Abstract
Marine diatoms are known to be responsible for about a quarter of global primary production and their photosynthesis is sustained by inorganic carbon-concentrating mechanisms and/or C4 metabolism. Activities of the inorganic carbon-concentrating mechanism are attenuated under enriched [CO2]; however, impacts of this factor on primary productivity and the molecular mechanisms of CO2 responses in marine diatoms are unknown. In this study, transgenic cells were generated of the marine diatom Phaeodactylum tricornutum by the introduction of a β-glucuronidase reporter gene under the control of an intrinsic CO2-responsive promoter, which is the sequence between −80 to +61 relative to the transcription start site of a chloroplastic-carbonic anhydrase gene, ptca1, obtained from P. tricornutum. The activity of the ptca1 promoter was effectively repressed in air-level CO2 by treating cells with a 1.0 mm cAMP analog, dibutyryl cAMP, or a cAMP phosphodiesterase inhibitor, 3-isobutyl-1-methylxanthine. Deletion of the intrinsic cAMP-response element from the ptca1 promoter caused a lack of repression of the reporter gene uidA, even under elevated [CO2] and a null phenotype to the strong repressive effects of dibutyryl cAMP and 3-isobutyl-1-methylxanthine on the ptca1 promoter. Deletion of the cAMP-response element was also shown to cause derepression of the uidA reporter gene in the dark. These results indicate that the cytosolic cAMP level increases under elevated [CO2] and represses the ptca1 promoter. This strongly suggests the participation of cAMP metabolism, presumably at the cytosolic level, in controlling CO2-acquisition systems under elevated [CO2] at the ocean surface in a marine diatom.
Marine diatoms are responsible for one-half of primary productivity in the ocean and hence play a key role in global cycles of carbon and other inorganic nutrients (Tréguer et al., 1995; Falkowski et al., 2000). [CO2] dissolved in seawater is limited under the present atmospheric pCO2 (below 15 μm at 20°C) that is much lower than the Km[CO2] of Rubisco in diatom species (Badger et al., 1998). This implies that marine diatoms need active uptake and accumulation systems for dissolved inorganic carbon (DIC) to support their photosynthesis. There is a substantial body of evidence that the operation of the inorganic carbon-concentrating mechanism (CCM) confers on marine diatom cells high-affinity photosynthesis for DIC (Colman and Rotatore, 1995; Johnston and Raven, 1996; Matsuda et al., 2001), which is due to the operation of active uptake of both CO2 and 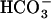 (Colman and Rotatore, 1995; Johnston and Raven, 1996; Matsuda et al., 2001). The activity of the CCM is suppressed under CO2-enriched conditions, whereas it is induced in CO2-limiting conditions; this regulation is due to CO2 sensing by algal cells and induction of the CCM facilitates an ample supply of CO2 to Rubisco even under extreme [CO2] limitation (Badger et al., 1980, 1998; Kaplan et al., 1980; Miller et al., 1990; Colman and Rotatore, 1995; Johnston and Raven, 1996; Tortell et al., 1997; Moroney and Somanchi, 1999; Matsuda et al., 2001).
(Colman and Rotatore, 1995; Johnston and Raven, 1996; Matsuda et al., 2001). The activity of the CCM is suppressed under CO2-enriched conditions, whereas it is induced in CO2-limiting conditions; this regulation is due to CO2 sensing by algal cells and induction of the CCM facilitates an ample supply of CO2 to Rubisco even under extreme [CO2] limitation (Badger et al., 1980, 1998; Kaplan et al., 1980; Miller et al., 1990; Colman and Rotatore, 1995; Johnston and Raven, 1996; Tortell et al., 1997; Moroney and Somanchi, 1999; Matsuda et al., 2001).
A number of physiological evidences have shown major differences in the mode of regulation in CCM expression between cyanobacteria and eukaryotic algae. The differences are particularly evident with respect to O2 dependency and the critical determinant for the extent of CCM expression. Decrease in [O2] from 21% to 2.6% during acclimation of the cyanobacterium Anabaena variabilis to low [CO2] caused a significant delay of CCM induction (Marcus et al., 1983). This is in complete agreement with the recent finding that transcription of most CCM components requires O2 for full induction in Synechococcus PCC7942 under CO2 limitation (Woodger et al., 2005). In contrast, it was clearly demonstrated in the green alga Chlamydomonas reinhardtii that inductions of CO2-regulated genes and the CCM are completely independent of O2 concentrations but that CO2 alone is important for this process (Vance and Spalding, 2005). The absence of O2 effect on CCM expression was also shown in the green alga Chlorella ellipsoidea (Matsuda et al., 1998). These investigations clearly revealed at least two distinct lines of the CO2-response system in CCM regulation. Close relations of the bacterial-type CCM regulation with [O2] probably reflect the metabolic state in photosynthesis induced by changing the CO2 to O2 ratio (Kaplan and Reinhold, 1999), whereas the data obtained in green algae suggest the occurrence of the relatively direct perception of CO2 signal in the eukaryotic CCM regulations. In cyanobacteria, it is assumed therefore that low CO2 signal will disappear or be diminished after cells have developed an effective level of the CCM during acclimation to low CO2. This is supported by the finding that transcription of CCM components occurs in response to a transient decrease in internal [DIC] at the initial stage of acclimation to low CO2 but was suppressed according to the development of the internal DIC pool at the late stage of acclimation to low CO2 in Synechococcus PCC7942 (Woodger et al., 2005). In sharp contrast, it was clearly demonstrated that [CO2] in the bulk medium is the critical determinant for the extent of CCM expression, but other DIC species and temporary decrease in internal DIC are not responsible for CCM regulation in the green alga C. ellipsoidea (Matsuda and Colman, 1995). The occurrence of [CO2] as a critical determinant for the CCM expression level has also been reported in the green algae C. reinhardtii (Bozzo and Colman, 2000; Vance and Spalding, 2005) and Chlorella kessleri (Bozzo et al., 2000), and the marine diatom Phaeodactylum tricornutum (Matsuda et al., 2001).
Light is a crucial but not absolute factor to develop a full expression of the CCMs in cyanobacteria and eukaryotic algae. Dependence on light seems to vary considerably among species, and in green algae it is often very weak (i.e. significant levels of CCM expression occurred in the dark in air; Matsuda and Colman, 1995; Bozzo and Colman, 2000). In C. reinhardtii, regulation of the CO2-responsive gene Cah1 was shown to be regulated by both silencer and enhancer in its promoter region in response to changing the ambient [CO2], and the absence of light constitutes a repressive signal to the Cah1 promoter via the silencer region (Kucho et al., 1999). These observations strongly suggest the occurrence of cross talk between light and CO2 signals in CCM regulation.
CO2-sensing mechanisms, as described above, have been studied in a limited number of algae, primarily freshwater and soil species (Matsuda and Colman, 1995; Rawat and Moroney, 1995; Badger et al., 1998; Eriksson et al., 1998; Kaplan and Reinhold, 1999; Kucho et al., 1999; Bozzo and Colman., 2000; Miura et al., 2004; Vance and Spalding, 2005; Woodger et al., 2005), but little molecular information is available on marine eukaryotic algae. CO2 is a redundant molecule that influences a variety of physiological processes in microbes, plants, and animals, and, as a precedent, the initial process of sensing [CO2] has been found in a mammalian tissue. In rat testis, the activity of soluble adenylyl cyclase (sAC) was shown to be stimulated directly by the addition of 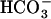 and Ca2+ (Chen et al., 2000; Jaiswal and Conti, 2003), with which ejaculated spermatozoa could undergo a series of
and Ca2+ (Chen et al., 2000; Jaiswal and Conti, 2003), with which ejaculated spermatozoa could undergo a series of 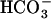 -induced initial activation processes (Chen et al., 2000). Since this gene for sAC was found to be evolutionally conserved from cyanobacteria to mammals (Chen et al., 2000), the CO2-sensing system mediated by cytosolic cAMP levels was proposed to be a general mechanism that might operate in the regulation of the CCM in cyanobacteria (Chen et al., 2000). In fact, several cyanobacterial sACs, CyaC, CyaB1, and Cya1, are stimulated or inhibited by the addition of
-induced initial activation processes (Chen et al., 2000). Since this gene for sAC was found to be evolutionally conserved from cyanobacteria to mammals (Chen et al., 2000), the CO2-sensing system mediated by cytosolic cAMP levels was proposed to be a general mechanism that might operate in the regulation of the CCM in cyanobacteria (Chen et al., 2000). In fact, several cyanobacterial sACs, CyaC, CyaB1, and Cya1, are stimulated or inhibited by the addition of 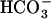 (Chen et al., 2000; Masuda and Ono, 2005). However, DIC species, critical for the stimulation of sACs, was not clear in these studies. Recently, a sAC, Cya1 in Synechocystis PCC6803, was shown to be stimulated by CO2 rather than
(Chen et al., 2000; Masuda and Ono, 2005). However, DIC species, critical for the stimulation of sACs, was not clear in these studies. Recently, a sAC, Cya1 in Synechocystis PCC6803, was shown to be stimulated by CO2 rather than 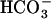 (Hammer et al., 2006). It is thus possible that CO2-responsive events, including CCM expression, might be controlled not only by
(Hammer et al., 2006). It is thus possible that CO2-responsive events, including CCM expression, might be controlled not only by 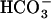 but also directly by [CO2], in some cases via the second-messenger cAMP. At present, however, no relation among sACs, cAMP, and CCM regulation has been reported either in cyanobacteria or in eukaryotic algae. In marine diatoms, some putative sACs are found in the genome database of Thalassiosira pseudonana, but these genes are not related to CO2/
but also directly by [CO2], in some cases via the second-messenger cAMP. At present, however, no relation among sACs, cAMP, and CCM regulation has been reported either in cyanobacteria or in eukaryotic algae. In marine diatoms, some putative sACs are found in the genome database of Thalassiosira pseudonana, but these genes are not related to CO2/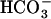 responses.
responses.
A β-carbonic anhydrase in the marine diatom P. tricornutum (PtCA1), thought to be one of the crucial chloroplastic components of the CCM (Satoh et al., 2001; Tanaka et al., 2005), is known to be regulated strictly by the ambient [CO2] and light (Harada et al., 2005). A promoter region of the PtCA1 gene (ptca1) was isolated previously, and it was demonstrated that the critical CO2-response sequence was located downstream −70 bp relative to the transcription start site (Harada et al., 2005). This relatively short core regulatory sequence comprises two putative cAMP-response elements, CRE1 at −70 to −63 and CRE2 at −21 to −14, and a putative p300-binding element at −52 to −46 relative to the transcription start site (Harada et al., 2005; Fig. 1). In this study, the activity of this core regulatory region was investigated by replacing the ptca1 gene with the β-glucuronidase (GUS) reporter gene uidA.
Figure 1.
The core regulatory region of Pptca1, which is −80 to +61 bp relative to the transcription start site. Three putative cis-elements, CRE1, p300-binding element, and CRE2, are boxed. The putative initiator sequence is underlined. The upstream and downstream sequences of the transcription start site are indicated with uppercase and lowercase letters, respectively.
RESULTS
Deletion and Substitution Assays of the ptca1 Promoter with the GUS Reporter Gene uidA
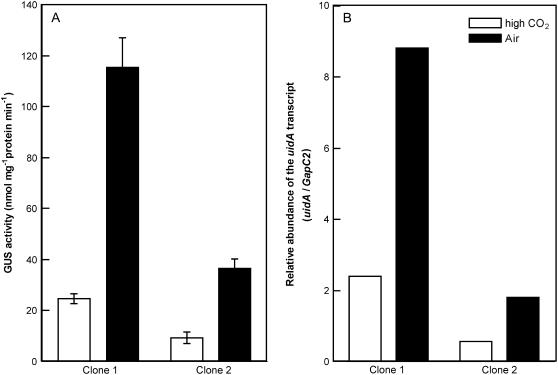
The upstream sequence from −80 to +61 relative to the transcription start site of the ptca1 gene (Fig. 1), which includes the CO2-responsive core regulatory region of the ptca1 promoter, Pptca1 (Harada et al., 2005), was ligated to the GUS reporter gene uidA. Prior to the deletion experiment, GUS activities in two independent P. tricornutum clones, which were transformed with the uidA reporter gene under the control of Pptca1, were compared with uidA transcript levels (Fig. 2). GUS activities increased 4.0- to 5.3-fold in the air-grown transformants as compared to those in high-CO2-grown transformants (Fig. 2A). Transcript levels of uidA were detected at levels corresponding to GUS activities irrespective of growth conditions and clones (Fig. 2B), indicating that GUS activity can be a direct indicator for activities of Pptca1.
Figure 2.
Levels of GUS activities and the GUS gene (uidA) transcript in two independent clones of transformants containing the uidA reporter. A, Levels of GUS activity in lysates of cells grown in high CO2 or acclimated to air. Values are means ± sd of three separate experiments. B, Relative levels of the uidA transcript in cells grown in high CO2 and acclimated to air. Transformant cells of P. tricornutum grown in 5% CO2 (white bars) were transferred to air and allowed to acclimate to air for 1 d (black bars) at 20°C under continuous illumination. GUS activity was measured as described in the text. Real-time PCR was carried out with the Smart Cycler thermal cycler system (Cepheid). Relative amount of the uidA product was quantified as ratios to levels of the GapC2 product as an internal standard.
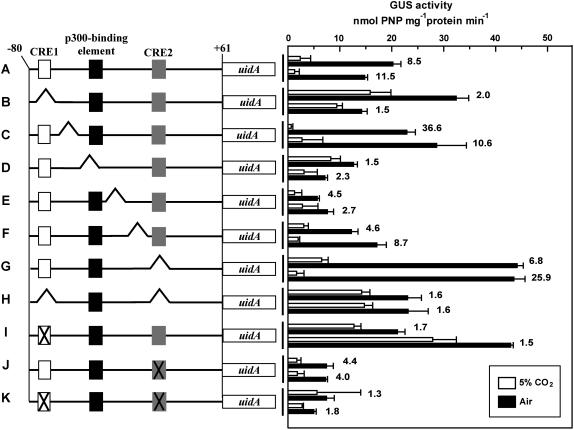
To identify the critical CO2-responsive cis-elements, a series of 10-bp deletions or base substitutions were carried out to the core regulatory region of the ptca1 promoter, Pptca1 (Fig. 3, left). Manipulated promoter constructs were fused with the reporter gene uidA (Fig. 3, left) and were introduced into P. tricornutum cells. A single deletion of the region −71 to −62 or the region −52 to −42, which, respectively, includes CRE1 or the putative p300-binding region, resulted in the apparent derepression of GUS expression in high-CO2-grown transformants (Fig. 3, B and D). GUS expression ratios (air/5% CO2) were reduced significantly to less than 2.0 (Fig. 3, B and D), and, thus, these transformants became weakly responsive to CO2 with nearly constitutive expressions of GUS. A similar regulatory profile of uidA was also seen in transformants by deleting both CRE1 and CRE2 (−21 to −13; Fig. 3H). Derepression of uidA was again the case when regions containing these elements were substituted with the respective antisense sequences (Fig. 3, I and K). A similar derepression of uidA under high CO2 was also observed by deleting the p300-binding element (Fig. 3D). On the other hand, a single deletion or substitution of the CRE2 region did not cause a reduction in the CO2 sensitivity of uidA expression (Fig. 3, G and J). Deletion of CRE2 appeared rather to enhance the expression levels of uidA in air, whereas the antisense substitution of CRE2 did not (Fig. 3, G and J). Deletions of sequences located between CREs and the p300-binding element did not cause significant reduction of CO2 sensitivity of Pptca1 (Fig. 3, C, E, and F).
Figure 3.
Activities of the intact and manipulated core regulatory regions of the ptca1 promoter, which were reported by GUS expression levels. Constructs A to K are depicted in the left half and GUS activities in transformants with each construct are shown in the right half. Two independent clones of each transformation were subjected to the GUS assay. Three putative cis-elements, CRE1, p300-binding element, and CRE2, are indicated by white, black, and gray boxes, respectively. A, The intact Pptca1 core regulatory region with 61 bp of the 5′-untranslated region, which is followed by the uidA reporter gene. B to G, A series of 10-bp deletions was carried out to construct A at −71 to −62, −61 to −52, −51 to −42, −41 to −32, −32 to −23, and −22 to −13 bp relative to the transcription start site. H, Regions including both CRE1 (−71 to −62) and 2 (−22 to −13) were deleted. I to K, Either or both CRE1 and 2 were substituted by their respective antisense sequences. Deleted and substituted portions are indicated as broken lines and cross marks, respectively. Values are means ± sd of three separate experiments. CO2-grown cells (white bars) were transferred to air and were acclimated to air in the light for 1 d (black bars). Values are means ± sd of three separate experiments. GUS activity ratios (air/5% CO2) are shown to the right of bars.
Pptca1 Activities Driven with Substituted Human Cytomegalovirus Minimal Promoter
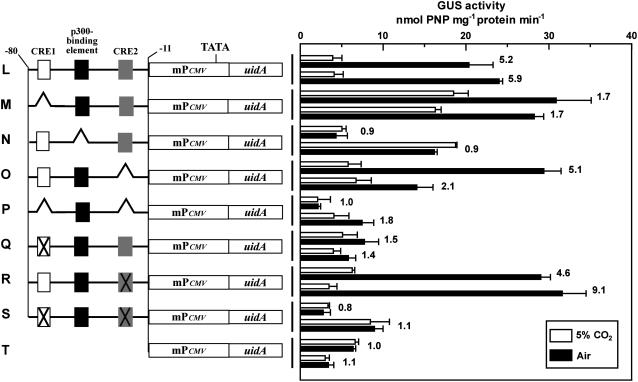
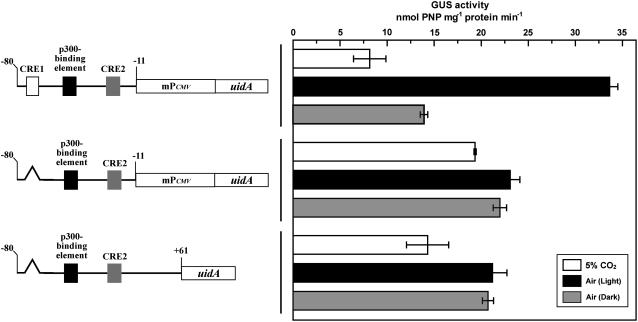
We further followed up the function of Pptca1 with a substituted minimal promoter. The region, −10 to +61 relative to the transcription start site of Pptca1, containing the initiator-like sequence (CACA) at −2 to +2 was substituted with the human cytomegalovirus (CMV) minimal promoter (mPCMV), which is a TATA-type promoter (Fig. 4, left). The CO2 responses of the core regulatory region of Pptca1 (−80 to −11) were well conserved (Fig. 4L) and were not changed significantly by deleting or substituting CRE2 (Fig. 4, O and R). Both deletion and/or antisense substitution of CRE1 and p300-binding element again revealed the apparent tendency of uidA derepression under 5% CO2, and uidA expression became constitutive by losing response to CO2 (Fig. 4, M, N, P, Q, and S). mPCMV alone did not show any CO2 response and was highly constitutive (Fig. 4T).
Figure 4.
Activities of the intact and manipulated core regulatory regions of Pptca1, which are driven by mPCMV. The putative minimal-promoter sequence of Pptca1 and the 5′-untranslated region (−11 to +61), which includes the putative initiator region, was substituted by the minimal-promoter region of CMV. The left and the right halves indicate constructs and GUS activities in each transformant, respectively. L to S, Substituted forms of the constructs A, B, D, G, H, I, J, and K in Figure 3; T, uidA ligated to mPCMV alone. The 5% CO2-grown cells (white bars) were transferred to air and were acclimated to air in the light for 1 d (black bars). Deleted and substituted portions are indicated as broken lines and cross marks, respectively. Values are means ± sd of three separate experiments. GUS activity ratios (air/5% CO2) are shown to the right of bars.
Responses of Pptca1 to the cAMP Analog and the cAMP Phosphodiesterase Inhibitor
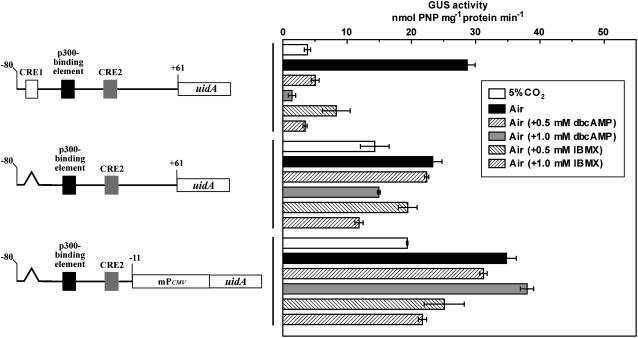
To clarify whether cAMP is involved in the CO2-responsive regulation of ptca1 expression, responses to an introduced Pptca1 core regulatory region, with or without CRE1, were analyzed using uidA as the reporter during acclimation of P. tricornutum cells from 5% CO2 to air, in the presence of 0.5 and 1.0 mm of the cAMP analog dibutyryl cAMP (dbcAMP) or the cAMP phosphodiesterase inhibitor 3-isobutyl-1-methylxanthine (IBMX). Interestingly, in the transformant with a complete Pptca1 core regulatory region, induction of GUS activity during acclimation to air was largely abolished by treatment of P. tricornutum cells with these drugs in a dose-dependent manner. That is, GUS activity was 18% and 5% of that in untreated air-acclimated cells in the presence of 0.5 and 1.0 mm dbcAMP, respectively, and, similarly, 28% or 12% in the presence of 0.5 and 1.0 mm IBMX, respectively (Fig. 5, top). In sharp contrast, uidA expression was insensitive to the strong repressive effects of these two drugs in the transformant with deleted CRE1, but was almost fully derepressed in air-acclimated cells under these drug treatments or in 5% CO2-grown cells without drugs (Fig. 5, middle). The same held true when CRE1-deleted Pptca1 was driven by mPCMV (Fig. 5, bottom).
Figure 5.
Activities of the CRE1-deleted Pptca1 during acclimation from 5% CO2 to air under illumination in the presence or the absence of dbcAMP or IBMX. The left and the right halves indicate constructs and GUS activities, respectively. Two different concentrations of dbcAMP and IBMX were used. Top, A transformant with construct A; middle, a transformant with construct B; bottom, a transformant with construct M. The 5% CO2-grown cells (white bars) were transferred to air and allowed to acclimate to air for 1 d in the absence (black bars) or the presence of drugs (striped and gray bars). Deleted portions are indicated as broken lines. Types of drug treatment are indicated in the inset. Values are means ± sd of three separate experiments.
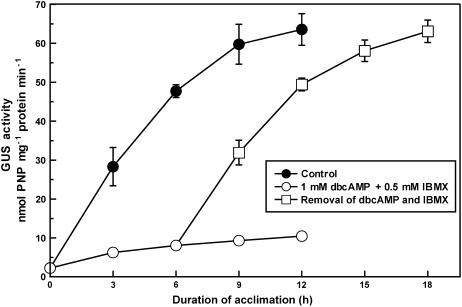
To examine the direct toxicity of dbcAMP and IBMX to P. tricornutum cells, 5% CO2-grown cells were transferred to air in the presence of both 1.0 mm dbcAMP and 0.5 mm IBMX, and these drugs were removed from the bulk medium after 6 h of acclimation to air. GUS expression was quickly derepressed upon removal of these drugs and increased for the next 12 h (Fig. 6, squares). The time course of the derepression was similar to that in air-acclimating cells without the drug treatment (Fig. 6, black circles and squares), whereas cells maintained under these drugs did not express GUS (Fig. 6, white circles).
Figure 6.
Recovery of GUS reporter expression after the removal of dbcAMP and IBMX in air. The 5% CO2-grown cells of P. tricornutum transformant containing the reporter construct A (Fig. 3A) were transferred to air in the presence (white circles) or the absence (black circles) of 1.0 mm dbcAMP and 0.5 mm IBMX. Cells were allowed to acclimate to air for 12 h, and GUS activity was measured at every 3 h after starting the acclimation. A part of air-acclimating cells under the drug treatment was washed at 6 h of acclimation with CO2-free F/2ASW to remove dbcAMP and IBMX from the medium and was allowed to acclimate to air for the next 12 h (white squares). GUS activity was measured as described in the text. Acclimation was carried out at 20°C under the photon-flux density of 50 μmol m−2 s−1. Values are means ± sd of three separate experiments.
Repression of the Endogenous ptca1 by dbcAMP and/or IBMX in Air
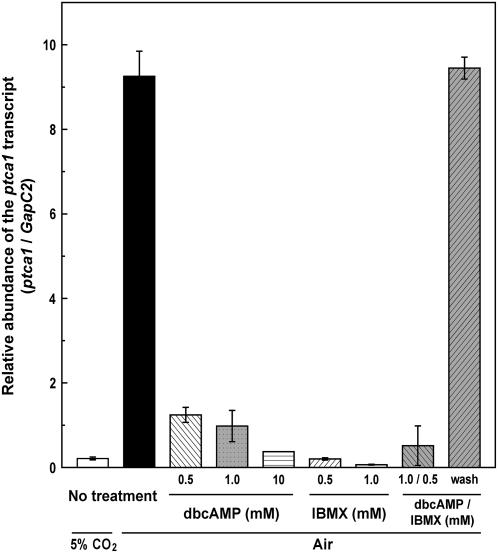
Responses of accumulations of the endogenous ptca1 transcript to the treatment with dbcAMP and/or IBMX were examined during acclimation of 5% CO2-grown cells to air (Fig. 7). As described by Harada and Matsuda (2005) previously, the ptca1 transcript was trace when grown in 5% CO2, whereas it accumulated about 40-fold of that in 5% CO2-grown cells after acclimation to air for 2 d (Fig. 7). Levels of the ptca1 transcript were found to be about 13%, 11%, and 4% of those in air-acclimated cells when 0.5, 1.0, and 10 mm dbcAMP, respectively, were added to the medium during acclimation to air (Fig. 7). Similarly, accumulation of the ptca1 transcript dropped to below 3% of that in the air-acclimated cells when 0.5 and 1.0 mm IBMX were added during acclimation to air (Fig. 7). The transcript level of the endogenous ptca1 was also suppressed to about 5% of that in air-acclimated cells during acclimation to air under the treatment with both 1.0 mm dbcAMP and 0.5 mm IBMX for 6 h, but increased to a level that was equivalent to that in air-acclimated cells within 12 h in air after removal of these two drugs from the bulk medium (Fig. 7).
Figure 7.
Effects of dbcAMP and IBMX treatments on transcript levels of the endogenous ptca1 during acclimation from 5% CO2 to air. Wild-type cells of P. tricornutum grown in 5% CO2 were transferred to air and allowed to acclimate to air for 2 d at 20°C in the absence or presence of 0.5, 1.0, or 10 mm dbcAMP and/or 0.5 or 1.0 mm IBMX as indicated in the diagram. In a part of the experiment, an aliquot of air-acclimating cells under the treatment with both 1.0 mm dbcAMP and 0.5 mm IBMX for 6 h (1.0/0.5) was washed with F/2ASW and allowed to acclimate to air for the next 12 h (wash). Total RNA was extracted from each cell and relative amounts of the ptca1 product were quantified as ratios to levels of the GapC2 product as an internal standard using the quantitative real-time PCR technique. Values are means ± sd of three separate experiments.
Responses of Pptca1 to Light
We previously showed that about 60% of PtCA1 expression in air condition was repressed in the absence of light (Harada et al., 2005). The possibility of participation of a cAMP-related element to Pptca1 regulation in response to light was examined using uidA-reporter constructs with and without CRE1 (Fig. 8, left). The Pptca1 core regulatory region with added mPCMV showed a clear repression to about 43% the maximum air-level uidA expression in the dark (Fig. 8, top), but repression was totally lost by deleting CRE1, irrespective of the type of minimal promoter (Fig. 8, middle and bottom). It was also noted that high-CO2 condition in the light always showed slightly stronger repressive effect on Pptca1 than that in air in the dark, irrespective of the existence of CRE1 and of types of minimal-promoter sequence (Fig. 8).
Figure 8.
Activities of the intact or the CRE1-deleted Pptca1 during acclimation from 5% CO2 to air under illumination or in the dark. The left and the right halves indicate constructs and GUS activities, respectively. Top, A transformant with the construct L shown in Figure 4; middle, a transformant with the construct M shown in Figure 4; bottom, a transformant with the construct B shown in Figure 3. The 5% CO2-grown cells (white bars) were transferred to air and were allowed to acclimate to air in the light (black bars) or the dark (gray bars) for 1 d. Deleted portions are indicated as broken lines. Values are means ± sd of three separate experiments.
DISCUSSION
The promoter region of ptca1 was previously isolated and it was demonstrated that the critical CO2-response sequence was located downstream −70 bp relative to the transcription start site (Harada et al., 2005). This relatively short core regulatory sequence comprises two putative cAMP-response elements, CRE1 at −70 to −63 and CRE2 at −21 to −14, and a putative p300-binding element at −52 to −46 relative to the transcription start site (Harada et al., 2005; Fig. 1). A rough truncation experiment, which was previously done on this region, suggested that these elements would participate in repressive regulation of the ptca1 expression in response to increase in [CO2] (Harada et al., 2005). A precisely targeted deletion or an antisense substitution of CRE1, conducted in this study, was clearly related to derepressions of Pptca1 under 5% CO2 condition (Figs. 3, B and I, and 4, M and Q). Although GUS expression levels varied depending on clones presumably due to the position effects on inserted fragments, GUS expression ratios were found to be relatively stable and revealed characteristics of each manipulated construct of Pptca1. This apparent tendency of the Pptca1 derepression in high CO2 was also held unchanged in the case of double deletions or antisense substitutions of both CRE1 and CRE2 (Figs. 3, H and K, and 4, P and S). CRE2 is, however, unlikely to be relevant to the repressive activity of the Pptca1 core regulatory region under high-CO2 conditions, since a single deletion or substitution of the CRE2 region did not cause a reduction in the CO2 sensitivity of uidA expression (Fig. 3, G and J). It is also likely that the p300-binding region would participate significantly in the CO2 responses of Pptca1 (Figs. 3D and 4N). These data clearly indicate that the CO2 response of the Pptca1 core regulatory region operates irrespective of the structures of the minimal-promoter region and that CRE1 and the p300-binding element are required for the repression of ptca1 in response to increases in the ambient [CO2].
In this study, mPCMV was shown to operate in cells of the marine diatom P. tricornutum. GUS expression driven by mPCMV alone without Pptca1 was highly constitutive and did not show any CO2 response (Fig. 4T). The activity of mPCMV did not disturb the CO2-responsive regulatory function of Pptca1 (−80 to −11). These data clearly indicate that CO2 response of Pptca1 is not governed by the sequence between −10 and +61 relative to the transcription start site. It should be also noted that a mammalian viral promoter could be used as a molecular tool for marine diatoms.
The occurrences of CREs and the p300-binding element on the CO2-responsive promoter are the striking features that indicate the participation of cAMP in the CO2 response of marine diatoms. Our preliminary attempts, however, failed to detect significant changes in cAMP concentration in response to growth [CO2] in a cell lysate of P. tricornutum. Cyclic AMP is a highly redundant molecule that operates in many different signal transduction pathways as a second messenger, and, therefore, concentration changes in cAMP in a particular physiological response are not precisely detected in a whole-cell lysate. This consideration prompted us to quantify relative changes in expression levels of the GUS reporter gene, which is driven with manipulated Pptca1, under treatments with modulator reagents for levels and activities of the cytosolic cAMP. The results obtained in this study clearly showed that either supply of the cAMP analog dbcAMP or an enrichment of the steady-state level of cAMP by inhibiting cAMP degradation with IBMX strongly abolished GUS expression in the transformant with a complete Pptca1 core regulatory region during acclimation to air (Fig. 5). These repressive effects by high CO2 or by these two drugs on Pptca1 largely disappeared in the absence of CRE1, and uidA was derepressed under these treatments (Fig. 5). Furthermore, dbcAMP and IBMX showed little toxicity to cells of P. tricornutum (Figs. 6 and 7). It is thus likely that cAMP is the key component in transmitting the high-CO2 signal to repress Pptca1 via the function of CRE1 in P. tricornutum. It is also noteworthy that increases in the dose of dbcAMP and IBMX from 0.5 to 1.0 mm exhibited trace repressive effects on the CRE1-deleted Pptca1 (Fig. 5). This probably indicates that CRE1 alone is not sufficient to account for the repressive effects of dbcAMP and IBMX but requires additional elements, such as the p300-binding element, for full function. It is also confirmed that dbcAMP and IBMX strongly repressed the transcription of the endogenous ptca1 even under air-level CO2 condition (Fig. 7), indicating clearly that increase in the cytosolic cAMP level plays a key role in repression of the native ptca1 promoter.
Responses of CCM components to light in eukaryotic algae are also diverse, but the absence of light is strongly or moderately repressive in many algal species (Matsuda and Colman, 1995; Villand et al., 1997; Kucho et al., 1999; Bozzo and Colman, 2000; Harada et al., 2005). However, the light signaling pathway in CCM regulation has not been elucidated in detail. In the marine diatom P. tricornutum, the ptca1 gene is repressed in the dark to about 40% of that under illumination (Harada et al., 2005), but the promoter sequence of ptca1 is not similar to any of known CO2-responsive promoters (Villand et al., 1997; Kucho et al., 1999; Harada et al., 2005). This indicates that the origins of CO2-response systems are also diverse. The Pptca1 core regulatory region with added mPCMV showed a clear repressive effect on uidA expression in the dark (Fig. 8, top), but the repression was totally lost by deleting CRE1 irrespective of the type of minimal promoter (Fig. 8, middle and bottom). It is strongly suggested from these results that cAMP pathways transmit both CO2 and light signals via CRE1. Derepressions of uidA expression by deleting CRE1 were almost 100% and GUS levels reached the maximum in these transformants grown in the dark (Fig. 8, middle and bottom), whereas trace repressions of uidA were always observed in high CO2 in the light in these CRE1-deleted transformants (Fig. 8, middle and bottom). It is probable from these results that the repression of Pptca1 in the absence of light is governed primarily by CRE1 but no other cis-element would be required, whereas the CO2 response, as described above, would require cooperation of other regulatory elements presumably related to cAMP. This consideration in turn makes us question whether light and CO2 signals might be transmitted to Pptca1 in different ways via transcription factors that interact either primarily with CRE1 or cooperatively with both CRE1 and other regulatory elements.
In animal cells, CRE has been reported as a cis-element that is targeted by the CRE-binding protein, CREB, whose phosphorylated form binds to the CREB-binding protein, CBP (Chrivia et al., 1993), and stimulates the formation of a basal transcriptional complex (Kwok et al., 1994). This process operates typically in transcriptional activations in response to hormones, neurotransmitters, and olfactants. The p300 is known as a functional homolog of CBP and also activates transcriptional processes in mammalian development by interacting with CREB (Rikitake and Moran, 1992; Tanaka et al., 1997). All of these animal systems, mediated by cAMP and CRE-related factors, function to activate transcriptions. In contrast, examples of repressive regulation of cAMP-mediated gene expression are limited. One of the best known cases is the catabolite-repression model in yeast, in which exposures to some favorable organic carbon sources, such as Glc, cause increases in cytosolic cAMP level and in turn repress genes controlling bioenergetic pathways, such as alcohol dehydrogenase and Fru-1,6-bisphosphatase (Cherry et al., 1989; Zaragoza et al., 1999). CCM repression in diatoms appears to be homologous to the catabolite-repression model as it is a down-regulation model by excess substrate for photosynthesis. However, catabolite repression in yeast has not been related to CRE or p300-binding element. Repression of Pptca1 is thus a new repression model by cAMP via a CRE-related promoter sequence.
Molecular research on acclimation of marine diatoms to changes in environmental factors is extremely important considering its ecophysiological significance, although it is still at a rudimentary stage. Responses of marine diatoms to changes in physicochemical factors, such as fluid motion, osmotic stress, iron, light, and [CO2], might be controlled by specific receptors and feedback mechanisms probably mediated, at the cytosolic level, by second messengers, such as Ca2+ (Falciatore et al., 2000) and cAMP (this study). It is also suggested in this study that there might be a fine discrimination of the CO2 signal from the light signal at the promoter level in a marine diatom. CO2 input at the ocean surface may thus act as a direct signal to the marine ecosystem as one of possible regulatory factors of primary productivity.
MATERIALS AND METHODS
Cells and Culture Conditions
The marine diatom Phaeodactylum tricornutum Bolin (UTEX 642) was obtained from the University of Texas Culture Collection and was grown in artificial seawater, which was supplemented with half-strength Guillard's “f” solution (F/2ASW; Guillard and Ryther, 1962; Harrison et al., 1980) under continuous illumination (50 μmol m−2 s−1) at 20°C with constant aeration with 5% CO2 or ambient air. Fifty milliliters of 5% CO2-grown cells were centrifuged at 3,500g at 20°C for 5 min, washed twice with 10 mL of CO2-free F/2ASW, resuspended in 50 mL of CO2-free F/2ASW, and allowed to acclimate to air by aeration with ambient air for 1 to 2 d. In some experiments, 0.5, 1.0, or 10 mm dbcAMP and/or IBMX were added during the acclimation to air. Prior to the acclimation to air in the dark, 5% CO2-grown cells were dark adapted for 1 h in 5% CO2.
Preparation of Chimeric Constructs
The upstream region of the ptca1 open reading frame was isolated as described previously (Harada et al., 2005). Using this region as a template, the core regulatory region, −80 to +61 bp relative to the transcription start site, was amplified by PCR. The core regulatory region was phosphorylated and inserted into the blunt-ended site of transformation vector pFcpApGUS (Harada et al., 2005; Fig. 3A). To create deletion constructs (Fig. 3, B–H), PCR primers were designed to perform an inverse elongation from the sequence neighboring each deletion site in the Pptca1 (Fig. 3, B–H) and PCR was done using construct A as a template. A double-deletion construct (Fig. 3H) was created in the same way as above using construct B as a template. To create constructs I to K (Fig. 3), PCR primers that contain antisense substitution at the 5′ termini were designed and amplified by PCR using construct A as a template. A double-substitution construct (Fig. 3K) was created by PCR on construct I as a template. To create constructs L to S (Fig. 4), the deleted or the substituted constructs of Pptca1 (−80 to −11), which lack the region containing the initiator and the omega sequence (−10 to +61), were amplified on templates of constructs A to K (Fig. 3) and fused with mPCMV by PCR. These fragments were then phosphorylated and inserted into the blunt-ended site of pFcpApGUS. As a control, uidA was fused with mPCMV alone and designated as construct T (Fig. 4).
Transformation of P. tricornutum
Transformation was carried out as described by Zaslavskaia et al. (2000). P. tricornutum cells grown in 5% CO2 under continuous illumination were harvested at the mid-logarithmic phase (OD730 = 0.3–0.4). Approximately 5 × 107 cells were spotted as a plaque of 2.5-cm diameter on the surface of the F/2ASW agar plate. Five hundred micrograms of tungsten microcarriers (1.1 μm particle size) were coated with approximately 1.0 μg of plasmid DNA containing 1.0 m CaCl2 and 16 mm spermidine. PDS-1000/He biolistic particle-delivery system (Bio-Rad) was used for microprojectile bombardments of microcarriers. The bombardment was done at 1,550 psi to the cells in the chamber under the pressure of −27 inches mercury with a target distance of 6 cm. Bombarded cells were cultured for 1 d under illumination and were suspended in 5 mL of F/2ASW. After the centrifugation at 3,000 rpm at 20°C for 5 min, cells were resuspended in 0.3 mL of F/2ASW, plated onto F/2ASW agar plate containing 100 μg mL−1 Zeocin.
GUS Assays
The transformants were cultured in 100 mL of F/2ASW in 5% CO2 or air. Five to 10 mL of cells were harvested and disrupted by a sonicator (Ultrasonic disruptor model UD-201; TOMY Seiko) at output level 3 in 0.5 mL of GUS extraction buffer [50 mm sodium phosphate, pH 7.0, 10 mm β-mercaptoethanol, 0.1% (w/v) sodium lauryl sarcosine, 0.1% (v/v) Triton X-100], and then centrifuged at 13,000 rpm for 5 min at 4°C. Twenty microliters of lysate was added to 980 μL of GUS assay buffer (10 mm p-nitrophenyl β-d-glucuronide in the GUS extraction buffer) and incubated at 37°C for 1 h. The reaction was terminated by the addition of 400 μL of 0.5 m Na2CO3 every 10 min after starting the reaction and the optical density of p-nitrophenol released from the substrate was measured at 405 nm.
Quantitative Real-Time PCR
Total RNA was extracted from 5% CO2-grown cells, air-acclimated cells, or cells treated with dbcAMP and/or IBMX during acclimation to air using RNeasy Plant Mini kit (QIAGEN) according to the protocol provided by the manufacturer. Total RNA (1 μg) was reverse transcribed using oligo(dT)20 primer (TOYOBO) and reverse transcriptase (Revertra Ace; TOYOBO) to form single-stranded cDNA. To amplify the uidA (accession no. S69414) and the ptca1 (accession no. AF414191) cDNAs, sets of forward and reverse primers, GUSRTF (5′-TTGCCAACGAACCGGATA-3′) and GUSRTR (5′-AATCGCCGCTTTGGACATAC-3′), and CA1F (5′-TCACAATTCCTAGCAGAAAATCATCG-3′) and CA1R (5′-ACGCATCCAATGTACAAGTACTTGGG-3′), were used for PCR. Quantitative PCR was standardized on known amounts of template using plasmids containing the uidA or the ptca1 cDNA. To normalize the quantification, the levels of the transcript of the cytosolic glyceraldehyde-3-P dehydrogenase gene (GapC2; accession no. AF063805), which is constitutively expressed under both 5% CO2 and air, were measured as an internal marker. Amplification of the GapC2 cDNA was done using the GapC2F (5′-TTTTTCGCCTTTCTAAACATCAGTT-3′) and GapC2R (5′-TACTCGGGCGTCAAGAAGG-3′) primer pair. The quantitative PCR was carried out with a Smart Cycler thermal cycler system (Cepheid) and Takara Ex Taq R-PCR Version 2.0 (Takara Bio) or SYBR Premix Ex Taq (Takara Bio) under PCR conditions as follows: for uidA, heating at 95°C for 10 s, followed by 45 cycles of denaturing at 95°C for 5 s, annealing at 64°C for 20 s, and extension at 72°C for 10 s; and for ptca1 and GapC2, same conditions, except annealing at temperatures of 63°C and 60°C, respectively.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers S69414, AF414191, AF063805, and M64944.
Acknowledgments
We thank Dr. Brian Colman (York University, Toronto) for critical reading and comments on this manuscript, Ms. Nobuko Higashiuchi for her technical assistance, and Ms. Miyabi Inoue for her skillful secretarial aid.
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology, Japan (MEXT; Grant-in-Aid for Scientific Research B no. 18310014 to Y.M.); by the Showa-Shell Sekiyu Environmental Research Foundation (to Y.M.); by the Program for Research on Halophilic Organism of the Salt Science Research Foundation (grant no. 05B02 to Y.M.); and by the University-Industry Joint Research Project from MEXT (to Kwansei-Gakuin University, Nano-Biotechnology Research Centre).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Yusuke Matsuda (yusuke@ksc.kwansei.ac.jp).
References
- Badger MR, Andrews TJ, Whitney SM, Ludwig M, Yellowlees DC, Leggat W, Price GD (1998) The diversity and co-evolution of Rubisco, plastids, pyrenoids and chloroplast-based CCMs in the algae. Can J Bot 76: 1052–1071 [Google Scholar]
- Badger MR, Kaplan A, Berry JA (1980) Internal inorganic carbon pool of Chlamydomonas reinhardtii: evidence for a carbon dioxide concentrating mechanism. Plant Physiol 66: 407–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzo GG, Colman B (2000) The induction of inorganic carbon and external carbonic anhydrase in Chlamydomonas reinhardtii is regulated by external CO2 concentration. Plant Cell Environ 23: 1137–1144 [Google Scholar]
- Bozzo GG, Colman B, Matsuda Y (2000) Active transport of CO2 and bicarbonate is induced in response to external CO2 concentration in the green alga Chlorella kessleri. J Exp Bot 349: 1341–1348 [PubMed] [Google Scholar]
- Chen Y, Cann MJ, Litvin TN, Iourgenko V, Sinclair ML, Levin LR, Buck J (2000) Soluble adenylyl cyclase as an evolutionarily conserved bicarbonate sensor. Science 289: 625–628 [DOI] [PubMed] [Google Scholar]
- Cherry JR, Johnson TR, Dollard JR, Shuster JR, Denis CL (1989) Cyclic AMP-dependent protein kinase phosphorylates and inactivates the yeast transcriptional activator ADR1. Cell 56: 409–419 [DOI] [PubMed] [Google Scholar]
- Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Gooman RH (1993) Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365: 855–859 [DOI] [PubMed] [Google Scholar]
- Colman B, Rotatore C (1995) Photosynthetic inorganic carbon uptake and accumulation in two marine diatoms. Plant Cell Environ 18: 919–924 [Google Scholar]
- Eriksson M, Villand P, Garderström P, Samuelsson G (1998) Induction and regulation of expression of a low-CO2-induced mitochondrial carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol 116: 637–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falciatore A, D'Alcalà MR, Croot P, Bowler C (2000) Perception of environmental signals by a marine diatom. Science 288: 2363–2366 [DOI] [PubMed] [Google Scholar]
- Falkowski P, Scholes RJ, Boyle E, Canadell J, Canfield D, Elser J, Gruber N, Hibbard K, Hogbeg P, Linder S, et al (2000) The global carbon cycle: a test of our knowledge of Earth as a system. Science 290: 291–296 [DOI] [PubMed] [Google Scholar]
- Guillard RRL, Ryther JH (1962) Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt and Detonula confervacea (Cleve) Gran. Can J Microbiol 8: 229–239 [DOI] [PubMed] [Google Scholar]
- Hammer A, Hodgson DRW, Cann MJ (2006) Regulation of prokaryotic adenylyl cyclases by CO2. Biochem J 396: 215–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Matsuda Y (2005) Identification and characterization of a new carbonic anhydrase in the marine diatom Phaeodactylum tricornutum. Can J Bot 83: 909–916 [Google Scholar]
- Harada H, Nakatsuma D, Ishida M, Matsuda Y (2005) Regulation of the expression of intracellular β-carbonic anhydrase in response to CO2 and light in the marine diatom Phaeodactylum tricornutum. Plant Physiol 139: 1041–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PJ, Waters RE, Taylor FJR (1980) A broad spectrum artificial seawater medium for coastal and open ocean phytoplankton. J Phycol 16: 28–35 [Google Scholar]
- Jaiswal BS, Conti M (2003) Calcium regulation of the soluble adenylyl cyclase expressed in mammalian spermatozoa. Proc Natl Acad Sci USA 100: 10676–10681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AM, Raven JA (1996) Inorganic carbon accumulation by the marine diatom Phaeodactylum tricornutum. Eur J Phycol 31: 285–290 [Google Scholar]
- Kaplan A, Badger MR, Berry JA (1980) Photosynthesis and intracellular inorganic carbon pool in the blue-green algae Anabaena variabilis: response to external CO2 concentration. Planta 149: 219–226 [DOI] [PubMed] [Google Scholar]
- Kaplan A, Reinhold L (1999) CO2 concentrating mechanisms in photosynthetic microorganisms. Annu Rev Plant Physiol Plant Mol Biol 50: 539–570 [DOI] [PubMed] [Google Scholar]
- Kucho K, Ohyama K, Fukuzawa H (1999) CO2-responsive transcriptional regulation of CAH1 encoding carbonic anhydrase is mediated by enhancer and silencer regions in Chlamydomonas reinhardtii. Plant Physiol 121: 1329–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok RP, Lundblad JR, Chrivia JC, Richards JP, Bachinger HP, Brennan RG, Roberts SG, Green MR, Goodman RH (1994) Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature 370: 223–226 [DOI] [PubMed] [Google Scholar]
- Marcus Y, Harel E, Kaplan A (1983) Adaptation of the cyanobacterium Anabaena variabilis to low CO2 concentration in their environment. Plant Physiol 71: 208–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S, Ono T (2005) Adenylyl cyclase activity of Cya1 from the cyanobacterium Synechocystis sp. strain PCC 6803 is inhibited by bicarbonate. J Bacteriol 187: 5032–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y, Bozzo GG, Colman B (1998) Regulation of dissolved inorganic carbon transport in green algae. Can J Bot 76: 1072–1083 [Google Scholar]
- Matsuda Y, Colman B (1995) Induction of CO2 and bicarbonate transport in green alga Chlorella ellipsoidea. Evidence for induction in response to external CO2 concentration. Plant Physiol 108: 253–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y, Hara T, Colman B (2001) Regulation of the induction of bicarbonate uptake by dissolved CO2 in the marine diatom Phaeodactylum tricornutum. Plant Cell Environ 24: 611–620 [Google Scholar]
-
Miller AG, Espie GS, Canvin DT (1990) Physiological aspects of CO2 and
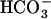 transport by cyanobacteria: a review. Can J Bot 68: 1291–1302 [Google Scholar]
transport by cyanobacteria: a review. Can J Bot 68: 1291–1302 [Google Scholar] - Miura K, Yamano T, Yoshioka S, Kohinata T, Inoue Y, Taniguchi F, Asamizu E, Nakamura Y, Tabata S, Yamato KT, et al (2004) Expression profiling-based identification of CO2-responsive genes regulated by CCM1 controlling a carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Physiol 135: 1595–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroney JV, Somanchi A (1999) How do algae concentrate CO2 to increase the efficiency of photosynthetic carbon fixation? Plant Physiol 119: 9–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat M, Moroney JV (1995) The regulation of carbonic anhydrase and ribulose-1,5-bisphosphate carboxylase/oxygenase activase by light and CO2 in Chlamydomonas reinhardtii. Plant Physiol 109: 937–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikitake Y, Moran E (1992) DNA-binding properties of the ElA-associated 300-kilodalton protein. Mol Cell Biol 12: 2826–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh D, Hiraoka Y, Colman B, Matsuda Y (2001) Physiological and molecular biological characterization of intracellular carbonic anhydrase from the marine diatom Phaeodactylum tricornutum. Plant Physiol 126: 1459–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Nakatsuma D, Harada H, Ishida M, Matsuda Y (2005) Localization of soluble β-carbonic anhydrase in the marine diatom Phaeodactylum tricornutum. Sorting to the chloroplast and cluster formation on the girdle lamellae. Plant Physiol 138: 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Naruse I, Maekawa T, Masuya H, Shiroishi T, Ishii S (1997) Abnormal skeletal patterning in embryos lacking a single Cbp allele: a partial similarity with Rubinstein-Taybi syndrome. Proc Natl Acad Sci USA 94: 10215–10220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortell PD, Reinfelder JR, Morel FMM (1997) Active uptake of bicarbonate by diatoms. Nature 390: 243–2449384376 [Google Scholar]
- Tréguer P, Nelson D, Van Bennekom A, DeMaster D, Leynaert A, Quéguiner B (1995) The silica balance in the world ocean: a reestimate. Science 268: 375–379 [DOI] [PubMed] [Google Scholar]
- Vance P, Spalding MH (2005) Growth, photosynthesis, and gene expression in Chlamydomonas over a range of CO2 concentrations and CO2/O2 ratios: CO2 regulates multiple acclimation states. Can J Bot 83: 796–809 [Google Scholar]
- Villand P, Eriksson M, Samuelsson G (1997) Carbon dioxide and light regulation of promoters controlling the expression of mitochondrial carbonic anhydrase in Chlamydomonas reinhardtii. Biochem J 327: 51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodger FJ, Badger MR, Price GD (2005) Sensing of inorganic carbon limitation in Synechococcus PCC7942 is correlated with the size of the internal inorganic carbon pool and involves oxygen. Plant Physiol 139: 1959–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O, Lindley C, Gancedo JM (1999) Cyclic AMP can decrease expression of genes subject to catabolite repression in Saccharomyces cerevisiae. J Bacteriol 181: 2640–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaslavskaia LA, Lippmeier JC, Kroth PG, Grossman AR, Apt KE (2000) Transformation of the diatom Phaeodactylum tricornutum (Bacillariophyceae) with a variety of selectable marker and reporter genes. J Phycol 36: 379–386 [Google Scholar]