Abstract
The release of organic anions from roots can protect plants from aluminum (Al) toxicity and help them overcome phosphorus (P) deficiency. Our previous findings showed that Al treatment induced malate and citrate efflux from rape (Brassica napus) roots, and that P deficiency did not induce the efflux. Since this response is similar to the malate efflux from wheat (Triticum aestivum) that is controlled by the TaALMT1 gene, we investigated whether homologs of TaALMT1 are present in rape and whether they are involved in the release of organic anions. We isolated two TaALMT1 homologs from rape designated BnALMT1 and BnALMT2 (B. napus Al-activated malate transporter). The expression of these genes was induced in roots, but not shoots, by Al treatment but P deficiency had no effect. Several other cations (lanthanum, ytterbium, and erbium) also increased BnALMT1 and BnALMT2 expression in the roots. The function of the BnALMT1 and BnALMT2 proteins was investigated by heterologous expression in cultured tobacco (Nicotiana tabacum) cells and in Xenopus laevis oocytes. Both transfection systems showed an enhanced capacity for malate efflux but not citrate efflux, when exposed to Al. Smaller malate fluxes were also activated by ytterbium and erbium treatment. Transgenic tobacco cells grew significantly better than control cells following an 18 h treatment with Al, indicating that the expression of BnALMT1 and BnALMT2 increased the resistance of these plant cells to Al stress. This report demonstrates that homologs of the TaALMT1 gene from wheat perform similar functions in other species.
Aluminum (Al) toxicity is the primary factor limiting crop production on acidic soils. When the soil pH falls below 5.0 the soluble Al in the soil solution exists predominantly as the toxic trivalent cation Al3+ that can inhibit root growth in many species at micromolar concentrations (Kochian et al., 2005). Some species have developed mechanisms to overcome Al-related stresses by either excluding Al from the root cells (resistance mechanisms) or by increasing their tolerance to Al once these cations have been absorbed by the roots (tolerance mechanisms). Exclusion mechanisms might involve the exudation of Al-chelating ligands, the binding of Al within the cell wall or mucilage, plant-induced pH changes in the rhizosphere that reduce the local concentration of Al3+ relative to other hydrolysis products, the selective permeability of the plasma membrane, or perhaps the efflux of Al itself from the root cells. Tolerance mechanisms might include the chelation of Al in the cytosol, the sequestration of Al in the vacuole or other organelles, or modifications to metabolism that allow cellular function to continue normally in the presence of Al (Foy et al., 1978; Taylor, 1991; Delhaize and Ryan, 1995; Horst, 1995; Kochian, 1995; Ma et al., 2001; Matsumoto, 2002).
The mechanism of Al resistance that has been observed most commonly in monocotyledonous and dicotyledonous species involves the release of organic anions such as malate, citrate, and oxalate from roots (Miyasaka et al., 1991; Delhaize et al., 1993; Pellet et al., 1995; Ryan et al., 1995b, 2001; Ma et al., 1997b, 2001; Zheng et al., 1998; Hoekenga et al., 2003; Kinraide et al., 2005). These anions bind the Al cations in the apoplasm and rhizosphere and minimize their harmful interactions with the root cells.
Phosphorus (P) deficiency is also reported to induce the release of organic anions in rape (Brassica napus; Hoffland et al., 1989), white lupin (Lupinus albus; Gardner et al., 1983), and alfalfa (Medicago sativa; Lipton et al., 1987). These organic anions can mobilize P; first by chelating metal ions that could otherwise immobilize P, and second by direct anion exchange. In contrast, P deficiency did not induce exudation in wheat (Triticum aestivum), taro (Colocasia esculenta), buckwheat (Fagopyrum esculentum), and soybean (Glycine max; Delhaize et al., 1993; Ma et al., 1997a; Ma and Miyasaka, 1998; Yang et al., 2000). Similarly, Ligaba et al. (2004) did not observe any induction of organic anion efflux under P-deficient conditions, but showed that Al-induced efflux is more pronounced in P-sufficient plants of rape.
Al resistance in wheat relies on the Al-dependent efflux of malate anions from the root apices and a strong correlation exists between relative Al resistance of different genotypes and the capacity for malate efflux (Delhaize et al., 1993; Ryan et al., 1995b; Raman et al., 2005). Pharmacological and electrophysiological studies suggest that malate efflux is facilitated by anion channels and this is consistent with the electrochemical gradient for anions across the plasma membrane (Ryan et al., 1997; Zhang et al., 2001). Maize (Zea mays) is another species showing Al-dependent organic anion efflux, and studies in this species also indicate a role for anion channels in malate and citrate export (Kollmeier et al., 2001; Piñeros and Kochian, 2001; Piñeros et al., 2002).
Recently the gene controlling the Al-dependent efflux of malate from wheat was isolated by Sasaki et al. (2004). This gene, designated TaALMT1, encodes a hydrophobic protein that localizes to the plasma membrane of root cells (Yamaguchi et al., 2005). TaALMT1 expression in Al-resistant genotypes of wheat is 5- to 10-fold higher than in Al-sensitive genotypes (Sasaki et al., 2004; Raman et al., 2005). Heterologous expression of TaALMT1 in cultured tobacco (Nicotiana tabacum) cells, Xenopus oocytes, and intact rice and barley (Hordeum vulgare) plants conferred an Al-activated malate efflux and increased the Al tolerance of tobacco suspension cells and intact barley plants (Delhaize et al., 2004; Sasaki et al., 2004).
Homologs of TaALMT1 have now been identified in other members of the poaceae (rice and barley) as well as in dicotyledons (see GenBank database). The Arabidopsis (Arabidopsis thaliana) genome contains at least 14 homologs with the predicted proteins showing an approximately 30% to 40% identity with TaALMT1. One of these homologs has recently been implicated in an Al-resistance mechanism of Arabidopsis that also relies on malate release (Hoekenga et al., 2003, 2006). Hoekenga et al. (2006) showed that plants carrying knockout mutations in At1g08430, a TaALMT1 homolog on chromosome 1, lose their capacity for Al-dependent malate efflux and are significantly more sensitive to Al treatment than wild-type plants.
Ligaba et al. (2004) demonstrated previously that Al treatment induced the release of malate and citrate from the roots of rape seedlings. The response could not be explained by Al-dependent changes in organic acid biosynthesis or by the cellular concentrations of citrate and malate in the roots (Ligaba et al., 2004). Therefore the aim of this study was to investigate whether TaALMT1 homologs are present in the important crop species rape and, if so, whether they are involved in the Al-induced efflux of organic acids. We isolated and characterized two ALMT1 homologs in rape designated BnALMT1 and BnALMT2 (B. napus Al-activated malate transporter). Expression studies in wild-type plants as well as transgenic studies in Xenopus oocytes and tobacco suspension cells suggest that these genes encode Al-activated malate transporters that can confer resistance to Al stress.
RESULTS
Isolation of ALMT1 Homologs in Rape
Previous investigation by Ligaba et al. (2004) demonstrated that treatment with Al induced malate and citrate efflux from the roots of +P and −P plants. Since this response in rape is similar to the Al-dependent malate efflux reported in Al-resistant wheat (Delhaize et al., 1993; Ryan et al., 1995a) we investigated whether a homolog of the wheat TaALMT1 gene was involved in the response in rape.
The Arabidopsis genome contains at least 14 genes that are homologous to the TaALMT1 gene in wheat (Sasaki et al., 2004). The phylogenetic similarity between rape and Arabidopsis was used to design degenerate primers from the consensus sequences of three homologs in Arabidopsis (At1g08430, At1g08440, and At2g27240) that are among the most similar to TaALMT1. About 600 bp fragment was amplified from cDNA prepared from the roots of rape seedlings. These fragments were cloned, sequenced, and RACE was used to obtain full-length cDNAs. Two different cDNAs, designated BnALMT1 and BnALMT2, were isolated using this procedure. Rape (B. napus AACC) is an amphidiploid species formed from the cross of Brassica oleracea (AA) and Brassica rapa (CC). Therefore, rape is a tetraploid and it is possible that BnALMT1 and BnALMT2 are homologs. The coding regions of BnALMT1 and BnALMT2 (DNA data bank of Japan AB194300, AB194301) were 1,494 and 1,479 bp long with the deduced proteins being comprised of 498 and 493 amino acid residues, respectively. BnALMT1 and BnALMT2 were 95% identical to one another, 80% identical to the At1g08430 gene from Arabidopsis, and 40% identical to the TaALMT1 gene from wheat (Fig. 1). Hydropathy plots (PSORT: http://psort.nibb.ac.jp/form.html) of the deduced proteins were similar to TaALMT1 with each having five to eight potential membrane-spanning domains located in the N-terminal half (data not shown).
Figure 1.
Multiple sequence alignment of the ALMT1-1 protein from wheat and the hypothetical proteins from rape (BnALMT1 and BnALMT2) and Arabidopsis (At1g08430, At1g08440, and At2g27240). Alignments were carried out using CLUSTAL (http://clustalw.genome.jp). Identical amino acids and similar amino acids were indicated by dark shading and light shading, respectively, using BOXSHADE software (http://www.ch.embnet.org/software/BOX_form.html). The two lines above the sequence indicate the position of degenerate primers.
Analysis of BnALMT Expression
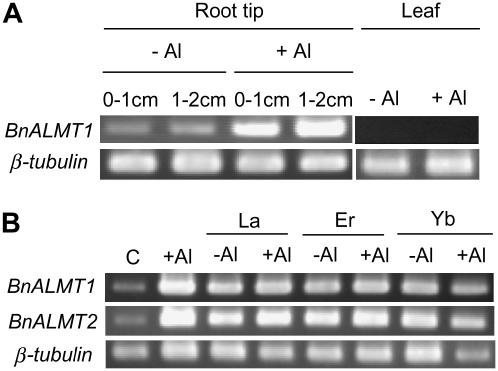
Semiquantitative reverse transcription (RT)-PCR was initially used to analyze BnALMT1 and BnALMT2 expression in rape seedlings. Expression in the apical 2 cm of roots was relatively low but increased by a 6 h treatment in 50 μm Al (Fig. 2). No expression of BnALMT1 was detected in the shoots with or without Al treatment (Fig. 2A). To determine the specificity of this response for Al ions, the effect of a number of lanthanides on BnALMT expression was tested. These elements, like Al, exist as trivalent cations in solution and are toxic to plants. Treatment with 50 μm lanthanum (La), erbium (Er), or ytterbium (Yb) increased expression of BnALMT1 and BnALMT2 in the roots in the presence and absence of Al (Fig. 2B).
Figure 2.
Analysis of BnALMT expression in rape seedlings. A, Tissue-specific expression of BnALMT1 seedlings grown in complete nutrient solution. Plants were exposed to 0.5 mm CaCl2 solution (pH 4.5) containing 0 or 50 μm Al for 6 h. RNA was isolated from the apical centimeter of the root (0–1 cm), the next centimeter of root (1–2 cm), and from young leaves. Semiquantitative RT-PCR was conducted on 5 μg total RNA as described in the “Materials and Methods.” β-tubulin was used as the reference gene. B, Effect of Al, La, Er, and Yb on BnALMT1 and BnALMT2 expression. Seedlings were exposed to 0.5 mm CaCl2 (pH 4.5) containing 0 or 50 μm AlCl3 in the presence or absence of 50 μm LaCl3, ErCl3, or YbCl3 for 6 h. RNA was isolated from the roots and semiquantitative RT-PCR was conducted on 5 μg total RNA. β-tubulin was used as the reference gene (C is control with 0.5 mm CaCl2).
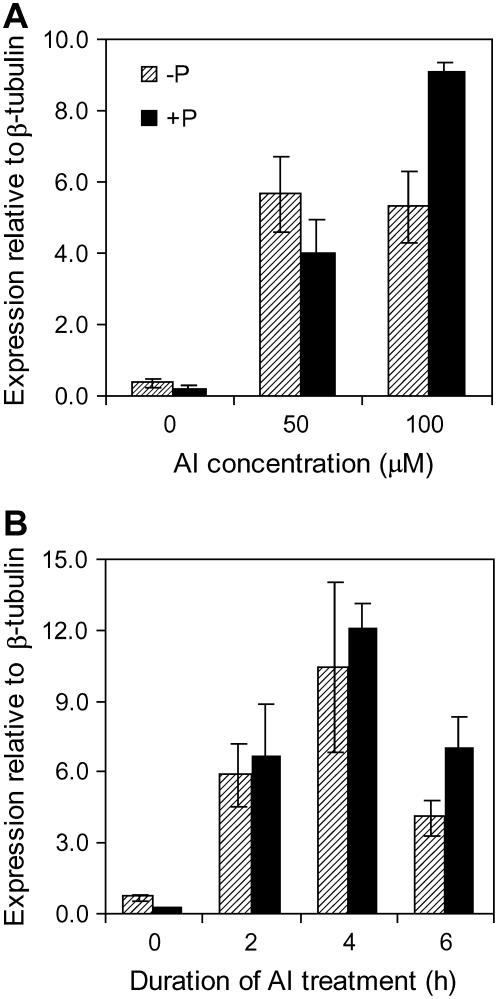
A second set of experiments used quantitative RT-PCR to examine BnALMT expression. Since the expression of BnALMT1 and BnALMT2 responded similarly to Al treatment these experiments used primers that amplified both genes together. Figure 3A shows that BnALMT expression was induced by Al treatment in a concentration-dependent manner. The expression of neither gene was increased by 8 d of P deprivation in the absence of Al. A time course indicates that the BnALMT genes are induced after 2 h exposure to Al with no further increases occurring after 6 h (Fig. 3B).
Figure 3.
Effect of Al concentration on BnALMT expression (A) and the time course of BnALMT expression levels after addition of 50 μm Al (B). P-sufficient (250 μm) or P-deficient (0 μm) seedlings were exposed to 0.5 mm CaCl2 solution (pH 4.5) containing 0, 50, or 100 μm AlCl3 for 6 h. RNA was isolated from the root and quantitative real-time RT-PCR was conducted on 1 μg total RNA. Bars represent means ± se of three replicates and independent experiments were performed at least three times. Expression of the BnALMT genes is shown relative to the β-tubulin reference gene using arbitrary units. Note that the primers used here amplified both BnALMT genes.
Expression of BnALMT1 and BnALMT2 in Xenopus Oocytes Confers Al-Activated Malate Efflux
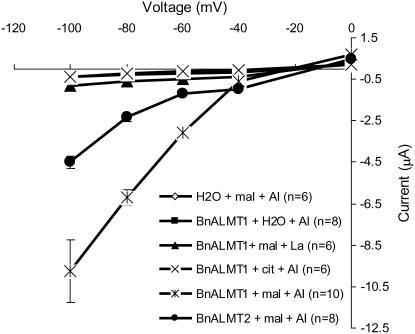
The function of the proteins encoded by BnALMT1 and BnALMT2 was examined by expressing their cRNA in Xenopus oocytes. The oocytes were first injected with the full-length cRNA of BnALMT1, BnALMT2, or with water (control), and later injected with malate or citrate before being exposed to 100 μm Al. For oocytes injected with the cRNA of either BnALMT1 or BnALMT2 as well as malate, treatment with 100 μm Al activated large inward or negative currents (Fig. 4). These inward currents were absent from control oocytes injected with water and malate and they were also absent from the oocytes injected with BnALMT1 cRNA and citrate (instead of malate). Oocytes injected with BnALMT1 cRNA and malate but treated with La (instead of Al) did not show the same currents either. The average current detected in oocytes expressing BnALMT1 was approximately 2-fold greater than the current measured in oocytes expressing BnALMT2. In summary, Al-dependent currents were only detected in cells expressing BnALMT1 or BnALMT2 that had been injected with malate. By convention, inward currents are generated either by the net uptake of cations or the net efflux of anions. Therefore, these inward currents are consistent with the Al-dependent efflux of malate2− or malate− anions from the oocytes.
Figure 4.
Electrophysiological characterization of BnALMT1 and BnALMT2 protein in Xenopus oocytes. Oocytes were injected with cRNA of BnALMT1, BnALMT2, or water. After an overnight incubation at 20°C the oocytes were injected with 50 nL of 100 mm Na malate or Na citrate before being exposed to 100 μm AlCl3 or LaCl3. From the holding potential 0 mV, the test voltage was clamped from −100 to 0 mV in 20 mV increments (3 s pulses). After clamping the membrane potential at a test voltage, the voltage was maintained at the holding potential for 5 s before being clamped at the next test voltage. The current-voltage curves were constructed from the current values measured at the end of the 3 s voltage pulses.
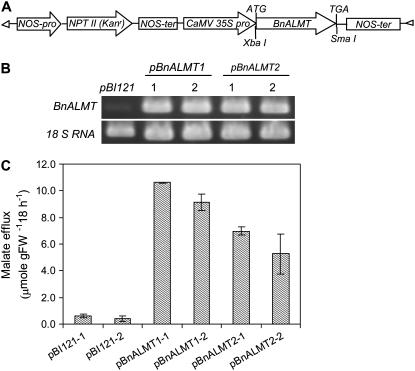
Expression of BnALMT1 and BnALMT2 in Tobacco Cultured Cells Confers Al-Activated Malate Efflux and Enhances Resistance to Al Stress
The function of proteins encoded by BnALMT1 and BnALMT2 was also investigated in transgenic plant cells using the constitutive 35S cauliflower mosaic virus promoter (Fig. 5A). To confirm expression of the genes, RNA was isolated from the transgenic calli and RT-PCR was conducted using gene-specific primers. Transcript of BnALMT1 and BnALMT2 was accumulated in the transgenic cells while no transcript was detected in the cells transformed with the empty vector (Fig. 5B). Al-dependent malate efflux was measured in two independent transgenic lines expressing each gene construct. Al treatment activated malate efflux from all the transgenic cell lines expressing BnALMT1 or BnALMT2 but not from the control lines containing the empty vector (Fig. 5C). No citrate was detected in the media with or without Al treatment (data not shown).
Figure 5.
Expression of BnALMT1 and BnALMT2 in tobacco cultured cells. A, Construct inserted into the vector pBI121 and introduced into Agrobacterium tumefaciens strain LBA 4404 for transforming tobacco cells. B, BnALMT1 and BnALMT2 expression in the transgenic cells analyzed by RT-PCR using tobacco 18 S RNA as control. C, Effect of Al on malate efflux from two independent transgenic lines (1 and 2) expressing BnALMT1, BnALMT2, or the empty vector. The transgenic cells (100 mg fresh weight 10 mL−1) were cultured in the dark condition at 25°C on a rotary shaker for 18 h in the Ca-Suc medium (pH 4.5) containing 0 or 100 μm AlCl3. Malate efflux was determined from 10 mL culture as described in the “Materials and Methods.” Bars represent means ± se of three replicates and the experiment was repeated four times.
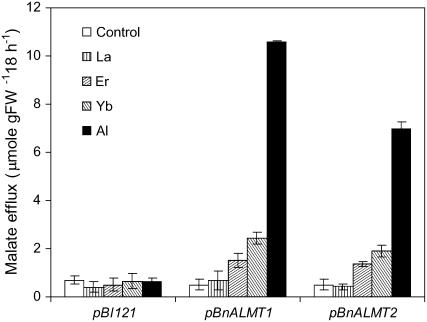
We determined whether malate efflux from the transgenic tobacco cells could also be induced by some of the elements from the lanthanide series. Figure 6 shows that Yb and Er, but not La, induced malate release from the transgenic cells but the magnitude was only 20% to 30% of the response induced by Al treatment. None of the treatments induced malate release from cells transformed with an empty vector. Malate efflux from cells expressing BnALMT1 was more than 50% greater than from cells expressing BnALMT2.
Figure 6.
The effect of Al, La, Er, and Yb on malate efflux from transgenic tobacco cells expressing BnALMT1, BnALMT2, and the vector (pBI121). The transgenic cells (100 mg fresh weight 10 mL−1) were cultured for 18 h in the Ca-Suc medium (pH 4.5) containing 0 or 100 μm AlCl3, LaCl3, ErCl3, or YbCl3 on a rotary shaker (110 rpm) under dark condition at 25°C. Malate efflux was determined from 10 mL culture as described in the “Materials and Methods.” Bars represent means ± se of three replicates from one experiment and the experiments were repeated four times.
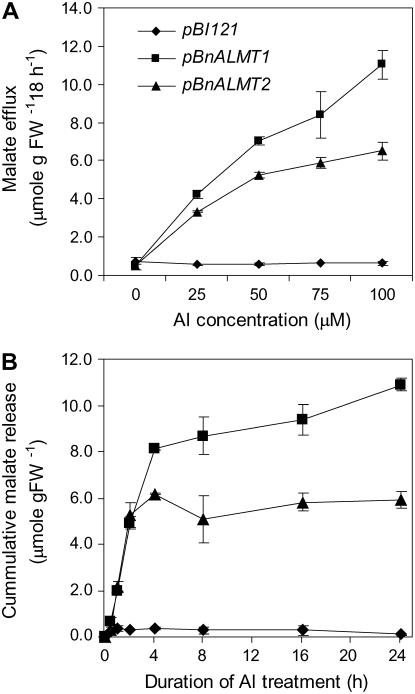
The concentration dependence of malate efflux was investigated by treating cells with a Ca-Suc medium containing 0, 25, 50, 75, or 100 μm Al. Figure 7A shows that cells transformed with the empty vector showed no increase in malate efflux with Al whereas the efflux from cells expressing BnALMT1 or BnALMT2 increased with Al concentrations up to 100 μm. The time course for Al-dependent malate efflux from transgenic tobacco cells was also determined by treating cells with 100 μm Al and measuring malate released to the media periodically over 24 h. Malate was detected in the media within 1 h of Al treatment in cells expressing either cDNA. Malate release continued at a constant rate of approximately 3 μmol g fresh weight−1 h−1 for approximately 4 h and then slowed (Fig. 7B). Since treatment solution contained only CaCl2 and Suc it is possible that nutrient deficiencies explain the decrease in efflux through time. Malate efflux from cells transformed with the empty vector remained negligible throughout the period.
Figure 7.
The effect of Al concentration (A) and time after Al addition of 100 μm Al (B) on malate efflux from transgenic tobacco cells expressing BnALMT1, BnALMT2, or transformed with the empty vector (pBI121). The transgenic cells were cultured in Ca-Suc medium, pH 4.5, containing 0, 25, 50, 75, or 100 μm AlCl3, at a cell density of 100 mg fresh weight 10 mL−1 for 18 h. The cells were cultured on a rotary shaker (110 rpm) in the dark condition at 25°C. Malate efflux was determined from the 10 mL culture as described in the “Materials and Methods.” Bars represent means ± se of three replicates and at least two independent experiments.
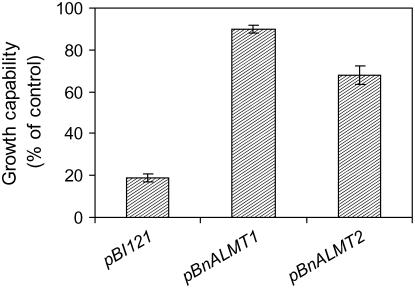
The effect of BnALMT expression on the Al resistance of tobacco cultured cells was investigated by measuring the recovery of growth following exposure to a toxic Al treatment. Transgenic cells expressing BnALMT1 or BnALMT2 were suspended in the standard Ca-Suc medium (pH 4.5) with or without 100 μm Al for 18 h. The cells were then washed with Ca-Suc medium (pH 5.8) and cultured in a modified Murashige and Skoog (MS) medium for an additional 7 d. The fresh weight of cells was then measured and compared with similar cells not treated with Al. Cells expressing BnALMT1 or BnALMT2 recovered better following the Al treatment than cells containing an empty vector. The fresh weight of cells expressing BnALMT1 and BnALMT2 were 90% and 75%, respectively, of the control cells not treated with Al compared to 20% for the vector controls (Fig. 8). These results indicate that cells expressing BnALMT1 or BnALMT2 show an enhanced resistance to Al toxicity compared to controls.
Figure 8.
Growth of the transgenic tobacco cells expressing BnALMT1, BnALMT2, and the vector (pBI121) following an 18 h treatment in control solution or 100 μm AlCl3. After Al treatment the cells were washed twice with Ca-Suc medium (pH 5.8) and transferred to a modified MS medium for an additional 7 d. Results show the cell growth relative to the Al-free controls of each construct. Bars represent means ± se of three replicates and the experiment was repeated three times.
DISCUSSION
The recent isolation of the TaALMT1 gene from wheat (Sasaki et al., 2004) enabled us to isolate two homologs from rape using conserved sequences from three TaALMT1 homologs in Arabidopsis. One of these Arabidopsis homologs, At1g08430, has recently been shown to facilitate malate efflux from roots (Hoekenga et al., 2006) but the rape homologs were cloned and their function was determined before this information was available. BnALMT1 and BnALMT2 are 95% identical to one another, 80% identical to At1g08430, and 40% identical to the TaALMT1 amino acid sequences (Fig. 1). Since rape (AACC) is an amphidiploid species formed from crossing B. oleracea (AA) with B. rapa (CC) it is possible that BnALMT1 and BnALMT2 are homologs of one another. However, addition mapping is required to verify this possibility. Both genes were expressed in the roots and induced by a 2 h treatment with Al in a concentration-dependent manner. This is similar to At1g08430 (Hoekenga et al., 2006) but contrasts with TaALMT1 that is constitutively expressed in wheat and unaffected by Al treatment (Sasaki et al., 2004). A number of Al-induced genes have been identified in wheat and other species but most of these encode stress-response proteins that are induced by a range of stresses including P deficiency for some (Ezaki et al., 1995; Snowden et al., 1995; Richards et al., 1998; Ermolayev et al., 2003). However, in the current study neither BnALMT1 nor BnALMT2 is induced by P deficiency (Fig. 3). Therefore, we conclude that P deficiency does not induce both BnALMT transcript accumulation (Fig. 3) and malate efflux (Ligaba et al., 2004) at least in rape.
The function of BnALMT1 and BnALMT2 was investigated by heterologous expression in Xenopus oocytes and tobacco suspension cells. When Xenopus oocytes injected with BnALMT cRNA and malate were exposed to Al, large inward currents were generated that were not observed in water controls or when malate was replaced with citrate (Fig. 4). These inward currents are consistent with the efflux of malate2− or malate− from the oocytes. Similarly, the expression of either BnALMT gene in tobacco suspension cells was associated with an Al-dependent efflux of malate that was not observed in control cells transformed with an empty plasmid (Fig. 5C). Al-dependent citrate efflux was not detected in these transgenic cells. Furthermore, BnALMT1 or BnALMT2 expression enhanced the Al resistance of the tobacco cells because cells expressing these genes grew 4-fold more rapidly than controls following Al treatment (Fig. 8). Previous reports have demonstrated that other trivalent cations such as many of the lanthanides (La, Er, and Yb) can activate malate release from the root apices of Al-tolerant wheat and rape plants but at significantly lower rates than observed for Al (Kataoka et al., 2002; Osawa and Matsumoto, 2002; Ligaba et al., 2004). Other reports also found that La was unable to activate an anion channel in wheat (Ryan et al., 1995a, 2001; Zhang et al., 2001; Sasaki et al., 2004) and maize root (Kollmeier et al., 2001), providing evidence that the Al response cannot be repeated by all trivalent cations. In this study we found that several lanthanides could induce BnALMT transcript in the roots of rape (Fig. 2B) but they could not activate malate efflux from transgenic tobacco or oocytes as effectively as Al, indicating some specificity for Al for protein activation but not for gene induction (Figs. 4 and 6). These results suggest that BnALMT expression and activation occur by separate mechanisms. It is possible that BnALMT expression is induced by a nonspecific stress response whereas the activation of the protein requires a specific interaction with Al. This hypothesis can be tested in the future by examining how BnALMT expression responds to other abiotic stresses.
The phenotypes conferred by BnAMLT1 and BnALMT2 expression in Xenopus oocytes and tobacco cultured cells were similar to those generated by TaALMT1 in the same transfection systems (see Sasaki et al., 2004) and by At1g08430 in Xenopus oocytes (Hoekenga et al., 2006). This is consistent with all four ALMT genes encoding malate permeases that are likely to be ion channels (Ryan et al., 1997; Zhang et al., 2001). While TaALMT1 is known to be targeted to the plasma membrane (Yamaguchi et al., 2005) the subcellular localization of the BnALMT genes is unknown. Nevertheless we predict that one or both of these proteins facilitate the malate efflux across the plasma membrane and contribute to the phenotype observed in rape plants. The malate efflux from the transgenic tobacco cells and the inward currents from oocytes were consistently larger from cells expressing BnALMT1 than from cells expressing BnALMT2. Whether these reflect real functional differences between the two genes requires further investigation.
Hoffland et al. (1992) reported citrate and malate efflux from P-deficient rape plants but that response could not be repeated in the previous study (Ligaba et al., 2004). However, they did confirm that Al treatment induces citrate efflux as well as malate efflux. Interestingly, BnALMT1 and BnALMT2 do not appear to be involved in the efflux of citrate from rape roots. Another member of the ALMT family could be involved or a different type of protein might be responsible. Magalhaes et al. (2004) identified single Al tolerance locus in sorghum that segregates with citrate release from roots. Once the gene responsible for this phenotype is isolated it will be possible to examine whether similar genes are involved in the release of citrate from rape.
A distinguishing feature between the ALMT gene from wheat with those from rape and Arabidopsis is that BnALMT1, BnALMT2, and At1g08430 expression is induced by Al, whereas TaALMT1 is constitutively expressed (Figs. 2 and 3; Sasaki et al., 2004; Hoekenga et al., 2006). Therefore, in rape and Arabidopsis, Al is required for two key steps: the induction of transcript and the activation of the proteins to transport malate. This is evidenced by the observations that malate efflux from the oocytes and transgenic tobacco cells constitutively expressing these genes only occurred when they were exposed to Al.
The Al-dependent efflux of organic anions from some plant species (e.g. wheat, buckwheat) occurs rapidly, suggesting all the necessary proteins are constitutively expressed and that exposure to Al activates the existing cellular machinery. In other species (e.g. Cassia tora), a delay of several hours occurs between the start of the Al treatment and the beginning of organic anion efflux and this has been interpreted as Al first triggering the induction of one or more proteins essential for organic anion efflux. These contrasting responses have been designated as pattern I and pattern II, respectively (Ma et al., 2001). Ligaba et al. (2004) previously demonstrated that the Al-dependent malate and citrate release from rape occurred rapidly and without any discernible delay. Therefore, prior to this study, gene induction was not considered necessary for this response in rape. Nevertheless, this study has demonstrated that the expression of two genes is increased from a low basal level by Al treatment. We have suggested that one or both of these genes are likely to be involved in malate efflux from roots. The induction of the BnALMT expression by Al was maximal after 2 h (Fig. 3B) but induction may have been completed much earlier. These results indicate that either: (1) the initial low level of BnALMT expression is sufficient to provide the rapid response; (2) pattern I-type response can involve gene expression if the induction is sufficiently rapid; or (3) the Al-dependent malate efflux from rape plants is facilitated by other constitutively expressed proteins.
The regulation of organic anion release from roots has been attributed, in some cases, to changes in organic anion synthesis (de la Fuente et al., 1997; Koyama et al., 2000; Tesfaye et al., 2001; Anoop et al., 2003) or degradation (Kispal et al., 1988; Neumann et al., 1999; Ligaba et al., 2004; Ryan et al., 2001). While changes to cellular metabolism might explain part of this response, the evidence presented here clearly demonstrates that anion efflux can be manipulated by the expression of genes that are unrelated to biosynthetic pathways. Instead, these findings support the hypothesis that the transport process is the crucial step in regulating organic anion efflux from plant roots. It remains unclear whether Al activates malate efflux by directly interacting with the ALMT1 proteins or whether intermediate steps are involved. However, the observation that Al is able to activate malate efflux in plant and animal expression systems indicates that a direct interaction between Al and the BnALMT proteins is the most likely mechanism.
This work describes the first isolation of TaALMT1 homolog genes from rape and the first functional characterization of a dicotyledonous member of this gene family in plant cells. BnALMT expression can increase malate efflux and enhance the Al resistance of tobacco cells. We conclude that BnALMT1 and BnALMT2 encode Al-activated malate transporters that function similarly to the TaALMT1 gene in wheat.
MATERIALS AND METHODS
Plant Cultivation
Rape (Brassica napus L. var. Natane nourin no. 20) seeds were sown on moist river sand and germinated in a dark chamber (LNC-131, Tabai) at 25°C. After 3 d, the seedlings were transferred to a cultivation chamber (CFH-405, Tomy) at a cycle of 14 h/25°C day and 10 h/20°C night and a light intensity of 40 μmol s−1 m−2. Six-day-old uniformly sized seedlings were transferred to a complete nutrient solution containing (in mmol L−1): Ca (NO3)2 (5), KNO3 (1.25), MgSO4 (2), KCl (0.25), and KH2PO4 (0.25); and (in μmol L−1): Fe (III)-EDTA (20), H3BO3 (25), MnSO4 (1.5), ZnSO4 (1.5), CuSO4 (0.5), and (NH4)6Mo7O24 (0.025), pH 5.0. The nutrient solution was renewed every 3 to 4 d to maintain optimum nutrition. The seedlings were grown in a cultivation chamber as described above. After 7 d in the complete nutrient solution, the seedlings were transferred to an aerated nutrient solution with (+P) or without (−P) 250 μm P in 3.5 L containers (16 seedlings per container). The plants were grown for an additional 8 d in a naturally illuminated phytotron at a day temperature of 25°C and night temperatures of 22°C. For Al treatment, all plants grown in the same pot were transferred to 1 L pot filled with 0.5 mm CaCl2, pH 4.5, containing 0, 50, or 100 μm AlCl3.
RNA Isolation
Roots of +P and −P plants were rinsed with deionized water and transferred to 1 L of 0.5 mm CaCl2, pH 4.5, containing 0, 50, or 100 μm AlCl3 for 6 h. After 6 h, the roots were carefully rinsed with deionized water and 1 g apical root tissue was excised and immediately frozen in liquid nitrogen. The frozen plant material was disrupted by mortar and pestle, and then lysed with Trizol reagent based on recommended procedure (Invitrogen/Life Technologies). After lyses of the plant material, RNA was isolated by a modified acid guanidinium thiocyanate-phenol-chloroform method of Chomezynski and Sacchi (1987). Contaminating genomic DNA was removed from the RNA samples using deoxyribonuclease (RT-grade, Wako). The RNA samples were stored at −80°C for further use.
Gene Cloning and Sequencing
To clone BnALMT1, RNA was isolated from Al-treated roots of +P plants. Degenerated PCR primers were designed from conserved sequences of three Arabidopsis (Arabidopsis thaliana) genes (At1g08430, At1g08440, and At2g27240) that show homology to the putative malate transporter from wheat (Triticum aestivum) TaALMT1. A fragment of about 600 bp was amplified using a pair of degenerated sense (5′-GCNGTNATGACNGTNGTNGTNGTNTTYG-3′) and antisense (5′-CANGGRTGNCKRAANCKRAAYTGNCCRTG-3′) primers. For sequencing, the PCR product was purified using microcon filters (Amicon Bioseparations, Millipore), and then cloned into the pGEM-T-Easy vector (Promega). The nucleotide sequence of the products was determined according to the enzymatic method of Sanger et al. (1977) using an ABI PRISM BigDye terminator cycle sequencing kit (Applied Biosystems) with a DNA sequencer model 310 Genetic Analyzer. The nucleotide and deduced amino acid sequence were analyzed by GENETYX software (Software Developing Company). Rapid amplification of cDNA ends (RACE) was used to clone 5′ and 3′ ends of the cDNA, and an adapter sequence was added to either 5′ or 3′ ends of the cDNA. The two PCR primers were either specific to the adapter or specific to a known sequence of the cDNA (5′-CACCCCCAAACTCTTGAAGGAAGTG-3′ for the 5′ and 5′-CTAGCACTTGGAGCTCATCAGCTAG-3′ for the 3′ RACE). For sequencing, the RACE product was purified from a gel using SUPREC-01 (TaKaRa), and then cloned into pGEM-T-Easy vector (Promega). All procedures were carried out using a SMART RACE cDNA amplification kit (CLONTECH).
Analysis of Gene Expression
Ligaba et al. (2004) showed that P supply affects the amount of Al-induced organic anions efflux. To investigate whether P affects an Al-induced transcript accumulation, plants were grown with or without 250 μm P 8 d prior to Al treatment. Gene expression was studied by using the semiquantitative RT-PCR and quantitative real-time RT-PCR techniques. For the semiquantitative RT-PCR, the first-strand cDNA synthesis was performed on 5 μg total RNA as a template in a 20 μL reaction mixture using Superscript II RT and Oligo(dT)12-18 following manufacturer's recommended protocol (Invitrogen/Life Technologies). After the first-strand cDNA synthesis, PCR was carried out using one-twentieth of the first-strand cDNA, 2.5 units of Ex-Taq DNA polymerase, 10× Ex-Taq buffer supplemented with MgCl2, and 200 μm dNTP mixture as recommended by the supplier (TaKaRa). About 400 bp BnALMT was amplified from the cDNA by using 5′-CTAGCACTTGGAGCTCATCAGCTAG-3′ and 5′-CACCCCCAAACTCTTGAAGGAAGTG-3′, primers that can amplify both BnALMT1 and BnALMT2. To amplify the BnALMT1 DNA, gene-specific sense (5′-ATGAATATTTTGAAGCCAGAACA-3′) and antisense (5′-AACTTCTTGAGTGACACGC-3′) primers were used. Similarly, DNA of BnALMT2 was amplified with gene-specific sense (5′-GTGAATATTTTGAAGCCAGAGAG-3′) and antisense (5′-GACTTCTTGAGTGACACGG-3′) primers. PCRs were performed on Thermal Cycler (GeneAmp System 9700, Applied Biosystems) set to give a temperature profile of 2 min at 94°C, followed by 30 cycles of 30 s at 94°C, 30 s at 57°C to 72°C (depending on primers annealing temperatures), 1 to 2 min at 72°C (depending on the length of the expected product), and a final 5 min extension at 72°C. PCR products were run on agarose gels and stained with EtBr. For the quantitative real-time PCR, first-strand cDNA synthesis was performed in a 20 μL reaction that contained 1 μg of total RNA using Superscript II RT as mentioned above. The 20 μL first-strand reaction was diluted to 40 μL and used for PCR. The PCR reactions were initiated by adding 1 μL of the first-strand cDNA to 10 μL mixture containing 1× SYBG Green PCR master mixture (Roche Diagnostics GmbH), 250 nm sense (5′-CACCCCCAAACTCTTGAAGGAAGTG-3′), and antisense (5′-CTAGCACTTGGAGCTCATCAGCTAG-3′) primers. Quantitative PCR was performed on the LightCycler instrument (Roche Diagnostics GmbH) with PCR conditions of 95°C for 10 min, 40 cycles of 95°C for 10 s, 57°C for 15 s, and 72°C for 20 s. The amount of transcript accumulation was normalized after determining the constitutive level of β-tubulin (accession no. AF258790) expression by using 5′-GACACTGTTGTTGAGCCATACAATGCAAC-3′ and 5′-GTTGTTCGGGATCCATTCACAAAGTAG-3′ primers. To amplify tobacco (Nicotiana tabacum) 18S RNA (accession no. AJ236016; Fig. 6B), sense (CTTAACGAGGATCCATTGGAG) and antisense (GGTATCTGATCGTCTTCGAG) primers were used.
Heterologous Expression of BnALMT1 and BnALMT2 in Tobacco Cells
The coding region of the BnALMT1 and BnALMT2 was amplified using a sense primer with a XbaI restriction site at the 5′ end (CGCGCTCTAGAATGGAGAAACTGAGAGAGATAGTG) and an antisense primer with a SmaI restriction site (CGCCCCGGGTCAAATCTGAAGTATACGAACACCC). The fragments were inserted into a binary vector pBI121 (CLONTECH) replacing the β-glucuronidase gene. The vector contains the cauliflower mosaic virus 35S promoter and the nopaline synthase terminator. This construct was introduced into Agrobacterium tumefaciens strain LBA 4404 (Invitrogene/Life Technologies). Tobacco cells (N. tabacum L. cv Samsun, a cell line SL) were transformed with the construct as previously described by Sasaki et al. (2004) with minor modifications. Briefly, 4 mL aliquots of a 4-d-old exponentially growing suspension of the SL cells were transferred to 90 mm petri dish and incubated with 120 μL overnight culture of A. tumefaciens LBA 4404 harboring the binary vector. After 48 h of cocultivation at 25°C under dark condition, the cells were washed five times with a modified MS liquid medium (Yamamoto et al., 1994) and plated on a modified MS solid medium containing 200 μg mL−1 kanamycin and 500 μg mL−1 carbenicillin. After 5 weeks of selection, the kanamycin-resistant calli were transferred to a fresh modified MS solid medium containing the antibiotics mentioned above. After an additional 1 month of selection, the transformants were transferred to a modified MS liquid medium containing 100 μg mL−1 kanamycin. The cells suspension of the transformants was subcultured every 7 d until uniform suspension was obtained.
Heterologous Expression of BnALMT1 and BnALMT2 in Xenopus Oocytes and Electrophysiological Assay
For complementary RNA (cRNA) preparation, the coding region of the BnALMT1 and BnALMT2 were cloned into the XbaI and SmaI sites of the pGEM-T Easy vector (Promega). After digesting with the XbaI and SmaI and blunting with T4 DNA polymerase, the fragments were inserted into the blunt-ended BglII site of vector pXBG-ev1, that is a pSP64T-derived Bluescript type vector into which Xenopus laevis oocytes β-globin 5′ and 3′ untranslated regions have been inserted (Preston et al., 1992). The construct was linearized with SmaI and cRNA was synthesized using the mMESSEGE mMACHINE in vitro transcription kit (Ambion), the synthesized cRNA was suspended in RNAse free water. Oocytes were obtained from ovarian lobes isolated from anesthetized adult female animals as described by Katsuhara et al. (2002). The oocytes were incubated overnight at 20°C in modified Barth's saline [MBS; 88 mm NaCl, 1 mm KCl, 2.4 mm NaHCO3, 15 mm Tris-HCl (pH 7.6), 0.3 mm Ca (NO3)24H2O, 0.41 mm CaCl24H2O, 0.82 mm MgSO47H2O, 10 mg mL−1 sodium penicillin, and 10 mg mL−1 streptomycin sulfate]. The oocytes were then injected with 50 nL of RNA solution (100 ng of RNA) or with 50 nL MilliQ water. A day after RNA injection, the oocytes were injected with 50 nL of 0.1 m Na malate or Na citrate. The oocytes were further incubated in MBS pH 4.5 for about 4 h. For the electrophysiological measurements, individual oocytes were transferred to MBS, pH 4.5, containing 100 μm AlCl3 or LaCl3. After approximately 10 min the two-electrode voltage-clamp system was used to measure net current across the oocyte membrane at different test voltages. The holding potential was set to 0 mV. The test voltage was clamped from −100 to 0 mV in 20 mV increments using an amplifier MEZ-7200, CEZ-1200 (Nihon Kohden). After clamping the membrane potential at a test voltage (3 s pulses), the voltage was maintained at the holding potential for 5 s before being clamped at the next test voltage. The current-voltage curves were constructed from the current values measured at the end of the 3 s voltage pulses.
Malate Assay
To determine the amount of malate efflux from tobacco cells, 300 mg fresh weight logarithmically growing cells were transferred to a centrifuge tube and pelleted at 2,000 rpm for 5 min. The cells were washed twice with Ca-Suc medium (3 mm CaCl2 and 3% Suc), pH 5.8. The cells were then suspended in Ca-Suc, pH 4.5, containing 100 μm AlCl3, LaCl3, ErCl3, or YbCl3. For the Al-dose experiment, the cells were suspended in Ca-Suc medium containing 0, 25, 50, 75, or 100 μm Al. After 18 h of incubation at dark on a rotary shaker (110 rpm) maintained at 25°C, 10 mL of the culture, corresponding to 100 mg fresh weight cells was centrifuged and the supernatant was used for malate assay. For the time-course experiment, the cells were suspended in Ca-Suc medium containing 100 μm Al and samples were taken at 0, 0.5, 1, 2, 4, 8, 16, and 24 h after treatment. The concentration of malate in the samples was determined using the method previously described by Delhaize et al. (1993).
Determination of Al Resistance
Transgenic cells (100 mg fresh weight) were treated with 100 μm Al in Ca-Suc (pH 4.5) for 18 h. The cells were then washed twice with Ca-Suc (pH 5.8) and suspended in 20 mL modified MS medium containing 100 μg mL−1 kanamycin for additional 7 d. The growth of the cells was determined as percentage of the control cells not treated with Al.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers BAE97280 and BAE97281.
Acknowledgments
The authors are very grateful to Dr. Takayuki Sasaki for help in cloning, Prof. Minoru Murata for help in sequence analysis, and Dr. Manny Delhaize for helpful comments on the manuscript. The authors also thank Ms. Shizuka Sasano and Mr. Hideki Nishimura for technical assistance.
This research was supported by a Grant-in-Aid for General Research (A) from the Ministry of Education, Science, Sports and Culture of Japan (grant no. 14206008 to H.M.); the Ohara Foundation for Agricultural Sciences; and a postdoctoral fellowship awarded by the Japan Society for the Promotion of Science (to A.L.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ayalew Ligaba (aligaba@rib.okayama-u.ac.jp).
References
- Anoop VM, Basu U, McCammon MT, McAlister-Henn L, Taylor GJ (2003) Modulation of citrate metabolism alters aluminum tolerance in yeast and transgenic canola overexpressing a mitochondrial citrate synthase. Plant Physiol 132: 2205–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomezynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159 [DOI] [PubMed] [Google Scholar]
- de la Fuente JM, Ramírez-Rodríguez V, Cabrera-Ponce JL, Herrera-Estrella L (1997) Aluminum tolerance in transgenic plants by alteration of citrate synthesis. Science 276: 1566–1568 [DOI] [PubMed] [Google Scholar]
- Delhaize E, Ryan PR (1995) Aluminum toxicity and tolerance in plants. Plant Physiol 107: 315–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Ryan PR, Hebb DM, Yamamoto Y, Sasaki T, Matsumoto H (2004) Engineering high-level aluminum tolerance in barley with the ALMT1 gene. Proc Natl Acad Sci USA 101: 15249–15254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Ryan PR, Randall P (1993) Aluminium tolerance in wheat (Triticum aestivum L.). II. Aluminium-stimulated excretion of malic acid from root apices. Plant Physiol 103: 695–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolayev V, Weschki W, Manteuffel R (2003) Comparison of Al-induced gene expression in sensitive and tolerant soybean cultivars. J Exp Bot 54: 2745–2756 [DOI] [PubMed] [Google Scholar]
- Ezaki B, Yamamoto Y, Matsumoto H (1995) Cloning and sequencing of the cDNAs induced by aluminium treatment and Pi starvation in tobacco cultured cells. Physiol Plant 93: 11–18 [Google Scholar]
- Foy CD, Chaney RL, White MC (1978) The physiology of metal toxicity in plants. Annu Rev Plant Physiol 29: 511–566 [Google Scholar]
- Gardner WK, Barber DA, Parbery DG (1983) The acquisition of phosphorus by Lupinus albus L. III. The probable mechanism by which phosphorus movement in the soil root interface is enhanced. Plant Soil 70: 107–124 [Google Scholar]
- Hoekenga OA, Maron LG, Cançado GMA, Piñeros MA, Shaff J, Kobayashi Y, Ryan PR, Dong B, Delhaize E, Sasaki T, et al (2006) AtALMT1 (At1g08430) is a novel, essential factor for aluminum tolerance in Arabidopsis thaliana and encodes an aluminum-activated malate transporter. Proc Natl Acad Sci USA 103: 9734–9743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekenga OA, Vision TJ, Shaff JE, Monforte AJ, Lee GP, Howell SH, Kochian LV (2003) Identification and characterization of aluminum tolerance loci in Arabidopsis (Landsberg erecta × Columbia) by quantitative trait locus mapping: a physiologically simple but genetically complex trait. Plant Physiol 132: 936–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffland E, Findenegg GR, Nelemans JA (1989) Utilization of rock phosphate by rape. Plant Soil 113: 155–160 [Google Scholar]
- Hoffland E, Van de Boogaard R, Nelemans JA, Findenegg GR (1992) Biosynthesis and root exudation of citric and malic acid in phosphate starved rape plants. New Phytol 122: 675–680 [Google Scholar]
- Horst WJ (1995) The role of the apoplast in aluminum toxicity and resistance of higher plants: a review. Z Pflanzenernaehr Bodenkd 158: 419–428 [Google Scholar]
- Kataoka T, Stekelenburg A, Nakanishi TM, Delhaize E, Ryan PR (2002) Several lanthanides activate malate efflux from roots of aluminium tolerant wheat. Plant Cell Environ 25: 453–460 [Google Scholar]
- Katsuhara M, Akiyama Y, Koshio K, Shibasaka M, Kasamo K (2002) Functional analysis of water channels in barley roots. Plant Cell Physiol 43: 885–893 [DOI] [PubMed] [Google Scholar]
- Kinraide TB, Parker DR, Zobel RW (2005) Organic acid secretion as a mechanism of aluminium resistance: a model incorporating the root cortex, epidermis, and the external unstirred layer. J Exp Bot 56: 1853–1865 [DOI] [PubMed] [Google Scholar]
- Kispal G, Rosenkrantz M, Guarente L, Srere PA (1988) Metabolic changes in Saccharomyces cerevisiae strains lacking citrate synthases. J Biol Chem 263: 11145–11149 [PubMed] [Google Scholar]
- Kochian LV (1995) Cellular mechanisms of aluminum toxicity and tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol 46: 237–260 [Google Scholar]
- Kochian LV, Piñeros MA, Hoekenga OA (2005) The physiology, genetics and molecular biology of plant aluminum resistance and toxicity. Plant Soil 274: 175–195 [Google Scholar]
- Kollmeier M, Dietrich P, Bauer CS, Horst WJ, Hedrich R (2001) Aluminum activates a citrate-permeable anion channel in the Al-sensitive zone of the maize root apex: a comparison between an Al-sensitive and an Al-tolerant cultivar. Plant Physiol 126: 397–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama H, Kawamura A, Kihara T, Hara T, Takita E, Shibata D (2000) Overexpression of mitochondrial citrate synthase in Arabidopsis thaliana improved growth on a phosphorus limited soil. Plant Cell Physiol 41: 1030–1037 [DOI] [PubMed] [Google Scholar]
- Ligaba A, Shen H, Shibata K, Yamamoto Y, Tanakamaru S, Matsumoto H (2004) The role of phosphorus in aluminum-induced citrate and malate exudation in rape (Brassica napus L.). Physiol Plant 120: 575–584 [DOI] [PubMed] [Google Scholar]
- Lipton D, Blanchar R, Blevins D (1987) Citrate, malate and succinate concentration in exudates from P sufficient and P stressed Medicago sativa L. seedlings. Plant Physiol 85: 315–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JF, Ryan PR, Delhaize E (2001) Aluminum tolerance in plants and the complexing role of organic acids. Trends Plant Sci 66: 273–278 [DOI] [PubMed] [Google Scholar]
- Ma JF, Zheng SJ, Matsumoto H (1997. a) Specific secretion of citric acid induced by Al stress in Cassia tora L. Plant Cell Physiol 38: 1019–1025 [Google Scholar]
- Ma JF, Zheng SJ, Matsumoto H, Hiradate S (1997. b) Detoxifying aluminum with buckwheat. Nature 390: 569–5709403684 [Google Scholar]
- Ma Z, Miyasaka SC (1998) Oxalate exudation by taro in response to Al. Plant Physiol 118: 861–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes JV, Garvin DF, Wang Y, Sorrells ME, Klein PE, Schaffert RE, Li L, Kochian LV (2004) Comparative mapping of a major aluminum tolerance gene in sorghum and other species in the poaceae. Genetics 167: 1905–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto H (2002) Metabolism of organic acids and metal tolerance in plants exposed to aluminum. In MNV Prasad, K Strzalka, eds, Physiology and Biochemistry of Metal Toxicity and Tolerance in Plants. Kluwer Academic Publisher, Dordrecht, The Netherlands, pp 95–109
- Miyasaka SC, Buta JG, Howell RK, Foy CD (1991) Mechanism of aluminium tolerance in snapbeans: root exudates of citric acid. Plant Physiol 96: 737–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Massonneau A, Maritinoia E, Roemheld V (1999) Physiological adaptation of phosphorous deficiency during proteoid root development in white lupin. Planta 208: 373–382 [Google Scholar]
- Osawa H, Matsumoto H (2002) Aluminum triggers malate independent potassium release via ion channels from the root apex in wheat. Planta 215: 405–412 [DOI] [PubMed] [Google Scholar]
- Pellet DM, Grunes DL, Kochian LV (1995) Organic acid exudation as an aluminium-tolerance mechanism in maize (Zea mays L.). Planta 196: 788–795 [Google Scholar]
- Piñeros M, Kochian LV (2001) A patch clamp study on the physiology of aluminum toxicity and aluminum tolerance in maize: identification and characterization of Al3+-induced anion channels. Plant Physiol 125: 292–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñeros MA, Magalhaes JV, Alve VMC, Kochian LV (2002) The physiology and biophysics of Al tolerance mechanism based on root citrate exudation in maize. Plant Physiol 129: 1194–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston GM, Carroll TP, Guggino WB, Agre P (1992) Appearance of water channels in Xenopus oocytes expressing red cell CHIP28 protein. Science 256: 385–387 [DOI] [PubMed] [Google Scholar]
- Raman H, Zhang K, Cakir M, Appels R, Garvin DF, Maron LG, Kochian LV, Moroni JS, Raman R, Imtiaz M, et al (2005) Molecular characterization and mapping of ALMT1, the aluminium-tolerance gene of bread wheat (Triticum aestivum L.). Genome 48: 781–791 [DOI] [PubMed] [Google Scholar]
- Richards KD, Schott EJ, Sharma YK, Davis KR, Gardner RC (1998) Aluminum induces oxidative stress genes in Arabidopsis thaliana. Plant Physiol 116: 409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan PR, Delhaize E, Jones DL (2001) Function and mechanism of organic anion exudation from plant roots. Annu Rev Plant Physiol Plant Mol Biol 52: 527–560 [DOI] [PubMed] [Google Scholar]
- Ryan PR, Delhaize E, Randall PJ (1995. a) Characterization of Al-stimulated efflux of malate from the apices of Al-tolerant wheat roots. Planta 196: 103–110 [Google Scholar]
- Ryan PR, Delhaize E, Randall PJ (1995. b) Malate efflux from root apices and tolerance to aluminum are highly correlated in wheat. Aust J Plant Physiol 22: 531–536 [Google Scholar]
- Ryan PR, Skerrett M, Findlay GP, Delhaize E, Tyerman S (1997) Aluminum activates an anion channel in the apical cells of wheat roots. Proc Natl Acad Sci USA 94: 6547–6552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74: 5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Yamamoto Y, Ezaki B, Katsuhara M, Ahn SJ, Ryan PR, Delhaize E, Matsumoto H (2004) A wheat gene encoding an aluminum-activated malate transporter. Plant J 37: 645–653 [DOI] [PubMed] [Google Scholar]
- Snowden KC, Richards KD, Gardner RC (1995) Aluminum-induced genes (induction by toxic metals, low calcium, and wounding and pattern of expression in root tips). Plant Physiol 107: 341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GJ (1991) Current views of the aluminum stress response; the physiological basis of tolerance. Curr Top Plant Biochem Physiol 10: 57–93 [Google Scholar]
- Tesfaye M, Temple SJ, Allan DJ, Vance CP, Samac DA (2001) Overexpression of malate dehydrogenase in transgenic alfalfa enhances organic acid synthesis and confers tolerance to aluminum. Plant Physiol 127: 1836–1844 [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Sasaki T, Sivaguru M, Yamamoto Y, Osawa H, Ahn SJ, Matsumoto H (2005) Evidence for the plasma membrane localization of Al-activated malate transporter (ALMT1). Plant Cell Physiol 46: 812–816 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Rikiishi S, Chang YC, Ono K, Kasai M, Matsumoto H (1994) Quantitative estimation of aluminum toxicity in cultured tobacco cells: correlation between aluminum uptake and growth inhibition. Plant Cell Physiol 35: 575–583 [Google Scholar]
- Yang ZM, Sivaguru M, Horst JW, Matsumoto H (2000) Aluminium tolerance is achieved by exudation of citric acid from roots of soybean (Glycine max). Physiol Plant 110: 72–77 [Google Scholar]
- Zhang W-H, Ryan PR, Tyerman S (2001) Malate-permeable channels and cation channels activated by aluminum in the apical cells of wheat roots. Plant Physiol 125: 1459–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SJ, Ma JF, Matsumoto H (1998) High aluminium resistance in buckwheat. I. Al-induced specific secretion of oxalic acid from root tips. Plant Physiol 117: 745–751 [DOI] [PMC free article] [PubMed] [Google Scholar]