Abstract
Proteinaceous aspartic proteinase inhibitors are rare in nature and are described in only a few plant species. One of them corresponds to a family of cathepsin D inhibitors (CDIs) described in potato (Solanum tuberosum), involving up to 15 isoforms with a high sequence similarity. In this work, we describe a tomato (Solanum lycopersicum) wound-inducible protein called jasmonic-induced protein 21 (JIP21). Sequence analysis of its cDNA predicted a putative function as a CDI. The JIP21 gene, whose protein has been demonstrated to be glycosylated, is constitutively expressed in flowers, stem, and fruit, and is inducible to high levels by wounding and methyl jasmonate in leaves of tomato plants. The genomic sequence of JIP21 shows that the gene is intronless and reveals the presence of both a methyl jasmonate box (TGACT) and a G-box (CACGT) in the promoter. In contrast to the presumed role of JIP21 based on sequence analysis, a detailed biochemical characterization of the purified protein uncovers a different function as a strong chymotrypsin inhibitor, which questions the previously predicted inhibitory activity against aspartic proteinases. Moreover, Egyptian cotton worm (Spodoptera littoralis) larvae fed on transgenic tomato plants overexpressing JIP21 present an increase in mortality and a delay in growth when compared with larvae fed on wild-type plants. These larvae belong to the Lepidoptera family whose main digestive enzymes have been described as being Ser proteases. All these results support the notion that tomato JIP21 should be considered as a chymotrypsin inhibitor belonging to the Ser proteinase inhibitors rather than a CDI. Therefore, we propose to name this protein tomato chymotrypsin inhibitor 21 (TCI21).
Plants respond to insect attack or wounding by transcriptional activation of a large number of genes. Proteins encoded by these wound-inducible genes perform different functions, such as repairing damaged tissues, participating in activation of the wound-signaling pathway, adjusting plant metabolism to the imposed nutritional demands, or inhibiting growth of the predator insect (Reymond et al., 2000; Ryan, 2000; León et al., 2001). Among these wound-induced antinutritional proteins, proteinase inhibitors (PINs) are the main components because they interfere with the digestive systems of the attacking herbivores, limiting their growth and development. Activation of PIN genes is mediated by jasmonic acid as a result of the activation of the octadecanoic pathway by insect attack. This activation not only occurs in the wounded leaves, but also in the distal ones (Farmer and Ryan, 1990; Ryan, 1990; Schaller, 2001).
PINs have been classified into four groups according to the protease they inhibit: Ser protease, Cys protease, aspartic protease, or metallocarboxy protease inhibitors. Ser proteinases are divided into two superfamilies: subtilisin and chymotrypsin families. The latter includes digestive enzymes, such as trypsin, chymotrypsin, and elastase. Inhibitors of these Ser proteinases have been described in many plant species and are widespread throughout the plant kingdom. The best studied is the soybean (Glycine max) trypsin inhibitor (STI), a representative member of the Kunitz-type Ser PIN family whose characterization has provided a basic understanding of the mechanism of action of the remaining inhibitors (Laskowski and Qasim, 2000). Other families of Ser PINs are represented by the soybean Bowman-Birk inhibitor (BBI) and PINs I and II of potato (Solanum tuberosum; Sánchez-Serrano et al., 1986; Cleveland et al., 1987; Birk, 1996).
In contrast to this broad distribution of the Ser PIN family, proteinaceous inhibitors of aspartic proteinases are less numerous and have been described in only a few plant species: potato (Keilova and Tomasek, 1976a, 1976b), tomato (Solanum lycopersicum; Werner et al., 1993), wheat (Triticum aestivum; Galleschi et al., 1993), Vicia sativa (Roszkowska-Jakimiec and Bankowska, 1998), Anchusa strigosa (Abuereish, 1998), and squash (Cucurbita pepo; Christeller et al., 1998). The biochemical characterization of the potato and V. sativa members showed that they were both cathepsin D inhibitors (CDIs), whereas the proteins from A. strigosa and squash inhibit pepsin, a digestive aspartic proteinase. In potato, a large family of CDIs has been described. The two first isoforms purified (PDI and NDI) were biochemically characterized, showing inhibitory activity against cathepsin D and Ser proteinases (Keilova and Tomasek, 1976a; Mares et al., 1989; Ritonja et al., 1990) and displaying sequence similarities with the STI. Since then, up to 15 isoforms have been found in potato and they have been classified as CDIs exclusively on the basis of sequence analysis. Potato CDIs constitutively accumulate in tubers and flower buds and are also induced by wounding in potato leaves (Strukelj et al., 1990; Hildmann et al., 1992; Maganja et al., 1992; Hannapel, 1993; Herbers et al., 1994; Ishikawa et al., 1994a; Kreft et al., 1997).
The use of PINs to transform crop plants for resistance to insect pests has been well documented (for reviews, see Jouanin et al., 1998; Schuler et al., 1998; Lawrence and Koundal, 2002). Selecting the appropriate PINs to generate this resistance implies taking into account the main digestive enzymes in the midgut of the target insect. In this sense, Ser PINs are effective against Lepidoptera (Hilder et al., 1987; Johnson et al., 1989; McManus et al., 1994; Duan et al., 1996; Li et al., 1998; De Leo et al., 2001). On the other hand, aspartic and Cys PINs have been used to generate resistance against coleopteran species (Orr et al., 1994; Leplé et al., 1995; Girard et al., 1998; Lecardonnel et al., 1999).
In this work, we describe the characterization of jasmonic-induced protein 21 (JIP21), a tomato wound- and jasmonate-inducible protein. Sequence analysis predicted a putative function as a CDI. A thorough biochemical study has been performed with the purified plant protein, which revealed a lack of activity against cathepsin D or other aspartic proteinases. Instead, JIP21 shows powerful activity as a chymotrypsin inhibitor. Tomato plants overexpressing JIP21 have been generated and resistance assays against larvae of the lepidopteran Egyptian cotton worm (Spodoptera littoralis) have been carried out, confirming the new proposed function.
RESULTS
Purification of JIP21 Protein and Cloning of cDNA and Genomic Sequences
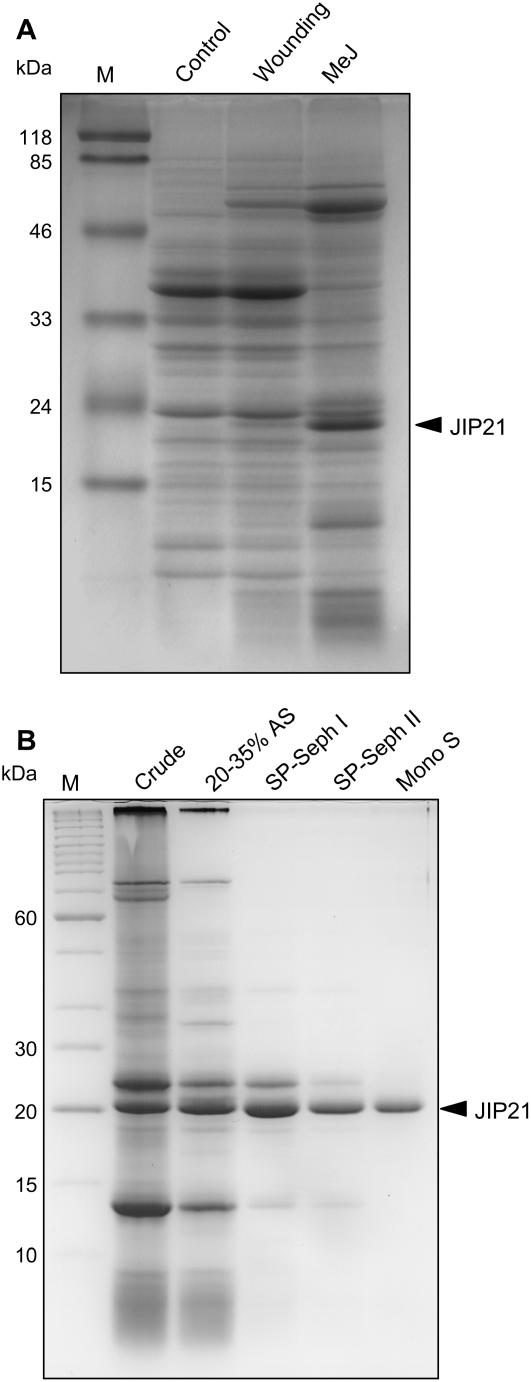
Comparing the electrophoretic profiles of protein extracts from control and wounded tomato leaves reveals the outstanding presence of a 21-kD polypeptide that accumulates upon wounding. Accumulation of this polypeptide is even higher when leaves are treated with 2 mm methyl jasmonate (MeJ; Fig. 1A). We purified this protein, named JIP21, from acidic protein extracts of tomato leaves treated with MeJ by ammonium sulfate fractionating by two serial chromatographies on a SP-Sephadex C25 and a final FPLC step through a Mono S HR 5/5 (Fig. 1B). Once purified, specific polyclonal antibodies were obtained and used to immunoscreen a cDNA library constructed from mRNAs of jasmonate-treated tomato leaves. Several identical clones were isolated and the longest one was used as a probe to obtain complete cDNA. This sequence proved to be very abundant in our library because about 7% of the cDNA clones corresponded to JIP21 cDNA. In addition, its sequence analysis revealed that it could be assumed as a possible CDI (Werner et al., 1993).
Figure 1.
SDS-PAGE (14%). A, Detection of tomato JIP21. Crude protein extracts of untreated control tomato leaves (control); wounded tomato leaves harvested upon 48 h (wounding); and tomato leaves harvested after 48 h of a 2 mm MeJ treatment (MeJ). B, Purification of JIP21 protein. Crude, Crude extract of MeJ-treated tomato leaves; 20% to 35% AS, proteins present after 20% to 35% ammonium sulfate fractionating; SP-Seph I and II, enrichment achieved by two consecutive chromatographies on SP-Sephadex C25; MonoS, final FPLC step using a Mono S HR 5/5 column. Lane M, Mr standards. Proteins were stained with Coomassie Brilliant Blue R-250. Bands corresponding to JIP21 are indicated with an arrowhead.
With the complete cDNA sequence, a genomic library constructed in the λ-EMBL vector was screened and the corresponding genomic sequence was isolated (GenBank accession no. AJ295638). The JIP21 gene is intronless as are the genomic sequences described for potato CDIs (Herbers et al., 1994; Ishikawa et al., 1994a, 1994b). The tomato genomic sequence contains a promoter of 1.4 kb, which is highly similar to the potato promoters described, and a 3′ region spanning more than 700 bp downstream from the open reading frame. An analysis of conserved motifs in the 5′-promoter sequence revealed the presence of a MeJ box (TGACT) between positions −384 and −380 and a putative inverted G-box (CACGT) at position −963. The G-box has been previously described in promoters of wound-inducible genes such as the potato pin II (Sánchez-Serrano et al., 1987; Thornburg et al., 1987), the soybean vspB (Mason et al., 1993), the tomato Thr deaminase (Samach et al., 1991), or the potato sporamin (Wang et al., 2002). In addition, an in silico analysis of the 5′-promoter regions of Arabidopsis (Arabidopsis thaliana) genes, identified as wound inducible by two-step microarray analysis, revealed G-box-related motifs in a significant proportion of the promoters (Delessert et al., 2004). All this supports the idea that the JIP21 gene is endowed with the structural and functional characteristics of a wound-inducible gene.
Pattern of Expression
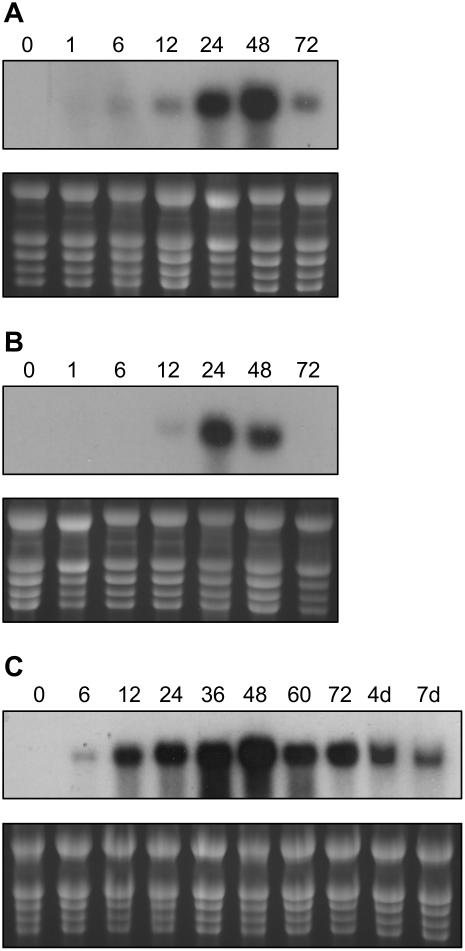
The expression of PINs in tomato is well known to be wound inducible (Graham et al., 1985a, 1985b; Martineau et al., 1991; Díez-Díaz et al., 2004). To follow the time course of JIP21 mRNA accumulation upon wounding, we performed northern-blot analysis from wounded and systemic leaves harvested 0, 1, 6, 12, 24, 48, and 72 h after wounding. In wounded leaves, mRNA accumulation can be detected as early as 6 h, whereas induction is not detected until 24 h after wounding in the immediately upper (distal) leaves. In the local response, the maximal accumulation takes place at 48 h and at 36 h in the distal one, where the mRNA levels decline faster (Fig. 2, A and B). The pattern of induction by MeJ is similar to that of wounding, but, in general terms, is much stronger and remains longer, even for 7 d after treatment (Fig. 2C). Similar to the rest of the PINs, JIP21 was not found to be induced by salicylic acid, ethylene, or pathogenic (citrus exocortis viroid and Pseudomonas syringae pv tomato) infections (data not shown).
Figure 2.
Time-course analysis of JIP21 mRNA accumulation in tomato leaves, in response to wounding or MeJ treatment by northern blot. A, Local response to wounding. mRNAs from tomato wounded leaves, harvested 0, 1, 6, 12, 24, 48, and 72 h after wounding. B, Distal response to wounding. mRNAs from immediately upper tomato leaves, harvested 0, 1, 6, 12, 24, 48, and 72 h after wounding. C, Response to MeJ treatment. mRNAs from tomato leaves, harvested 0, 6, 12, 24, 36, 48, 72 h, 4 d, and 7 d after spraying plants with a 2 mm solution.
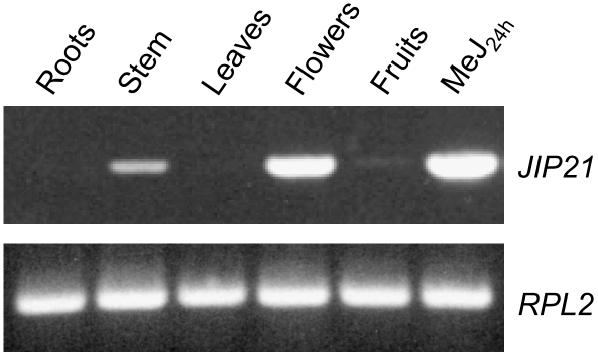
Regarding the JIP21 levels in other tissues, we could detect the constitutive presence of JIP21 in flowers by northern-blot analysis. Moreover, we detected constitutive levels of JIP21 not only in flowers, but also in stem and fruits by reverse transcription (RT)-PCR using specific primers against the cDNA sequence (Fig. 3). All PCR products have been cloned and sequenced and they all correspond to the original cDNA. These results appear to indicate that constitutive levels are not due to the expression of JIP21 isoforms in these tissues.
Figure 3.
Constitutive mRNA levels of JIP21 in different tissues (roots, stem, leaves, flowers, and fruits) of tomato, as analyzed by RT-PCR. mRNA from tomato leaves harvested 24 h after a 2 mm MeJ treatment is included as a positive control. Top, PCR performed with specific JIP21 primers. Bottom, PCR performed with specific primers of the RPL2 gene (Fleming et al., 1993) used as a control for RT.
N-Glycosylation of JIP21
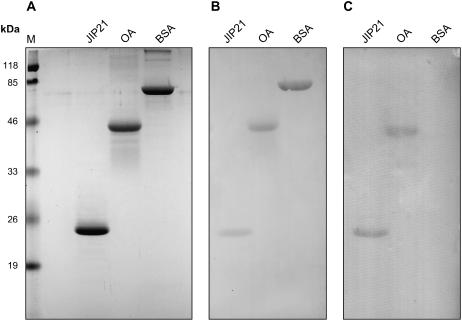
Sequence analysis of the JIP21 protein shows the presence of a putative N-glycosylation site at Asn 51. This site has also been described in some of the CDIs characterized in potato, but no glycosylation studies have been performed to date. Using the periodic acid-Schiff staining technique associated with western-blot analysis (Strömqvist and Gruffman, 1992), we found that JIP21 is effectively a glycoprotein (Fig. 4). In this assay, ovoalbumin protein has been used as a positive control of periodic acid-Schiff staining, and bovine serum albumin as a negative one.
Figure 4.
N-glycosylation of JIP21. Two micrograms of JIP21 purified protein, ovoalbumin (OA), used as a positive control, and bovine serum albumin (BSA), used as a negative control, were separated in 14% SDS-PAGE. A, Proteins stained with Coomassie Brilliant Blue R-250. B, Proteins transferred onto Immobilon membrane and stained with Ponceau-S. C, Proteins transferred onto Immobilon membrane and stained with periodic acid-Schiff reagent. Lane M, Mr standards.
Biochemical Characterization of the Purified Protein
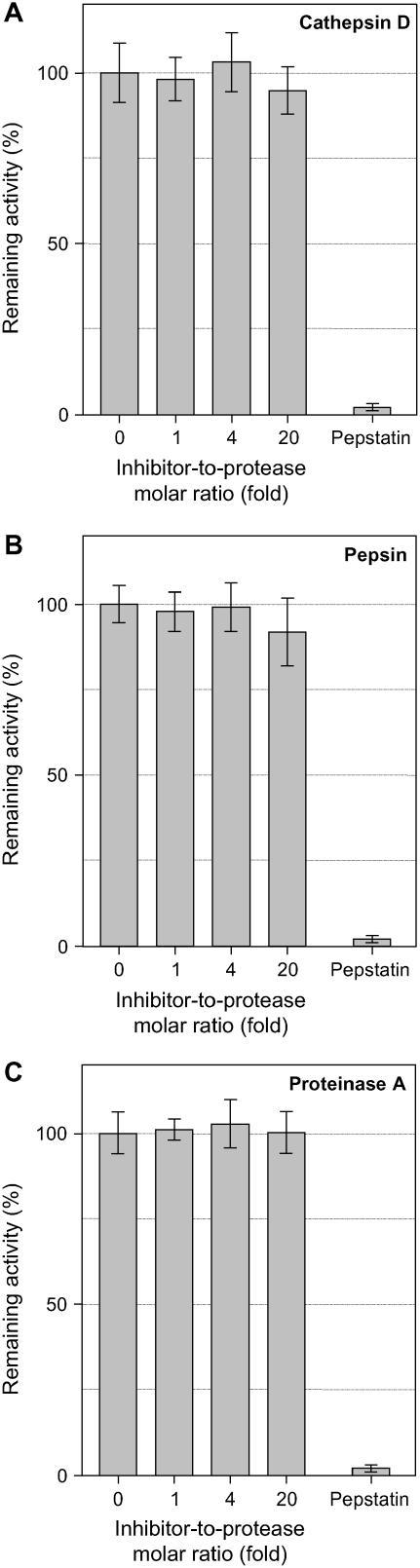
To characterize the predicted CDI activity of tomato JIP21, we performed an inhibition assay using hemoglobin labeled with fluorescein as a substrate for the proteinase. Endopeptidase inhibitors directly interact with the active center of the protease at a 1:1 molar ratio (Laskowski et al., 2000). Unexpectedly, the purified JIP21 protein showed no inhibitory activity against cathepsin D at pH 2.8, even at the inhibitor:protease weight ratio (w/w) of 10:1 (equivalent to 20:1 in the molar ratio). As shown in Figure 5A, an excess of JIP21 does not seem to affect cathepsin D activity, whereas pepstatin, used as a positive control, totally blocked the activity of this protease. To discard the possibility that the lack of activity might be due to a shift in the optimal pH for inhibition, the same studies were performed at a pH range from 2 to 8 and no inhibition was observed (data not shown). Because cathepsin D is a lysosomal protein and is not, therefore, exactly related to digestive processes, we decided to test JIP21 activity against a digestive aspartic proteinase such as pepsin. As seen in Figure 5B, excess amounts of purified JIP21 displayed no activity against pepsin, unlike the complete inhibition exerted by pepstatin. It has been described (Cater et al., 2002) that CDIs are also active against proteinase A, a yeast (Saccharomyces cerevisiae) aspartic proteinase. We decided to check JIP21 inhibitory activity against the yeast enzyme even though it is not an herbivore digestive peptidase. Once again, JIP21 did not inhibit this proteinase (Fig. 5C). These results appear to indicate that tomato JIP21 might not be an aspartic PIN, which prompted us to search for other possibilities.
Figure 5.
Inhibition assay of aspartic proteinases by JIP21. Reactions containing 50 ng of cathepsin D (A), pepsin (B), or proteinase A (C) were preincubated with increasing amounts of JIP21 in a final volume of 50 μL, then 10 μL of fluoresceinated hemoglobin (0.5% [w/v]) were added as a substrate. The proteolytic activity was determined as soluble fluorescence measured at 525 nm. Enzymatic activity is expressed in relative terms as the net emitted fluorescence with respect to the control reaction. Ten nanograms of pepstatin were used as a positive inhibition control in all assays. Results are the means of three independent assays.
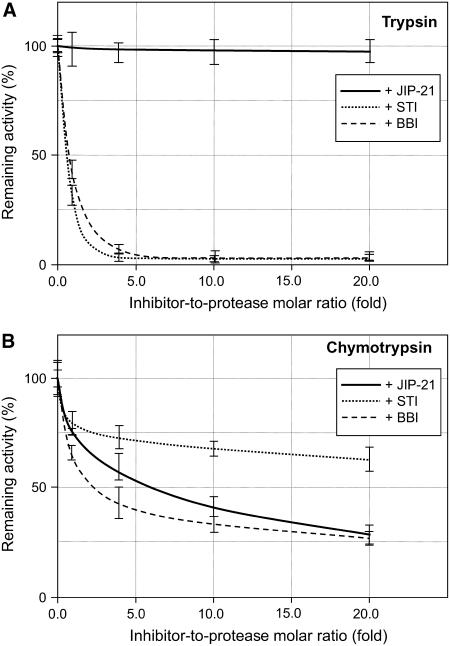
In this regard, we went on to test the activity of the purified protein against digestive proteinases belonging to the Ser protease family. We used the STI and the bifunctional trypsin and chymotrypsin BBI (Birk, 1996) as controls for these assays. Figure 6B shows that JIP21 is a potent inhibitor of chymotrypsin, where this inhibitory activity is comparable with the BBI inhibitor itself. On the other hand, inhibition against trypsin was negligible (Fig. 6A) when compared with its antichymotrypsin activity. Other Ser proteinases, such as elastase, proteinase K, or subtilisin were also unaffected by JIP21 (data not shown). The same negative results were obtained when assayed against chymosin (an aspartic proteinase), papain (a Cys proteinase), or metallocarboxypeptidase A. Together, these results suggest that JIP21 appears to be a Ser protease inhibitor with a high specificity against chymotrypsin, which is in contrast to the previously presumed function.
Figure 6.
Inhibitory activity of JIP21 on trypsin and chymotrypsin. Reactions containing 50 ng of trypsin (A) or chymotrypsin (B), and increasing amounts of JIP21 (continuous line), STI (dotted line), or BBI (dashed line) were preincubated in a final volume of 50 μL, and then 10 μL of fluoresceinated hemoglobin (0.5% [w/v]) were added as a substrate. The proteolytic activity was determined as soluble fluorescence measured at 525 nm. Enzymatic activity is expressed in relative terms as the net emitted fluorescence with respect to the control reaction. Three independent assays were performed for each protease experiment.
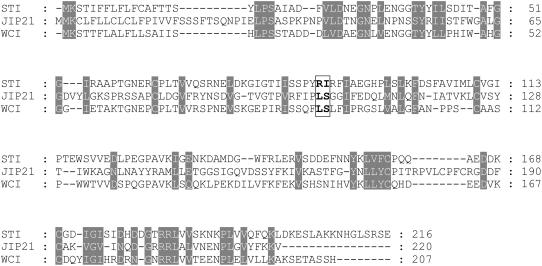
The chymotrypsin inhibitory activity described herein contrasts with the expected CDI activity deduced from sequence analysis. Apart from the obvious similarity to potato CDI, the JIP21 amino acid sequence diverges in relation to the rest of the aspartic PINs described. On the contrary, JIP21 is homologous to the Kunitz-type Ser PINs. Figure 7 shows a comparative analysis between JIP21 and two members of this family: the STI and the winged bean chymotrypsin inhibitor (WCI; Shibata et al., 1988). The putative inhibitory sites are highlighted in this figure. JIP21 contains the residues Leu-Ser, previously proposed as the binding site to chymotrypsin of the WCI, unlike the STI, with Arg-Ile as active residues for trypsin (Song and Suh, 1998). In fact, it has been described that a single mutation (Leu → Arg) in the reactive site converted WCI into a strong inhibitor of trypsin (Khamrui et al., 2005). Interestingly, the inhibitory site for chymotrypsin in the BBI also contains the residues Leu-Ser (Werner and Wemmer, 1991). These data further explain the observed activity of JIP21 against chymotrypsin.
Figure 7.
Comparison of the deduced amino acid sequence of JIP21 with the STI (GenBank accession no. S45092) and the WCI (GenBank accession no. D13976). Identical or closely related amino acids are shaded in gray. Residues corresponding to the active sites of both STI and WCI, and the putative active site of JIP21, are boxed and highlighted in bold.
Generation and Characterization of Tomato Plants Overexpressing JIP21
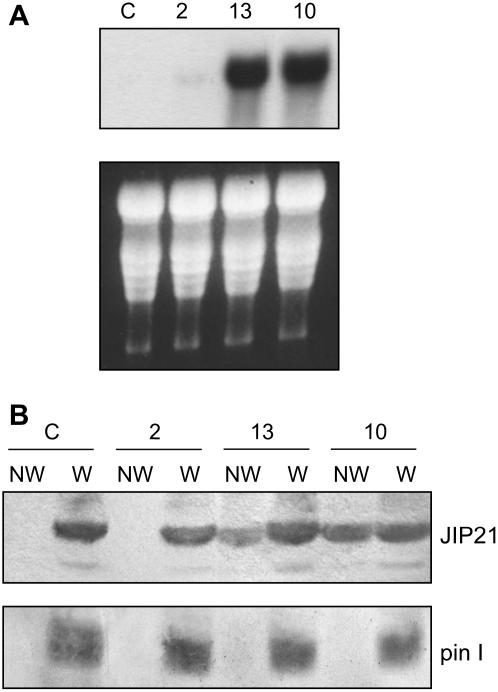
To study the biological function of JIP21, we generated tomato plants overexpressing the protein. For this purpose, a DNA cassette consisting of JIP21 cDNA, driven by a double cauliflower mosaic virus (CaMV) 35S promoter and a nos terminator, was ligated into a pBin19 plasmid. The resulting construction was used to generate tomato transgenic plants via Agrobacterium-mediated transformation. Insertion of the transgene was detected by Southern-blot hybridization of genomic DNA digested with BamHI, a restriction enzyme that does not cut the cDNA sequence. The transgenic lines obtained displayed additional bands to the single endogenous one found in wild-type plants. Levels of expression of the transgene were detected by northern and western blot, and both analyses showed good correlation (Fig. 8, A and B). Transgenic line number 2 shows a very low level of the JIP21 transcript and no detectable protein. Transgenic lines 10 and 13, with high constitutive levels of transcript, accumulate JIP21 protein at levels that are comparable with a wounded control leaf, as shown in Figure 8B. These levels are not due to any wound or pest on the transgenic plants, as indicated by the absence of pin I (Graham et al., 1985a) in nonwounded leaves (Fig. 8B, bottom). Moreover, when transgenic plants were wounded, the constitutive levels of JIP21 did not appear to be altered, whereas pin I accumulation was as apparent as in wounded wild-type plants. This indicates that CaMV-mediated overexpression of JIP21 does not affect the pattern of expression of other PINs.
Figure 8.
Characterization of different JIP21-overexpressing tomato lines (2, 13, and 10). A, Northern-blot analysis. Total RNA was extracted from transgenic lines and control plants, separated in formaldehyde-agarose gels, and transferred onto Nytran membranes. Hybridization was performed at 65°C using JIP21 cDNA as a probe. B, Western-blot analysis. Crude protein extracts were obtained, separated in 14% SDS-PAGE, and transferred onto nitrocellulose membranes. The membranes were then incubated with JIP21 antibody (top) or with the tomato pin I antibody (bottom). NW, Nonwounded leaves; W, wounded leaves; C, control tomato plants.
Homozygous plants were obtained for line 10, which integrated one single copy of the transgene. JIP21 represents about 3% of the total soluble protein in crude protein extracts from this line, as estimated by gel densitometry. These crude extracts displayed strong antichymotrypsin activity, unlike the wild-type control plants (data not shown). Consequently, this line was used for insect feeding bioassays.
Effect of Overexpression of JIP21 on Egyptian Cotton Worm Larvae
Because JIP21 is a powerful Ser PIN, we decided to test its biological effect on insects whose main digestive enzymes belong to this family of proteases. Thus, we performed insect feeding assays with Egyptian cotton worm larvae, which belong to the Lepidoptera family. Neonate larvae were placed on detached transgenic or control leaves. Leaves were replaced daily with fresh ones to avoid accumulation of the endogenous PINs caused by larval feeding, as pointed out by Abdeen et al. (2005). At the end of the assay (7 d), surviving larvae were counted and weighed. As Table I indicates, larvae fed with JIP21-overexpressing tomato leaves presented a percentage of mortality of 20%, whereas only 6% mortality was observed in the larvae fed on wild-type plants. Besides, we observed a mean weight reduction of approximately 40% in larvae fed on transgenic tomato leaves (Table I). Our results clearly indicate that JIP21 exerts an antinutritional effect on Egyptian cotton worm larvae, which demonstrates its defensive function and supports the novel role of JIP21 as a chymotrypsin inhibitor against insects and herbivores.
Table I.
Effect of overexpression of JIP21 on the mortality and growth of Egyptian cotton worm larvae
Neonate larvae were placed on daily renewed detached tomato control or transgenic leaves. Sufficient humidity was provided by damp absorbing paper. Seven days later, the surviving larvae were counted and weighed. Bioassays were repeated three times.
| Ratio of Mortality | Weight | |
|---|---|---|
| mg | ||
| Control | 6% | 51.5 ± 5.1 |
| Line 10 | 20% | 30.1 ± 4.6 |
DISCUSSION
In this work, we have purified and characterized JIP21, a defensive protein with a novel function as a chymotrypsin inhibitor in tomato. This is in contrast to its role as a CDI previously predicted on the grounds of its sequence analysis.
JIP21 strongly accumulates after wounding and treatment with MeJ at levels perfectly detectable by Coomassie Blue staining. Purification of the protein allowed us to obtain its corresponding antibodies and the cDNA sequence by immunoscreening. Sequence analysis revealed that the cDNA corresponds to a possible CDI (Werner et al., 1993). In fact, the JIP21-deduced amino acid sequence presents an elevated identity with the family of CDIs described in potato (Mares et al., 1989; Ritonja et al., 1990; Strukelj et al., 1990; Hildmann et al., 1992; Maganja et al., 1992; Hannapel, 1993; Herbers et al., 1994; Ishikawa et al., 1994a; Kreft et al., 1997). Apart from this large family in potato, proteinaceous aspartic PINs are uncommon and are described in only a few plant species, yeast (Schu and Wolf, 1991), and the nematode Ascaris lumbricoides (Abu-Erreish and Peanasky, 1974). None of these inhibitors seems to be related to potato CDIs.
The deduced amino acid sequence of JIP21 cDNA reveals the presence of a putative N-glycosylation site, which is also described in some of the CDIs characterized in potato. We herein demonstrate that such glycosylation actually occurs. It is well established, at least in animal glycoproteins, that glycosylation participates in important processes, such as maintenance of protein conformation and solubility, stabilization of the polypeptide against uncontrolled proteolysis, intracellular sorting and externalization of glycoproteins, or mediation of its biological activity (Olden et al., 1985; Dwek, 1995). In silico folding simulation indicates that the N-glycosylation site falls close to the putative inhibition site; thus deglycosylation of the purified protein or mutagenesis of the Asn-Ser-Ser site might yield information about the role of the glycan on either the stability or activity of JIP21.
JIP21 has a pattern of expression similar to the first identified tomato PINs, pin I and pin II (Graham et al., 1985a, 1985b). That is, it is locally and systemically induced by both wounding and MeJ treatment. When compared to wounding, MeJ induction is stronger, more rapid, and long lasting. JIP21 is not induced by pathogens (citrus exocortis viroid and P. syringae pv tomato). However, the cDNA described by Werner et al. (1993) was obtained from a library from RNAs of tomato leaves infected with potato spindle tuber viroid. This might be due to any injury on the material used for the potato spindle tuber viroid library.
We have detected constitutive levels of JIP21 in tomato plant flowers. Accumulation in these organs has also been described for other PINs, such as potato pin II (Peña-Cortés et al., 1991), CDI (Hildmann et al., 1992; Ishikawa et al., 1994a), tomato metallocarboxypeptidase inhibitor (Martineau et al., 1991), and a BBI from pea (Pisum sativum; Domoney et al., 2002). A possible role of this localization could be the protection of vulnerable tissues of flowers. Moreover, we have detected an accumulation of JIP21 mRNA in the stem and fruit of tomato control plants by RT-PCR. The fact that all the PCR products have been shown to correspond to the original cDNA suggests that the constitutive levels are not due to the expression of isoforms of JIP21 in these tissues, unlike potato, where a family of 15 members has been described to date. Hence, whereas the promoter of one isoform of the CDI in potato has been shown to direct the expression of the β-glucuronidase reporter gene in tubers alone and not in leaves upon wounding (Herbers et al., 1994), the JIP21 promoter sequence, which we have isolated by screening a genomic library, could be responsible for the whole pattern of expression observed.
Detailed biochemical characterization of JIP21 has allowed us to uncover a novel function that differs from that predicted by its comparative sequence analysis. Although the JIP21-deduced amino acid sequence presents an elevated identity with the family of CDIs of potato, we have not detected any inhibitory activity either against this protease or other proteases of the aspartic family assayed. It is also interesting to note that, among the large number of putative CDIs described in potato over recent years, biochemical characterization was only performed in early studies when the two first potato isoforms (PDI and NDI) were purified. In those studies, a weak affinity of PDI for cathepsin D was described when compared with the affinity shown by soybean and potato trypsin inhibitors to their proteinase targets (Keilova and Tomasek, 1976a). Since then, potato isoforms have been identified only by means of sequence analysis. In a more recent study using a recombinant protein obtained in Pichia pastoris from a tomato cDNA clone with a high identity to that described by Werner et al. (1993), a revision of the name is proposed given the weak activity detected against human cathepsin D (Cater et al., 2002).
In this work, the biochemical activity of the purified tomato protein is tested against a number of proteinases. By doing so, we observed a strong inhibitory activity of JIP21 against chymotrypsin, which showed no effect against other proteinases. Thus, we propose JIP21 to be a member of the Ser PIN family, acting specifically against chymotrypsin. This is consistent with the fact that the first study of the primary structure of the CDI from potato considered its structure homologous to that of the STI, which belongs to the Kunitz-type Ser PIN family (Mares et al., 1989). Tomato JIP21, along with all the potato CDIs, presents strong sequence similarity to potato Kunitz-type Ser PINs (Walsh and Twitchell, 1991; Valueva et al., 1998; Heibges et al., 2003). Besides, comparative sequence analysis of JIP21 identifies three domains in JIP21 according to functional and structural protein domains using the PRODOM database (http://www.toulouse.inra.fr/prodom.html). The main central domain is Kunitz like, whose member of reference is precisely the STI. In fact, we have conducted in silico structural studies using the SWISS-MODEL server (http://www.expasy.ch/swissmod/SWISS-MODEL.html) and we observed good folding compatibility between JIP21 and the STI. All of this points to the Kunitz-like nature of JIP21 and accounts for its inhibitory activity to Ser proteinases.
All our data question the formerly proposed activity of JIP21 against cathepsin D. Moreover, cathepsin D is a lysosomal aspartic protease implicated in cancer, apoptosis, and Alzheimer's disease (for reviews, see Callahan et al., 1998; Liaudet-Coopman et al., 2006). It is synthesized in the endoplasmic reticulum as preprocathepsin D and, once in the lysosome, the single-stranded procathepsin (52 kD) is activated to cathepsin D to finally constitute the mature double-stranded cathepsin D (31 and 14 kD, respectively; Yamamoto, 1995). Its functions are related to the degradation or activation of proteins inside the lysosome. Therefore, it is not an extracellular protein and it is unlikely to have a digestive role.
If JIP21 effectively is a Ser PIN, it should have a biological effect on insects of which the main digestive proteases belong to this family. To verify this point, we have generated tomato plants overexpressing JIP21 at levels comparable to a wounded tomato plant and we have evaluated the effect of this overexpression on the mortality and growth of Egyptian cotton worm larvae. Egyptian cotton worm belongs to the Lepidoptera family whose main digestive enzymes are Ser proteases (Houseman et al., 1989; for review, see Terra and Ferreira, 1994). The biochemical characterization of our protein and the antinutritional effect of the overexpressed protein on these larvae strongly support our proposal that JIP21 is a Ser protease inhibitor instead of the function assumed to date. All our data suggest that the formerly called tomato CDI could be referred to henceforth as tomato chymotrypsin inhibitor 21 (TCI21).
MATERIALS AND METHODS
Plant Material and Treatments
Tomato (Solanum lycopersicum L. cv Rutgers) plants were grown under standard greenhouse conditions (20°C–25°C and 16-h light/8-h dark photoperiods).
Wounding and MeJ treatments were performed with 3- to 4-week-old plants. Wounding was performed by crushing one compound leaf per plant with forceps. To study the local response, wounded leaves were harvested at different times and the immediate upper leaves were used to analyze the systemic response. MeJ was applied by spraying a 2 mm solution and treated leaves were harvested at different times. Plant material was used immediately or stored frozen at −80°C.
Protein Analysis
Protein extracts of tomato leaves were performed by homogenization in acidic extraction buffer (84 mm citric acid, 32 mm sodium phosphate, pH 2.8) as described in Rodrigo et al. (1993). Proteins were separated by SDS-PAGE and stained with Coomassie Brilliant Blue R-250 following the method described by Conejero and Semancik (1977).
Purification of JIP21 Protein
Crude extracts from tomato leaves, sprayed with 2 mm MeJ and harvested after 48 h, were subjected to fractionated precipitation using ammonium sulfate. Proteins precipitating between 20% to 30% (w/v) saturation were sedimented, dialyzed against 50 mm sodium acetate buffer (pH 5.5), and chromatographed in a SP-Sephadex C25 (Pharmacia) column using a linear salt gradient (0–0.5 m NaCl in acetate buffer). Fractions enriched in JIP21 protein (eluted around 0.2 m NaCl) were collected, concentrated by lyophilization, and rechromatographed in SP-Sephadex C25 under the same conditions. Finally, fractions containing JIP21 were applied to a FPLC system (Pharmacia) using a Mono S HR 5/5 column and eluted with a linear NaCl gradient (0–0.5 m NaCl in acetate buffer). The protein peak corresponding to JIP21 was collected, concentrated, and equilibrated in 50 mm Tris-HCl (pH 7.5) to be stored at −20°C.
Antibodies and Immunoblots
Anti-JIP21 serum was obtained by injecting female New Zealand rabbits with purified preparations of JIP21 following standard procedures. For western-blot immunoassay, proteins were separated by SDS-PAGE, transferred onto nitrocellulose membranes using semidry electrotransfer equipment, and immunodetected using a 1:5,000 dilution of anti-JIP21 serum or a 1:500 dilution of anti-pin I serum previously obtained in our laboratory from the recombinant protein (Graham et al., 1985a). Membranes were incubated with goat anti-rabbit IgG conjugated to alkaline phosphatase (Promega) as a secondary antibody. Immunodetection was carried out with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate according to standard protocols.
N-Glycosylation Assay
For the N-glycosylation assay, we followed the method described by Strömqvist and Gruffman (1992), which combines the protein transfer to polyvinyl difluoride membranes with periodic acid-Schiff sugar staining. Samples containing purified JIP21, ovoalbumin as a positive control, and bovine serum albumin as a negative control were separated in SDS-PAGE and electrotransferred onto Immobilon membranes (Millipore). The homogeneous transfer was checked by using reversible Ponceau staining. After washing with distilled water, membranes were treated with 1% (w/v) periodic acid in 3% acetic acid for 5 min and then washed with distilled water for 15 min. The Schiff reagent (Sigma) was added and membranes were kept in darkness for 15 min. Successive washings were performed, first with sodium bisulfite 0.5% (w/v) for 5 min, and later with distilled water again. Finally, membranes were rinsed in methanol for a few seconds to eliminate the background and enhance the contrast of the bands.
PIN Assay
Proteinase activity was assayed using hemoglobin labeled with fluorescein as a substrate based on the method described by Twining (1984) for casein. Proteolytic activity was determined as soluble fluorescence in TCA, originating from the hydrolysis of the hemoglobin. Reaction buffers were citrate-phosphate buffer (84 mm citric acid, 32 mm sodium phosphate, pH 2.8) for the aspartic proteinases and 50 mm Tris-HCl (pH 7.5) buffer for the rest of the proteinases assayed. To perform inhibition analyses through a pH range from 2 to 8, McIlvaine buffers were used. These buffers were prepared by mixing the proper volumes of 0.1 m citric acid and 0.2 m disodium phosphate to achieve the desired pH values. Reactions were performed in Eppendorf tubes containing 50 μL of the enzyme to a final concentration of 1 to 10 μg/mL, and increasing amounts of the different inhibitors. Control reactions contained no inhibitor. Reactions were preincubated for 15 min at 4°C and then 10 μL of fluoresceinated hemoglobin (0.5% [w/v]) were added. After 1 h at 37°C, digestion was stopped by adding 1 volume of 20% (w/v) TCA, and the precipitate was removed by centrifugation. Supernatants were added to 2.5 mL 0.5 m Tris-HCl (pH 8.5) and fluorescence at 525 nm was measured using an excitation wavelength of 490 nm in a Perkin-Elmer LS 50 B luminescence spectrophotometer. Enzymatic activity is expressed in relative terms as the net emitted fluorescence (without the background) in relation to the control reaction. Three independent assays were performed for each protease experiment. Cathepsin D was purchased from Calbiochem. Trypsin and chymotrypsin were obtained from Roche. The rest of the proteinases (pepsin, proteinase A, chymosin, elastase, subtilisin, papain, and carboxypeptidase A), as well as the PINs used (STI, BBI, and pepstatin), were obtained from Sigma.
cDNA Library, Screenings, and DNA Sequence Analysis
A cDNA library was constructed from mRNAs of tomato leaves harvested after 48 h of a 2 mm MeJ treatment in a Uni-ZAP XR vector (Stratagene), following the manufacturer's instructions. Phagemid-infected Escherichia coli cells were grown in the presence of 10 mm isopropyl-β-d-thiogalactopyranoside to induce the synthesis of the β-galacturonase fusion protein, and upon plaque formation, proteins were transferred to nitrocellulose membranes. Clones expressing JIP21 fused to β-galacturonase were revealed by immunostaining as indicated above. As a control, membranes containing protein extracts of tomato leaves, which were either treated or not with 2 mm MeJ and separated by SDS-PAGE, were processed simultaneously.
The cDNA obtained in the immunoscreening was used as a probe to screen a tomato genomic DNA library constructed in λ-EMBL (CLONTECH), and the positive clones were isolated, purified, and characterized, as described in Sambrook et al. (1989).
DNA sequencing was performed on an ABI PRISM DNA sequencer 377 (Perkin-Elmer). Computer-assisted analyses of DNA sequences were carried out using the University of Wisconsin Genetics Computer Group package (Genetics Computer Group) and the online services available at the National Center of Biotechnology Information (http://www.ncbi.nlm.nih.gov).
Nucleic Acids
Total RNA was prepared by using the TRIzol reagent (Invitrogen) following the manufacturer's instructions. For northern analysis, 30 μg of RNA were separated in formaldehyde-agarose gels and transferred onto Nytran (Schleicher & Schuell) membranes. 32P-labeled probes were prepared using the Rediprime labeling kit (Amersham) as recommended by the manufacturer. Hybridization and washing conditions were performed as described in Church and Gilbert (1984).
RT-PCR and Cloning of PCR Products
For RT reactions, we used 5 μg of total RNA obtained from different tomato tissues and Moloney murine leukemia virus reverse transcriptase (Promega). Five microliters of the reverse transcriptase reaction were used for PCR, employing the following oligonucleotide primers to specifically amplify JIP21: JIP21F (5′-CCGAATTCATATGATGAAGTGTTTATTT-3′) and JIP21R (5′-CCAATTTTATTAAGAAAGACATGC-3′). The primers used to amplify RPL2 (Fleming et al., 1993) were RPL2F (5′-GGTGACCGTGGTGTCTTTGC-3′) and RPL2R (5′-ACCAACGTTTTGTCCAGGAGGT-3′). PCR reactions were performed in a Perkin-Elmer thermocycler under the following conditions: 30 cycles of 94°C for 30 s, 50°C for 1 min, and 72°C for 1 min, followed by a final extension of 72°C for 15 min.
JIP21 PCR products were purified by elution from agarose gels cloned into the vector pGEM-T Easy (Promega) and sequenced on both strands.
Generation of Transgenic Plants
To generate the overexpression construct, JIP21 cDNA was prepared by digestion of the plasmid pBlue-JIP21 cDNA, obtained in the immunoscreening, with the enzymes EcoRI and XhoI. The cDNA insert was blunt-end ligated between a double CaMV 35S promotor and the nos terminator signal in a modified pBlueScript vector. The correct sense orientation of the cDNA was checked, and then the cassette CaMV 35S 2X:JIP21cDNA:nos was digested with HindIII and finally cloned into the vector pBIN19 to give the plasmid called pBin19-JIP21sense.
The pBin19-JIP21sense construct was introduced into the Agrobacterium tumefaciens strain LBA 4404 and used for tomato transformation as described by Ellul et al. (2003). Transformants were selected in kanamycin-containing medium and propagated in soil for subsequent analysis.
Feeding Bioassays
Neonate larvae of the Egyptian cotton worm (Spodoptera littoralis) were kindly provided by Koppert Biological Systems. These larvae were placed in 140-mm diameter petri dishes containing freshly detached tomato leaves. Plates were kept at 22°C with an 8-h light/16-h dark photoperiod. Damp absorbing paper provided sufficient humidity to the plates. Leaves were replaced daily and surviving larvae were weighed every day throughout the assay. At the end of the test (7 d), mortality was evaluated and surviving insects were weighed.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AJ295638.
Acknowledgments
We thank Asunción Saurí (Universidad Politécnica de Valencia) and Dr. Joaquín Fayos (Productos Citrosol S.A.) for their assistance with purification of JIP21 and obtaining anti-JIP21 and anti-pin I polyclonal antibodies. We also thank Dr. Fernando García-Marí (Universidad Politécnica de Valencia) for his technical advice and Koppert Biological Systems for providing the Spodoptera littoralis larvae. We are grateful to Prof. C.A. Ryan (Washington University, Pullman) for kindly providing us with tomato pin I cDNA.
This work was supported by Comisión Interministerial de Ciencia y Tecnología, Spanish Ministry of Science and Technology (grant no. BMC2003–07837).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ismael Rodrigo (irodrig@ibmcp.upv.es).
References
- Abdeen A, Virgos A, Olivella E, Villanueva J, Aviles X, Gabarra R, Prat S (2005) Multiple insect resistance in transgenic tomato plants over-expressing two families of plant proteinase inhibitors. Plant Mol Biol 57: 189–202 [DOI] [PubMed] [Google Scholar]
- Abu-Erreish GM, Peanasky RJ (1974) Pepsin inhibitors from Ascaris lumbricoides: isolation, purification, and some properties. J Biol Chem 249: 1558–1565 [PubMed] [Google Scholar]
- Abuereish GM (1998) Pepsin inhibitor from roots of Anchusa strigosa. Phytochemistry 48: 217–221 [DOI] [PubMed] [Google Scholar]
- Birk Y (1996) Protein proteinase inhibitors in legume seeds. Arch Latinoam Nutr 44: 26S–30S [PubMed] [Google Scholar]
- Callahan LM, Chow N, Cheetham JE, Cox C, Coleman PD (1998) Analysis of message expression in single neurons of Alzheimer's disease brain. Neurobiol Aging 19: S99–S105 [DOI] [PubMed] [Google Scholar]
- Cater SA, Lees WE, Hill J, Brzin J, Kay J, Phylip LH (2002) Aspartic proteinase inhibitors from tomato and potato are more potent against yeast proteinase A than cathepsin D. Biochim Biophys Acta 1596: 76–82 [DOI] [PubMed] [Google Scholar]
- Christeller JT, Farley PC, Ramsay RJ, Sullivan PA, Laing WA (1998) Purification, characterization and cloning of an aspartic proteinase inhibitor from squash phloem exudate. Eur J Biochem 254: 160–167 [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland TE, Thornburg RW, Ryan CA (1987) Molecular characterization of wound inducible inhibitor I gene from potato and the processing of its mRNA and protein. Plant Mol Biol 8: 199–207 [DOI] [PubMed] [Google Scholar]
- Conejero V, Semancik JS (1977) Analysis of the proteins in crude plant extracts by polyacrylamide gel electrophoresis. Phytopathology 67: 1424–1426 [Google Scholar]
- Delessert C, Wilson IW, Van Der Straeten D, Dennis ES, Dolferus R (2004) Spatial and temporal analysis of the local response to wounding in Arabidopsis leaves. Plant Mol Biol 55: 165–181 [DOI] [PubMed] [Google Scholar]
- De Leo F, Bonade-Bottino M, Ceci LR, Gallerani R, Jouanin L (2001) Effects of a mustard trypsin inhibitor expressed in different plants on three lepidopteran pests. Insect Biochem Mol Biol 31: 593–602 [DOI] [PubMed] [Google Scholar]
- Díez-Díaz M, Conejero V, Rodrigo I, Pearce G, Ryan CA (2004) Isolation and characterization of wound-inducible carboxypeptidase inhibitor from tomato leaves. Phytochemistry 65: 1919–1924 [DOI] [PubMed] [Google Scholar]
- Domoney C, Welham T, Ellis N, Mozzanega P, Turner L (2002) Three classes of proteinase inhibitor gene have distinct but overlapping patterns of expression in Pisum sativum plants. Mol Genet Genomics 267: 359–369 [DOI] [PubMed] [Google Scholar]
- Duan X, Li X, Xue Q, Abo-Ei-Saad M, Xu D, Wu R (1996) Transgenic rice plants harboring an introduced potato proteinase inhibitor II gene are insect resistant. Nat Biotechnol 14: 494–496 [DOI] [PubMed] [Google Scholar]
- Dwek RA (1995) Glycobiology: more functions for oligosaccharides. Science 269: 1234–1235 [DOI] [PubMed] [Google Scholar]
- Ellul P, Garcia-Sogo B, Pineda B, Rios G, Roig LA, Moreno V (2003) The ploidy level of transgenic plants in Agrobacterium-mediated transformation of tomato cotyledons (Lycopersicon esculentum Mill.) is genotype and procedure dependent. Theor Appl Genet 106: 231–238 [DOI] [PubMed] [Google Scholar]
- Farmer EE, Ryan CA (1990) Interplant communication: airborne methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proc Natl Acad Sci USA 87: 7713–7716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AJ, Mandel T, Roth I, Kuhlemeier C (1993) The patterns of gene expression in the tomato shoot apical meristem. Plant Cell 5: 297–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galleschi L, Friggeri M, Repiccioli R, Come D (1993) Aspartic proteinase inhibitor from wheat: some properties. Proceedings of the Fourth International Workshop on Seeds. Angers, France, pp 207–211
- Girard C, Le Metayer M, Bonade-Bottino M, Pham-Delegue MH, Jouanin L (1998) High level of resistance to proteinase inhibitors may be conferred by proteolytic cleavage in beetle larvae. Insect Biochem Mol Biol 28: 229–237 [DOI] [PubMed] [Google Scholar]
- Graham JS, Pearce G, Merryweather J, Titani K, Ericsson L, Ryan CA (1985. a) Wound-induced proteinase inhibitors from tomato leaves. I. The cDNA-deduced primary structure of pre-inhibitor I and its posttranslational processing. J Biol Chem 260: 6555–6560 [PubMed] [Google Scholar]
- Graham JS, Pearce G, Merryweather J, Titani K, Ericsson L, Ryan CA (1985. b) Wound-induced proteinase inhibitors from tomato leaves. II. The cDNA-deduced primary structure of pre-inhibitor II. J Biol Chem 260: 6561–6564 [PubMed] [Google Scholar]
- Hannapel DJ (1993) Nucleotide and deduced amino acid sequence of the 22-kD cathepsin D inhibitor protein of potato (Solanum tuberosum L.). Plant Physiol 101: 703–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heibges A, Salamini F, Gebhardt C (2003) Functional comparison of homologous members of three groups of Kunitz-type enzyme inhibitors from potato tubers (Solanum tuberosum L.). Mol Genet Genomics 269: 535–541 [DOI] [PubMed] [Google Scholar]
- Herbers K, Prat S, Willmitzer L (1994) Cloning and characterization of a cathepsin D inhibitor gene from Solanum tuberosum L. Plant Mol Biol 26: 73–83 [DOI] [PubMed] [Google Scholar]
- Hilder VA, Gatehouse AMR, Sheerman SE, Barker RF, Boulter D (1987) A novel mechanism for insect resistance engineered into tobacco. Nature 330: 160–163 [Google Scholar]
- Hildmann T, Ebneth M, Peña-Cortés H, Sánchez-Serrano JJ, Willmitzer L, Prat S (1992) General roles of abscisic and jasmonic acids in gene activation as a result of mechanical wounding. Plant Cell 4: 1157–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman JG, Downe AER, Philogene BJR (1989) Partial characterization of proteinase activity in the larval midgut of the European corn borer Ostrinia nubilalis Hubner (Lepidoptera: Pyralidae). Can J Zool 67: 864–868 [Google Scholar]
- Ishikawa A, Ohta S, Matsuoka K, Hattori T, Nakamura K (1994. a) A family of potato genes that encode Kunitz-type proteinase inhibitors: structural comparisons and differential expression. Plant Cell Physiol 35: 303–312 [PubMed] [Google Scholar]
- Ishikawa A, Yoshihara T, Nakamura K (1994. b) Jasmonate-inducible expression of a potato cathepsin D inhibitor-GUS gene fusion in tobacco cells. Plant Mol Biol 26: 403–414 [DOI] [PubMed] [Google Scholar]
- Johnson R, Narvaez J, An G, Ryan CA (1989) Expression of proteinase inhibitors I and II in transgenic tobacco plants: effects on natural defense against Manduca sexta larvae. Proc Natl Acad Sci USA 86: 9871–9875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouanin L, Bonadé-Bottino M, Girard C, Morrot G, Giband M (1998) Transgenic plants for insect resistance. Plant Sci 131: 1–11 [Google Scholar]
- Keilova H, Tomasek V (1976. a) Isolation and some properties of cathepsin D inhibitor from potatoes. Collect Czech Chem Commun 41: 489–497 [Google Scholar]
- Keilova H, Tomasek V (1976. b) Further characteristics of cathepsin D inhibitor from potatoes. Collect Czech Chem Commun 41: 2440–2447 [Google Scholar]
- Khamrui S, Dasgupta J, Dattagupta JK, Sen U (2005) Single mutation at P1 of a chymotrypsin inhibitor changes it to a trypsin inhibitor: x-ray structural (2.15 A) and biochemical basis. Biochim Biophys Acta 1752: 65–72 [DOI] [PubMed] [Google Scholar]
- Kreft S, Ravnikar M, Mesko P, Pungercar J, Umek A, Kregar I, Strukelj B (1997) Jasmonic acid inducible aspartic proteinase inhibitors from potato. Phytochemistry 44: 1001–1006 [DOI] [PubMed] [Google Scholar]
- Laskowski M Jr, Qasim MA (2000) What can the structures of enzyme-inhibitor complexes tell us about the structures of the enzyme substrate complexes? Biochim Biophys Acta 1477: 324–337 [DOI] [PubMed] [Google Scholar]
- Laskowski M Jr, Qasim MA, Lu SM (2000) Interaction of standard mechanism, canonical protein inhibitors with serine proteinases. In C Kleanthous, ed, Protein-Protein Recognition: The Frontiers in Molecular Biology Series. Oxford University Press, Oxford, pp 228–279
- Lawrence PK, Koundal KR (2002) Plant protease inhibitors in control of phytophagous insects. Electronic J Biotechnol 5: 93–109 [Google Scholar]
- Lecardonnel A, Chauvin L, Jouanin L, Beaujean A, Prévost G, Sangwan-Norreel B (1999) Effects of rice cystatin I expression in transgenic potato on Colorado potato beetle larvae. Plant Sci 140: 87–98 [Google Scholar]
- León J, Rojo E, Sánchez-Serrano JJ (2001) Wound signalling in plants. J Exp Bot 52: 1–9 [DOI] [PubMed] [Google Scholar]
- Leplé JC, Bonadé-Bottino M, Augustin S, Pilate G, Dumanois Lê Tân V, Delplanque A, Cornu D, Jouanin L (1995) Toxicity to Chrysomela tremulae (Coleoptera: Crysomelidae) of transgenic poplars expressing a cysteine proteinase inhibitor. Mol Breed 1: 319–328 [Google Scholar]
- Li YE, Zhu Z, Chen ZX, Wu X, Wang W, Li SJ (1998) Obtaining transgenic cotton plants with cowpea trypsin inhibitor. Acta Gossypii Sinica 10: 237–243 [Google Scholar]
- Liaudet-Coopman E, Beaujouin M, Derocq D, Garcia M, Glondu-Lassis M, Laurent-Matha V, Prebois C, Rochefort H, Vignon F (2006) Cathepsin D: newly discovered functions of a long-standing aspartic protease in cancer and apoptosis. Cancer Lett 237: 167–179 [DOI] [PubMed] [Google Scholar]
- Maganja DB, Strukelj B, Pungercar J, Gubensek F, Turk V, Kregar I (1992) Isolation and sequence analysis of the genomic DNA fragment encoding an aspartic proteinase inhibitor homologue from potato (Solanum tuberosum L.). Plant Mol Biol 20: 311–313 [DOI] [PubMed] [Google Scholar]
- Mares M, Meloun B, Pavlik M, Kostka V, Baudys M (1989) Primary structure of cathepsin-D inhibitor from potatoes and its structural relationship to trypsin inhibitor family. FEBS Lett 251: 94–98 [DOI] [PubMed] [Google Scholar]
- Martineau B, McBride KE, Houck CM (1991) Regulation of metallocarboxypeptidase inhibitor gene expression in tomato. Mol Gen Genet 228: 281–286 [DOI] [PubMed] [Google Scholar]
- Mason HS, DeWald DB, Mullet JE (1993) Identification of a methyl jasmonate-responsive domain in the soybean vspB promoter. Plant Cell 5: 241–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus MT, White DWR, McGregor PG (1994) Accumulation of a chymotrypsin inhibitor in transgenic tobacco can affect the growth of insect pests. Transgenic Res 3: 50–58 [Google Scholar]
- Olden K, Bernard BA, Humphries MJ, Yeo T-K, Yeo K-T, White SL, Newton SA, Bauer HC, Parent JB (1985) Function of glycoprotein glycans. Trends Biochem Sci 12: 78–82 [Google Scholar]
- Orr GL, Strickland JA, Walsh TA (1994) Inhibition of Diabrotica larval growth by a multicystatin from potato tubers. J Insect Physiol 40: 893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Cortés H, Willmitzer L, Sánchez-Serrano JJ (1991) Abscisic acid mediates wound induction but not developmental-specific expression of the proteinase inhibitor II gene family. Plant Cell 3: 963–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE (2000) Differential gene expression in response of mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritonja A, Krizaj I, Mesko P, Kopitar M, Lucovnik P, Strukelj B, Pungercar J, Buttle DJ, Barrett AJ, Turk V (1990) The amino acid sequence of a novel inhibitor of cathepsin D from potato. FEBS Lett 267: 13–15 [DOI] [PubMed] [Google Scholar]
- Rodrigo I, Vera P, Tornero P, Hernandez-Yago J, Conejero V (1993) cDNA cloning of viroid-induced tomato pathogenesis-related protein P23: characterization as a vacuolar antifungal factor. Plant Physiol 102: 939–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszkowska-Jakimiec W, Bankowska A (1998) Cathepsin D inhibitor from Vicia sativa L. Rocz Akad Med Bialymst 43: 245–249 [PubMed] [Google Scholar]
- Ryan CA (1990) Proteinase inhibitors in plants: genes for improving defenses against insects and pathogens. Annu Rev Phytopathol 28: 425–449 [Google Scholar]
- Ryan CA (2000) The systemin signaling pathway: differential activation of plant defensive genes. Biochim Biophys Acta 1477: 112–121 [DOI] [PubMed] [Google Scholar]
- Samach A, Hareven D, Gutfinger T, Ken-Dror S, Lifschitz E (1991) Biosynthetic threonine deaminase gene of tomato: isolation, structure, and upregulation in floral organs. Proc Natl Acad Sci USA 88: 2678–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sánchez-Serrano JJ, Keil M, O'Connor A, Schell J, Willmitzer L (1987) Wound-inducible expression of a potato proteinase inhibitor II in transgenic tobacco plants. EMBO J 6: 303–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Serrano JJ, Schmidt R, Schell J, Willmitzer L (1986) Nucleotide sequence of proteinase inhibitor II encoding cDNA of potato (Solanum tuberosum) and its mode of expression. Mol Gen Genet 203: 15–20 [Google Scholar]
- Schaller F (2001) Enzymes of the biosynthesis of octadecanoid-derived signalling molecules. J Exp Bot 52: 11–23 [PubMed] [Google Scholar]
- Schu P, Wolf DH (1991) The proteinase yscA-inhibitor, IA gene: studies of cytoplasmic proteinase inhibitor deficiency on yeast physiology. FEBS Lett 283: 78–84 [DOI] [PubMed] [Google Scholar]
- Schuler TH, Poppy GM, Kerry BR, Denholm I (1998) Insect-resistant transgenic plants. Trends Biotechnol 16: 168–175 [DOI] [PubMed] [Google Scholar]
- Shibata H, Hara S, Ikenaka T (1988) Amino acid sequence of winged bean (Psophocarpus tetragonolobus (L.) DC.) chymotrypsin inhibitor, WCI-3. J Biochem (Tokyo) 104: 537–543 [DOI] [PubMed] [Google Scholar]
- Song HK, Suh SW (1998) Kunitz-type soybean trypsin inhibitor revisited: refined structure of its complex with porcine trypsin reveals an insight into the interaction between a homologous inhibitor from Erythrina caffra and tissue-type plasminogen activator. J Mol Biol 275: 347–363 [DOI] [PubMed] [Google Scholar]
- Strömqvist M, Gruffman H (1992) Periodic acid/Schiff staining of glycoproteins immobilized on a blotting matrix. Biotechniques 13: 744–746 [PubMed] [Google Scholar]
- Strukelj B, Pungercar J, Ritonja A, Krizaj I, Gubensek F, Kregar I, Turk V (1990) Nucleotide and deduced amino acid sequence of an aspartic proteinase inhibitor homologue from potato tubers (Solanum tuberosum L.). Nucleic Acids Res 18: 4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terra WR, Ferreira C (1994) Insect digestive enzymes: properties, compartmentalization and function. Comp Biochem Physiol 109: 1–62 [Google Scholar]
- Thornburg RW, An G, Cleveland TE, Johnson R, Ryan CA (1987) Wound-inducible expression of a potato inhibitor II-chloramphenicol acetyltransferase gene fusion in transgenic tobacco plants. Proc Natl Acad Sci USA 84: 744–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twining SS (1984) Fluorescein isothiocyanate-labeled casein assay for proteolytic enzymes. Anal Biochem 143: 30–34 [DOI] [PubMed] [Google Scholar]
- Valueva TA, Revina TA, Kladnitskaya GV, Mosolov VV (1998) Kunitz-type proteinase inhibitors from intact and Phytophthora-infected potato tubers. FEBS Lett 426: 131–134 [DOI] [PubMed] [Google Scholar]
- Walsh TA, Twitchell WP (1991) Two Kunitz-type proteinase inhibitors from potato tubers. Plant Physiol 97: 15–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SJ, Lan YC, Chen SF, Chen YM, Yeh KW (2002) Wound-response regulation of the sweet potato sporamin gene promoter region. Plant Mol Biol 48: 223–231 [DOI] [PubMed] [Google Scholar]
- Werner MH, Wemmer DE (1991) 1H assignments and secondary structure determination of the soybean trypsin/chymotrypsin Bowman-Birk inhibitor. Biochemistry 30: 3356–3364 [DOI] [PubMed] [Google Scholar]
- Werner R, Guitton MC, Muhlbach HP (1993) Nucleotide sequence of a cathepsin D inhibitor protein from tomato. Plant Physiol 103: 1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K (1995) Cathepsin E and cathepsin D: biosynthesis, processing and subcellular location. Adv Exp Med Biol 362: 223–229 [DOI] [PubMed] [Google Scholar]