Abstract
It is widely accepted that gene duplication is a primary source of genetic novelty. However, the evolutionary fate of duplicated genes remains largely unresolved. The classical Ohno's Duplication-Retention-Non/Neofunctionalization theory, and the recently proposed alternatives such as subfunctionalization or duplication-degeneration-complementation, and subneofunctionalization, each can explain one or more aspects of gene fate after duplication. Duplicated genes are also affected by epigenetic changes. We constructed a phylogenetic tree using Dof (DNA binding with one finger) protein sequences from poplar (Populus trichocarpa) Torr. & Gray ex Brayshaw, Arabidopsis (Arabidopsis thaliana), and rice (Oryza sativa). From the phylogenetic tree, we identified 27 pairs of paralogous Dof genes in the terminal nodes. Analysis of protein motif structure of the Dof paralogs and their ancestors revealed six different gene fates after gene duplication. Differential protein methylation was revealed between a pair of duplicated poplar Dof genes, which have identical motif structure and similar expression pattern, indicating that epigenetics is involved in evolution. Analysis of reverse transcription-PCR, massively parallel signature sequencing, and microarray data revealed that the paralogs differ in expression pattern. Furthermore, analysis of nonsynonymous and synonymous substitution rates indicated that divergence of the duplicated genes was driven by positive selection. About one-half of the motifs in Dof proteins were shared by non-Dof proteins in the three plants species, indicating that motif co-option may be one of the forces driving gene diversification. We provided evidence that the Ohno's Duplication-Retention-Non/Neofunctionalization, subfunctionalization/duplication-degeneration-complementation, and subneofunctionalization hypotheses are complementary with, not alternative to, each other.
Darwin's positive selection theory cannot adequately explain the rapid rise and early diversification of more than 250,000 flowering plant species (Darwin and Seward, 1903; Davies et al., 2004; De Bodt et al., 2005). The theory that gene duplication events are the primary source of genetic novelty leading to speciation, first postulated by Ohno (1970), has gained wide acceptance (Lynch and Conery, 2000; Gu et al., 2003; Moore and Purugganan, 2003, 2005; Blanc and Wolfe, 2004; Li et al., 2005). However, the specific evolutionary route(s) of duplicated genes has remained largely unresolved (Force et al., 1999; Lynch and Conery, 2000; He and Zhang, 2005b; Moore and Purugganan, 2005). According to Ohno (1970), after a duplication event, one daughter gene retains the preduplication function, while the other one, in the majority of cases, accumulates deleterious mutations and is eliminated, or, in the minority of cases, survives by gaining a new function. This hypothesis, referred to here as Ohno's Duplication-Retention-Non/Neofunctionalization (DRNNF), has been the subject of intensive debate (Taylor and Raes, 2004). Hughes (1994) proposed the subfunctionalization (SF) model for proteins, under which duplicated genes share the same functions for a period of time and then evolve into functionally distinct proteins with each daughter gene specialized in a subset of functions of the ancestral gene. Force et al. (1999) summarized three observations on genome-wide duplication events that are contradictory to DRNNF, including (1) a higher proportion of the duplicated genes retained than expected by chance alone, (2) nucleotide substitution patterns reflective of purifying selection on both copies of the duplicated genes, and (3) a relative paucity of null allele for loci that have avoided nonfunctionalization. They then proposed a model similar to SF, called duplication-degeneration-complementation (DDC), to also include regulatory element functions (Force et al., 1999). Under this model, the majority of duplicated genes accumulates degenerative mutations for a period of time and then undergoes functional specialization by complementary partition of ancestral functions. Therefore, preservation of duplicated genes is through complementary subfunctionalization of the progenitor gene rather than the evolution of new functions. Recently, He and Zhang (2005b) further extended the DDC model, termed subneofunctionalization (SNF), under which a large proportion of duplicate genes undergo rapid subfunctionalization, and the subfunctionalized genes may later also evolve new functions not in the ancestral gene. A fundamental assumption of the SF/DDC model is that each ancestral gene had at least two functions that could subsequently be partitioned between two daughter genes. However, this assumption could be valid only if the genes of ancestral species had almost all the functions that extant relatives contain, and, to our knowledge, this is unlikely the case. Moreover, if the ancestral genes possessed multiple functions, where were their origins? To answer this question, a subfunction co-option concept has also been put forth (Raff, 1996; Carroll, 2001; Cameron et al., 2005), which suggests that a gene evolves by co-opting a new function not found in the ancestral gene. Recently, it was suggested that epigenetic changes might play important roles in the evolution of duplicated genes (Rodin and Riggs, 2003; Rapp and Wendel, 2005; Rodin et al., 2005). The term epigenetics can be applied to mean alteration of phenotype, morphological or molecular, without change in either gene coding sequence or promoter region (Rapp and Wendel, 2005). Epigenetic changes in gene expression include promoter methylation, DNA packaging, repositioning, microRNA, and small interfering RNA (Bender, 2002; Rapp and Wendel, 2005; Rodin et al., 2005). Epigenetic changes in proteins include posttranslational modification of proteins such as methylation (Chen et al., 2006).
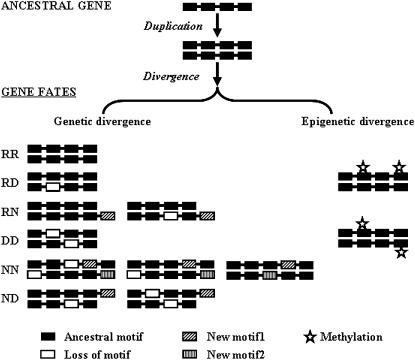
Based on the above information, we hypothesized that both genetic and epigenetic changes are involved in the evolution of duplicated genes (Fig. 1). Genetic changes in proteins include retention (R), indicating that a copy retains the original motif organization and function; degeneration (D), indicating that a copy degenerates or loses one or more motifs and functions; and neofunctionalization (N), indicating that a copy acquires one or more motifs and functions. There are six possible combinations of these three types of genetic changes in coding regions for duplicated genes: RR, RD, RN, DD, NN, and ND. Epigenetic changes in proteins (i.e. protein methylation) or in promoter regions (i.e. DNA methylation) can cause functional diversification in duplicated genes that share the same motif structure and change them from RR into RD or DD type. RD and RN correspond to Ohno's hypothesis (Ohno, 1970), DD corresponds to the SF or DDC model (Hughes, 1994; Force et al., 1999), and NN corresponds to the SNF model (He and Zhang, 2005b). We consider that gene function consists of both protein function conferred by the coding region, as suggested by Hughes (1994), and the pattern of gene expression mostly controlled by regulatory elements, as suggested by Force et al. (1999).
Figure 1.
Possible evolutionary modes of paralogs in the protein coding region after gene duplication.
In this study, we compared genes of a plant-specific gene family, the DNA binding with one finger domain (Dof) transcription factor, in three angiosperm plants, poplar (Populus trichocarpa) Torr. & Gray ex Brayshaw, Arabidopsis (Arabidopsis thaliana), and rice (Oryza sativa), all of which have been completely sequenced and have undergone at least one round of genome-wide duplication (Bowers et al., 2003; Raes et al., 2003; Sterck et al., 2005; Tuskan et al., 2006). We selected the Dof gene family because of its diverse biological functions (Yanagisawa, 2004). Dof proteins typically consist of multiple domains, including a highly conserved N-terminal DNA-binding domain and a C-terminal domain for transcriptional regulation (Yanagisawa, 2002). The high degree of conservation of the Dof domain and the diversity of the remaining portion of the protein provide rich materials for studying the fates of gene diversification. Our purpose was to search for footprints of genes evolution; to determine the validity of DRNNF, SF, DDC, or SNF in describing the fates of duplicated genes; and to examine the forces that have driven the divergence of duplicated genes. We first constructed a maximum-likelihood phylogenetic tree using full-length protein sequences of the Dof genes. From the phylogenetic tree, we identified 27 pairs of paralogous Dof genes. Then we predicted the ancestral protein sequences for the paralogs. Analysis of protein motif structure of the Dof paralogs and their ancestors revealed six different genetic changes in coding region after gene duplication. We also investigated potential epigenetic changes in the proteins of duplicated genes that have the same protein motif structure. Prediction of methylation in protein sequences indicates that epigenetics is also involved in gene evolution. We also examined expression of the paralogs by reverse transcription (RT)-PCR, massively parallel signature sequencing (MPSS), and microarray data analysis. To examine the driving force for the gene evolution, we performed nonsynonymous and synonymous substitution rate (ka and ks) analysis of the duplicated genes. We also searched the non-Dof genes in Arabidopsis, poplar, and rice for the motifs in Dof proteins and revealed that motif co-option may be one of the forces driving gene diversification. We provided evidence that the previously proposed DRNNF, SF/DDC, and SNF hypotheses are complementary with, not alternative to, each other. Our study also suggested that epigenetic changes might be involved in evolution of duplicated genes.
RESULTS AND DISCUSSION
The Dof Gene Family and Conservative Motifs in the Dof Proteins
Using the 66 Dof protein sequences from Arabidopsis and rice to query the recently sequenced poplar genome, we identified 41 poplar Dof genes (Supplemental Table S1) and manually verified their uniqueness. These genes were analyzed along with the 36 and 30 Dof genes from Arabidopsis and rice, respectively (Supplemental Table S1).
A total of 41 conserved motifs were identified in all 107 Dof protein sequences (Table I). The motif 1 was identified to be the Dof domain using the Conserved Domain Search Service (Marchler-Bauer and Bryant, 2004). Although many motifs are shared by poplar, Arabidopsis, and rice, species-specific motifs were also found (motifs 18, 20, 25, 33, and 37 in poplar and 36 in Arabidopsis; Supplemental Table S2). We also found two motifs (17 and 29) shared by rice and poplar only, one (34) shared by rice and Arabidopsis only, and three (12, 23, and 28) shared by Arabidopsis and poplar only (eudicot specific; Supplemental Table S2). In addition, several motifs were commonly linked (e.g. 8, 2, and 3). These results suggest that motif acquisition/divergence had continued to occur in poplar, Arabidopsis, and rice after the monocot/eudicot split and the eurosids I/eurosids II split.
Table I.
Conserved motifs identified by MEME using amino acid sequences of the 107 Dof genes from poplar, Arabidopsis, and rice
| Motif No. | Multilevel Consensus Sequence |
|---|---|
| 1 | EILKCPRCDSMNTKFCYYNNYNLSQPRHFCKTCRRYWTKGGALRNVPVGGGCRKNKR |
| 2 | MEEEKTEKCVWVPKTLRIDDPDEAAKSSIWTTLGIKNDKK |
| 3 | KEEKHHVIETSPVLQANPAALSRSMNFQE |
| 4 | KDPAIKLFGKTIPVP |
| 5 | SMAERARLAKIPLPE |
| 6 | HVMNGVHHPPIKNNGTVLKFGSDAPLCESMASVLNLGEKT |
| 7 | GRLLFPFEDLKQQVSS |
| 8 | SDNNSPTLGKHSRDE |
| 9 | MDTAQWPQEIVVKPIEEIVTNTCPKPP |
| 10 | SSIESLSCINQDLHWKLQQQRLAMLF |
| 11 | GYWTGMLGGGSW |
| 12 | LTISDFSTKVPLSDNDHLMYYYSLDSAHKHQDHQDRTKQCTSHETSSFHLPPLPGQDTVSQEILWSNSHMMDNHNLEMSQQPVLGPETQDPNLLFGNWSPFDMSSDDTFSR |
| 13 | DSSRVSQLAPVKTEGNQGLNLSKPYLGIPGNDQY |
| 14 | HIDLALVYAKFLNHH |
| 15 | MVFSSIPVYLD |
| 16 | PHQIPCFPGVPWPYPWNPA |
| 17 | VQLSHLHNILGSQETIANPNFMESKYNIGMLENPRPIDFMDSKFEALVGSSRNYDFMGNGDLGMVSGLGDMSHHHGLAPNFSDICSPFGMSLDGNSGTFMETCQRLMLPYDQ |
| 18 | ENSRLQQQLQQQPHLLQHNFATPQNILATNNSGNLVSPALRNKESGNLVLPPAPGMSTMGSYFPGDGFFTSLEAIQSLNNNQPPIQSFPFQPLNQPVNLGGDLGETSNLGLLHGFNAVPAFGSQNQQQRQFYHVDYRDNKSIEHSFYPHDQESLIQSSRPA |
| 19 | HEGQDLNLAFP |
| 20 | RFQELIESQDMNAFGLQDLLIDEIVQDALWSDDATLPHFPNWQPMVQLQDFDSFSVDDRLKISANFISDDNWSSFDLSGFEVFPRP |
| 21 | DIIGHMPQPQPQLPILPPLHHLGDYNSGDIGLDFGGIQ |
| 22 | FYPVPAYWGCTVP |
| 23 | NHRLDFGEEDFEQDYYDVGSDDLIENQEI |
| 24 | EKTLKKPD |
| 25 | FQPLNVYNYTGESMEDSTITTIMPPTSSTIAHPWQVPNTSSGMDMTNYWNWDDIENYVSADLNVPWDDSEIK |
| 27 | IERKARPQ |
| 28 | IENHVQKPQPIMFENLEISKPQVCAAGNSRKEGAASGDPATEWFFGNSYDQVTATPTNRSNNGNNDNTGNWNGVQAWGDL |
| 29 | WNNEASMAAAQHSTGQACITNIPNQVQLCPTPMLAVPSICPPNIPLQFVPASYWGCMPTWAAGTRNVPLCGSNGCL |
| 30 | HHHHHHM |
| 31 | YHMNTVDQYYWSQSQWNDMDM |
| 32 | GFPLQEFKPTLSFSLDGLGS |
| 33 | NMINWVNPQQPIQAQQQKQNLPDLVLGDQKDLSEILYQAMINPPSSVLQQNSISCNNFDTKSFNVNNNGVLL |
| 34 | MDNLNVFANEDNQVN |
| 35 | HLATTHGGFRHDFPVKRRRCY |
| 36 | KIDQPSVAQMVSVEIQPGNHQPFKNVQENIDFVGSF |
| 37 | TSVSASVGKSGTNKIKTIASEIGRSGFGNGFEHELSSSPIMWASPQNSHIFALLRATQNPNPSTPCNSIFVKEEGFLIGKHFI |
| 38 | IDVKPNTKLLSLDWQDQGCYDVGKDTFGY |
| 39 | SASHYRHITIPEALQ |
| 40 | EPLAKGTCSEITKVETKGPSEEISEEPEMFSGLGQGEEEIQAAMRVNEAEVIAKHKE |
| 41 | VSNNLLNGIVESKIFPRGDMNPSFEPALLEQGSDCGIFSEIGSFTSLITSTNDL |
Gene Duplicates and Ancestral Protein Sequences
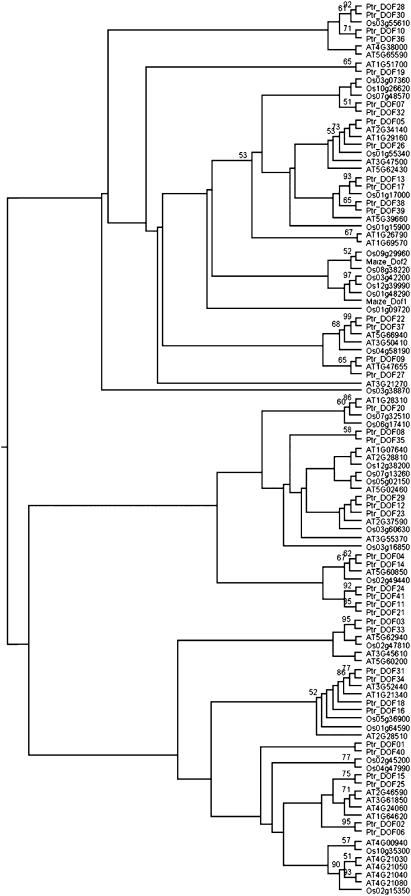
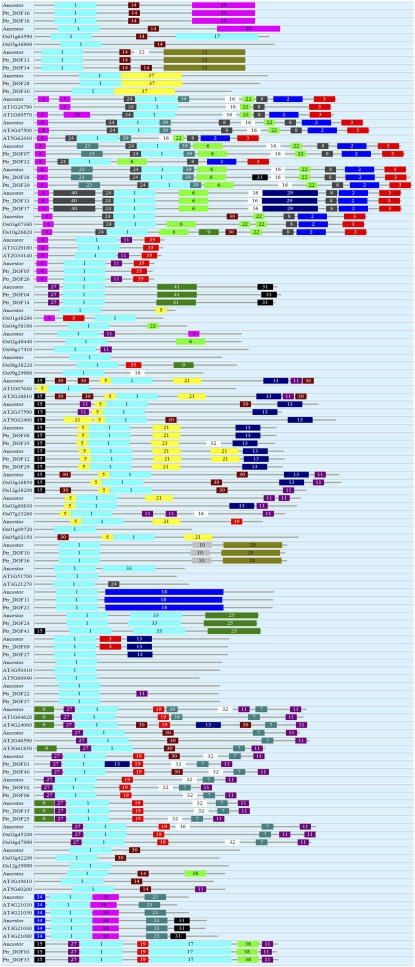
We constructed a maximum-likelihood phylogenetic tree using full-length protein sequences of the Dof genes (Fig. 2). From the phylogenetic tree, we identified 27 pairs of paralogous genes in the terminal nodes, which were well supported by bootstrap analysis. We predicted the immediate ancestral protein sequences of the 27 pairs of paralogs and identified protein motif structure of the duplicated genes and their ancestors (Fig. 3).
Figure 2.
Phylogenetic analysis of full-length protein sequences of 107 Dof genes in poplar, Arabidopsis, and rice. Two maize Dof proteins, Dof1 (GenBank accession no. CAA46875) and Dof2 (CAA56287), were also included to validate the tree topology. Maximum-likelihood tree was built using 100 bootstrap replicates.
Figure 3.
Motif structure of gene duplicates located in the terminals of the phylogenetic tree in Figure 2 and their ancestors. Boxes labeled with numbers are protein motifs.
Genetic Divergence of the Dof Paralogs after Gene Duplication
All of the six evolutionary outcomes resulted from genetic changes (Fig. 1) that existed in the 27 Dof paralogs (Table II), and there were 10 RR, six RN, two RD, three NN, three DN, and three DD evolutionary outcomes. According to Ohno's hypothesis (Ohno, 1970), there are two potential fates of duplicated genes equivalent to RD and RN. However, these types of gene fates (two RD + six RN) account for only 30% of the 27 pairs of Dof paralogs. According to the DDC model (Force et al., 1999), there are three potential fates equivalent to DD, RD, and DN, respectively. However, these types of gene fates (three DD + two RD + three DN) account for only 30% of the 27 pairs of paralogs. According to the SNF model (He and Zhang, 2005b), there are three potential fates of duplicate gene pairs equivalent to NN, DD, and RN, respectively. However, these types of gene fates (three NN + three DD + six RN) account for only 44% of the 27 pairs of paralogs. These results indicate that none of the three existing hypotheses (Ohno, DDC, and SNF) could explain the majority (≥50%) of the genetic changes in the coding region of the Dof paralogs. Although some proteins evolved following the DDC model, such as hemoglobin (Hughes, 2005) and the tRNA endonucleases in the hyperthermophilic, sulfate-reducing Archaeoglobus fulgidus and the thermophilic, methane-producing Methanococcus jannaschii (Tocchini-Valentini et al., 2005), we found only three typical DDC cases (DD type) in the coding regions of the Dof paralogs (Table II). Recently, He and Zhang (2005a) reported that duplicate genes in yeast generally have longer protein sequences and more functional domains than singleton genes. Therefore, we suggest that DDC is not a major route for protein sequence evolution after gene duplication.
Table II.
Evolutionary modes of the Dof duplicates in poplar, Arabidopsis, and rice after recent gene duplication
| Gene 1 | Gene 2 | Gene Fate | ks |
|---|---|---|---|
| Ptr_DOF02 | Ptr_DOF06 | RR | 0.14 |
| Ptr_DOF03 | Ptr_DOF33 | RR | 0.15 |
| Ptr_DOF24 | Ptr_DOF41 | RD | 0.18 |
| Ptr_DOF15 | Ptr_DOF25 | RR | 0.18 |
| Ptr_DOF13 | Ptr_DOF17 | RR | 0.19 |
| Ptr_DOF10 | Ptr_DOF36 | RR | 0.20 |
| Ptr_DOF08 | Ptr_DOF35 | RN | 0.20 |
| Ptr_DOF28 | Ptr_DOF30 | RR | 0.22 |
| Ptr_DOF12 | Ptr_DOF29 | RN | 0.22 |
| AT4G21040 | AT4G21080 | RR | 0.22 |
| Ptr_DOF04 | Ptr_DOF14 | RR | 0.23 |
| Ptr_DOF38 | Ptr_DOF39 | RN | 0.24 |
| Ptr_DOF11 | Ptr_DOF21 | RR | 0.24 |
| Ptr_DOF07 | Ptr_DOF32 | DD | 0.25 |
| Ptr_DOF31 | Ptr_DOF34 | RN | 0.25 |
| Ptr_DOF01 | Ptr_DOF40 | NN | 0.26 |
| Ptr_DOF22 | Ptr_DOF37 | RN | 0.28 |
| AT4G21030 | AT4G21050 | RR | 0.37 |
| Os02g45200 | Os04g47990 | DN | 0.54 |
| Os03g42200 | Os12g39990 | RN | 0.55 |
| AT2G46590 | AT3G61850 | RD | 0.68 |
| AT1G26790 | AT1G69570 | DN | 0.72 |
| Os07g13260 | Os05g02150 | NN | 0.89 |
| AT1G07640 | AT2G28810 | DN | 0.95 |
| AT3G45610 | AT5G60200 | DD | 0.95 |
| Os03g07360 | Os10g26620 | NN | 1.04 |
| AT4G38000 | AT5G65590 | DD | 1.49 |
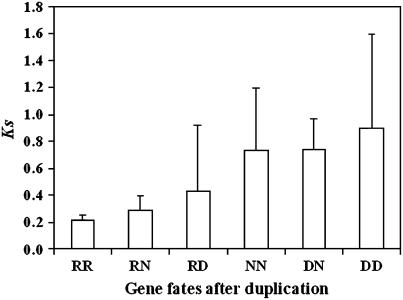
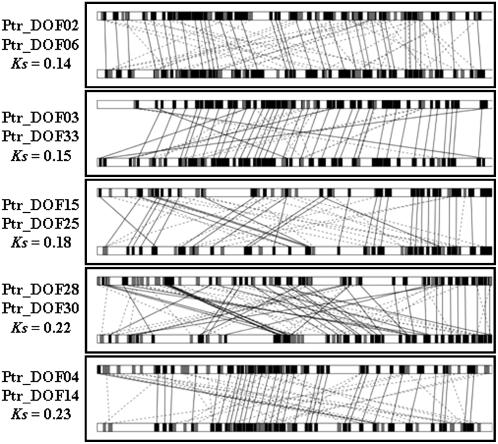
About 37% (10 RR) of the 27 pairs of paralogs retained the ancestral motif organization in protein sequences. It is possible that these genes are still in the process of evolving. This possibility is supported by the fact that the average ks of RR paralogs is lower than that of NN, DN, or DD paralogs (Fig. 4). Another explanation for the RR paralogs is that Dof genes are transcriptional factors, which were preferentially retained in duplicate form, as shown in Arabidopsis (Seoighe and Gehring, 2004). This notion is also supported by the fact that most of the RR paralogs are found in poplar (Table II) in which mutation rate is lower compared with herbaceous annual plants because poplar has a much longer generation time (Sterck et al., 2005; Tuskan et al., 2006).
Figure 4.
The synonymous substitution rate (ks) of different gene fate categories. Values shown represent the average ± 95% confidence interval.
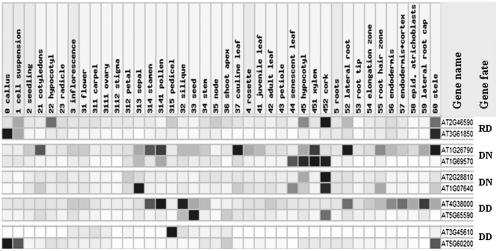
Although the evolutionary fates of the promoter region of RR paralogs are not clear, the expression patterns of the sampled duplicate genes in poplar clearly suggest that some of the duplicates have diverged in functions after duplication. For example, the expression levels of Ptr_DOF02 and Ptr_DOF15 are generally stronger in all of the six tissues examined than those of Ptr_DOF 06 and Ptr_DOF25, respectively, and the expression of Ptr_DOF28 is stronger in shoot tip but weaker in leaf than that of Ptr_DOF 30 (Fig. 5). Diversification in expression of duplicate genes was also revealed in other types of paralogs (Table III; Figs. 5 and 6). This indicates that the regulatory mechanism of the Dof paralogs might experience rapid evolution. Because changes in the promoter regions of duplicate genes can result in subfunctionalization (Force et al., 1999), we compared the 1,000-bp region upstream of the translation start codon (ATG) of the RR paralogs in poplar. We found divergence in upstream regions of the RR paralogs although there are conserved regions and microsynteny (Fig. 7). Further experiments, such as promoter deletions or mutations, will be needed to pinpoint the changes in the cis-elements that are responsible for the expression diversity in the duplicated genes. If experiments reveal that duplicate genes share the same set of cis-elements in the promoter regions, epigenetic aspects should be explored, such as DNA methylation, microRNA, and small interfering RNA.
Figure 5.
Expression of Dof duplicates in poplar revealed by RT-PCR analysis. 18S rRNA was used as an internal standard.
Table III.
Expression of rice Dof duplicates revealed by MPSS analysis
| Gene Fate | Gene Name | Young Leaves Stressed in Cold | Young Roots Stressed in Cold | Young Leaves Stressed in Drought | Young Roots Stressed in Drought | Germinating Seedlings | Germinating Seed | Immature Panicle | Mature Leaves: Replicate A | Mature Leaves: Replicate B | Vegetative Meristematic Tissue | Ovary and Mature Stigma | Mature Pollen | Mature Roots: Replicate A | Mature Roots: Replicate B | Young Leaves Stressed in NaCl | Young Roots Stressed in NaCl | Stem | Young Leaves | Young Roots |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RN | Os03g42200 | 0 | 0 | 0 | 0 | 0 | 68 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Os12g39990 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| NN | Os07g13260 | 0 | 0 | 0 | 0 | 18 | 0 | 5 | 0 | 0 | 0 | 43 | 0 | 0 | 0 | 2 | 0 | 0 | 15 | 0 |
| Os05g02150 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| NN | Os03g07360 | 235 | 0 | 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 70 | 0 | 0 | 41 | 0 |
| Os10g26620 | 296 | 76 | 3 | 0 | 10 | 0 | 24 | 0 | 29 | 5 | 0 | 0 | 10 | 35 | 366 | 0 | 0 | 85 | 30 | |
| DN | Os02g45200 | 14 | 21 | 0 | 5 | 0 | 80 | 34 | 0 | 0 | 0 | 0 | 5 | 8 | 0 | 0 | 0 | 19 | 0 | 0 |
| Os04g47990 | 0 | 0 | 0 | 96 | 0 | 0 | 9 | 0 | 0 | 15 | 5 | 0 | 0 | 82 | 6 | 5 | 0 | 0 | 37 |
Figure 6.
Expression of Dof duplicates in Arabidopsis revealed by analysis of microarray data using GENEVESTIGATOR (Zimmermann et al., 2004). All gene-level profiles were normalized for coloring such that for each gene the highest signal intensity obtains value 100% (dark) and absence of signal obtains value 0% (white).
Figure 7.
Comparative analysis of the 1,000-bp region upstream of the translation start codon (ATG) of the RR paralogs. Solid dark lines connect similar regions and gray broken lines connect matched regions in reversed orientation.
Driving Forces for Genetic Divergence
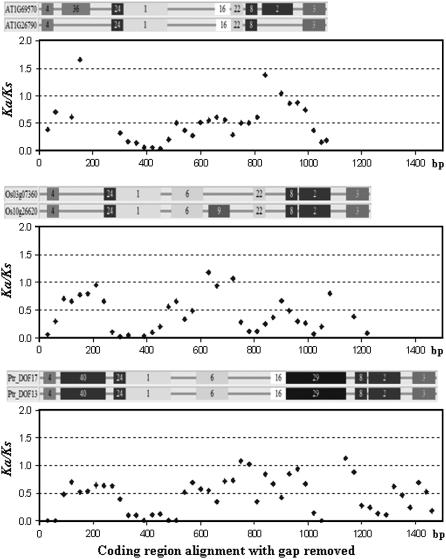
To explore whether Darwinian positive selection was involved in driving gene divergence after duplication, we calculated the nonsynonymous/synonymous substitution ratio (ka/ks) for the coding region of some recently duplicated paralogs using a sliding window of 20 amino acids. Generally, a ka:ks ratio > 1 indicates positive selection, a ratio < 1 indicates negative or purifying selection, and a ratio = 1 indicates neutral evolution (Wang et al., 2005). For the Dof paralogs, ka:ks ratios were always near zero for motif 1, the conserved Dof domain, suggesting strong purifying selection on this motif. In contrast, much higher ka:ks ratios were generally found in the regions outside the motif 1, especially in the intermotif regions (Fig. 8). Such positive selections have also been observed in the RLK/Pelle gene family in Arabidopsis and rice (Shiu et al., 2004). A higher proportion of new exons had ka:ks ratios > 1 and a higher frequency of insertions/deletions (indels) than did the old exons, implying that positive selection played an important role in the evolution of new domains (Wang et al., 2005). Therefore, positive selection is one of the major driving forces for the emergence of new motifs/functions in protein after gene duplication.
Figure 8.
The nonsynonymous to synonymous substitution ratio (ka:ks) for the coding region of several recently duplicated paralogs with a sliding window of 20 amino acids and a step size of 10 amino acids. Boxes labeled with numbers are protein motifs.
Furthermore, more than one-half (21) of the 41 motifs in Table I was also found in non-Dof genes in the three plant species (Table IV). We hypothesize that co-option of new motifs from non-Dof genes might be an important source of domain expansions in Dof genes. Such domain fusion or co-option was also observed in the other gene families (Raff, 1996; Carroll, 2001; Cameron et al., 2005; Force et al., 2005). In our study, the co-oped motifs appear to have undergone local tandem duplications, minor insertions and deletions, and translocations (Fig. 3). These local events contributed to further gene diversification in the Dof family.
Table IV.
Motifs in Dof proteins of poplar, Arabidopsis, and rice identified in non-Dof proteins
| Non-Dof Proteins | E Value | Motifs | ||||||
|---|---|---|---|---|---|---|---|---|
| eugene3.00700193 | 2.40E-11 | 11 | ||||||
| eugene3.00290199 | 7.60E-10 | 14 | ||||||
| fgenesh1_pg.C_LG_VII001151 | 6.80E-09 | 14 | 23 | 11 | 11 | |||
| eugene3.00180470 | 7.00E-09 | 10 | 24 | 23 | ||||
| fgenesh1_pg.C_LG_V000738 | 7.50E-09 | 30 | ||||||
| estExt_fgenesh1_pm_v1.C_LG_I0692 | 8.90E-09 | 35 | ||||||
| fgenesh1_pg.C_LG_III001633 | 2.50E-08 | 30 | 30 | 30 | 30 | 34 | 30 | 30 |
| fgenesh1_pg.C_scaffold_18421000001 | 3.00E-08 | 40 | ||||||
| fgenesh1_pg.C_scaffold_3342000001 | 6.00E-08 | 39 | 24 | 31 | ||||
| fgenesh1_pg.C_LG_VI001314 | 8.90E-08 | 34 | 39 | 9 | ||||
| eugene3.00180651 | 9.90E-08 | 15 | ||||||
| At2g04495 | 2.80E-12 | 4 | 10 | 2 | 23 | |||
| At1g56170 | 6.70E-09 | 26 | 30 | 26 | 6 | 26 | ||
| Os02g13580 | 2.70E-08 | 20 | 13 | 40 | 40 | 40 | 37 | 2 |
Epigenetic Divergence
To investigate possible involvement of epigenetic changes, we performed computer prediction of potential methylation sites in protein sequences of duplicated poplar genes (RR) that share the same motif structure. Our analysis revealed two Arg methylation sites within the motif regions in Ptr_DOF14, whereas another copy of the duplicate, Ptr_DOF04, does not have the methylation sites (Fig. 9). It is interesting that the expression pattern of Ptr_DOF04 is very similar to that of Ptr_DOF14 (Fig. 5). Because Ptr_DOF04 and Ptr_DOF14 are similar in terms of expression and protein motif structure, we suggest that the diversification of these two genes resulted from epigenetic changes such as Arg methylation. It has been reported that Arg methylation plays important roles in RNA processing, transcriptional regulation, and signal transduction (Bedford and Richard, 2005; Boisvert et al., 2005; Chen et al., 2006). Arg methylation has been observed on a variety of proteins associated with gene regulation, including DNA-binding transcriptional activators, transcriptional coactivators, and many RNA-binding proteins involved in RNA processing, transport, and stability (Lee et al., 2005). Experiments need to be done to confirm the predicted methylation and to investigate its effect on protein function.
Figure 9.
Methylation sites (indicated by arrows) predicted in RR paralogs suggest epigenetic divergence. Boxes labeled with numbers are protein motifs. The ruler indicates the size of the proteins in amino acids.
In spite of the aforementioned possibility that sequence difference in the promoter regions accounts for the expression difference between duplicated genes (Table III; Figs. 5 and 6), it is still possible that epigenetics plays a role in expression diversification of these genes.
CONCLUSION
Our results show that the duplication and subsequent divergence of the Dof gene family in three plant species do not fit Onho's classical DRNNF model, or the more recently proposed alternatives SF/DDC or SNF alone, in terms of gene functions conferred by the coding regions. We conclude that the existing models are complementary with, not alternative to, one another. We anticipate that the six gene fates (RR, RD, RN, DD, NN, and ND) may also fit other gene families at variable ratios among them. We also suggest that epigenetics may play an important role in gene diversification after duplication. Based on our analysis of the Dof gene families in poplar, Arabidopsis, and rice, we also conclude that after a gene duplication event, the evolution of the duplicated genes is driven by purifying selection, Darwinian positive selection, local duplication and translocations, and domain co-option. The divergent expression may also be affected by epigenetic regulations.
MATERIALS AND METHODS
Sequences
The Arabidopsis (Arabidopsis thaliana) Dof gene name list was obtained from two Arabidopsis transcription factor databases (http://Arabidopsis.med.ohio-state.edu/AtTFDB/ and http://datf.cbi.pku.edu.cn). The corresponding coding and protein sequences were downloaded from http://www.arabidopsis.org/ (The Institute for Genomic Research [TIGR] annotation release 5). The 5′ end of AT5G62430 was found truncated and it was corrected according to http://datf.cbi.pku.edu.cn. The rice (Oryza sativa) Dof gene name list was obtained from http://ricetfdb.bio.uni-potsdam.de/ and the corresponding coding and protein sequences were downloaded from http://www.tigr.org/ (TIGR rice pseudomolecules release 3). Os03g42200 was manually corrected. The 5′ end of Os07g48570 was found truncated after searching the expressed sequence tag database and was corrected according to http://ricetfdb.bio.uni-potsdam.de/. 9640.m03713 in TIGR rice pseudomolecules release 2 was assigned as Os12g38200 in TIGR rice pseudomolecules release 3. The sequence of Os12g38200 was found to be incorrect and was replaced by the sequence of 9640.m03713, which was confirmed by examining the pseudomolecules of the rice genome (Build 4.0) released recently by The International Rice Genome Sequencing Project (International Rice Genome Sequencing Project, 2005). To obtain populus Dof gene sequences, Arabidopsis and rice Dof protein sequences were used to search the populus genome annotation (http://genome.jgi-psf.org/Poptr1/Poptr1.home.html) using tBLASTn (Altschul et al., 1990). Pt_DOF15, Pt_DOF27, and Pt_DOF29 were found to be 3′ truncated and were manually corrected.
ks and ka Calculations
Pairwise alignments of the paralogous nucleotide sequences were made using ClustalW (Thompson et al., 1994), with the corresponding protein sequences as the alignment guides. Gaps in the alignments were removed. ks (synonymous substitution rate) and ka (nonsynonymous substitution rate) analysis was carried out using the K-Estimator program (Comeron, 1999).
Tree Construction
A multiple alignment analysis was performed with M-Coffee (Wallace et al., 2006). Phylogeny was created by maximum-likelihood analysis using PHYML (Guindon and Gascuel, 2003) with protein model JTT+I+G, which was selected by modelGenerator (Keane et al., 2006). Bootstrap analysis of 100 replicates was then performed, and the consensus tree was then displayed with bootstrap values.
Prediction of Ancestral Protein Sequences
Gapped Ancestral Sequence Prediction (Edwards and Shields, 2004) was used to predict ancestral sequences from phylogenetic trees and the corresponding multiple sequence alignments. The input tree was unrooted and it was rooted automatically by midpoint rooting in Gapped Ancestral Sequence Prediction analysis.
Motif Identification
Protein motifs of the Dof genes were identified statistically using MEME (Bailey and Elkan, 1994) with motif length set as 6 to 200, motif sites 2 to 107, and e value < 1 × 10−10. The MAST program (Bailey and Gribskov, 1998) was used to search protein motifs of the extant Dof proteins in non-Dof genes as well as the ancestral Dof protein sequences. The motifs were further characterized using the Conserved Domain Search Service (Marchler-Bauer and Bryant, 2004).
Gene Expression
The tissue-specific expression analysis of Arabidopsis genes was performed using Meta-Analyzer in GENEVESTIGATOR (Zimmermann et al., 2004) with ATH1-22k array. The tissue-specific expression data of rice genes were obtained from the MPSS database (20-bp signatures; http://mpss.udel.edu/rice/?).
For multiple-tissue RT-PCR analysis of gene expression in poplar (Populus trichocarpa) “Nisqually-1,” stem and leaf tissues were taken from plants grown in vitro on media containing Murashige and Skoog salts (Murashige and Skoog, 1962), 3% Suc, and 0.25% Gelrite (PhytoTechnology Laboratories) at 23°C ± 1°C under cool-white fluorescent light (approximately 125 μmol m−2 s−1, 16-h photoperiod). Root, shoot tip, petiole, and bark tissues were taken from plants grown in a greenhouse in Knoxville, TN, under natural lighting and temperatures ranging from 25°C to 35°C. Total RNA was extracted from root, stem, shoot tip, petiole, leaf, and bark using the Spectrum Plant Total RNA kit (Sigma-Aldrich) and then treated with AMPD1 DNase I (Sigma-Aldrich) to eliminate DNA, according to the manufacturer's instructions. RNA purity was determined spectrophotometrically, and quality was determined by examining rRNA bands on agarose gels. cDNA was synthesized from 2 μg of RNA using the PowerScript PrePrimed Single Shots with random hexamers as primer (CLONTECH Laboratories) in a 20-μL reaction. For PCR reactions using gene-specific primers (Supplemental Table S3), the cDNA was diluted 50-fold, and 2.0 μL was used for a 20-μL PCR reaction. For PCR reactions using 18S rRNA-specific primers (QuantumRNA Universal 18S Internal Standard; Ambion), the cDNA was diluted 20,000 times, and 2.0 μL was used for a 20-μL PCR reaction containing 0.5 units TaKaRa Taq HS (Takara Mirus Bio), 1× PCR buffer, 200 μm of each dNTP, and 0.5 μm of each gene-specific primer (or 0.25 μm of 18S rRNA-specific primers). PCR was performed as follows: one round at 94°C for 2 min; 35 cycles: 94°C for 30 s, 60°C for 30 s, 72°C for 1 min; and a final step at 72°C for 7 min. The amplified products (18 μL each) were separated on a 1% agarose gel, stained with ethidium bromide, and documented with Gel Doc 2000 (Bio-Rad).
Comparative Analysis of Promoter Sequences
Comparative analysis of the 1,000-bp region upstream of the translation start codon (ATG) was performed using the GATA program (Nix and Eisen, 2005), with window size of 7 and lower cutoff score of 12 bit.
Protein Methylation Prediction
Protein sequence alignment was performed with M-Coffee (Wallace et al., 2006). Protein methylation prediction was performed with the MeMo Web server (Chen et al., 2006).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers CAA46875 (Dof1) and CAA56287 (Dof2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Dof gene list in Arabidopsis, rice, and poplar.
Supplemental Table S2. Summary of motif distributions.
Supplemental Table S3. Primers for RT-PCR.
Supplementary Material
Acknowledgments
We thank F. Chen and R.C. Moore for reviewing the manuscript and valuable comments. We also thank the anonymous reviewers for the inspiring comments on the manuscript.
This work was supported by the National Science Foundation (grant no. 0421743 to G.A.T. and Z.-M.C.), by the U.S. Department of Energy/Oak Ridge National Laboratory (subcontract to Z.-M.C.), and by the Tennessee Agricultural Experiment Station.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: (Max) Zong-Ming Cheng (zcheng@utk.edu).
The online version of this article contains Web-only data.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Bailey TL, Gribskov M (1998) Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14: 48–54 [DOI] [PubMed] [Google Scholar]
- Bedford MT, Richard S (2005) Arginine methylation an emerging regulator of protein function. Mol Cell 18: 263–272 [DOI] [PubMed] [Google Scholar]
- Bender J (2002) Plant epigenetics. Curr Biol 12: R412–R414 [DOI] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH (2004) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, Chenard CA, Richard S (2005) Protein interfaces in signaling regulated by arginine methylation. Sci STKE 2005: re2. [DOI] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438 [DOI] [PubMed] [Google Scholar]
- Cameron RA, Chow SH, Berney K, Chiu TY, Yuan QA, Kramer A, Helguero A, Ransick A, Yun M, Davidson EH (2005) An evolutionary constraint: strongly disfavored class of change in DNA sequence during divergence of cis-regulatory modules. Proc Natl Acad Sci USA 102: 11769–11774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB (2001) Chance and necessity: the evolution of morphological complexity and diversity. Nature 409: 1102–1109 [DOI] [PubMed] [Google Scholar]
- Chen H, Xue Y, Huang N, Yao X, Sun Z (2006) MeMo: a web tool for prediction of protein methylation modifications. Nucleic Acids Res 34: W249–W253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron JM (1999) K-Estimator: calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics 15: 763–764 [DOI] [PubMed] [Google Scholar]
- Darwin F, Seward AC, editors (1903) More Letters of Charles Darwin, Vol 2. John Murray, London
- Davies TJ, Barraclough TG, Chase MW, Soltis PS, Soltis DE, Savolainen V (2004) Darwin's abominable mystery: insights from a supertree of the angiosperms. Proc Natl Acad Sci USA 101: 1904–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S, Maere S, Van de Peer Y (2005) Genome duplication and the origin of angiosperms. Trends Ecol Evol 20: 591–597 [DOI] [PubMed] [Google Scholar]
- Edwards RJ, Shields DC (2004) GASP: gapped ancestral sequence prediction for proteins. BMC Bioinformatics 5: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, Cresko WA, Pickett FB, Proulx SR, Amemiya C, Lynch M (2005) The origin of subfunctions and modular gene regulation. Genetics 170: 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li WH (2003) Role of duplicate genes in genetic robustness against null mutations. Nature 421: 63–66 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704 [DOI] [PubMed] [Google Scholar]
- He X, Zhang J (2005. a) Gene complexity and gene duplicability. Curr Biol 15: 1016–1021 [DOI] [PubMed] [Google Scholar]
- He X, Zhang J (2005. b) Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 169: 1157–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL (1994) The evolution of functionally novel proteins after gene duplication. Proc Biol Sci 256: 119–124 [DOI] [PubMed] [Google Scholar]
- Hughes AL (2005) Gene duplication and the origin of novel proteins. Proc Natl Acad Sci USA 102: 8791–8792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Keane TM, Creevey CJ, Pentony MM, Naughton TJ, McInerney JO (2006) Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol 6: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, Teyssier C, Strahl BD, Stallcup MR (2005) Role of protein methylation in regulation of transcription. Endocr Rev 26: 147–170 [DOI] [PubMed] [Google Scholar]
- Li WH, Yang J, Gu X (2005) Expression divergence between duplicate genes. Trends Genet 21: 602–607 [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH (2004) CD-Search: protein domain annotations on the fly. Nucleic Acids Res 32: W327–W331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RC, Purugganan MD (2003) The early stages of duplicate gene evolution. Proc Natl Acad Sci USA 100: 15682–15687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RC, Purugganan MD (2005) The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol 8: 122–128 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nix DA, Eisen MB (2005) GATA: a graphic alignment tool for comparative sequence analysis. BMC Bioinformatics 6: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S (1970) Evolution by Gene Duplication. Springer-Verlag, Heidelberg
- Raes J, Vandepoele K, Simillion C, Saeys Y, Van de Peer Y (2003) Investigating ancient duplication events in the Arabidopsis genome. J Struct Funct Genomics 3: 117–129 [PubMed] [Google Scholar]
- Raff R (1996) The Shape of Life. University of Chicago Press, Chicago
- Rapp RA, Wendel JF (2005) Epigenetics and plant evolution. New Phytol 168: 81–91 [DOI] [PubMed] [Google Scholar]
- Rodin SN, Parkhomchuk DV, Rodin AS, Holmquist GP, Riggs AD (2005) Repositioning-dependent fate of duplicate genes. DNA Cell Biol 24: 529–542 [DOI] [PubMed] [Google Scholar]
- Rodin SN, Riggs AD (2003) Epigenetic silencing may aid evolution by gene duplication. J Mol Evol 56: 718–729 [DOI] [PubMed] [Google Scholar]
- Seoighe C, Gehring C (2004) Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet 20: 461–464 [DOI] [PubMed] [Google Scholar]
- Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, Li WH (2004) Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16: 1220–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterck L, Rombauts S, Jansson S, Sterky F, Rouze P, Van de Peer Y (2005) EST data suggest that poplar is an ancient polyploid. New Phytol 167: 165–170 [DOI] [PubMed] [Google Scholar]
- Taylor JS, Raes J (2004) Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet 38: 615–643 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocchini-Valentini GD, Fruscoloni P, Tocchini-Valentini GP (2005) Structure, function, and evolution of the tRNA endonucleases of Archaea: an example of subfunctionalization. Proc Natl Acad Sci USA 102: 8933–8938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Wallace IM, O'Sullivan O, Higgins DG, Notredame C (2006) M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res 34: 1692–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zheng H, Yang S, Yu H, Li J, Jiang H, Su J, Yang L, Zhang J, McDermott J, et al (2005) Origin and evolution of new exons in rodents. Genome Res 15: 1258–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S (2002) The Dof family of plant transcription factors. Trends Plant Sci 7: 555–560 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S (2004) Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol 45: 386–391 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.