Abstract
In this study, we show that both arginine decarboxylase (ADC) protein and mRNA transcript are present at different phases of mitosis in Scots pine (Pinus sylvestris) zygotic embryogenesis. We also examined the consistency of polyamine (PA) profiles with the effective temperature sum, the latter indicating the developmental stage of the embryos. PA metabolism was analyzed by fitting statistical regression models to the data of free and soluble conjugated PAs, to the enzyme activities of ADC and ornithine decarboxylase (ODC), as well as to the gene expression of ADC. According to the fitted models, PAs typically had the tendency to increase at the early stages but decrease at the late stages of embryogenesis. Only the free putrescine fraction remained stable during embryo development. The PA biosynthesis strongly preferred the ADC pathway. Both ADC gene expression and ADC enzyme activity were substantially higher than putative ODC gene expression or ODC enzyme activity, respectively. ADC gene expression and enzyme activity increased during embryogenesis, which suggests the involvement of transcriptional regulation in the expression of ADC. Both ADC mRNA and ADC protein localized in dividing cells of embryo meristems and more specifically within the mitotic spindle apparatus and close to the chromosomes, respectively. The results suggest the essential role of ADC in the mitosis of plant cells.
The polyamines (PAs), found in bacteria, fungi, animals, and plants, are evolutionary ancient small polycations that are implicated in various physiological and developmental processes. In higher plants, these processes include stimulation of cell division, response to environmental stresses and regulation of rhizogenesis, embryogenesis, senescence, floral development, and fruit ripening (Kakkar and Sawhney, 2002). Both in prokaryotes and in eukaryotes, PAs play important roles in a number of cellular processes, such as DNA conformation, chromatin condensation, RNA processing, translation, and protein activation (Childs et al., 2003), which is reflected in a strict regulatory control of their intracellular levels. The intracellular free-PA pool is affected both by PA synthesis and degradation, as well as cellular influx, efflux, and conjugation mechanisms (Tiburcio et al., 1997). In plants, however, the molecular mechanisms of the action of PAs are still in most cases unknown, and hardly anything is known about the role of PAs at the cellular level in conifers.
Diamine putrescine (Put) and triamine spermidine (Spd) are present in most organisms, but tetra-amine spermine (Spm) is predominantly found in eukaryotes (Cohen, 1998). The first step in PA biosynthesis is the formation of Put from Orn and Arg via the rate-limiting enzymes Arg decarboxylase (ADC; EC 4.1.1.19) and Orn decarboxylase (ODC; EC 4.1.1.17), respectively. Mammalian and fungal cells utilize mainly ODC to synthesize Put (Coleman et al., 2004), whereas an additional indirect route from Arg to Put is prevalent in plant and bacterial cells (Bagni and Tassoni, 2001). Put is converted into Spd and Spm by Spd synthase (EC 2.5.1.16) and Spm synthase (EC 2.5.1.22), respectively, which add aminopropyl groups generated from S-adenosylmethionine by S-adenosylmethionine decarboxylase (EC 4.1.1.50; Tiburcio et al., 1997). In plant cells, PAs often occur as free molecules (soluble PAs), but they can also be conjugated to low-molecular-mass compounds (conjugated soluble PAs, SH) or bound to different macromolecules (conjugated insoluble PAs, PH; Martin-Tanguy, 1997; Tiburcio et al., 1997). PAs are catabolized by the action of amino oxidases, which include diamino oxidase, oxidizing the diamines Put and cadaverine, and PA oxidase, which oxidizes Spd and Spm (Tiburcio et al., 1997).
Different physiological roles have been proposed for ADC and ODC because their production in plants is often tissue specific and developmentally regulated (Kumar et al., 1997). It has been suggested that ODC is involved in the regulation of cell division in actively growing plant tissues (Cohen et al., 1984; Acosta et al., 2005). ADC activity has been found in elongating cells, embryonic cells, and cells exposed to various stress conditions (Flores, 1991). Bortolotti et al. (2004) showed that ADC protein is located in the nuclei of nonphotosynthetic tissue of tobacco (Nicotiana tabacum), whereas ADC in photosynthetic tissues is located in chloroplasts. Recently, ODC and ADC gene transcripts were colocalized in developing primary and lateral roots and hypocotyls of etiolated soybean (Glycine max) seedlings (Delis et al., 2005).
The importance of PAs in embryogenesis has been documented in many angiosperm species (Lin et al., 1984; Yadav and Rajam, 1998), and recently the knockout mutants of PA biosynthesis have shown that Spd synthase (Imai et al., 2004) and S-adenosylmethionine decarboxylase (Ge et al., 2006) are essential for embryo development in Arabidopsis (Arabidopsis thaliana). However, the intracellular localization of PAs and their function at the molecular level during embryogenesis have not been elucidated. The role of PAs has been under active investigation also in gymnosperms, but most of the studies have focused on somatic embryogenesis, while relatively little is known about PA biosynthesis during zygotic embryo development. Thus, based on the previous studies on PAs during somatic embryo development in conifers and during zygotic embryo development in angiosperms, we hypothesized that PAs also have a role in zygotic embryo development of gymnosperms. The role of PAs might be specific depending on the developmental stage of embryos, which could appear as a variation at metabolite, protein, or transcript level. In this work, we were able to construct the statistical regression models of the free and conjugated Put, Spd, and Spm concentrations, and show the yearly recurrent PA concentration profiles during the zygotic embryo development of Scots pine (Pinus sylvestris). Furthermore, because ODC activity is commonly connected with cell division in plants (Galston and Sawhney, 1990), we also hypothesized that, in Scots pine developing embryos, ODC mRNA transcripts or proteins should be found in dividing cells. It showed up, however, that the ADC pathway was the prevalent route to form Put in Scots pine embryos, and instead of ODC both ADC mRNA transcripts and ADC protein were specifically localized in mitotic cells of primary meristems in embryos.
RESULTS
Zygotic Embryo Development and Effective Temperature Sum
In gymnosperms including Scots pine, the time of fertilization and, consequently, embryo development are known to vary between years in the same locality according to the effective temperature sum (d.d.; i.e. the heat sum unit based on the daily mean temperatures minus the adapted +5°C base temperature; Sarvas, 1962; Krasowski and Owens, 1993; Sirois et al., 1999; Owens et al., 2001). Therefore, in this work, the effective temperature sum rather than the sampling date was used as the main explanatory variable in the statistical analyses. The zygotic embryos of the two Scots pine clones (K818 and K884) were derived from cone collections started after fertilization and repeated four times during the period of embryo development in 2001 and 2003. On sampling dates I, II, III, and IV, the effective temperature sums were 569.7, 667.3, 775.6, and 876.0 d.d., respectively, in 2001, and 495.1, 594.0, 702.8, and 819.2 d.d. in 2003.
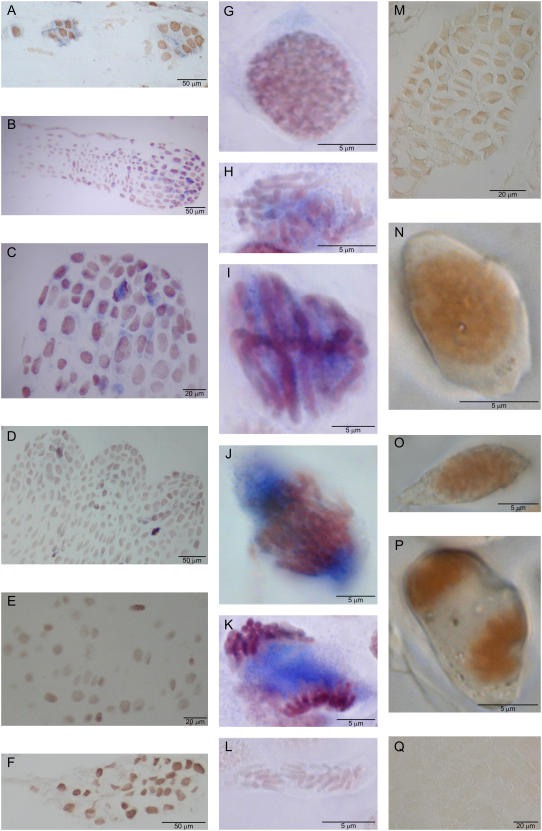
The anatomical preparates of the developing embryos from the year 2003 indicate the following developmental pattern. On sampling date I, when the effective temperature sum was approximately 500 d.d., around 50% of the embryos were still at the proembryogeny stage and 50% at the early embryogeny stage. On sampling dates II and III, when the effective temperature sum was between 600 and 700 d.d., nearly 100% of the embryos were at the early embryogeny stage. On sampling date IV, when the effective temperature sum was over 800 d.d., approximately 85% of the embryos had reached the late embryogeny stage while the remaining 15% were still at the early embryogeny stage. In Figure 1, both early embryos from sampling dates II (Fig. 1A) and III (Fig. 1F) and late embryos from sampling date IV (Fig. 1, B–D) have been presented.
Figure 1.
The localization of ADC and ODC mRNAs and ADC enzyme in developing zygotic embryos of Scots pine. ADC gene expression was localized by in situ hybridization with DIG-labeled RNA probes (blue signal) and ADC protein by immunolocalization (brown signal). The embryo in A was from sampling date II and the embryos in E, F, and M were from sampling date III. The embryos in B to D, G to L, N to P, and Q were from sampling date IV. A, ADC gene expression was localized in the cytoplasm of the cells in both dominant and subordinate early embryos. B and C, ADC gene expression localized in the cells of primary shoot and root meristem of late embryo. D, ADC gene expression in the mitotic cells of shoot meristem in the late embryo at the cotyledonary stage. E, No label was detected in the sections hybridized with the sense ADC probe. F, No mRNAs of a putative ODC gene were found in the developing early embryo. G, H, I, J, and K, ADC mRNA transcripts during the mitotic stages in the cells of late embryos. L, No label was detected in the mitotic cells in the sections hybridized with the ADC sense probe. M and N, ADC protein was localized in the nuclei of embryos. O and P, ADC protein was located with the chromosomes in the mitotic cells. Q, No signal was detected in the control sections incubated with preimmune serum.
Changes in PA Content during Zygotic Embryo Development
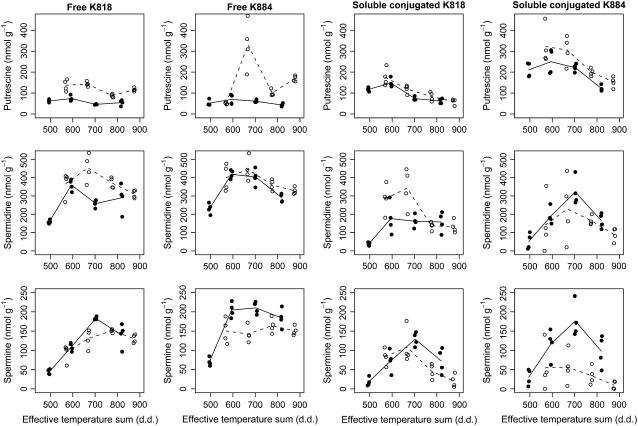
By presenting PA concentrations as quadratic (second-order polynomial) functions of the effective temperature sum, we were able to combine results from different clones, years, and PA fractions to the same regression model and to show that the PA concentrations followed consistent profiles, at least for Spd and Spm (Fig. 2; Table I).
Figure 2.
The fluctuation of free and soluble conjugated PAs (Put, Spd, and Spm) in developing zygotic embryos of Scots pine. Immature embryos were collected four times in 2001 and 2003 throughout the period of embryo development from two Scots pine clones, K818 and K884. The free and soluble conjugated PA concentrations are presented relative to the effective temperature sum (d.d.) and the year (• = 2003, ○ = 2001). The mean values of the four replicates pertaining to the same clone, year, and sampling date are connected by solid lines for 2003 and dashed lines for 2001.
Table I.
Estimated regression coefficients and their 95% EMs from the models fitted for the PA concentrations associated with explanatory variables and with their mutual interactions
The explanatory variables are C (clone; 0 = K818, 1 = K884), F (fraction; 0 = free, 1 = soluble conjugated), V (year; 1 = 2001, 0 = 2003), T1 (centered and scaled [divided by 100] linear term of the effective temperature sum [d.d.]), and T2 (quadratic term of the effective temperature sum). The residual degrees of freedom are 102 for Put and 106 for Spd and Spm. Coef., Regression coefficient.
| Put
|
Spd
|
Spm
|
||||
|---|---|---|---|---|---|---|
| Coef. | 95% EM | Coef. | 95% EM | Coef. | 95% EM | |
| Intercept | 61.5 | 23.2 | 326.4 | 53.7 | 164.4 | 21.2 |
| F | 36.7 | 31.5 | −141.9 | 72.9 | −49.3 | 28.7 |
| C | −0.4 | 31.5 | 99.9 | 72.9 | 62.5 | 28.7 |
| V | 56.7 | 31.6 | 92.1 | 72.9 | −27.4 | 28.7 |
| T1 | −10.4 | 13.6 | 8.9 | 31.6 | 19.6 | 12.4 |
| T2 | −4.0 | 12.1 | −35.7 | 28.0 | −24.4 | 11.0 |
| F × C | 136.9 | 40.8 | 6.3 | 93.7 | 13.8 | 36.9 |
| F × V | −42.9 | 41.2 | −4.5 | 94.4 | 1.5 | 37.2 |
| C × V | −53.6 | 44.6 | −94.6 | 94.4 | −44.7 | 37.2 |
| F × T1 | −11.0 | 17.4 | 0.0 | 40.4 | −16.8 | 15.9 |
| F × T2 | 0.4 | 15.5 | 5.4 | 35.7 | −1.5 | 14.1 |
| C × T1 | 8.1 | 17.4 | −38.2 | 40.4 | −11.5 | 15.9 |
| C × T2 | 0.3 | 15.5 | −28.1 | 35.7 | −11.5 | 14.1 |
| V × T1 | −0.7 | 19.5 | −19.6 | 45.2 | −0.2 | 17.8 |
| V × T2 | 8.5 | 17.4 | 8.5 | 39.8 | 12.4 | 15.7 |
| F × C × V | 79.4 | 46.5 | −81.5 | 104.5 | −43.4 | 41.2 |
| F × C × T1 | −37.3 | 18.4 | 35.3 | 42.5 | 11.9 | 16.7 |
| F × C × T2 | −29.0 | 16.7 | −8.6 | 37.4 | 1.2 | 14.8 |
| F × V × T1 | −25.9 | 22.5 | −34.2 | 52.2 | −15.8 | 20.6 |
| F × V × T2 | 4.7 | 20.5 | −0.7 | 46.0 | −0.4 | 18.1 |
| C × V × T1 | 26.4 | 22.5 | 42.9 | 52.2 | −1.3 | 20.6 |
| C × V × T2 | 10.8 | 20.5 | 28.7 | 46.0 | 20.0 | 18.1 |
The free Put content was not observed to change during the study period apart from clone K884 in 2001, when we found a slightly increasing trend (Fig. 2, top, left side). The latter exception to the rule was supported by the estimated value [+26.4 nmol g−1 (100 d.d.)−1] of the interaction term C × V × T1 between clone, year, and the linear term of effective temperature sum being larger than its error margin [EM; 95% EM 22.5 nmol g−1 (100 d.d.)−1; Table I]. The observation of no trend elsewhere in the free Put was consistent with the estimates and EMs for the main effects of effective temperature sum T1 (slope) and T2 (curvature), and with those of the interaction effects C × T1 and C × T2 between clone and effective temperature sum, as well as interactions V × T1 and V × T2 between year and effective temperature sum, in which the estimates were all well within their EMs. In clone K884, the free Put observations on sampling date II were excluded from statistical analysis due to the wide scattering of the observations. The soluble conjugated fraction of the Put appeared in both clones to increase at the beginning of embryo development and to decrease during embryo maturation (Fig. 2, top, right side). However, the results on relevant interaction terms F × T1 and F × T2 were not sufficiently supportive about this for clone K818, whereas for clone K884 strong evidence for this observed pattern was provided by the estimates and EMs of the interaction effects F × C × T1 and F × C × T2. The insoluble conjugated Put fraction was considerable and maximally contained nearly 35% of the total Put in the sample, but there was no trend in the insoluble conjugated Put content (data not shown).
Spd was the most abundant PA during Scots pine zygotic embryogenesis. Both free and soluble conjugated Spd increased at the beginning of embryo development, reached their maximum when the effective temperature sum was between 600 and 750 d.d., and decreased thereafter (Fig. 2, middle). There was somewhat more random fluctuation in 2001 than in 2003 and more fluctuation in clone K818 than in clone K884. The main quadratic effect of T2 [estimate −35.7, 95% EM 28.0, both in nmol g−1 (100 d.d.)−1; Table I] conveyed strong evidence for the observed downward curvature in the PA concentrations by the effective temperature sum together with the “nonsignificance” of the main linear effect T1. This observation was qualitatively consistent for both the free and the soluble conjugated forms over the 2 years and in the two clones, as can be judged from the result for all the interaction terms involving T1 and T2, the estimates of these being all within their EMs (Table I). The insoluble conjugated Spd fraction always contained less than 11% of the total Spd fraction in the sample (data not shown).
Both the free and the soluble conjugated forms of Spm fluctuated in a consistent way in the two pine clones (Fig. 2, bottom). The concentrations appeared to increase typically when the level of the effective temperature sum was 500 d.d. until it was around 700 d.d., after which the concentrations started to go down. There was convincing evidence for the observed overall downward curvature provided by the fitted regression model, in which the main linear and quadratic terms of effective temperature sums [T1: estimate 19.6, 95% EM 12.4; T2: estimate −24.4, 95% EM 11.0; all values in nmol g−1 (100 d.d.)−1; Table I] were well supporting the observed pattern, and none of the estimated interaction terms involving T1 and T2 modified this appreciably. The content of insoluble conjugated Spm was very low or under the HPLC detection level in most samples.
ADC and ODC Expression at Transcript and Protein Levels
ADC gene expression and ADC enzyme activity were clearly preferred compared to putative ODC gene expression and ODC enzyme activity, respectively. This suggests that the ADC pathway is the main route to produce Put in developing zygotic embryos.
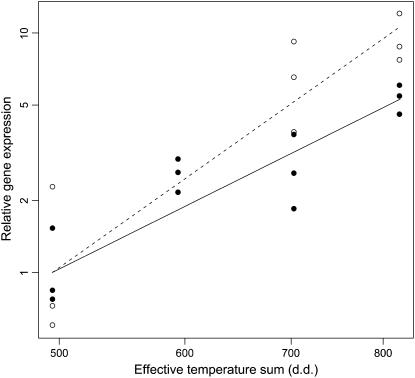
Relative ADC gene expression showed an increasing trend over time and by the effective temperature sum in both of the clones on the double logarithmic scale. The slope appeared steeper in clone K818 (slope 4.7, 95% EM 0.80) than in K884 (slope 3.3, 95% EM 0.75; Fig. 3). An ascending trend was also evident in ADC gene expression during embryo development, when the gene expression was normalized by another housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (data not shown). The mRNA levels of the putative ODC gene were very low. The fluorescence signal showed up after 35 cycles in real-time reverse transcription (RT)-PCR and therefore could not be reliably distinguished from spurious background.
Figure 3.
The expression of ADC gene in developing zygotic embryos of Scots pine in 2003. The expression was quantified by real-time PCR and normalized by the expression of the ACT gene. Observed relative ADC gene expression is presented relative to the effective temperature sum (d.d.) in clones K884 (•) and K818 (○) with the fitted regression lines (solid for K884, dashed for K818). There are three replicates per clone and per sampling date, except the missing data set of clone K818 on sampling date II.
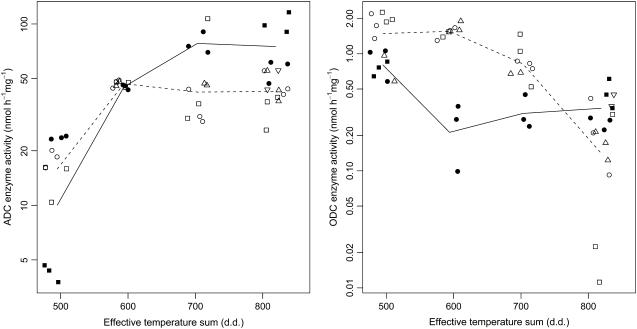
ADC enzyme activity seemed to increase in both clones during early embryo development, when the effective temperature sum rose from 500 to 600 d.d. Thereafter, the activity still seemed to increase in clone K884 but not in K818 (Fig. 4). The evidence for an increasing trend was weak and for curvature even weaker, though, as reflected by the estimate to EM ratio of the linear and quadratic terms T1 and T2 (Table II). Also, the results on interaction terms C × T1 and C × T2 did not provide sufficient basis to conclude that the slope and curvature were different for the two clones.
Figure 4.
ADC and ODC enzyme activities in developing zygotic embryos of Scots pine presented in relation to the effective temperature sum (d.d.) in the four sampling dates in 2003. Enzyme activities were analyzed by measuring the 14CO2 evolution of decarboxylated radiolabeled Arg and Orn. The black symbols (• and ▪) represent the activity measurements from clone K884 and the open symbols (○, □, ▵, and ▿) those from clone K818. At each sampling date, the replicate measurements (two or three) done from the same protein extraction are marked with the same symbol. Protein extractions per clone and per sampling date varied from one to four. The locations of the symbols are horizontally jittered about the exact x coordinate to separate overlapping values. The geometric mean values for each clone at different sampling dates are connected with solid line for K884 and dashed line for K818.
Table II.
Estimated regression coefficients and their 95% EMs from the models fitted for the natural logarithms of ADC and ODC enzyme activities associated with the fixed explanatory variable C (clone; 0 = K818, 1 = K884) and the linear and quadratic terms T1 and T2 of the effective temperature sum (centered and scaled by 100), as well as their interactions
The residual degrees of freedom are 12 for ADC and 13 for ODC. Coef., Regression coefficient.
| ADC
|
ODC
|
|||
|---|---|---|---|---|
| Coef. | 95% EM | Coef. | 95% EM | |
| Intercept | 3.90 | 0.43 | 0.10 | 0.53 |
| C | 0.39 | 0.85 | −1.58 | 1.03 |
| T1 | 0.22 | 0.22 | −0.76 | 0.25 |
| T2 | −0.22 | 0.24 | −0.36 | 0.27 |
| C × T1 | 0.33 | 0.36 | 0.58 | 0.42 |
| C × T2 | −0.13 | 0.42 | 0.67 | 0.50 |
ODC enzyme activity showed a decreasing trend with downward curvature in clone K818 throughout the sampling period, as suggested by the estimated values −0.76 (95% EM 0.25) of the linear term T1 and −0.36 (95% EM 0.27) of the quadratic term T2. In clone K884, ODC enzyme activity was initially lower and decreased further during the early developmental stages of the embryos, when the effective temperature sum was between 500 and 600 d.d., after which it remained at that level (Fig. 4). The estimated coefficients and EMs for the C × T1 and C × T2 interactions supported the existence of a less steep overall slope but an upward curvature for clone K884 than for K818 (Table II). ADC and ODC activities were also measured in pellets, in which the enzyme activities were clearly lower than in supernatants but no trend could be detected (data not shown).
Localization of ADC Gene Expression and ADC Protein in Dividing Cells
ADC gene expression was localized in the dominant embryo at the early embryogeny stage and also in two subordinate embryos (Fig. 1A). In late embryos, ADC expression was found to be located in the regions undergoing cell division, especially in the shoot apical and axillary meristems, but also in the dividing cells of the root meristem (Fig. 1, B–D). The ADC probe specificity was confirmed by the absence of signals in the sections hybridized with the sense ADC probe (Fig. 1, E and L). The transcripts of a putative ODC gene were not detected in embryos (Fig. 1F).
ADC gene expression occurred during mitosis and was found in cells throughout the mitotic stages (Fig. 1, G–K). In early prophase, when chromatids became visible in the nucleus, ADC expression was located in the cytoplasm (Fig. 1G). In late prophase and metaphase, ADC transcripts were also in the cytoplasmic region (Fig. 1, H and I), but in anaphase and telophase cells ADC transcripts clearly accumulated within the area of the mitotic spindle apparatus (Fig. 1, J and K).
The immunocytochemical localization showed the presence of ADC protein in the nuclei of late embryos (Fig. 1, M and N). High level of ADC protein was detected especially in mitotic cells, but, in contrast to ADC transcripts, ADC protein was found to locate close to the chromosomes and not within the mitotic spindle as the mRNA (Fig. 1, O and P). No positive signal was found in the control sections incubated with preimmune serum (Fig. 1Q).
DISCUSSION
PA Concentrations Show Consistent Profiles during Zygotic Embryogenesis
In this study, we found high PA concentrations in developing zygotic embryos of Scots pine, in accordance with the high PA contents reported in tissues undergoing rapid cell division, active metabolism, and somatic embryogenesis (Egea-Cortines and Mizrahi, 1991; Kakkar and Sawhney, 2002). Spd, which was the most abundant PA in this study, also was the main PA in the zygotic and somatic embryos of Pinus radiata D. Don. (Minocha and Minocha, 1995; Minocha et al., 1999b). We found that the individual PA levels differed between the pine clones within 1 year and between years when studying the same pine clone. This might be due to the variation reported in PA metabolism between genotypes (Sarjala and Savonen, 1994) and/or due to the environmental factors affecting PA levels (Bouchereau et al., 1999).
The zygotic embryo development of Scots pine takes 2 years. Generally, wind pollination occurs at the beginning of the growing season, late May or early June in Scandinavia, after which the pollen tube germination gradually ceases to be continued during the following growing season about 1 year later. Fertilization occurs usually at late June or early July, depending on the effective temperature sum (Sarvas, 1962; Krasowski and Owens, 1993). When the embryo development of Picea mariana Mill. was described by the effective temperature sum, the developmental stages, although unequal in duration, succeeded each other according to the monotonically increasing sigmoid function of the effective temperature sum (Sirois et al., 1999). In this work, anatomical studies also confirmed that Scots pine embryo development occurred along with the increasing effective temperature sum. Therefore, to be able to compare PA metabolism during early embryo development at two different years, the effective temperature sum instead of the sampling date was used as the main explanatory variable in statistical analyses. In most of the cases, the PA metabolism in developing embryos followed consistent profiles in different years, which suggested that PAs have an important role in embryo development and that individual PAs may have different roles at different developmental stages. The profiles of PAs seemed to show an increasing trend at the early stages but a decreasing trend at the late stages of embryo development. Only the free Put fraction remained stable throughout the period of embryo development. In this study, an increase in the ratio of free Spd and Put as well as in the ratio of free Spm and Put was found at the beginning of the zygotic embryo development. This is in accordance with the reports on somatic embryogenesis in Picea abies L. Karst. (Santanen and Simola, 1992), Scots pine (Niemi et al., 2002), and P. radiata (Minocha et al., 1999b), as well as in the zygotic embryos of P. radiata (Minocha et al., 1999b).
ADC Pathway Is Preferred during Zygotic Embryogenesis
In this study, both ADC gene expression at the mRNA transcript level and the activity of ADC enzyme were detected in developing zygotic embryos. The higher rate of ADC activity compared to ODC was prominent, which is in agreement with the reports of somatic embryo development in Picea rubens Sarg. (Minocha et al., 1996) as well as the zygotic embryogenesis of P. abies (Santanen and Simola, 1999). We found the highest ADC and ODC activities in the supernatant fraction containing soluble proteins, but some were also detected in the pellet.
In this work, ADC gene expression and ADC enzyme activity showed a steady increase throughout the embryo development. This might be due to the increasingly important role of the more developed embryo in PA metabolism compared to the senescing megagametophyte. We found that ADC expression increased during embryo development at both the mRNA and the enzyme activity levels. This suggests an involvement of transcriptional regulation in ADC gene expression but does not rule out posttranscriptional regulation of the gene. ADC mRNA has been found to accumulate under various stress conditions in Oryza sativa (Chattopadhyay et al., 1997), G. max L. Merr. (Nam et al., 1997), Brassica juncea (Mo and Pua, 2002), Arabidopsis (Urano et al., 2003), and Malus sylvestris L. Mill. (Hao et al., 2005). However, ADC has been reported to be regulated at the posttranslational level in Avena sativa (Malmberg and Cellino, 1994; Borrell et al., 1996) and also in potassium deficiency-stressed Arabidopsis (Watson and Malmberg, 1996). This indicates that different regulation mechanisms may be involved in ADC expression in different gene family members, plant species, tissues, and physiological conditions, as well as at different developmental stages.
The Role of ADC in Cell Division
In this study, we were able to localize ADC gene expression and ADC protein specifically in the mitotic cells of developing zygotic embryos. There is evidence that PAs are involved in the regulation of cell division in animals (Bello-Fernandez et al., 1993), microorganisms (Theiss et al., 2002), and plants (Kaur-Sawhney et al., 1980). In mammalian cells, the progression of the normal cell cycle is dependent on PAs (Oredsson, 2003), and PA contents have been reported to vary throughout the cell cycle in Helianthus tuberosus (Serafini-Fracassini, 1991). However, the exact intracellular localizations of ADC, ODC, and PAs during mitosis are hardly known, which has prevented more precise interpretation of the biological role of PAs in cell division. In this study, the specific localization of ADC protein in the cell nuclei and in the chromosomal area of mitotic cells may indicate that PAs synthesized via ADC are involved in chromatin condensation during cell division. Interestingly, in synchronized human cervix and prostatic carcinoma cell cultures, ODC protein was localized in perinuclear site at the beginning of mitosis and in the nucleoplasm surrounding the chromosomes during metaphase, anaphase, and telophase (Schipper et al., 2004). PAs have been implicated in the formation of higher-order chromosomal fibers in vitro and in vivo (Belmont et al., 1989). In mammalian cells, PA depletion causes major chromosomal alterations, such as decondensation and fragmentation of mitotic chromosomes (Pohjanpelto and Knuutila, 1982). Pollard et al. (1999) suggested that in yeasts PAs are repressors of transcription in vivo and that histone hyperacetylation antagonizes the ability of PAs to stabilize highly condensed states of chromosomal fibers.
ADC mRNAs, in this study, were located within the mitotic spindle apparatus of dividing cells, whereas ADC protein was close to the chromosomes. This may suggest that the transport of ADC mRNA during mitosis is mediated by microtubules and that ADC mRNA is translationally repressed while en route. Studies in Drosophila melanogaster Meigen and Xenopus laevis Daudin embryos, as well as neurons, have implicated microtubules in localizing mRNA (Nasmyth and Jansen, 1997), but to our knowledge this has not been reported earlier in higher plants. However, RNA localization appears to be an ancient cellular mechanism, while RNA transport and local translation have been documented in vertebrates, invertebrates, and unicellular organisms (Kindler et al., 2005). First evidence for the intracellular localization of specific mRNAs was discovered with maternal mRNAs within eggs or oocytes (Jeffery et al., 1983; Gavis and Lehmann, 1994), but, through improved sensitivity of in situ hybridization techniques, localized transcripts have been found in an increasing number of somatic cell types, including fibroblast, myoblast, neurons, oligodendrocytes, and epithelial cells (St Johnston, 1995).
ADC mRNA and ADC protein seem to take place particularly at the mitotic phase of the cell cycle, when most protein synthesis is inhibited. Pyronnet et al. (2000) found that mammalian ODC mRNA contains a cap-independent internal ribosome entry site that functions exclusively at the G2/M phase of the cell cycle. Commonly, ODC activity is also connected with cell division in plant cells (Galston and Sawhney, 1990), and recently ODC mRNA was in situ localized in the dividing cells of shoots and roots in Lycopersicon esculentum Mill., whereas ADC transcripts were not found in the dividing cells (Acosta et al., 2005). In tobacco cv Xanthi, ADC was found to be responsible for Put synthesis in old hypergenous vascular tissues, whereas ODC expression coincided with early cell divisions and ODC catalyzed Put synthesis in hypogenous tissues (Paschalidis and Roubelakis-Angelakis, 2005). Because plants are capable of using both the ODC and the ADC pathways in PA synthesis, it could be hypothesized that ADC may have taken, at least in some cells, the functions that belong to ODC in animal cells. Hanfrey et al. (2001) did not find a genomic sequence homologous to known ODC genes or ODC enzyme activity in Arabidopsis. Although conserved or similar basic mechanisms are operational at the core of the cell division cycle of all eukaryotes, plants show novel features, especially in molecules involved in the regulation of cell cycle control (Dewitte and Murray, 2003).
CONCLUSION
In this study, we report PA metabolism to follow yearly consistent profiles during zygotic embryo development in Scots pine. There is typically an increasing trend in PA concentrations at the early developmental stages and a decreasing trend during late embryo development, except in the case of the free Put fraction, which remains relatively stable throughout the embryo development. Our results show that the ADC pathway is the main PA route and that ADC expression is regulated, at least in part, at the mRNA level. We also show that ADC gene expression and ADC protein are present specifically in mitotic cells of embryo meristems, suggesting the essential role of ADC in the mitosis of plant cells.
MATERIALS AND METHODS
Collection of Immature Seeds
During the growing seasons 2001 and 2003, 1-year-old immature seed cones were collected from two open-pollinated elite Scots pine (Pinus sylvestris) clones, K818 and K884, growing in the Scots pine clone collection in Punkaharju, Finland (61°48′ N; 29°17′ E). In both years, the same grafts, one per pine clone, were used to collect the cones. The cone collection was repeated four times in July throughout the period of embryo development. In 2001 cones were collected on July 9 (sampling date I), July 16 (sampling date II), July 23 (sampling date III), and July 30 (sampling date IV), and in 2003 cones were collected on July 8 (sampling date I), July 15 (sampling date II), July 22 (sampling date III), and July 29 (sampling date IV). Immature zygotic embryos surrounded by the immature megagametophyte, called zygotic embryos, were dissected from the developing cones and fixed immediately for the microscopic examination as described below. Dissected zygotic embryos for PA analysis, enzyme analysis, and real-time RT-PCR were stored at −80°C until use.
Microscopic Observations
Developing embryos were fixed for study of the anatomical features of the embryos, for in situ hybridization and for immunolocalization. For all these purposes, the following protocol from fixation to coverslip mounting was used. The tissues were fixed in 4% (w/v) p-formaldehyde in 1× phosphate-buffered saline (PBS) buffer (10 mm phosphate, 150 mm NaCl, pH 7.4). After dehydration with a graded series of ethanol, ethanol was replaced by tertiary butanol and then gradually by paraffin. Sections (5 and 7 μm) were cut from the embedded samples with a microtome, mounted on SuperFrostPlus slides (Menzel-Glaser), and fixed by drying overnight at 40°C.
To study the developmental stage of the embryos, the preparates were stained with toluidine blue (0.05% toluidine blue in water) and the sections were examined under a light microscope. In 2003 the developmental stage was specified from six to 15 embryos per sampling date, totaling 85 embryos. The sequence of embryo development was divided into three phases according to Singh (1978). These stages include proembryogeny (i.e. the stages before suspensor elongation), early embryogeny (i.e. the stages after suspensor elongation and before the establishment of the root meristem), and late embryogeny (i.e. the establishment of root and shoot meristems and the further development of the embryo).
Measurement of PAs by HPLC
In addition to the free PAs, soluble and insoluble conjugated PAs were also examined. Four samples per sampling date and each sample consisting of 250 mg of embryos were extracted in 5% (w/v) perchloric acid. Crude extract for free PAs, hydrolyzed supernatant for perchloric acid-soluble conjugated PAs, and hydrolyzed pellet for perchloric acid-insoluble conjugated PAs were dansylated and separated by HPLC according to Sarjala and Kaunisto (1993) and Fornalé et al. (1999). The embryonic PA concentrations were expressed as nmol g−1 initial fresh weight of embryos.
Analysis of ADC and ODC Activities
The proteins of Scots pine zygotic embryos were extracted for enzyme activity measurements from one to four independent samples per sampling date. Embryos (400 mg fresh weight) were homogenized in an ice-cold mortar with liquid nitrogen and dissolved in 4 mL of extraction buffer containing 50 mm Tris-HCl, pH 8.4, 0.5 mm pyridoxal-5-phosphate, 0.1 mm EDTA, and 5.0 mm dithiothreitol. The solution was centrifuged at 15,500g for 20 min at +4°C, and the supernatant as well as the pellet resuspended with 4 mL of extraction buffer were used for both ADC and ODC enzyme assays according to Minocha et al. (1999a), with some modifications. The ADC and ODC activity reaction mixtures (300 μL) contained 200 μL of protein extract, 50 μL of buffer, and 50 μL of either 12 mm Arg containing 9.3 kBq l-[U-14C]Arg (specific activity 11.1 GBq/mmol; Amersham) or 12 mm Orn containing 3.7 kBq dl-[1-14C]Orn hydrochloride (specific activity 2.07 GBq/mmol; Amersham). The presence of true ADC activity was confirmed by adding 20 μL of 16 mm unlabeled l-Orn into the reaction mixture according to Tassoni et al. (2000) to inhibit arginase activity. In the inhibition assay for ODC, 12.5 mm dl-α-difluoromethylornithine (Sigma) was used. Blank samples were made by adding 200 μL of perchloric acid. Three activity measurements were performed from each protein sample and the activities were expressed as nmol CO2 h−1 (mg protein)−1. Protein content was measured with the Bradford method (Quick Start Bradford Protein Assay; Bio-Rad) using bovine serum albumin (BSA) as a standard.
PCR Primers
The primers 5′-AGAAATTGGGGATGCTGGAT-3′ and 5′-GCCATCACGATTGTATTCACC-3′ were designed to amplify a 466-bp fragment of the Scots pine ADC gene (AF306451). For amplifying a 420-bp fragment of a putative ODC gene, the primers 5′-AAGCGGTGAAGCCATTAAAA-3′ and 5′-TTGCGTTGCAGACGTATTTC-3′ were used. The primers for ODC were designed using GenBank Pinus taeda EST sequence CO176452. The selection of a putative ODC sequence was based on similarity at amino acid level with the ODC genes isolated from other plant species. The designing of the ACT primers 5′- GCTTGCTTATGTAGCCCTTGA-3′ and 5′-GGTCTTGGCAATCCACATCT-3′ was based on the Pinus contorta Dougl. ex Loud. actin (ACT) gene sequence M36171. The ACT primers were designed to contain an intron in the sequence between the primers to reveal possible genomic DNA contamination. The functioning of ACT primers and the primer pair against Scots pine glyceraldehyde-3-phosphate dehydrogenase sequence (L07501) was shown in Jaakola et al. (2004). All primers were chosen to have equal annealing temperatures close to the optimal temperature of 58°C.
RNA Isolation, RT-PCR, and cDNA Cloning
The total RNA of Scots pine zygotic embryos was extracted for the gene expression studies date as described in our previous paper (Vuosku et al., 2004), using the automated magnetic-based KingFisher mL method (Thermo Electron Corporation) and the manufacturer's KingFisher total RNA purification kit. Three independent RNA extractions were done per clone and per sampling date. However, the extractions from the embryos of clone K818 on sampling date II were unsuccessful and could not be repeated because of the shortage of the embryos. cDNA was prepared from 3 μg of total RNA, which was reverse transcribed by SuperScript II reverse transcriptase (Invitrogen) from an anchored oligo(dT) primer using standard methods in a reaction volume of 20 μL. Fragments of the PA biosynthesis genes and the housekeeping gene ACT were amplified from cDNA by standard PCR using DyNAzymeEXT polymerase (Finnzymes). PCR fragments were cloned into the pDrive Cloning Vector of Qiagen PCR Cloning kit (Qiagen). DNA sequencing was performed using an automated sequencer (model 377; PE Applied Biosystems) and dye terminator sequencing reagents (PE Applied Biosystems).
Real-Time PCR Analyses of ADC, ODC, and ACT mRNA Transcripts
The quantification of mRNA was done with real-time PCR. A first-strand cDNA template was synthesized from 3 μg of total RNA as described above. Three independent cDNA preparations were made for every sampling date, and every PCR reaction was done in duplicate to control for the variability of PCR amplification. The real-time PCR was performed in 50 μL of reaction mixture composed of 2 μL of cDNA, Brilliant SYBR Green QPCR Master Mix, and 150 nm gene-specific primers using the Mx3000P real-time PCR system (Stratagene). PCR amplification was initiated by incubation at 95°C for 10 min followed by 40 cycles: 30 s at 95°C, 1 min at 58°C, and 1 min at 72°C. The PCR conditions were optimized for high amplification efficiency >95% for all primer pairs used. ADC and ODC gene expressions were normalized by nonregulated reference gene expression derived from the housekeeping gene ACT. Q-Gene software (Muller et al., 2002) was used for the calculation of mean normalized expressions of the target genes. Mean normalized expressions are the linear expression values of a target gene relative to a reference gene, calculated from the average CT value, the PCR cycle where the fluorescence intensity of an amplicon crosses a threshold line, of the two replicates for the target gene and for the reference gene (Simon, 2003).
RNA Probes for in Situ Hybridization
To prepare RNA probes, we used a PCR-based technique, in which a T7 polymerase promoter sequence (TAATACGACTCACTATAGGG) was introduced at the 5′ ends of the gene-specific primers (Young et al., 1991). Plasmid DNA containing the cDNA in question served as the PCR template. The gene-specific downstream primer contained an artificially introduced T7 promoter at its 5′ end, which enabled the synthesis of antisense transcripts. The upstream primer containing T7 promoters at the 5′ ends was used for the synthesis of sense transcripts, i.e. as negative control. A PCR fragment representing the coding region of ADC was produced under standard PCR conditions using DyNAzymeEXT polymerase (Finnzymes). The PCR fragment was gel purified using the DNA gel extraction kit (Millipore Corporation), and 250 ng was subsequently used as a template DNA for in vitro transcription by T7 RNA polymerase (Invitrogen), incorporating dig-UTP via DIG RNA labeling mix (Roche Molecular Biochemicals). Template DNA was digested with four units of RNAse-free DNAse (Invitrogen) in a reaction volume of 20 μL for 10 min at 37°C, and the probe was precipitated and hydrolyzed in 1× carbonate buffer (80 mm NaHCO3, 120 mm Na2CO3). The yield of the DIG-labeled RNA probe was estimated by comparing the intensity of the sample to the defined controls made with DIG-labeled control RNA (Roche Molecular Biochemicals).
In Situ Hybridization Analysis
The sections for in situ hybridization were done as described above. In situ hybridization was done according to Mähönen et al. (2000) with some modifications. The sections were dewaxed in xylol, hydrated in an ethanol dilution series, and treated sequentially with 0.2 m HCl, proteinase K (10 μg mL−1), 4% (w/v) p-formaldehyde, and 0.5% acetic anhydride in 0.1 m triethanolamine. The samples were hybridized in a solution containing 50% (v/v) formamide, 300 mm NaCl, 10 mm Tris, pH 7.0, 10 mm Na3PO4, pH 7.0, 50 mm EDTA, 10% dextran sulfate, 200 μg mL−1 tRNA, 1× Denhardt's solution, and 10 units mL−1 RNase inhibitor overnight at 55°C in a water atmosphere. The amount of RNA probe used was about 200 ng per slide. After hybridization, the slides were washed in 0.2× SSC buffer (30 mm NaCl, 3 mm sodium citrate, pH 7.0) at 55°C for 60 min and treated with RNase A (10 μg mL−1) in NTE buffer (500 mm NaCl, 10 mm Tris, pH 8.0, 5 mm EDTA). The hybridized probe was detected using alkaline phosphatase-conjugated anti-DIG antibodies and NBT/BCIP as substrates (Roche Molecular Biochemicals).
Immunolocalization
The IgGs obtained against the tobacco (Nicotiana tabacum) ADC protein (Bortolotti et al., 2004) were used for immunolocalization. The sections were deparaffinized, rehydrated in an ethanol dilution series, and treated with 3% H2O2 in 1× PBS to destroy endogenous peroxidase activity. The sections were washed with PBS followed by a 0.3% Triton X-100/5% BSA in 0.1% phosphate buffer (PBTBSA), and then they were incubated with anti-ADC antiserum at concentration 1:250 in PBTBSA for 30 min. After rinsing in PBTBSA, the avidin-biotin complex method (Vector Labs), using diaminobenzidine as a substrate for peroxidase, was carried out to detect the antigen-antibody complex according to the vendor's instructions.
Statistical Methods
The PA concentrations were analyzed by normal linear regression models in which T (effective temperature sum) was the main explanatory variable. C (clone; values 0 = K818, 1 = K884), F (fraction; 0 = free, 1 = soluble conjugated), and V (year; 1 = 2001, 0 = 2003) were binary covariates. A second-order polynomial curve for the response was assumed, including both T1 = the centered and scaled linear term (original values minus the overall mean value divided by 100) and T2 = the quadratic (square of T1) term of the effective temperature sum. Interactions between both terms of effective temperature sum with the three binary factors were allowed for by including appropriate product terms in the model.
The development of ADC gene expression as a function of effective temperature sum in 2003 was analyzed by fitting a regression line through the origin for the logarithm of relative expression as a linear function of the logarithm of effective temperature sum (LET). The slopes were estimated for both clones separately, and the common slope assumption was evaluated by the appropriate interaction term C × LET.
ADC and ODC enzyme activities by effective temperature sum in 2003 were analyzed by linear mixed regression models. Here, the natural logarithm of the enzyme activity measurement was taken as the response variable. As in the above PA models, the linear and quadratic terms T1 and T2 of effective temperature sum were the interesting quantitative covariates, and C = clone was a binary covariate. The interactions of effective temperature sum terms with clone were also included in the models. In addition, the variability of the response over the separate isolation runs performed at each measurement occasion was allowed for by including an appropriate random effect term.
The PA models and the gene expression models were fitted using function lm() and the models for the enzyme activities by function lme() in the R statistical environment (http://www.r-project.org/). Assessment of the model assumptions were based on an informal inspection of the four diagnostic graphs provided by the default plot methods for the fitted model objects. The results were tabulated by giving the point estimates and the 95% EMs for the coefficients in each model, so that estimate ± EM provides the 95% confidence interval for the coefficient. The estimate to EM ratio multiplied by 2 can be used as an approximate t statistic to perform a test of significance for any coefficient of interest, but, in light of recent recommendations, we prefer to avoid explicit reporting of P values.
Acknowledgments
We thank Dr. Anneli Kauppi, Dr. Antti Pajunen, and Dr. Marja Nissinen from the University of Oulu, Dr. Eila Tillman-Sutela from the Finnish Forest Research Institute, Dr. Alexandra C. Baumgartner from the University of Göttingen, and MA Merja Lappalainen for help, enthusiasm, and interest during the work. We are also grateful to Ms. Eeva Pihlajaviita, Finnish Forest Research Institute, for technical help, and to the personnel of the Finnish Forest Research Institute at Punkaharju Research Unit for conducting the collections of the research material.
This work was supported by the Academy of Finland (project no. 53440 to T.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Hely Häggman (hely.haggman@oulu.fi).
References
- Acosta C, Pérez-Amador MA, Carbonell J, Granell A (2005) The two ways to produce putrescine in tomato are cell-specific during normal development. Plant Sci 168: 1053–1057 [Google Scholar]
- Bagni N, Tassoni A (2001) Biosynthesis, oxidation and conjugation of aliphatic polyamines in higher plants. Amino Acids 20: 301–317 [DOI] [PubMed] [Google Scholar]
- Bello-Fernandez C, Packham G, Cleveland JL (1993) The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc Natl Acad Sci USA 90: 7804–7808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont AS, Braunfeld MB, Sedat JW, Agard DA (1989) Large-scale chromatin structural domains within mitotic and interphase chromosomes in vivo and in vitro. Chromosoma 98: 129–143 [DOI] [PubMed] [Google Scholar]
- Borrell A, Besford RT, Altabella T, Masgrau C, Tiburcio AF (1996) Regulation of arginine decarboxylase by spermine in osmotically-stressed oat leaves. Physiol Plant 98: 105–110 [DOI] [PubMed] [Google Scholar]
- Bortolotti C, Cordeiro A, Alcázar R, Borrell A, Culiañez-Maciá FA, Tiburcio AF, Altabella T (2004) Localization of arginine decarboxylase in tobacco plants. Physiol Plant 120: 84–92 [DOI] [PubMed] [Google Scholar]
- Bouchereau A, Aziz A, Larher F, Martin-Tanguy J (1999) Polyamines and environmental challenges: recent development. Plant Sci 140: 103–125 [Google Scholar]
- Chattopadhyay MK, Gupta S, Sengupta DN, Ghosh B (1997) Expression of arginine decarboxylase in seedlings of indica rice (Oryza sativa L.) cultivars affected by salinity stress. Plant Mol Biol 34: 477–483 [DOI] [PubMed] [Google Scholar]
- Childs AC, Mehta DJ, Gerner EW (2003) Polyamine-dependent gene expression. Cell Mol Life Sci 60: 1394–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen E, Arad SM, Heimer YH, Mizrahi Y (1984) Polyamine biosynthetic enzymes in the cell cycle of Chlorella. Plant Physiol 74: 385–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S (1998) A Guide to the Polyamines. University Press, Oxford
- Coleman CS, Hu G, Pegg A (2004) Putrescine biosynthesis in mammalian tissues. Biochem J 379: 849–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delis C, Dimou M, Efrose RC, Flemetakis E, Aivalakis G, Katinakis P (2005) Ornithine decarboxylase and arginine decarboxylase gene transcripts are co-localized in developing tissues of Glycine max etiolated seedlings. Plant Physiol Biochem 43: 19–25 [DOI] [PubMed] [Google Scholar]
- Dewitte W, Murray JAH (2003) The plant cell cycle. Annu Rev Plant Biol 54: 235–264 [DOI] [PubMed] [Google Scholar]
- Egea-Cortines M, Mizrahi Y (1991) Polyamines in cell division, fruit set and development, and seed germination. In RD Slocum, HE Flores, eds, Biochemistry and Physiology of Polyamines in Plants. CRC Press, Boca Raton, FL, pp 143–158
- Flores HE (1991) Changes in polyamine metabolism in response to abiotic stress. In RD Slocum, HE Flores, eds, Biochemistry and Physiology of Polyamines in Plants. CRC Press, Boca Raton, FL, pp 213–225
- Fornalé S, Sarjala T, Bagni N (1999) Endogenous polyamine content and metabolism in the ectomycorrhizal fungus Paxillus involutus. New Phytol 143: 581–587 [DOI] [PubMed] [Google Scholar]
- Galston AW, Sawhney RK (1990) Polyamines in plant physiology. Plant Physiol 94: 406–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavis ER, Lehmann R (1994) Translational regulation of nanos by RNA localization. Nature 369: 315–318 [DOI] [PubMed] [Google Scholar]
- Ge C, Cui X, Wang Y, Hu Y, Fu Z, Zhang D, Cheng Z, Li J (2006) BUD2, encoding an S-adenosylmethionine decarboxylase, is required for Arabidopsis growth and development. Cell Res 16: 446–456 [DOI] [PubMed] [Google Scholar]
- Hanfrey C, Sommer S, Mayer MJ, Burtin D, Michael AJ (2001) Arabidopsis polyamine biosynthesis: absence of ornithine decarboxylase and the mechanism of arginine decarboxylase activity. Plant J 27: 551–560 [DOI] [PubMed] [Google Scholar]
- Hao YJ, Kitashiba H, Honda C, Nada K, Moriguchi T (2005) Expression of arginine decarboxylase genes in apple cells and stressed shoots. J Exp Bot 56: 1105–1115 [DOI] [PubMed] [Google Scholar]
- Imai A, Matsuyama T, Hanzawa Y, Akiyama T, Tamaoki M, Saji H, Shirano Y, Kato T, Hayashi H, Shibata D, et al (2004) Spermidine synthase genes are essential for survival of Arabidopsis. Plant Physiol 135: 1565–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakola L, Pirttilä AM, Vuosku J, Hohtola A (2004) Method based on electrophoresis and gel extraction for obtaining genomic DNA-free cDNA without DNase treatment. Biotechniques 37: 744–748 [DOI] [PubMed] [Google Scholar]
- Jeffery WB, Tomlinson CR, Brodeur RD (1983) Localization of actin messenger RNA during early ascidian development. Dev Biol 99: 408–417 [DOI] [PubMed] [Google Scholar]
- Kakkar RK, Sawhney VK (2002) Polyamine research in plants—a changing perspective. Physiol Plant 116: 281–292 [Google Scholar]
- Kaur-Sawhney R, Flores HE, Galston AW (1980) Polyamine-induced DNA synthesis and mitosis in oat leaf protoplasts. Plant Physiol 65: 368–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler S, Wang H, Richter D, Tiedge H (2005) RNA transport and local control of translation. Annu Rev Cell Dev Biol 21: 223–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasowski MJ, Owens JN (1993) Ultrastructural and histochemical postfertilization megagametophyte and zygotic embryo development of white spruce (Picea glauca) emphasizing the deposition of seed storage products. Can J Bot 71: 98–112 [Google Scholar]
- Kumar A, Altabella T, Taylor MA, Tiburcio AF (1997) Recent advances in plant polyamine research. Trends Plant Sci 2: 124–130 [Google Scholar]
- Lin PPC, Egli DB, Li GM, Meckel L (1984) Polyamine titer in the embryonic axis and cotyledons of Glycine max (L.) during seed growth and maturation. Plant Physiol 76: 366–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen AP, Bonke M, Kauppinen L, Riikonen M, Benfey PN, Helariutta Y (2000) A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev 14: 2938–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmberg RL, Cellino ML (1994) Arginine decarboxylase of oats is activated by enzymatic cleavage into two polypeptides. J Biol Chem 28: 2703–2706 [PubMed] [Google Scholar]
- Martin-Tanguy J (1997) Conjugated polyamines and reproductive development: biochemical, molecular and physiological approaches. Physiol Plant 100: 675–688 [Google Scholar]
- Minocha R, Long S, Maki H, Minocha SC (1999. a) Assays for the activities of polyamine biosynthetic enzymes using intact tissues. Plant Physiol Biochem 37: 597–603 [Google Scholar]
- Minocha R, Shortle WC, Coughlin DJ, Minocha SC (1996) Effects of aluminum on growth, polyamine metabolism, and inorganic ions in suspension cultures of red spruce (Picea rubens). Can J For Res 26: 550–559 [Google Scholar]
- Minocha R, Smith DR, Reeves C, Steele K, Minocha S (1999. b) Polyamine levels during the development of zygotic and somatic embryos of Pinus radiata. Physiol Plant 105: 155–164 [Google Scholar]
- Minocha SC, Minocha R (1995) Role of polyamines in somatic embryogenesis. In YPS Bajaj, ed, Biotechnology in Agriculture and Forestry, Vol 30: Somatic Embryogenesis and Synthetic Seed I. Springer-Verlag, Heidelberg, pp 55–72
- Mo H, Pua EC (2002) Up-regulation of arginine decarboxylase gene expression and accumulation of polyamines in mustard (Brassica juncea) in response to stress. Physiol Plant 114: 439–449 [DOI] [PubMed] [Google Scholar]
- Muller PY, Janovjak H, Miserez AR, Dobbie Z (2002) Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 32: 1372–1379 [PubMed] [Google Scholar]
- Nam KH, Lee SH, Lee JH (1997) Differential expression of ADC mRNA during development and upon acid stress in soybean (Glycine max) hypocotyls. Plant Cell Physiol 38: 1156–1166 [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Jansen RP (1997) The cytoskeleton in mRNA localization and cell differentiation. Curr Opin Cell Biol 9: 396–400 [DOI] [PubMed] [Google Scholar]
- Niemi K, Sarjala T, Chen X, Häggman H (2002) Spermidine and methylglyoxal bis(guanylhydrazone) affect maturation and endogenous polyamine content of Scots pine embryogenic cultures. J Plant Physiol 159: 1155–1158 [Google Scholar]
- Oredsson SM (2003) Polyamine dependence of normal cell-cycle progression. Biochem Soc Trans 31: 366–370 [DOI] [PubMed] [Google Scholar]
- Owens JN, Øystein J, Dæhlen OG, Skrøppa T (2001) Potential effects of temperature on early reproductive development and progeny performance in Picea abies (L.) Karst. Scand J For Res 16: 221–237 [Google Scholar]
- Paschalidis KA, Roubelakis-Angelakis KA (2005) Spatial and temporal distribution of polyamine levels and polyamine anabolism in different organs/tissues of the tobacco plant. Correlations with age, cell division/expansion, and differentiation. Plant Physiol 138: 142–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohjanpelto P, Knuutila S (1982) Polyamine deprivation causes major chromosomal alterations in polyamine-dependent Chinese hamster ovary cell line. Exp Cell Res 141: 333–339 [DOI] [PubMed] [Google Scholar]
- Pollard KJ, Samuels ML, Crowley KA, Hansen JC, Peterson CL (1999) Functional interaction between GCN5 and polyamines: a new role for core histone acetylation. EMBO J 18: 5622–5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyronnet S, Pradayrol L, Sonenberg N (2000) A cell cycle-dependent internal ribosome entry site. Mol Cell 5: 607–616 [DOI] [PubMed] [Google Scholar]
- Santanen A, Simola LK (1992) Changes in polyamine metabolism during somatic embryogenesis in Picea abies. J Plant Physiol 140: 475–480 [Google Scholar]
- Santanen A, Simola LK (1999) Metabolism of L[U-14C]-arginine and L[U-14C]-ornithine in maturating and vernalized embryos and megagametophytes of Picea abies. Physiol Plant 104: 433–440 [Google Scholar]
- Sarjala T, Kaunisto S (1993) Needle polyamine concentrations and potassium nutrition in Scots pine. Tree Physiol 13: 87–96 [DOI] [PubMed] [Google Scholar]
- Sarjala T, Savonen EM (1994) Seasonal fluctuation in free polyamines in Scots pine needles. J Plant Physiol 144: 720–725 [Google Scholar]
- Sarvas R (1962) Investigations on the flowering and seed crop of Pinus sylvestris. Commun Inst For Fenn 53: 1–198 [Google Scholar]
- Schipper RG, Cuijpers VMJI, de Groot LHJM, Thio M, Verhofstad AAJ (2004) Intracellular localization of ornithine decarboxylase and its regulatory protein, antizyme-1. J Histochem Cytochem 52: 1259–1266 [DOI] [PubMed] [Google Scholar]
- Serafini-Fracassini D (1991) Cell cycle-dependent changes in plant polyamine metabolism. In RD Slocum, HE Flores, eds, Biochemistry and Physiology of Polyamines in Plants. CRC Press, Boca Raton, FL, pp 159–173
- Simon P (2003) Q-gene: processing quantitative real-time RT-PCR data. Bioinformatics 19: 1439–1440 [DOI] [PubMed] [Google Scholar]
- Singh H (1978) Embryology of Gymnosperms. Borntrager, Berlin
- Sirois L, Bégin Y, Parent J (1999) Female gametophyte and embryo development of black spruce along a shore-hinterland climatic gradient of a recently created reservoir, northern Quebec. Can J Bot 77: 61–69 [Google Scholar]
- St Johnston D (1995) The intracellular localization of messenger RNAs. Cell 81: 161–170 [DOI] [PubMed] [Google Scholar]
- Tassoni A, van Buuren M, Franceschetti M, Fornalé S, Bagni N (2000) Polyamine content and metabolism in Arabidopsis thaliana and effect of spermidine on plant development. Plant Physiol Biochem 38: 383–393 [Google Scholar]
- Theiss C, Bohley P, Voigt J (2002) Regulation by polyamines of ornithine decarboxylase activity and cell division in the unicellular green alga Chlamydomonas reinhardtii. Plant Physiol 128: 1470–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiburcio AF, Altabella T, Borrell A, Masgrau C (1997) Polyamine metabolism and its regulation. Physiol Plant 100: 664–674 [Google Scholar]
- Urano K, Yoshiba Y, Nanjo T, Igarashi Y, Seki M, Sekiguchi F, Yamaguchi-Shinozaki K, Shinozaki K (2003) Characterization of Arabidopsis genes involved in biosynthesis of polyamines in abiotic stress responses and developmental stages. Plant Cell Environ 26: 1917–1926 [Google Scholar]
- Vuosku J, Jaakola L, Jokipii S, Karppinen K, Kämäräinen T, Pelkonen VP, Jokela A, Sarjala T, Hohtola A, Häggman H (2004) Does extraction of DNA and RNA by magnetic fishing work for diverse plant species? Mol Biotechnol 27: 209–215 [DOI] [PubMed] [Google Scholar]
- Watson MB, Malmberg RL (1996) Regulation of Arabidopsis thaliana (L.) Heynh. arginine decarboxylase by potassium deficiency stress. Plant Physiol 111: 1077–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav JS, Rajam MV (1998) Temporal regulation of somatic embryogenesis by adjusting cellular polyamine content in eggplant. Plant Physiol 116: 617–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young IY, Ailles L, Deugau K, Kisilevsky R (1991) Transcription of cRNA for in situ hybridization from polymerase chain reaction-amplified DNA. Lab Invest 64: 709–712 [PubMed] [Google Scholar]