Abstract
Physiological and genetic studies with the ramosus (rms) mutants in garden pea (Pisum sativum) and more axillary shoots (max) mutants in Arabidopsis (Arabidopsis thaliana) have shown that shoot branching is regulated by a network of long-distance signals. Orthologous genes RMS1 and MAX4 control the synthesis of a novel graft-transmissible branching signal that may be a carotenoid derivative and acts as a branching inhibitor. In this study, we demonstrate further conservation of the branching control system by showing that MAX2 and MAX3 are orthologous to RMS4 and RMS5, respectively. This is consistent with the long-standing hypothesis that branching in pea is regulated by a novel long-distance signal produced by RMS1 and RMS5 and that RMS4 is implicated in the response to this signal. We examine RMS5 expression and show that it is more highly expressed relative to RMS1, but under similar transcriptional regulation as RMS1. Further expression studies support the hypothesis that RMS4 functions in shoot and rootstock and participates in the feedback regulation of RMS1 and RMS5 expression. This feedback involves a second novel long-distance signal that is lacking in rms2 mutants. RMS1 and RMS5 are also independently regulated by indole-3-acetic acid. RMS1, rather than RMS5, appears to be a key regulator of the branching inhibitor. This study presents new interactions between RMS genes and provides further evidence toward the ongoing elucidation of a model of axillary bud outgrowth in pea.
Lateral branching structures exist in many forms throughout the plant kingdom, with the outgrowth of these branches being a dynamic process responsive to a number of genetic and environmental cues (for review, see McSteen and Leyser, 2005). Among the highly branched mutants that exist in dicotyledonous plants, there are five in pea (Pisum sativum; ramosus [rms]), four in Arabidopsis (Arabidopsis thaliana; more axillary shoots [max]), and three in petunia (Petunia hybrida; decreased apical dominance [dad]) of particular interest. In rice (Oryza sativa), increased tillering dwarf mutants have been isolated (e.g. high tillering dwarf [htd]; Ishikawa et al., 2005; Zou et al., 2005). In contrast with the pleiotropic bushy mutant of pea (Symmons et al., 2002) none of the rms, max, dad, and htd branching mutants have decreased auxin concentration (Beveridge et al., 1994; Napoli et al., 1999; Ishikawa et al., 2005), nor do they have the profound effects associated with auxin deficiency, such as those of the axr collection of mutants (Lincoln et al., 1990). Other branching mutants, like bushy (bus1-1) in Arabidopsis and teosinte branched 1 (tb1) in maize (Zea mays) and rice, have pleiotropic effects relating to vasculature and inflorescence development, respectively (Reintanz et al., 2001; Takeda et al., 2003). The relative specificity of the rms phenotype has led to the conclusion that these genes have a specialized role in the control of axillary bud outgrowth.
In most species, branching in a wild-type plant is actively suppressed by the shoot tip, with early and continuing work defining apically derived auxin as a key regulator (Leyser, 2005; Bennett et al., 2006). Buds that are receptive to auxin undergo outgrowth during conditions of reduced auxin supply from the shoot tip. Other long-distance signals are involved in determining whether the buds are receptive to a depleted auxin supply (Morris et al., 2005). Extensive physiological characterization of branching mutants is defining the network of signals that relay information between the apex, root, stem, and bud, including a novel and as yet unidentified plant growth signal that we have temporarily named shoot multiplication signal (SMS; Beveridge, 2006). In this article, we will use the term SMS to refer to the molecular signal, deficient in particular mutants, that acts as a secondary messenger for auxin to inhibit the outgrowth of buds into branches.
In pea, mutations rms1 through rms5 increase basal and aerial branching (Blixt, 1976; Arumingtyas et al., 1992; Rameau et al., 1997). Grafting a wild-type rootstock to an rms1, rms2, or rms5 mutant shoot can almost completely restore the mutant branching phenotype to wild type (Beveridge et al., 1994, 1996; Morris et al., 2001). Further grafting has shown that RMS1 and RMS5 are required to act together either in the shoot or rootstock for production of the same graft-transmissible signal, SMS (Morris et al., 2001). Evidence that SMS is a novel signal comes from the observation that rms1 and rms5 mutant plants are not auxin deficient nor do they contain excess cytokinin (Beveridge et al., 1997; Morris et al., 2001), and from molecular evidence indicating involvement of carotenoids (Sorefan et al., 2003).
The finding that RMS1, one of the genes responsible for the production of this inhibitory signal of bud outgrowth, was homologous with MAX4 and DAD1 (Sorefan et al., 2003; Snowden et al., 2005) indicated that the pathways of bud inhibition are conserved across species at this level. Conservation of the branching control network between monocots and dicots has been revealed by the discovery that D3/OsMAX2 (Ishikawa et al., 2005) and HTD1/OsMAX3 (Zou et al., 2005) control tillering in rice. The MAX4, RMS1, and DAD1 genes encode for a member of the carotenoid cleavage dioxygenase (CCD) family of proteins, CCD8 (Sorefan et al., 2003). CCD enzymes oxidatively cleave carotenoids at specific positions and some are required for abscisic acid biosynthesis and drought response (Tan et al., 2003). MAX3 is a candidate for an ortholog of RMS5 because, like RMS1 and RMS5, MAX3 and MAX4 require coexpression in tissues to confer graft-transmissible suppression of branching (Morris et al., 2001; Booker et al., 2005). Consistent with a related function to MAX4, MAX3 encodes a divergent member of the CCD family, CCD7, and can cleave multiple carotenoid substrates (Booker et al., 2004). If SMS is carotenoid derived, the most likely candidates for precursors are zeaxanthin and β-carotene because mutant plants defective in α-carotene or epoxycarotenoid biosynthesis do not show a branching phenotype (Schwartz et al., 2004). In vitro coexpression of CCD7 and CCD8 with β-carotene gives rise to 10′-apo-β-carotenol (C27) and β-ionone (C13) plus a C18 product that has been identified as 13-apo-β-carotenone (Schwartz et al., 2004). Presently, this is the most likely chain of events leading to the synthesis of SMS, although it has recently been shown that CCD8 is also able to act directly on carotenoid substrates (Auldridge et al., 2006).
Two-shoot grafts showed that SMS can only move acropetally in shoots (Foo et al., 2001). Similar results have been obtained in grafting studies with Arabidopsis mutants max4 and max3, also indicating acropetal action of the substance in this species (Turnbull et al., 2002; Booker et al., 2005). The RMS pathway may regulate cytokinins because rms1, rms3, rms4, and rms5 mutants have significantly reduced export of cytokinins identified from xylem sap (Beveridge, 2000). Studies in pea and Arabidopsis suggest that SMS acts as an inhibitor of branching (Foo et al., 2001; Turnbull et al., 2002), rather than the branching genes inactivating a branching stimulus, which is the argument put forward in petunia based on the effects of adventitious roots (Snowden et al., 2005).
Whereas application of indole-3-acetic acid (IAA) can partially compensate for shoot tip removal and suppress branching in wild-type pea, this is not the case for rms mutants (Beveridge et al., 1994; Cline, 1994) and axillary buds from isolated max mutant stem segments also have reduced auxin response (Sorefan et al., 2003). Consequently, the role of auxin in shoot branching in these mutants may be mediated with high specificity to the SMS system. In pea, RMS1 is highly expressed in roots of vegetative plants and, in stem tissue, is up-regulated in response to IAA and strongly down-regulated after decapitation or treatment with auxin transport inhibitors. RMS1 transcript levels are greatly elevated in the epicotyl of rms3, rms4, and rms5 plants when compared to wild type, indicating feedback regulation. This feedback appears to be auxin independent, yet involves a shoot-to-rootstock signal (Foo et al., 2005; Beveridge, 2006). Mutant rms4 scions cause up-regulation of RMS1 in wild-type rootstocks and, conversely, rms4 rootstocks show weakened up-regulation of RMS1 transcript levels when grafted to wild-type scions. Such expression studies show that RMS1 transcription is also regulated by RMS4 action in the rootstock (Foo et al., 2005). The feedback process is directly or indirectly influenced by RMS2 because rms2 mutant plants have lower RMS1 expression than wild-type plants (Foo et al., 2005; for review, see Beveridge, 2006). RMS1 expression is therefore controlled by a long-distance signaling network involving IAA and an IAA-independent feedback signal that is modulated by the outcome of the interaction between SMS and RMS4 gene action. In Arabidopsis, recent work to examine the hormonally controlled regulation of MAX4 has shown that feedback regulation of the branching inhibitor occurs, but at a very low level in comparison to pea (Bainbridge et al., 2005).
Perception of SMS may be controlled by RMS3 and RMS4 in pea and MAX2 in Arabidopsis because grafting studies show that these genes act mostly in the shoot to inhibit branching (Booker et al., 2005). Consistent with this, MAX2 is postulated to work via the action of targeted protein degradation because it is an F-box protein (Woo et al., 2001; Stirnberg et al., 2002). The F-box is a degenerate motif of 40 amino acids in the N-terminal region of the protein, which predominantly binds to SKP1 as a part of the Skip Cullin F-box (SCF) E3 ligase complex (Kuroda et al., 2002). It is the C-terminal region of the F-box protein that recruits target proteins to provide substrate specificity to the SCF E3. Studies in Arabidopsis have revealed SCF E3 ligases form a component in the regulation of many hormone pathways, including ethylene (ethylene-insensitive 3-binding F-box protein [EBF1/2]; Stepanova and Alonso, 2005), jasmonates (coronatine-insensitive 1 [COI1]; Deveto et al., 2003), and gibberellins (SLEEPY1; Dill et al., 2004). Transport inhibitor response 1 (TIR1) is involved in the regulation of auxin pathways via targeted degradation of multiple AUX/IAA transcriptional repressor proteins and also acts as an auxin receptor, thus increasing affinity for target proteins (Dharmasiri et al., 2005; Kepinski and Leyser, 2005). Among its known functions, SCF participates in meristem activation in the context of mitotic cycling by playing an essential role in the controlled degradation of various cell cycle proteins (Vodermaier, 2004).
We confirm here that RMS4 and RMS5 are orthologous to MAX2 and MAX3, respectively. We provide evidence that RMS1 and RMS5 are coregulated by a long-distance signal and that RMS4 (PsMAX2), which may play a role in the response to SMS, is also involved in the regulation of RMS1 and RMS5 in the shoot and rootstock.
RESULTS
The Pea RMS4 Branching Gene Is Orthologous to the Arabidopsis MAX2 Gene
Degenerate primers were designed for PsMAX2 amplification based on the Arabidopsis MAX2 sequence and three partial expressed sequence tags (ESTs) from Medicago truncatula found in the database, which showed strong homology at the amino acid level: bf633414 from a drought response cDNA library (71% identical), bg448180 (70% identical), and bf648046 (62% identical) from an elicited cell culture cDNA library. PCR products showing homology with MAX2 were isolated in pea and completion of the full-length sequence was obtained by PCR walking and 5′- and 3′-RACE techniques. The coding region is 2,127 bp and, like MAX2, contains no introns. The PsMAX2 sequence hypothetically codes for a protein sequence of 708 amino acids that has highest sequence identity with MAX2 (59%) and rice OsMAX2 (45%).
In silico analyses show that the PsMAX2 and MAX2/ORE9 proteins contain an N-terminal F-box motif with homology to the consensus motif (Patton et al., 1998). The presence of 18 Leu-rich repeats (LRRs) containing Leus, or other aliphatic residues (Fig. 1), places PsMAX2 in the LRR family of F-box proteins.
Figure 1.
Predicted amino acid sequence of the PsMAX2/RMS4 protein and alignment with Arabidopsis and rice MAX2 predicted protein sequences. The F-box is shown with one of the degenerate F-box consensus sequences above (Patton et al., 1998). The LRRs conserved between the three proteins are underlined and highlighted in gray. Numbers corresponding to the different mutant alleles denote relative positions of mutations with a list of the different mutant alleles for rms4 indicating the change to the nucleotide and the resulting amino acid.
We developed a cleaved amplified polymorphic sequence marker corresponding to PsMAX2 to map it in the JI281 × JI399 recombinant inbred line population (Ellis et al., 1992). PsMAX2 was found to locate to linkage group VII close to the markers Tps 1/153 and G11/2+ that are in the same region as RMS4 (Rameau et al., 1998). Sequencing of PsMAX2 in the six rms4 mutant alleles generated by ethyl methanesulfonate mutagenesis compared with wild type showed point mutations leading to amino acid substitutions or early stop codons, confirming PsMAX2 is RMS4 (Fig. 1). Two rms4 lines, rms4-4 (M2/716) and rms4-5 (XV/23), could not be amplified using primers designed across the length of the gene. These mutants were generated by γ-radiation and fast neutrons, respectively, and are therefore likely to be deletion mutants.
We created a phylogenetic tree to compare MAX2 sequences with other F-box proteins to support our case for RMS4/MAX2 orthology. A branch of the tree with uniform support clusters the RMS4 and MAX2 protein sequences (Fig. 2) when compared with other F-box proteins known to be associated with regulation of plant hormonal pathways and a Medicago cyclin-like F-box sequence, which all have between 18% to 25% sequence identity at the amino acid level to PsMAX2.
Figure 2.
Neighbor-joining tree of well-characterized F-box proteins and calculated sequence identity with PsRMS4. The bootstrap value for 1,000 trials is indicated for each branch. Accession numbers for protein sequences: M. truncatula cyclin-like F-box (ABD32619); AtTIR1 (NP567135); AtCOI1 (O04197); HsSKP2 (NP005974); OsD3 (BAD69288); PsRMS4 (ABD67495); AtMAX2 (NP565979); AtFBL4 (AAM60829); LeEBF2 (ABC24972); and AtEBF1(NP565597).
The Pea RMS5 Branching Gene Is Orthologous to the Arabidopsis MAX3 Gene
Using the Arabidopsis MAX3 and rice sequences and a partial sequence from M. truncatula (phosphate-starved) root EST library (AW126158), degenerate primers were designed to make a probe for use in screening a bacterial artificial chromosome (BAC) library constructed from the pea germplasm line (PI 26918; Coyne et al., 2000). Screening yielded a positive clone and 5 kb of the BAC were sequenced, including approximately 700 bp upstream and 500 bp downstream of the hypothetical start and stop codons. The genomic DNA from start to stop codon is 3,880 bp and contains six introns (Fig. 3). Exon-intron boundaries are conserved between the CCD7 orthologs examined apart from two differences in gene structure: rice contains an extra intron in the first exon when compared with Arabidopsis and pea, and both Arabidopsis and rice lack the fifth intron found in pea in the Arabidopsis fifth exon. The length of the coding region and the exon-intron boundaries were determined by the sequencing of cDNAs in two varieties of pea, Porta and Paloma. The RMS5 cDNA sequences in these varieties shared 100% identity at the nucleotide and amino acid level and differed from PI 26918 by several nucleotides, which translated into differences at the amino acid level. The deduced protein of 620 amino acids showed highest homology with MAX3 (59% identical) and OsMAX3 (56% identical), and all three proteins form a well-supported clade, such as the one shown in the phylogenetic tree of the CCD gene family with MAX3 and OsMAX3 (Snowden et al., 2005; data not shown). Lignostilbene-α,β-dioxygenase (lsd) enzymes from cyanobacteria are the next closest relatives (28% to Nostoc sp. PCC7120/BAB7593). AtCCD7/MAX3 is targeted to the stroma of the chloroplast (Booker et al., 2004); PsMAX3 also bears an N-terminal peptide sequence that is predicted for plastid targeting with a probability of 90.2% (http://www.inra.fr/predotar).
Figure 3.
Alignment of the predicted amino acid sequence of PsCCD7 cv PI 26918 (DQ403160) compared with Arabidopsis MAX3 (NP_182026), rice HTD1 (AL663000.4), Nostoc sp. PCC7120/BAB75983, Synechocystis sp. SynACO (2BIX_B), pea CCD1 (BAC10549), and pea CCD8/RMS1 (AAS66907). Intron positions corresponding to the genomic DNA sequence are denoted by triangles and positions of rms5 mutations by a shaded residue and asterisk (*). Conserved His (H) residues implicated in binding of Fe2+ in the active site are highlighted in green; residues highlighted in gray are implicated in the active site of the structurally characterized Synechocystis sp. apocarotenoid-15,15′-oxygenase (Kloer et al., 2005).
PsMAX3 was sequenced in three rms5 mutant lines of pea (rms5-1, rms5-2, and rms5-3) and compared to the corresponding wild-type lines. The mutant rms5-2 (Wt10852) has a G-to-A base change at nucleotide 510, generating an early stop codon at amino acid 170. The mutants rms5-1 (Wt15244) and rms5-3 (Wt15241), obtained independently from two different genetic backgrounds (Porta and Paloma, respectively), have a T-to-G nucleotide substitution at nucleotide 1,779, leading to an amino acid change of Tyr (593Y) to Asp (D). This residue is particularly conserved between the CCD7 and lsd enzymes (Fig. 3). The similarities in functionality as evidenced by mutant grafting experiments, taken together with the base changes found in the three rms5 alleles, provide strong evidence that PsMAX3 is RMS5.
RMS4 Is Constitutively Expressed
Expression studies were carried out using real-time PCR to examine the expression profile of RMS4 in wild-type (cv Torsdag) plant tissues when the plants were 15 d old (five leaves expanded). RMS4 was expressed in all tissues, with the highest expression in the stipule (Fig. 4). RMS4 expression was also examined by real-time PCR in basal (epicotyl) and apical (apex) parts of the plant during development in response to IAA and decapitation and in the epicotyls and stipules of the rms1, rms2, rms3, and rms5 mutants (data not shown). The RMS4 transcript was expressed ubiquitously under all of these conditions.
Figure 4.
A, RMS4 gene expression in the different tissues of wild-type pea plants (cv Torsdag) determined by real-time PCR. RNA was extracted from the dissected plants at the five-node stage. Values are average ± se of two biological replicates (except for roots; n = 1) of pools of eight plants. B, Scheme of a node showing the different parts of the pea compound leaf.
RMS5 Gene Expression
RMS5 transcript levels were analyzed in various tissues of young wild-type plants (Fig. 5A). In this experiment, expression of RMS5 was detected in roots and stems: highest in roots, 3- to 6-fold less in the epicotyl, internodes, and apex. To examine the location of RMS1 and RMS5 gene expression within the stem, we dissected vascular tissue from the remainder of the stem using fine dissecting needles under a dissecting microscope (Fig. 5B). RMS5 was more highly expressed than RMS1 and both transcripts were more highly abundant in the vasculature than in the remainder of the stem. In all analyses performed in the wild type to date, RMS5 expression has been higher than RMS1.
Figure 5.
RMS5 and RMS1 gene expression. A, Tissue profile of RMS5 gene expression. B, RMS5 and RMS1 gene expression in the vascular tissue and remaining stem tissue from the upper internode (2.5–3.0 cm from apex) of wild-type plants. RNA was extracted from the dissected plant tissues at the five-node stage (14 to 15 d old). Values are average ± se of two biological replicates of pools of 12 plants, except for leaves (n = 8) and roots (n = 4). RMS1 and RMS5 expression shown is relative to RMS1 in the vascular bundle.
RMS5 Expression Responds to Variations in Auxin Levels
Decapitation, IAA, and the auxin transport inhibitor, 1-N-naphthylphthalamic acid (NPA), were used to test the response of RMS5 and RMS1 expression to treatments that modify IAA content (Fig. 6). In this experiment, RMS5 expression was found to be nearly 6 times higher than RMS1 expression in node 5 of intact plants (data not shown). Twenty-four hours after auxin was applied in a ring around the stem of intact plants, expression of RMS1 and RMS5 was enhanced up to 10-fold below the site of treatment. After decapitation, expression decreased more than 100-fold for RMS1 and 4-fold for RMS5 over the 24-h period. Application of IAA to the stump of decapitated plants restored RMS1 and RMS5 transcript levels to that of intact plants. NPA, which reportedly inhibits polar auxin transport (Rubery, 1990), was applied as a ring just below the shoot tip as a less destructive means than decapitation of blocking apical auxin supply to tissues below (Morris et al., 2005). Twenty-four hours after NPA application, both RMS5 and RMS1 transcript levels had decreased to a magnitude similar to that for decapitation. Furthermore, when IAA was applied as a ring just below the ring of NPA, intact levels of RMS5 and RMS1 transcripts were restored. The response of RMS5 to auxin application and depletion is proportional, whereas RMS1 expression has a larger decrease in expression when auxin flow is reduced than the increase after IAA application. These results provide evidence that RMS1 and RMS5 expression is under the control of IAA, although the magnitude of the responses appears to differ.
Figure 6.
RMS1 and RMS5 gene expression after 24 h at node 5 of intact, decapitated IAA (3,000 mg L−1) and NPA-treated plants relative to RMS1 and RMS5 gene expression in intact plants, respectively. RNA was extracted from the dissected plant tissues at the five-node stage (14 to 15 d old). Values are average ± se of two biological replicates of pools of eight plants. RMS1 and RMS5 expression shown is relative to RMS1 and RMS5 expression in intact plants, independently.
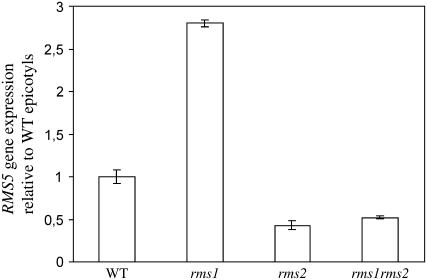
RMS5 Expression Is Feedback Regulated
RMS1 transcript levels are known to be feedback up-regulated in many rms mutants (Foo et al., 2005). To determine whether this feedback may also regulate RMS5, we investigated RMS5 expression in epicotyls of wild-type, rms1, rms2, and rms1 rms2 double-mutant plants. The rms2 mutant was included in this experiment because it lacks feedback regulation of RMS1. Small, but significant, differences in RMS5 expression were observed in the epicotyl of these lines (Fig. 7). RMS5 expression in rms1 is significantly higher than in wild type (t test; P < 0.01), and RMS5 expression is significantly lower in rms2 and rms1 rms2 than in rms1 (t test; P < 0.001 for both). RMS5 expression is also up-regulated in rms4 mutant plants, although to a lesser extent than RMS1 (Fig. 8A).
Figure 7.
RMS5 gene expression in epicotyls of wild-type (cv Torsdag), rms1, rms2, and rms1 rms2 double-mutant plants. RNA was extracted from the dissected plant tissues at the five-node stage (14 to 15 d old). Values are average ± se of three biological replicates of pools of six plants.
Figure 8.
Gene expression and branching phenotype in reciprocal grafts between wild type and rms4 grown under short-day conditions (12-h photoperiod) measured 40 d after grafting. A, RMS1 and RMS5 gene expression in the epicotyls of rootstocks relative to the wild-type self grafts. Values are average ± se of two biological replicates of five to six plants. B, Number of buds or branches >2 mm in length of reciprocal grafts between wild type and rms4 (n = 10–12). C, Total lateral length (mm) of branches of reciprocal grafts between wild type and rms4 (n = 10–12).
We used grafts to study the effect of rms4 scions and rootstocks on the expression of RMS5 because a similar experiment has previously shown that RMS1 transcript levels in rootstocks are affected by a graft-transmissible signal from scions (Foo et al., 2005). In these experiments, the epicotyl of the rootstock just below the graft union was harvested. The expression of RMS5 is highly elevated in the rootstock epicotyl of rms4/rms4 grafts compared to a wild-type/wild-type graft (Fig. 8A). When comparing a nonbranching graft combination (wild type/wild type) with a branching graft combination (rms4/wild type), expression of RMS5 was enhanced up to 25-fold in rootstock epicotyls as observed for RMS1 expression. This shows that RMS5 in rootstocks is responsive to feedback regulation by a shoot-derived signal. Comparing RMS5 expression in rootstock epicotyls of grafts between rms4 shoots and different rootstocks, it is clear that the wild-type rootstock can decrease RMS5 expression about 5-fold, as it does for RMS1. In contrast, where the scion is wild type, the genotype of the rootstock does not affect RMS5 expression levels, whereas it does for RMS1 (Fig. 8A).
RMS4 Acts in Shoot and Rootstock to Regulate SMS Biosynthesis
Although it is onerous to produce transgenic pea plants, grafts between different genotypes of pea are easily performed. We used grafts between rms4 and wild-type plants to observe whether the feedback up-regulation of RMS genes might cause reduced branching. In particular, we tested whether mutant rms4 rootstocks may be more effective than wild type at inhibiting shoot branching. Grafts of wild type/rms4 and wild type/wild type on a dwarf background were grown under natural short-day conditions to enhance branching in wild type, mostly at the basal nodes. To measure whether the buds were more or less inhibited in their outgrowth, branches in the different grafts were measured, showing that branching in the wild-type self grafts was greater than in the wild-type scions grafted to rms4 rootstocks. Total lateral length was reduced in wild-type scions grafted to rms4 rootstocks, as was the number of branches greater than 2 mm in length (Fig. 8, B and C). This demonstrates that rms4 rootstocks are indeed inhibitory to branching in wild-type shoots, supporting the hypothesis that feedback regulation occurring in rms4 rootstocks has physiological consequences for shoot branching.
DISCUSSION
Genes Involved in SMS Biosynthesis and Response Are Conserved across Species
In this article, we report that one of the enzymes already shown in physiological studies to be responsible for production of SMS, RMS5, is the pea ortholog of MAX3 and RMS4, putatively involved in SMS signal transduction, PsMAX2. rms mutants represent the largest collection of nonpleiotropic increased branching mutants available in one species, with mutagenesis for increased branching in pea being at or near saturation (Rameau et al., 1997). Five loci have been identified in pea where mutations cause increased branching at basal and aerial nodes. Part of the same network of genes regulating branching is found in other species, including Arabidopsis, petunia, and rice (Table I). Mutants have been identified at two OsMAX loci in rice and other OsMAX homologs exist in the database (Booker et al., 2005; Ishikawa et al., 2005; Snowden et al., 2005; Zou et al., 2005). We thus demonstrate remarkable conservation of branching control between Arabidopsis, pea, and rice. This discovery will further the quest for finding the identity of SMS and presents an ideal situation to explore interspecies similarities and variations in the control of shoot branching.
Table I.
Homologous branching genes isolated in different species and putative function
References for the characterization of these genes: MAX4 (At4g32810) and RMS1 (AY557341.1; Sorefan et al., 2003); DAD1 (AY743219), DAD2, DAD3 (Napoli, 1996; Snowden et al., 2005); MAX3 (At2g44990; Booker et al., 2004); RMS5 (DQ403160; Morris et al., 2001); HTD1 (AL663000.4; Zou et al., 2005); MAX1 (NP_565617; Booker et al., 2005; Lazar and Goodman, 2006); MAX2/ORE9 (AC007087; Woo et al., 2001; Stirnberg et al., 2002); RMS4 (DQ403159; Beveridge et al., 1996); D3 (AP006533; Ishikawa et al., 2005); DAD1 (AY743219; Snowden et al., 2005); RMS2 (Beveridge et al., 1994; Foo et al., 2005); RMS3 (Beveridge, 2000); MAX1 rice homolog (http://drnelson.utmem.edu/rice.html) full-length sequences: NP_917096, NP_917099, NP_917100, BAD17629; MAX4 rice homologs AP003296, AP003376.
| Species | Biosynthesis of SMS | Signal Reception | Unknown Function | ||
|---|---|---|---|---|---|
| Arabidopsis | MAX4 | MAX3 | MAX1 | MAX2/ORE9 | |
| Pea | RMS1 | RMS5 | RMS4 | RMS2, RMS3 | |
| Rice | Two homologs | HTD1 | Five homologs | D3 | |
| Petunia | DAD1 | DAD2, DAD3 | |||
Differences have been reported recently in the transcriptional control of RMS1, MAX4, and DAD1 between pea, Arabidopsis, and petunia (Sorefan et al., 2003; Bainbridge et al., 2005; Foo et al., 2005; Snowden et al., 2005). RMS1 expression in pea is highly regulated by auxin and by a feedback signal. Evidence for this regulation has been reported for Arabidopsis based on MAX4-β-glucuronidase reporter studies, although the differences observed were considerably reduced compared with pea. We continue by observing the influence of these signals on the gene expression of another member of the SMS synthesis pathway, RMS5, with the aim of analyzing these results in the context of RMS1 to better understand the point of regulation of SMS.
We have identified RMS5 as orthologous to CCD7/MAX3, a member of the same family as CCD8/RMS1/MAX4. In a phylogenic tree, RMS5 and MAX3 have highest homology to cyanobacterial lsd (data not shown; Snowden et al., 2005), which specifically cleaves interphenyl double bonds of stilbenes leading to lignin degradation and are known to function as homo- and heterodimers (Kamoda and Saburi, 1993). It is not yet known whether CCD7 and CCD8 act as either homo- or heterodimers; however, biochemical activity has been shown when expressed individually in bacteria (Schwartz et al., 2004). Two of the rms5 mutants, generated in independent mutagenesis events, are mutated at the same Tyr residue, which is conserved between the CCD7 and LSD enzymes. Due to the high identity between these two proteins and mutation leading to replicate loss of function, this Tyr residue may have an important function in both enzymes (Fig. 3).
RMS4 encodes the pea ortholog of MAX2/ORE9 (Fig. 2), a member of the LRR group of the F-box protein family (Woo et al., 2001; Stirnberg et al., 2002). Proof of association with the SCF complex has been carried out for the ORE9 (identical to MAX2) protein by yeast (Saccharomyces cerevisiae) two-hybridization studies demonstrating the interaction with ASK1 (Woo et al., 2001). By association, we assume the same functionality for the RMS4 protein. The LRR and associated region has been shown to facilitate protein-protein interactions with target proteins and may require phosphorylation or other forms of modification before protein interaction, such as hormone binding (Kepinski and Leyser, 2002, 2005; Dharmasiri et al., 2005). rms4 mutants appear to have few pleiotropic phenotypes, except for a slightly rounded leaflet phenotype and reduced stem length (Beveridge et al., 1996). This is in contrast to max2, which has multiple phenotypic traits associated with the mutations, including rounded leaves, fasciated stems, and elongated hypocotyls (Stirnberg et al., 2002). Furthermore, max2 is allelic with ore9, which shows delayed senescence (Woo et al., 2001). So far, no obvious delayed senescence trait has been observed in rms4 mutants (data not shown).
RMS4 Is Expressed in All Plant Tissues and Is More Highly Abundant in Stipules
Expression of RMS4 was detected in all tissues tested, the highest expression being in stipules (Fig. 4A). Stipules in pea are well-developed appendages at the leaf base and are outgrowths of the leaf primordium (Fig. 4B). Early in development, stipules in Arabidopsis are very strong sources of free auxin, possibly functioning to retard young leaf development and shoot tip growth (Aloni et al., 2003). Dormant buds in pea have very minimal, if any, connection to the main vasculature (Tepper, 1993), and it is only after growth to a certain size combined with auxin production by its own young leaves that the elongating bud starts to form connections to the primary vascular bundles in the stem (Gould et al., 1987). The higher expression of RMS4 in stipules, which lie in close proximity to axillary buds, could translate to a role for RMS4 in response to SMS, enabling local suppression of axillary bud outgrowth. OsMAX2 gene expression shows evidence of multiple forms of the transcript, with leaves expressing a long transcript and other organs expressing a shorter transcript (Ishikawa et al., 2005). In our analysis, real-time PCR primers are designed to the 3′ end of the sequence and we found evidence for the presence of this full-length sequence in all tissues tested.
Based on grafting studies demonstrating that the major effect of RMS4 is in the shoot, RMS4 is the postulated receptor of SMS in the shoot (Beveridge et al., 1996; Beveridge, 2000, 2006). However, widespread expression of RMS4 and the effect of rms4 rootstocks on RMS1 and RMS5 transcript levels and shoot branching (Figs. 4A and 8; Beveridge et al., 1996) show that it may act in all tissues and be involved in monitoring SMS levels for feedback regulation.
Transcriptional Analyses Show Coregulation of RMS1 and RMS5
Because activity of RMS1/MAX4 and RMS5/MAX3 is required in the same location, such as stem or roots, to produce SMS and branching inhibition, it is proposed that they act in the same pathway (Morris et al., 2001; Booker et al., 2005). RMS5 expression studies were carried out with the aim of comparing with RMS1, whose transcript levels are highly responsive to branching regulators (Foo et al., 2005). The higher than wild-type level of RMS5 transcript in rms1 and rms4 mutant stem and epicotyl tissues indicates that RMS5 may be under the same feedback regulation as RMS1 (Figs. 7 and 8A; Foo et al., 2005). The fact that RMS5 expression is lower in rms2 and rms1 rms2 than in rms1 and wild-type plants suggests that this process is affected in rms2 plants. Relative to RMS1, RMS5 is a more highly abundant transcript (Fig. 5B) that shows a lesser magnitude of fluctuations in transcript level (Figs. 5 and 6; Foo et al., 2005). In contrast, in Arabidopsis, MAX3 expression is similar to MAX4 expression in all tissues, except in the roots, where its expression is reported to be much lower (Auldridge et al., 2006). Similar to previous RMS1 expression data, RMS5 has the characteristic acropetal expression profile, greatest in the roots, but relatively strong along the stem (Fig. 5). This is consistent with rms5 mutant grafting studies that show RMS5 acts in the shoot and rootstock (Morris et al., 2001).
RMS1 and RMS5 are highly expressed in the vasculature of stem tissue (Fig. 5B). This is in keeping with predictions that SMS moves via the xylem (Foo et al., 2001) and that auxin acts indirectly in branch inhibition. One suggestion is that the specific role of auxin may be to regulate the loading or unloading of xylem-borne secondary messengers required for inhibition of bud outgrowth (Booker et al., 2003). A more recent hypothesis is that SMS is a regulator of auxin transport in the stem: Arabidopsis max mutants have increased accumulation of PIN1 efflux carriers in the xylem parenchyma leading to increased auxin transport (Bennett et al., 2006). The polarity of auxin efflux carriers in the stem and axillary buds requires further investigation because this polarity may affect critical signaling between the bud and the main stem.
If transcript abundance is reflective of downstream regulation of the SMS signal, it appears that the RMS1 enzyme is more highly regulated than the RMS5 enzyme and may possibly control a rate-limiting step in the SMS biosynthesis pathway. Evidence at the transcriptional level for allosteric control is obtained where earlier steps in the pathway are inhibited by the ultimate product of the pathway (Slocum, 2005). In this case, low levels of RMS1 in wild-type plants may be due to feedback inhibition, whereas RMS5 may retain a higher or standby level of transcription (Figs. 5 and 6). This type of pathway control can be almost instantaneous and may indeed be the case here because the transcriptional response of RMS1 to auxin depletion after decapitation or NPA treatment indicates that the RMS1 transcript may have a very short half-life (Foo et al., 2005). Furthermore, the level of response of RMS1 and RMS5 to IAA application and/or depletion differs with RMS1 showing a greater response to IAA depletion than RMS5 (Fig. 6). It is likely that the rapid turnover of the RMS1 transcript may enable this relatively quick response to auxin depletion while allowing a comparatively greater accumulation of RMS5 transcript levels in the presence of normal auxin levels.
What Is the Link between Depletion of IAA, Transcription of RMS1 and RMS5, and Bud Outgrowth?
Depletion of IAA in the stem by application of auxin transport inhibitors causes a decrease in RMS1 and RMS5 expression in a similar time frame (Fig. 6) to that observed for auxin depletion after decapitation, but does not always cause bud outgrowth even at upper nodes (Morris et al., 2005). This could indicate that, in an intact plant, SMS depletion may not always be sufficient to cause outgrowth of buds at a particular stage of bud development or under particular conditions and that other stimuli or events, such as decapitation, need to occur to enable buds to be responsive to a reduction in SMS level. Because bud outgrowth and inhibition is a dynamic process that is believed to cycle between various stages from dormancy to activity, we propose that a specific bud stage is vital for sensitivity to changes in SMS via auxin in pea (Shimizu-Sato and Mori, 2001; Beveridge, 2006). For example, SMS may function mostly on axillary buds forming at nodes above the highest expanded leaf, nodes that are not tested by our decapitation and auxin transport experiments. We draw attention to these findings to fuel future experiments to understand where and when SMS acts on bud inhibition and outgrowth.
The rms4 Mutant May Produce More SMS Than Wild Type
We have produced a higher than wild-type level of RMS1 expression in the rootstock by grafting an rms4 rootstock to a wild-type shoot and have linked this result with reduced shoot branching and, presumably, a higher level of SMS (Fig. 8). Up-regulation of RMS1 expression in the epicotyls of rms4 rootstocks grafted to wild-type scions showed that RMS4 is required in the rootstock to regulate RMS1 expression. RMS1 is up-regulated in the rootstock epicotyl tissue of rms4 rootstocks grafted to wild-type scions compared to the same tissue of wild-type self grafts (Fig. 8A; Foo et al., 2005), whereas RMS5 expression is unaffected (Fig. 8A). Besides RMS5 transcripts being consistently more abundant, this is the only experiment that has revealed a difference in gene expression regulation between RMS1 and RMS5. It is possible that specific up-regulation of RMS1 transcripts in rootstocks might affect levels of SMS and cause the inhibition of branching observed (Fig. 8, B and C). Again, this is suggestive of RMS1 controlling a rate-limiting step in the SMS biosynthesis pathway. This may also be the case in Arabidopsis because the two genotypes resulting from a cross between max3 and max4 heterozygous for MAX4 (max3/MAX3 max4/MAX4 and MAX3/MAX3 max4/MAX4) have a slightly higher inflorescence number compared to wild type. This quantitative dosage effect of MAX4 on inflorescence number suggests that MAX4 may control the rate-limiting step in the synthesis of SMS (Auldridge et al., 2006). However, it has been reported that overexpression using 35S-driven constructs of MAX3 and MAX4 show no novel phenotypes, suggesting that if MAX4 does control the rate-limiting step, it is unlikely to be at the level of transcription in Arabidopsis (Sorefan et al., 2003; Booker et al., 2004; Bainbridge et al., 2005). Similarly, 35S-driven overexpression of DAD1 in dad1 mutants led to plants that could not be discriminated from wild type (Snowden et al., 2005). Either the SMS pathway is regulated differently in different species or posttranscriptional regulation may be more important in SMS biosynthesis. It is also possible that simultaneous up-regulation of as yet unidentified genes or changes in protein stability, translation, or interactions with novel effectors not monitored in our study in pea or modified in transgenic experiments is required for enhanced SMS synthesis.
We did, however, use dwarf basal branching rms and wild-type lines under short-day conditions so our phenotypic study was carefully poised to enhance any possible chance of observing branching suppression in wild-type pea, and it is possible that such inhibition may not have been as easily observed in the other systems. Previous results show that mutant rms4 rootstocks on other genetic backgrounds are more effective than wild type at inhibiting branching in other mutant scions, such as rms1, rms2, and rms5 (Beveridge et al., 1996, 1997; Morris et al., 2001). This demonstrates that RMS4 may act locally and distally to regulate RMS1 and RMS5. Because this regulation can occur directly in roots, it supports evidence that auxin-independent feedback activation of RMS1 and RMS5 does not require growing axillary shoots (Foo et al., 2005).
CONCLUSION
This study demonstrates that IAA regulation and long-distance feedback regulation of RMS genes extends to at least two members of the SMS synthesis pathway. To date, pea is the only species where a clear demonstration of this long-distance feedback regulation has been made. The transcriptional coregulation of RMS1 and RMS5 highlights the possibility that if a similar control network occurs in other species, it may involve regulation of different genes in the network or may act under conditions yet to be revealed. A precise understanding of the regulation of SMS biosynthesis in pea will be useful in future efforts to isolate and identify the nature of the SMS molecule.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Plants described in Figure 4 were grown in a heated glasshouse (mean 15°C night/22°C day) under a 16-h photoperiod (the natural day length was extended or supplemented during the day, when necessary, with halogen lamps). Pots (5 L) contained one to three plants and were filled with peat and soil (1:1) and supplied regularly with nutrient solution. For Figures 5 to 8, two plants were sown per 2-L pot using potting mix supplemented weekly with Osmocote (producer) and Flowfeed EX7 (Grow Force). Plants were grown under a 12-h photoperiod (Fig. 8) and an 18-h photoperiod (Figs. 5–7) with the light during daylight hours supplemented by GRO-LUX lights (Fig. 8) and the natural day length extended by incandescent light (Figs. 5–7). Plants for the auxin treatments were grown in 14-cm growth bags with potting mix, one plant per bag, and grown under an 18-h photoperiod (natural day length extended by incandescent light), 18°C ± 2°C night, 26°C ± 4°C day. The wild-type cultivar used is always Torsdag, unless otherwise stated, and a description of the rms genotypes used can be found in Arumingtyas et al. (1992).
Auxin Treatments
Plants were decapitated or left intact and treated with lanolin containing IAA (dissolved in ethanol) at a final concentration of 0 or 3,000 mg L−1, applied to the cut stump or as a ring at internode 5, the oldest unexpanded internode. NPA was applied in lanolin at the same concentration and in the same manner as for IAA (Beveridge, 2000).
Epicotyl-to-Epicotyl Wedge Grafts
The grafting experiment was carried out on a cv Térèse background, using wild-type Térèse and rms4-3 (M3T-946) under short-day conditions. Grafts with 6- to 7-d-old plants were performed epicotyl to epicotyl (Beveridge et al., 1994). Cotyledonary shoots were regularly removed because they tend to weaken the growth of the scion.
Isolation of PsMAX2 and Mapping
Two degenerate primers, m1523 (5′-GAGAATTTTTGACNCMIYTNCC-3′) and m2005 (5′-CAACTCTCATCTCTGTNSWCATRTC-3′) were designed in consensus regions between the Arabidopsis (Arabidopsis thaliana) MAX2 protein and the deduced amino sequences of Medicago truncatula ESTs. These primers were tested on pea (Pisum sativum) genomic DNA and the amplified products were separated, using electrophoresis and a fragment of 550 bp of the 5′ region was identified. This was followed by two rounds of PCR walking, using the two specific primers M1626 (5′-GCTCAGACCAAATTCACTCACAGCC-3′) and M1579 (5′-TCTCCTTCCATCTTTATGCGAATCTC-3′) and the restriction enzyme EcoRI (Fermentas) and, in the second round of PCR walking, the primers M98 (5′-CCTGAATCTCAAACCTTACCAACCG-3′) and M55 (5′-TCTCAATCAACCCCATATCATCAAGG-3′) with the enzyme PvuII (Fermentas). A sequence of 1,547 bp was obtained. Several rounds of 5′ RACE (RACE system; BRL) and 3′ RACE were used followed by a last PCR walking in 5′ to get a pea sequence of 2,907 bp with 642 bp before the ATG and 141 bp of the 3′-untranslated region end.
PsMAX2 was mapped in the recombinant inbred lines mapping population from the John Innes Centre (kindly provided by Noel Ellis). PsMAX2 was sequenced for the two parents, JI281 and JI399, and a marker was designed that distinguished a size difference of 31 bp in the PsMAX2 PCR product generated using genomic DNA for the two parents. The primers used were forward (5′-ACATACAGATTTCATTACCACCTTT-3′) and reverse (5′-AGTGAGGATTTATTGAAGAGTAGAA-3′).
Southern hybridization analysis using a radiolabeled partial-length probe and a standard protocol was used to identify RMS4 copy number in pea (Sorefan et al., 2003).
Sequencing of PsMAX2 in rms4 Mutant Lines
The PsMAX2 gene was amplified from genomic DNA in the rms4-1 to rms4-8 mutants and their corresponding parental lines to discover point mutations associated with the phenotype of rms4 mutants (Arumingtyas et al., 1992). Sequence alignments and single-nucleotide protein detection were performed using Genalys software, developed at the Centre National de Génotypage (http://software.cng.fr).
Isolation of PsMAX3
Degenerate primers were designed in consensus regions of the MAX3 protein in Arabidopsis, rice (Oryza sativa), and a truncated protein from M. truncatula. One set of primers, forward (5′-GATGCCNCCAAAGAGACTCTTGTC-3′) and reverse (5′-GTATCCGTGAAWCCCAATCATG-3′), amplified a 420-bp region of pea genomic DNA showing highest homology with the Arabidopsis MAX3 protein. This was used as a probe to screen a 1× pea BAC library (Coyne et al., 2000), spotted in duplicate on nylon filters. Replicate clones were identified and, after PCR analysis, the BAC was found to contain the PsMAX3 sequence. The genomic open reading frame was sequenced by walking and 1.5 kb of the upstream 5′ region. The cDNA PsMAX3 sequence was amplified from cDNA generated from epicotyls and the sequence products used to confirm exon-intron boundaries for PsMAX3 genomic sequence.
Sequencing of PsMAX3 in rms5 Mutant Lines
PsMAX3 cDNA was amplified from three rms5 mutants, Wt10852 (rms5-2), Wt15241 (rms5-3), and Wt15414 (rms5-1) and their corresponding parental lines, Paloma and Porta. They were sequenced and analyzed by the aforementioned software to identify point mutations.
Phylogenetic Analysis
ClustalX was used to align the sequences, excluding positions for gaps and correcting for multiple substitutions. Neighbor-joining trees were constructed from these alignments with ClustalX and consisted of 1,000 trials with bootstrap. Global alignments for sequence identity analysis in Figure 1B and in the text were calculated with the program Align, using the default analysis options.
RMS4 Expression Analysis
Pea plants were grown using an aeroponic culture system with 16-h light for 15 d and tissues were harvested at a stage of development of five expanded nodes and divided into different tissue types before isolation of RNA (bioreplicate 1, n = 40; bioreplicate 2, n = 8). Epicotyls were used as a reference for comparison to other tissue types. Pea plants were also grown in pots with 16-h light until they reached the five expanded nodes stage of development (nodes 1 and 2 include buds and internode 1; nodes 4 and 5 include only a small amount of stem at nodes 4 and 5 and the axillary buds). Total RNA was isolated from tissues using TRIzol reagent (Sigma) and followed by DNAse treatment using the Qiagen kit. Concentration and purity of total RNA was checked by spectrophotometer (Jasco V-530) and gel electrophoresis. Reverse transcription reactions were carried out in a 20-μL volume (containing 2–5 μg RNA, 5 μm poly(dT), 0.125 mm dNTPs, 10 mm dithiothreitol, 1× first-strand reverse transcriptase buffer, and 200 units revert Aid H-minus Maloney murine leukemia virus reverse transcriptase [Fermentas]) and incubated at 42°C for 1 h. Primers were designed to RMS4 sequence using the LC probe design software for the Roche Lightcycler. Three sets were tested for efficiency before choosing forward (5′-GCATAGCAATGGTAACAGTAGCGG-3′), reverse (5′-CCGGTTTCGGTTGCCCT-3′), and the constitutive controls elongation factor (EF) 1α forward (5′-GATGCACCTGGACATCGTGAC-3′) and reverse (5′-CTTAGGGGTGGTAGCATCCATCT-3′). To calculate relative transcript levels, the comparative cycle method based on nonequal efficiencies was used (Pfaffl, 2001). Transcript levels of RMS4 were evaluated against EF1α, which was found to be constitutively expressed in these tissues under these conditions. Numbers represent the relative differences in steady-state gene expression of RMS4 when compared against EF1α expression. PCR reactions were performed in triplicate as technical replicates. The results shown are the average of the two biological replicates, each taken as an average of the three technical replicates. Standard error bars were generated based on biological replicates.
RMS5 and RMS1 Gene Expression Analysis
Total RNA was isolated using NucleoSpin RNA plant kits (Machery-Nagel). RNA was quantified using NanoDrop 1000. cDNA was generated in a 20-μL volume with 1.6 to 5 μg total RNA, 50 to 250 ng random hexamers (Invitrogen), 0.5 mm dNTPs, 5 mm dithiothreitol (Invitrogen), 1.25 × first-strand buffer, 40 units RnaseOut (Invitrogen), and 200 units SuperScript III reverse transcriptase (Invitrogen) incubated at 50°C for 1 h. cDNA was checked via PCR (25 cycles) and gel electrophoresis with actin primers forward (5′-CAACTATGTTTCCCGGTATTG-3′), and reverse (5′-AAGTCTGTGCCTCGACATCC-3′). RMS1 and RMS5 expression was measured against actin and 18S using Taqman real-time PCR with Platinum Quantitative PCR SuperMix-UDG (Invitrogen). Primers and probe for RMS1, RMS5, actin, and 18S were as follows: RMS1 forward (5′-AAGGAGCTGTGCCCTCAGAA-3′), RMS1 probe [5′-(6-Fam)-CATTCTTTGTGCCTCGACCAGGAGCA-(Tamra)-3′], RMS1 reverse (5′-ATTATGGAGATCACCACACCATCA-3′; Foo et al., 2005). RMS5 primers and probe were designed using Primer Express software: RMS5 forward (5′-CGGCATCTTAAAGACTCCGTACA-3′), RMS5 probe [5′-(6-Fam)-CGTGATCCG-CCAAATCACAACCGT-(Tamra)-3′], RMS5 reverse (5′-TGGATACGATCGGGAAGTTCA-3′); 18S forward (5′-ACGTCCCTGCCCTTTGTACA-3′), 18S probe [5′-(6-Fam)-ACCGCCCGTCGCTCCTACCG-(Tamra)-3′, 18S reverse (5′-CACTTCACCGGACCATTCAAT-3′; Ozga et al., 2003); actin forward (5′-GTGTCTGGATTGGAGGATCAATC-3′), actin probe [5′-(6-Fam)-CACCTTCCAGCAGATGTGGATATCTAAGGC-(Tamra)-3′], actin reverse (5′-GGCCACGCTCATCATATTCA-3′). Relative expression was calculated based on equal efficiency. For all experiments, two or three biological replicates each consisting of at least six to nine plants were used, with error bars representing different biological replicates.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: DQ403159 (RMS4) and DQ403160 (RMS5).
Acknowledgments
We would like to thank Dr. Clarice Coyne for BAC library filters, Dr. Noel Ellis for contributing the mapping population, Beate Hoffman for technical assistance, and Dr. Ian Small for assistance with interpretation of sequence motifs; we also want to acknowledge Patrick Grillot for preparation of plant materials and plant care.
This work was supported by FP6 Project Grain Legumes (FOOD–CT–2004–506223), Institut National de la Recherche Agronomique, and Région Ile de France, by the Australian Research Council, and by Australian postgraduate awards (to T.B. and E.A.D.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Catherine Rameau (rameau@versailles.inra.fr).
References
- Aloni R, Schwalm K, Langhans M, Ullrich CI (2003) Gradual shifts in sites of free-auxin production during leaf-primordium development and their role in vascular differentiation and leaf morphogenesis in Arabidopsis. Planta 216: 841–853 [DOI] [PubMed] [Google Scholar]
- Arumingtyas EL, Floyd RS, Gregory MJ, Murfet IC (1992) Branching in Pisum: inheritance and allelism in tests with 17 ramosus mutants. Pisum Genet 24: 17–31 [Google Scholar]
- Auldridge M, Block A, Vogel J, Dabney-Smith C, Mila I, Bouzayen M, Magallanes-Lundback M, DellaPenna D, McCarty D, Klee H (2006) Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J 45: 982–993 [DOI] [PubMed] [Google Scholar]
- Bainbridge K, Sorefan K, Ward S, Leyser O (2005) Hormonally controlled expression of the Arabidopsis MAX4 shoot branching regulatory gene. Plant J 44: 569–580 [DOI] [PubMed] [Google Scholar]
- Bennett T, Sieberer T, Willett B, Booker J, Lusching C, Leyser O (2006) The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr Biol 16: 553–563 [DOI] [PubMed] [Google Scholar]
- Beveridge CA (2000) Long-distance signalling and a mutational analysis of branching in pea. Plant Growth Regul 32: 193–203 [Google Scholar]
- Beveridge CA (2006) Advances in the control of axillary bud outgrowth: sending a message. Curr Opin Plant Biol 9: 35–40 [DOI] [PubMed] [Google Scholar]
- Beveridge CA, Ross JJ, Murfet IC (1994) Branching mutant rms-2 in Pisum sativum: grafting studies and endogenous indole-3-acetic acid levels. Plant Physiol 104: 953–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge CA, Ross JJ, Murfet IC (1996) Branching in pea: action of genes Rms3 and Rms4. Plant Physiol 110: 859–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge CA, Symons GM, Murfet IC, Ross JJ, Rameau C (1997) The rms1 mutant of pea has elevated indole-3-acetic acid levels and reduced root-sap zeatin riboside content but increased branching controlled by graft-transmissible signal(s). Plant Physiol 115: 1251–1258 [Google Scholar]
- Blixt S (1976) Linkage studies in Pisum. XV. Establishing the rms gene and linkage of rms and fas in chromosome 3. Agric Hort Genet 34: 83–87 [Google Scholar]
- Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O (2004) MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signalling molecule. Curr Biol 14: 1232–1238 [DOI] [PubMed] [Google Scholar]
- Booker J, Chatfield S, Leyser O (2003) Auxin acts in xylem-associated or medullary cells to mediate apical dominance. Plant Cell 15: 495–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker J, Sieberer T, Wright W, Williamson L, Willett B, Stirnberg P, Turnbull C, Srinivasen M, Goddard P, Leyser O (2005) MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3 and MAX4 to produce a carotenoid-derived branch inhibiting hormone. Dev Cell 8: 443–449 [DOI] [PubMed] [Google Scholar]
- Cline MG (1994) The role of hormones in apical dominance; new approaches to an old problem in plant development. Plant Physiol 90: 220–237 [Google Scholar]
- Coyne CJ, Meksem K, Lightfoot DA, Keller KE, Martin RR, McClendon MT, Inglis DA, Storlie EW, McPhee KE (2000) Construction of a bacterial artificial chromosome library for pea (Pisum sativum L.). Pisum Genet 32: 23–26 [DOI] [PubMed] [Google Scholar]
- Deveto A, Muskett PR, Shirasu K (2003) Role of ubiquitination in the regulation of plant defence against pathogens. Curr Opin Plant Biol 6: 307–311 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun TP (2004) The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin induced degradation. Plant Cell 16: 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis TH, Turner L, Hellens RP, Lee D, Harker CL, Enard C, Domoney C, Davies DR (1992) Linkage maps in pea. Genetics 130: 649–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Bullier E, Goussot M, Foucher F, Rameau C, Beveridge CA (2005) The branching gene RAMOSUS1 mediates interactions among two novel signals and auxin in pea. Plant Cell 17: 464–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Turnbull C, Beveridge CA (2001) Long-distance signaling and the control of branching in the rms1 mutant of pea. Plant Physiol 126: 203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould KS, Cutter EG, Young JPW, Charlton WA (1987) Positional differences in size, morphology, and in vitro performance of pea axillary buds. Can J Bot 65: 406–411 [Google Scholar]
- Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J (2005) Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol 46: 79–86 [DOI] [PubMed] [Google Scholar]
- Kamoda S, Saburi Y (1993) Structure and enzymatical comparison of lignostilbene-α,β-dioxygenase isozymes, I, II and III, from Pseudomonas paucimobolis TMY1009. Biosci Biotechnol Biochem 57: 931–934 [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2002) Ubiquitination and auxin signalling: a degrading story. Plant Cell (Suppl) 14: S81–S95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Kloer DP, Ruch S, Al-Babili S, Beyer P, Schulz GE (2005) The structure of a retinal-forming carotenoid oxygenase. Science 308: 267–269 [DOI] [PubMed] [Google Scholar]
- Kuroda H, Takahashi N, Shimada H, Seki M, Shinozaki K, Matsui M (2002) Classification and expression analysis of Arabidopsis F-box-containing protein genes. Plant Cell Physiol 43: 1073–1085 [DOI] [PubMed] [Google Scholar]
- Lazar G, Goodman HM (2006) MAX1, a regulator of the flavonoid pathway, controls vegetative axillary bud outgrowth in Arabidopsis. Proc Natl Acad Sci USA 103: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser O (2005) The fall and rise of apical dominance. Curr Opin Genet Dev 15: 468–471 [DOI] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M (1990) Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2: 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P, Leyser O (2005) Shoot branching. Annu Rev Plant Biol 56: 353–374 [DOI] [PubMed] [Google Scholar]
- Morris SE, Cox MCH, Ross JJ, Kristantini S, Beveridge CA (2005) Auxin dynamics after decapitation are not correlated with the initial growth of axillary buds. Plant Physiol 138: 1665–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SE, Turnbull CGN, Murfet IC, Beveridge CA (2001) Mutational analysis of branching in pea: evidence that Rms1 and Rms5 regulate the same novel signal. Plant Physiol 126: 1205–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli CA (1996) Highly branched phenotype of the petunia dad1-1 mutant is reversed by grafting. Plant Physiol 111: 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli CA, Beveridge CA, Snowden KC (1999) Reevaluating concepts of apical dominance and the control of axillary bud outgrowth. Curr Top Dev Biol 44: 127–169 [DOI] [PubMed] [Google Scholar]
- Ozga JA, Yu J, Reinecke DM (2003) Pollination-, development-, and auxin-specific regulation of gibberellin 3β-hydroxylase gene expression in pea fruit and seeds. Plant Physiol 131: 1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Tyers M (1998) Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet 14: 236–243 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rameau C, Bodelin C, Cadier D, Grandjean O, Miard F, Murfet IC (1997) New ramosus mutants at loci Rms1, Rms3, and Rms4 resulting from the mutation breeding program at Versailles. Pisum Genet 29: 7–12 [Google Scholar]
- Rameau C, Dénoue D, Fraval F, Haurogné K, Josserand J, Laucou V, Batge S, Murfet IC (1998) Genetic mapping in pea. 2. Identification of RAPD and SCAR markers linked to genes affecting plant architecture. Theor Appl Genet 97: 916–928 [Google Scholar]
- Reintanz B, Lehnen M, Reichelt M, Gershenzon J, Kowalczyk M, Sandberg G, Godde M, Uhl R, Palme K (2001) bus, a bushy Arabidopsis CYP79F1 knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates. Plant Cell 13: 351–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubery P (1990) Phytotropins: receptors and endogenous ligands. Symp Soc Exp Biol 44: 119–146 [PubMed] [Google Scholar]
- Schwartz SH, Qin X, Loewen MC (2004) The biochemical characterisation of two carotenoid cleavage enzymes from Arabidopsis indicates that a carotenoid-derived compound inhibits lateral branching. J Biol Chem 279: 46940–46945 [DOI] [PubMed] [Google Scholar]
- Shimizu-Sato S, Mori H (2001) Control of outgrowth and dormancy in axillary buds. Plant Physiol 127: 1405–1413 [PMC free article] [PubMed] [Google Scholar]
- Slocum RD (2005) Genes, enzymes and regulation of arginine biosynthesis in plants. Plant Physiol Biochem 43: 729–745 [DOI] [PubMed] [Google Scholar]
- Snowden KC, Simkin AJ, Janssen BJ, Templeton KR, Loucas HM, Simons JL, Karunairetnam S, Gleave AP, Clark DG, Klee HJ (2005) The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE 8 gene affects branch production and plays a role in leaf senescence, root growth and flower development. Plant Cell 17: 746–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K, Booker J, Haurogné K, Goussot M, Bainbridge K, Foo E, Chatfield S, Ward S, Beveridge C, Rameau C, et al (2003) MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and Pea. Genes Dev 17: 1469–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Alonso JM (2005) Arabidopsis ethylene signalling pathway. Sci STKE 276: cm4. [DOI] [PubMed] [Google Scholar]
- Stirnberg P, van den Sande K, Leyser HMO (2002) MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129: 1131–1141 [DOI] [PubMed] [Google Scholar]
- Symmons GM, Ross JJ, Murfet IC (2002) The bushy pea mutant is IAA deficient. Plant Physiol 128: 734–74111842176 [Google Scholar]
- Takeda T, Suwa Y, Suzuki M, Kitano H, Ueguchi-Tanaka M (2003) The ostb1 gene negatively regulates lateral branching in rice. Plant J 33: 513–520 [DOI] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng W, Liu L, Li Q, Cline K, McCarty DR (2003) Molecular characterisation of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35: 44–56 [DOI] [PubMed] [Google Scholar]
- Tepper HB (1993) Developmental features accompanying the imposition and release of apical dominance in pea. J Plant Physiol 142: 722–729 [Google Scholar]
- Turnbull CGN, Booker JP, Leyser HMO (2002) Micro grafting techniques for testing long-distance signalling in Arabidopsis. Plant J 32: 255–262 [DOI] [PubMed] [Google Scholar]
- Vodermaier HC (2004) APC/C and SCF: controlling each other and the cell cycle. Curr Biol 14: R787–R796 [DOI] [PubMed] [Google Scholar]
- Woo HE, Chung KM, Park J, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG (2001) ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13: 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Zengxiang C, Zhang S, Zhang W, Jiang G, Zhao X, Zhai W, Pan X, Zhu L (2005) Characterisation and fine mapping of a mutant gene for high tillering and dwarf in rice (Oryza sativa L.). Planta 222: 604–612 [DOI] [PubMed] [Google Scholar]