes indicate exons; intervening lines indicate introns. fon4-1 and fon4-3 mutations are attributed to an approximately 200-kb deletion and an approximately 20-kb deletion, respectively, whereas a G-to-A base change at the 3′ end of the first intron was found in fon4-2.
We further confirmed this CLV3-like gene as FON4 by a complementation test in which a 2,969-bp wild-type genomic DNA fragment, including the entire gene and 1,912 bp upstream of the start cordon, was transformed into the fon4-1 mutant. The abnormality of the inflorescence and flower was rescued in the transgenic plants (Supplemental Fig. S1). Furthermore, application of the synthetic 14-amino acid CLE motif peptides of FON4, FON4p, was able to rescue the SAM defect of the fon4-1 mutants (see below), indicating that FON4 is an important mediator of the CLV pathway, which confirmed that the CLV3-like gene was FON4 in rice.
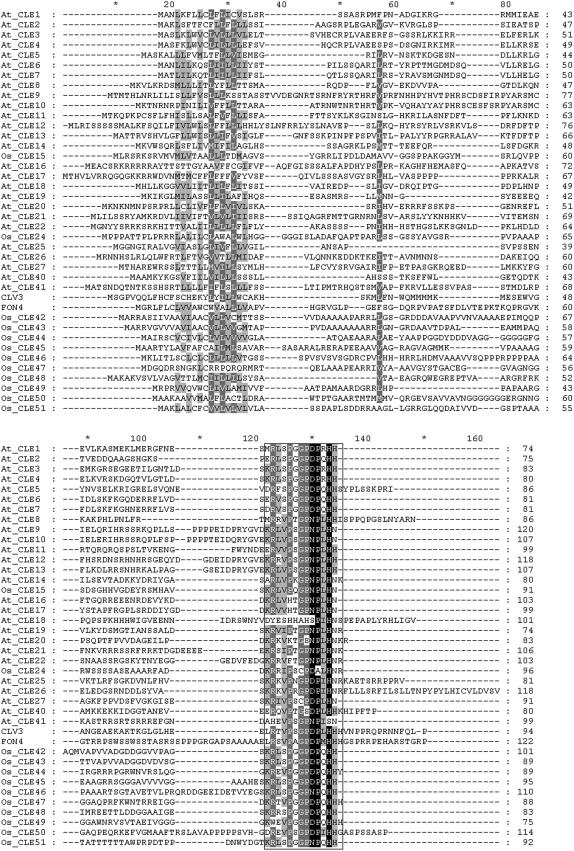
To compare the CLE motif of FON4 with those of other CLE members in both Arabidopsis and rice, we first searched the rice genomic database using the CLE motif of FON4 and identified a total of 13 members of the rice CLE family. Together with the 27 members in Arabidopsis (Cock and McCormick, 2001; Fiers et al., 2005), we carried out the alignment analysis of all known and predicted CLE proteins in rice and Arabidopsis. The result indicates that FON4 has little sequence similarity to other CLE proteins outside the CLE motif near the C termini, as is true for other CLE proteins (Fig. 7). The amino acid sequence identity and similarity of the FON4 and CLV3 CLE motifs were 78% (11/14) and 100%, respectively. In contrast, other predicted rice CLE proteins have CLE motifs with eight or nine residues identical to CLV3 and often contain dissimilar amino acid replacements. To elucidate the evolutionary relationship between FON4 and CLV3, a neighbor-joining phylogenetic tree was generated by using multiple sequence alignments of the CLE motifs of these 40 CLE family members from rice and Arabidopsis with bootstrap analysis (1,000 replicates; Supplemental Fig. S2). Even though there are only 14 amino acid residues in the CLE motif, we subdivided the 40 members of the CLE family into eight subfamilies, designated group 1 to group 8, according to clades with at least 50% bootstrap support. In this neighbor-joining tree, we grouped CLV3 and FON4 in group 3. The sequence conservation and the function similarity in meristem regulation support the hypothesis that the FON4 gene is the rice ortholog of CLV3.
Figure 7.
Alignment of CLE family members in rice and Arabidopsis. CLE motifs are boxed.
Expression Patterns of FON4
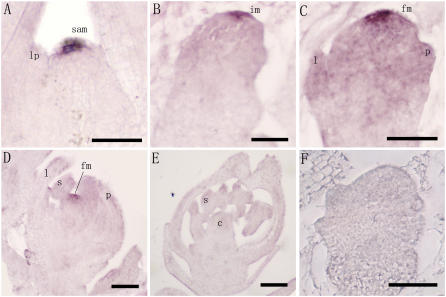
To study the function of FON4, the expression patterns of FON4 were analyzed using RNA in situ hybridization with FON4 antisense RNA as a probe. FON4 transcripts were detected in small groups of cells at the apex of the vegetative SAM, the inflorescence meristem, and the floral meristem (Fig. 8, A–D).These results indicate that FON4 is likely active in all rice vegetative and reproductive shoot meristems that are responsible for generating aerial organs. After the carpel has formed, expression of FON4 was no longer detectable (Fig. 8E). The region of FON4 expression probably represents the stem cell pool in SAMs, consistent with its function in controlling meristem size. The FON4 expression pattern is very similar to that of CLV3, which is mainly expressed in the overlying L1 and L2 cells in Arabidopsis (Fletcher et al., 1999).
Figure 8.
In situ analysis of FON4 expression in wild-type rice. A to D, FON4 transcripts are detected in the central zone of the meristems. A, Vegetative SAM. B, Inflorescence meristem. C, Floret at initiation of palea primordium. D, Floret before carpel initiation. E, After the carpel developed, no FON4 transcripts were detected. F, Sense probe as control. lp, Leaf primordia; im, inflorescence meristem; fm, floral meristem; l, lemma; p, palea; lo, lodicule; s, stamen; c, carpel. Bars = 50 μm.
The FON4 CLE Motif Affects the Formation of Aerial Organs by Inhibiting SAM Size
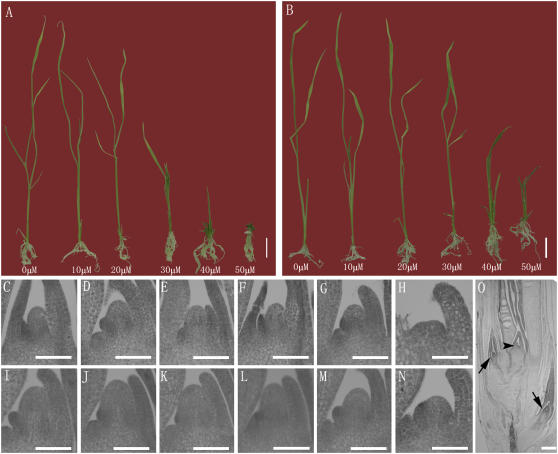
In Arabidopsis, exogenous application of three synthetic 14-amino acid CLE peptides, corresponding to the conserved CLE motifs of CLV3, CLE19, and CLE40, leads to consumption of root meristems by organ formation (Fiers et al., 2005). In addition, CLV3p (the CLE motif of CLV3) can restrict shoot meristem size (Fiers et al., 2006; Ni and Clark, 2006). To test whether the FON4 CLE motif also represents the primary functional domain of FON4, we obtained a synthetic 14-amino acid peptide, FON4p (Fig. 7), corresponding to the CLE motif of FON4. Then, wild-type and fon4-1 mutant rice seeds were germinated in cuvettes containing agar media with different concentrations of FON4p, ranging from 0 to 50 μm for 15 d. Although no obvious effect on root growth was observed at 15 d when treated with 0 to 50 μm FON4p, an inhibitory effect on aerial organ growth of the wild type was observed when treated with 20 μm FON4p. At 30 μm and higher concentrations, FON4p caused significant inhibition of apical growth in the wild type. Interestingly, visible inhibition of fon4-1 shoot growth was first detected at the FON4p concentration of 30 μm, and the reduction of growth was less for fon4-1 than the wild type at each concentration (Fig. 9, A and B). Fiers et al. (2006) recently reported that the synthetic CLV3p peptide was not stable in liquid media and was no longer present after 8 d. To rule out the possibility that the failure of FON4p to cause root meristem consumption was due to the late observation time, we repeated the experiment and observed the root meristem of wild-type rice treated with 50 μm FON4p at 4, 8, and 12 d, respectively; again, we did not observe obvious defects in root meristems (Supplemental Fig. S3).
Figure 9.
Effect of CLE peptides of FON4 (FON4p) on rice SAMs. Wild type and fon4-1 mutants treated with FON4p for 15 d. A, Wild-type rice treated with different concentrations of FON4p. The plants from left to right were treated with 0, 10, 20, 30, 40, and 50 μm FON4p, respectively. B, fon4-1 mutants treated with different concentrations of FON4p. Plants from left to right were treated with 0, 10, 20, 30, 40, and 50 μm FON4p, respectively. C to H, SAMs of wild-type plants treated with 0, 10, 20, 30, 40, and 50 μm FON4p, respectively. Note the minishing of the SAM while the concentration of FON4p increased. I to N, SAMs of fon4-1 treated with 0, 10, 20, 30, 40, and 50 μm FON4p, respectively. Note the minishing of the SAM while the concentration of FON4p increased. O, Wild-type plant treated with 50 μm FON4p. Arrowhead indicates the culm SAM; arrows indicated the tiller SAM. Bars = 2 mm (A and B); 100 μm (C–N); and 200 μm (O).
Consistent with shoot growth phenotypes, longitudinal sections of the shoot apices of the wild type and fon4-1 mutants after treatment with FON4p revealed that the application of FON4p caused the reduction of SAM size, probably due to an imbalance of the stem cell maintenance and organ primordia initiation (Fig. 9, C–N). It was clear that higher concentrations of FON4p caused greater meristem size reductions (Fig. 9, C–N). The fact that SAMs of wild type could be reduced more severely by FON4p than the fon4 SAMs suggests that, in the absence of endogenous FON4 function, more FON4p is needed to achieve the same effect. In addition, secondary apices could be observed near the base of the terminated shoots and short, bushy shoots were formed in both wild type and fon4-1 (Fig. 9O; data not shown). Overall, these results suggest that the CLE motif of FON4 could negatively regulate SAM size in rice. Strikingly, unlike the inhibitory effect of the CLV3 CLE peptide on Arabidopsis root growth, there were no obvious defects observed in root growth and root apical meristems treated with FON4p (Fig. 10, B and D). It is possible that regulation of rice root meristem development depends on other genes distinct from FON4.
Figure 10.
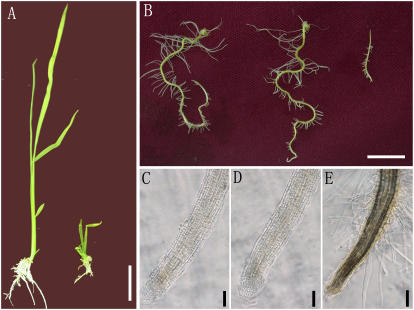
Effect of CLE peptides of CLV3 (CLV3p) in wild-type rice. A, Treatment with CLV3p delayed the development of rice. Right is treated with 50 μm CLV3p; left is control. Bar = 2 cm. B, Root morphology of wild-type rice treated with no peptide, 50 μm FON4p, and CLV3p, respectively (from left to right). Bar = 1 cm. C to E, Lateral root tips of wild-type rice treated with no peptide (C), 50 μm FON4p (D), and CLV3p (E). Note consumption of the root meristem after treatment with CLV3p. Bars = 100 μm.
Exogenous CLV3 CLE Peptide Leads to Developmental Defects in Both Rice Shoot and Root
To further investigate whether the function of FON4 is similar to that of CLV3, we treated Arabidopsis seeds with 25 and 50 μm FON4p and rice seeds with the CLV3 CLE motif (CLV3p) at 50 μm, respectively. After 4-, 8-, 12-, and 15-d treatments, we did not detect any abnormality of root and above-ground organs in Arabidopsis treated with FON4p compared with the control (data not shown); however, CLV3p could reduce Arabidopsis root growth at a concentration of 10 μm (Fiers et al., 2005), implying that FON4p is functionally distinct from CLV3p and unable to change Arabidopsis root development. It was shown that the Arabidopsis CLE19 and CLE40 peptides with only 50% to 64% similarity to CLV3p were able to inhibit root growth in Arabidopsis (Hobe et al., 2003; Fiers et al., 2004, 2005; Wang et al., 2005). In addition, transgenes encoding the CLE domains of CLE1 and CLE6 have the ability to rescue clv3-1 (Ni and Clark, 2006), suggesting these CLE motifs can be recognized by the receptors to activate the CLV-signaling pathway. Our results with FON4p suggest that it is different from CLV3p at key residues, preventing this rice CLE motif from being recognized by the Arabidopsis receptors. Intriguingly, treatment of rice seeds with CLV3p caused not only the defect in aerial parts resembling that of wild-type rice treated with FON4p (Fig. 10A), but also the consumption of root meristems (Fig. 10, B, C, and E). This suggests that CLV3p can be recognized by receptors in both rice and Arabidopsis.
DISCUSSION
FON4 Regulates Apical Meristem Size and Determinacy of Floral Meristems
We isolated and characterized three rice mutant alleles, fon4-1, fon4-2, and fon4-3. These mutants had increased floral organ numbers, especially in the inner whorls, similar to those of the fon1-2 and fon1-3 mutants, which contain a point mutation and a T-DNA insertion, respectively, in the FON1 gene (Suzaki et al., 2004; Moon et al., 2006). The increased floral organ numbers in fon4 mutants can be explained by enlarged floral meristems, which allowed the initiation of a greater than normal number of floral organ primordia. In addition, the fon4 floral meristem was not consumed by the formation of the carpel and ovule primordia, but instead maintained a persistent meristem even after producing a few carpel primordia. This likely contributes to a more dramatic increase of stamen and carpel numbers in the fon4 mutants than those of the outer whorl organs. The abnormal persistence of the fon4 floral meristem suggested that FON4 is needed for normal determinacy of the floral meristem. The similarity of phenotypes further suggests that FON1 and FON4 probably are involved in the same pathway for the regulation of floral meristem development.
In addition to the similarities between fon1 and fon4 mutants, we also observed some apparent differences between fon1 and fon4 mutants. Besides the floral defects, fon4 mutants also had enlarged shoot apical and inflorescence meristems, resulting in an increase of culm thickness and primary rachis branch number. Therefore, we propose that FON4 normally restricts the sizes of the SAM, the inflorescence meristem, and the floral meristem, thereby regulating both vegetative and reproductive development in rice. Unlike fon4 mutants, no obvious abnormalities in the vegetative and inflorescence meristems were observed in fon1-2 (Suzaki et al., 2004), whereas reduced tiller number and increased apical dominance were found in fon1-3 and fon1-4 (Moon et al., 2006). fon4 mutants also had abnormal floral organ morphology, including homeotic conversion of glumes and stamens. Such phenotypes were limited to lodicules and stamens in fon1 and fon2 mutants (Nagasawa et al., 1996). Therefore, it is possible that FON4 has a more extensive function than FON1 and FON2. Alternatively, FON1 and FON2 might have a greater degree of functional redundancy with genes other than FON4.
FON4 Is the Putative Ortholog of Arabidopsis CLV3
The CLV pathway is one of the best-characterized signaling mechanisms in the regulation of meristem size. It is thought that CLV1 and CLV2 form a disulfide-linked heterodimer of approximately 185 kD (Trotochaud et al., 1999). CLV3 is a putative peptide ligand that interacts with a disulfide-linked CLV1/CLV2 receptor complex. The presumed extracellular domain of CLV1 is composed almost exclusively of LRRs, which can potentially bind to ligands (Clark et al., 1997).
To support the hypothesis that the CLV pathway is conserved in monocots, rice genes FON1 and OsLRK1 were recently studied (Kim et al., 2000; Suzaki et al., 2004). The FON1 gene encoding a putative transmembrane receptor kinase is highly similar to CLV1. In addition, the fon1-2 mutant with a point mutation in the LRR domain also causes a severe phenotype. The similarity between FON1 and CLV1 in the LRRs suggests that they may interact with related ligands. In maize, fea2 and td1 encode putative orthologs of CLV2 and CLV1, respectively (Taguchi-Shiobara et al., 2001; Bommert et al., 2005). Our results presented here indicate that the rice gene FON4 encodes a putative small secreted protein, which has a CLE motif that is highly similar in sequence to that of CLV3. The presence of a 25-amino acid putative signal sequence at the N terminus of FON4 is consistent with the hypothesis that FON4 is secreted to the extracellular space for binding with its receptors. Taken together, we propose that FON4 is a putative ligand of FON1 and functions to limit meristem size in rice. At the same time, the differences in defects caused by fon1 and fon4 mutations suggest that FON1 is probably not the only receptor of FON4.
FON4 is expressed at the apex of shoot meristems, very similar to that of CLV3 in Arabidopsis (Fletcher et al., 1999). However, FON1 is expressed throughout the whole meristem and also in lateral organ primordia (Suzaki et al., 2004). This expression pattern is markedly different from that of the Arabidopsis CLV1 gene, which is expressed in the L3 layer of the meristems (Clark et al., 1997). Nevertheless, the domain of FON1 expression is larger than that of FON4. Recent evidence indicated that the CLE motif of CLV3 can regulate the WUS expression pattern (Fiers et al., 2006). Thus, we hypothesize that the conserved feedback loop regulatory systems consisting of the CLV ligand-receptor system and the homeodomain protein WUS may exist in rice and play a central role in regulating stem cell number. There might be an organizing center within the FON1 expression region, where FON4 cannot reach, possibly due to some sequestration mechanism (Casanova and Struhl, 1993; Chen and Struhl, 1996; Hajnal et al., 1997). The organizing center may be marked by a stem cell identity gene, homologous to Arabidopsis WUS, which is negatively regulated by FON4. In agreement with this hypothesis, we demonstrated that treatment with the CLE motif of FON4 caused termination of the SAM.
Functional Divergence of the CLE Motifs of FON4 and CLV3
Overexpression of three CLE family genes, CLV3, CLE19, and CLE40, caused consumption of the root meristems in Arabidopsis (Hobe et al., 2003; Fiers et al., 2004; Wang et al., 2005). Treatment with synthetic 14-amino acid CLE peptides corresponding to the conserved CLE motif of CLV3, CLE19, and CLE40 were able to mimic these overexpression phenotypes; in addition, some expressed CLE motifs can nearly replace CLV3 function (Fiers et al., 2005, 2006; Ni and Clark, 2006), these results indicate that CLE motifs of CLE family members are the major functional regions of these CLE genes. Similarly, treatment with the 14-amino acid CLE peptide of FON4 could reduce the SAM size, suggesting that the FON4 CLE motif is also the main functional domain of FON4.
However, no obvious effect was found in the rice root apical meristem when treated with the FON4p. There are two possible explanations for this; one is that there is a CLV-like signaling pathway regulating rice root apical meristem size, but the ligand is different from FON4 such that the CLE motif of FON4 could not be recognized by the root receptors. In contrast, the CLE functional motif of CLV3 can be recognized by the receptors in both shoot and root apical meristems in Arabidopsis. Another possibility is that there is no CLV-like signaling pathway in regulating rice root apical meristem size. This is unlikely because treatment of rice roots with CLV3p caused consumption of the root meristem, suggesting that a CLV-like signal pathway is involved in regulation of rice root meristem development. Thus, the functions of CLE genes in regulating rice root apical meristem development have diverged from those of FON4.
MATERIALS AND METHODS
Plant Materials
Three rice (Oryza sativa) mutants, fon4-1, fon4-2, and fon4-3, with an increased number of floral organs, were used in this study. fon4-1 was isolated from the M2 populations of 9522, a cultivar of japonica, by mutagenesis with γ-rays (Chu et al., 2005; Liu et al., 2005). M2 progeny tests of heterozygotes yielded a segregation of 299 normal and 101 mutant plants (χ2 [3:1] = 0.013; P > 0.9), indicating monofactorial recessive inheritance of the mutant characteristics. The other two spontaneous mutants, fon4-2 and fon4-3, whose genetic backgrounds were japonica and indica, respectively, had similar phenotypes with fon4-1. Allelism tests indicated that fon4-2 and fon4-3 were alleles of fon4-1. The 9522 cultivar was used as a wild-type strain for observation of phenotypes and for RNA in situ analysis. All plants were grown in the paddy field of the Shanghai Academy of Agriculture Sciences.
Morphological Analysis and Measurement of Meristem Size
The sample was fixed according to the method of Itoh et al. (2000). SEM observation was performed as described previously (Nagasawa et al., 2003). Observation by Nomarski microscopy was performed according to the protocol outlined by Suzaki et al. (2004). The width and height of the meristems were measured by using the methods of Nagasawa et al. (1996).
Map-Based Cloning of FON4
The fon4 locus was first mapped to a region between simple sequence repeat marker PSM415 (5′-CTCCCTCCTGCTCGTTTTCTC-3′; 5′-ACCTAGTTAGGTAGCGCCCAT-3′) and RM6094 on the long arm of chromosome 11 by using 96 F2 plants of fon4-1 and Guang-lu-ai 4 (spp. indica). Then, by using 2,100 F2 plants, the FON4 locus was narrowed to a region between two insertion/deletion markers, CH1142 (5′-TGTAGCTCAGAGGTGCTGTGT-3′; 5′-TGCTTGGTGGCAATCGT-3′) and CH1143 (5′-CAAAAATGAGTACACTCCCCTT-3′; 5′-TCATCACACCATCACCCATAC-3′), which were designed as described previously (Shen et al., 2004).
We then used the amino acid sequences of CLV1, CLV2, and CLV3 of Arabidopsis (Arabidopsis thaliana) as queries, and carried out tBLASTn (Altschul et al., 1997) searches in the rice genome database. The open reading frame of the putative gene was predicted by using the GeneMark gene prediction program (Lukashin and Borodovsky, 1998; Lomsadze et al., 2005). Mutations in fon4-1, fon4-2, and fon4-3 were determined by PCR amplification and sequence analysis using the primers designed based on the rice genome sequence. cDNA of FON4 was amplified and total RNAs were purified from young panicles by reverse transcription-PCR by using the FON4-specific primers: FP1 (5′-GTGTGTTTGCTTGACATGGGCCG-3′) and RP1 (5′-GATTTGCACCGTCCGTCCGTTC-3′). The nucleotide sequence for the cDNA of FON4 can be found in GenBank (accession no. DQ836359) and the gene structure of FON4 was deduced by comparing the sequence of cDNA and genomic sequence. A 2,969-bp DNA fragment containing the coding region, 1,912 bp of the sequence upstream of the start codon and 412 bp downstream of the stop codon, was amplified from genomic DNA and cloned into a binary vector pCAMBIA 1301, then transformed into fon4-1 by Agrobacterium-mediated transformation (Hiei et al., 1994).
Database Search and Multiple Sequence Alignments
To search for rice CLE genes, tBLASTn (Altschul et al., 1997) provided by The Institute for Genomic Research (http://tigrblast.tigr.org/tgi) was performed. Using the CLE motif of FON4 as a query sequence, we found many candidates. These new identified sequences were then used as queries to repeat the above search until no novel sequences were revealed. Multiple sequence alignment using the MUSCLE program (version 3.52; http://www.drive5.com/muscle; Edgar and Robert, 2004) with the default parameters were performed on the rice CLE sequences and the reported Arabidopsis CLE genes (Cock and McCormick, 2001; Fiers et al., 2005). A phylogenetic tree was constructed with the aligned CLE protein sequences of rice and Arabidopsis using the MEGA program (version 3.0; http://www.megasoftware.net/index.html; Kumar et al., 2004) using the neighbor-joining method with the following parameters: Poisson correction, pairwise deletion, and interior branch test (1,000 replicates; random seed).
In Situ Hybridization
FON4-specific probes were generated by inserting the cDNA fragment of FON4 in pMD18-T (TaKaRa); then this fragment, digested with EcoRI and HindIII, was subcloned into pBluescript SK(+) and sequenced to confirm the orientation. Sense and antisense probes were transcribed in vitro from the T7 or T3 promoter with respective RNA polymerases using the digoxigenin RNA-labeling kit (Roche). OSH1 and DL probes were prepared as described previously (Sato et al., 1996; Yamaguchi et al., 2004).
Samples were fixed in formaldehyde acetic acid (5% acetic acid, 50% ethanol, and 3.7% formaldehyde in water) and embedded in Paraplast Plus (Sigma). Microtome sections, 8-μm thick, were applied to glass slides (Sigma). RNA hybridization and immunological detection of the hybridized probes were performed according to the protocol of Kouchi and Hata (1993).
In Vitro Peptide Treatment
Treatment of rice and Arabidopsis seeds with CLE peptides was performed according to the method described by Fiers et al. (2005). The peptides were synthesized by GL Biochem. The sterilized rice and Arabidopsis seeds were planted on media containing different concentrations of peptide, Murashige and Skooge microelements and macroelements (Duchefa), and 3% (w/v) Suc, pH 5.8, with 1.5% (w/v) agar. Rice seeds were cultured in a greenhouse with a temperature of 26°C, 10 h light per day, and Arabidopsis seeds were cultured in a greenhouse with a temperature of 23°C, 16 h light per day.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number DQ836359.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Complementation of the fon4-1 mutant by FON4 genomic DNA.
Supplemental Figure S2. The phylogenetic tree constructed using the CLE motifs of 27 and 13 CLE family genes from Arabidopsis and rice, respectively.
Supplemental Figure S3. Effect of FON4p on rice root meristems.
Supplementary Material
Acknowledgments
We gratefully acknowledge Z.-J. Luo and M.-J. Chen for mutant screening and generation of F2 populations for mapping, C.-M. Zhang for rice transformation, and X.-S. Gao for SEM.
This work was supported by the National Key Basic Research Developments Program of the Ministry of Science and Technology, People's Republic of China (grant nos. 2006CB101700, 2005CB120802, and 2001CB109002), the National “863” High-Tech Project (grant no. 2005AA2710330), the Shanghai Municipal Committee of Science and Technology (grant no. 03JC14061), the Program for New Century Excellent Talents in University (grant no. NCET–04–0403), the Shuguang Scholarship (grant no. 04SG15), the Shanghai Institutes of Biological Sciences (Reproductive Development Project), and the U.S. Department of Energy (grant no. DE–FG02–02ER15332).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Dabing Zhang (zhangdb@sjtu.edu.cn).
The online version of this article contains Web-only data.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton MK, Poethig RS (1993) Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and shoot meristemless mutant. Development 119: 823–831 [Google Scholar]
- Bendtsen JD, Nielsen H, Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783–795 [DOI] [PubMed] [Google Scholar]
- Bommert P, Lunde C, Nardmann J, Vollbrecht E, Running M, Jackson D, Hake S, Werr S (2005) Thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development 132: 1235–1245 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Eshed Y (2000) Formation and maintenance of the shoot apical meristem. Trends Plant Sci 5: 110–115 [DOI] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Casanova J, Struhl G (1993) The torso receptor localizes as well as transduces the spatial signal specifying terminal body pattern in Drosophila. Nature 362: 152–155 [DOI] [PubMed] [Google Scholar]
- Chen Y, Struhl G (1996) Dual roles for patched in sequestering and transducing hedgehog. Cell 87: 553–563 [DOI] [PubMed] [Google Scholar]
- Chu HW, Liu HS, Li H, Wang HM, Wei JL, Li N, Ding SY, Huang H, Ma H, Huang CF, et al (2005) Genetic analysis and mapping of the rice leafy-hull mutant Oslh. J Plant Physiol Mol Biol 31: 594–598 [PubMed] [Google Scholar]
- Clark SE (2001) Meristems: start your signaling. Curr Opin Plant Biol 4: 28–32 [DOI] [PubMed] [Google Scholar]
- Clark SE, Jacobsen SE, Levin J, Meyerowitz EM (1996) The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 122: 1567–1575 [DOI] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM (1993) CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119: 397–418 [DOI] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM (1995) CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121: 2057–2067 [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585 [DOI] [PubMed] [Google Scholar]
- Cock JM, McCormick S (2001) A large family of genes that share homology with CLAVATA3. Plant Physiol 126: 939–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, Robert C (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endrizzi K, Moussain B, Haecker A, Levin JZ, Laux T (1996) The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J 10: 967–979 [DOI] [PubMed] [Google Scholar]
- Fiers M, Golemiec E, van der Schors R, van der Geest L, Li KW, Stiekema WJ, Liu CM (2006) The CLV3/ESR motif of CLV3 is functionally independent from the nonconserved flanking sequences. Plant Physiol 141: 1284–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M, Golemiec E, Xu J, van der Geest L, Heidstra R, Stiekema W, Liu CM (2005) The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 17: 2542–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M, Hause G, Boutilier K, Casamitjana-Martinez E, Weijers D, Offringa D, van der Geest L, van Lookeren Campagne M, Liu CM (2004) Mis-expression of the CLV3/ESR-like gene CLE19 in Arabidopsis leads to a consumption of root meristem. Gene 327: 37–49 [DOI] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Hajnal A, Whitfield CW, Kim SK (1997) Inhibition of Caenorhabditis elegans vulval induction by gap-1 and by let-23 receptor tyrosine kinase. Genes Dev 11: 2715–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hobe M, Muller R, Grunewald M, Brand U, Simon R (2003) Loss of CLE40, a protein functionally equivalent to the stem cell restricting signal CLV3, enhances root waving in Arabidopsis. Dev Genes Evol 213: 371–381 [DOI] [PubMed] [Google Scholar]
- Itoh JI, Kitano H, Matsuoka M, Nagato Y (2000) SHOOT ORGANIZATION genes regulate shoot apical meristem organization and the pattern of leaf primordium initiation in rice. Plant Cell 12: 2161–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Trotochaud AE, Clark SE (1999) The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11: 1925–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Qian Q, Mao L, Zhou QY, Zhai WX (2005) Characterization of the rice floral organ number mutant fon3. J Integ Plant Biol 47: 100–106 [Google Scholar]
- Kayes JM, Clark SE (1998) CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125: 3843–3851 [DOI] [PubMed] [Google Scholar]
- Kim C, Jeong DH, An G (2000) Molecular cloning and characterization of OsLRK1 encoding a putative receptor-like protein kinase from Oryza sativa. Plant Sci 152: 17–26 [Google Scholar]
- Kouchi H, Hata S (1993) Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol Gen Genet 238: 106–119 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5: 150–163 [DOI] [PubMed] [Google Scholar]
- Laux T, Mayer KFX, Berger J, Jürgens G (1996) The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122: 87–96 [DOI] [PubMed] [Google Scholar]
- Liu HS, Chu HW, Li H, Wang HM, Wei JL, Li N, Ding SY, Huang H, Ma H, Huang CF, et al (2005) Genetic analysis and mapping of rice (Oryza sativa L.) male-sterile (OsMS-L) mutant. Chin Sci Bull 2: 122–125 [Google Scholar]
- Lomsadze A, Ter-Hovhannisyan V, Chernoff Y, Borodovsky M (2005) Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res 33: 6494–6506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashin AV, Borodovsky M (1998) GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res 26: 1107–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Moon S, Jung KH, Lee DE, Lee DY, Lee J, An K, Kang HG, An G (2006) The rice FON1 gene controls vegetative and reproductive development by regulating shoot apical meristem size. Mol Cells 21: 147–152 [PubMed] [Google Scholar]
- Muller R, Borghi L, Kwiatkowska D, Laufs P, Simon R (2006) Dynamic and compensatory responses of Arabidopsis shoot and floral meristems to CLV3 signaling. Plant Cell 18: 1188–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa N, Miyoshi M, Kitano H, Satoh H, Nagato Y (1996) Mutations associated with floral organ number in rice. Planta 198: 627–633 [DOI] [PubMed] [Google Scholar]
- Nagasawa N, Miyoshi M, Sano Y, Satoh H, Hirano HY, Sakai H, Nagato Y (2003) SUPERWOMAN 1 and DROOPING LEAF genes control floral organ identity in rice. Development 130: 705–718 [DOI] [PubMed] [Google Scholar]
- Ni J, Clark SE (2006) Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiol 140: 726–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ride JP, Davies EM, Franklin FC, Marshall DF (1999) Analysis of Arabidopsis genome sequence reveals a large new gene family in plants. Plant Mol Biol 39: 927–932 [DOI] [PubMed] [Google Scholar]
- Sato Y, Hong SK, Tagiri A, Kitano H, Yamamoto N, Nagato Y, Matsuoka M (1996) A rice homeobox gene, OSH1, is expressed before organ differentiation in a specific region during early embryogenesis. Proc Natl Acad Sci USA 93: 8117–8122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jürgens G, Laux T (2000) The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Shen YJ, Jiang H, Jin JP, Zhang ZB, Xi B, He YY, Wang G, Wang C, Qian L, Li X, et al (2004) Development of genome-wide DNA polymorphism database for map-based cloning of rice genes. Plant Physiol 135: 1198–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T, Sato M, Ashikari M, Miyoshi M, Nagato Y, Hirano HY (2004) The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development 131: 5649–5657 [DOI] [PubMed] [Google Scholar]
- Taguchi-Shiobara F, Yuan Z, Hake S, Jackson D (2001) The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev 15: 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotochaud AE, Hao H, Wu G, Yang ZB, Clark SE (1999) The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell 11: 393–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoosthuyse V, Miege C, Dumas C, Cock JM (2001) Two large Arabidopsis thaliana gene families are homologous to the Brassica gene superfamily that encodes pollen coat proteins and the male component of the self-incompatibility response. Plant Mol Biol 46: 17–34 [DOI] [PubMed] [Google Scholar]
- Wang X, Mitchum MG, Gao B, Li C, Diab H, Baum TJ, Hussey RS, Davis EL (2005) A parasitism gene from a plant parasitic nematode with function similar to CLAVATA3/ESR (CLE) of Arabidopsis thaliana. Mol Plant Pathol 6: 187–191 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Nagasawa N, Kawasaki S, Matsuoka M, Nagato Y, Hirano HY (2004) The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16: 500–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.