Abstract
Despite much study of the role of S-adenosylmethionine (SAM) in the methylation of DNA, RNA, and proteins, and as a cofactor for a wide range of biosynthetic processes, little is known concerning the intracellular transport of this essential metabolite. Screening of the Arabidopsis (Arabidopsis thaliana) genome yielded two potential homologs of yeast (Saccharomyces cerevisiae) and human SAM transporters, designated as SAMC1 and SAMC2, both of which belong to the mitochondrial carrier protein family. The SAMC1 gene is broadly expressed at the organ level, although only in specialized tissues of roots with high rates of cell division, and appears to be up-regulated in response to wounding stress, whereas the SAMC2 gene is very poorly expressed in all organs/tissues analyzed. Direct transport assays with the recombinant and reconstituted SAMC1 were utilized to demonstrate that this protein displays a very narrow substrate specificity confined to SAM and its closest analogs. Further experiments revealed that SAMC1 was able to function in uniport and exchange reactions and characterized the transporter as highly active, but sensitive to physiologically relevant concentrations of S-adenosylhomocysteine, S-adenosylcysteine, and adenosylornithine. Green fluorescent protein-based cell biological analysis demonstrated targeting of SAMC1 to mitochondria. Previous proteomic analyses identified this protein also in the chloroplast inner envelope. In keeping with these results, bioinformatics predicted dual localization for SAMC1. These findings suggest that the provision of cytosolically synthesized SAM to mitochondria and possibly also to plastids is mediated by SAMC1 according to the relative demands for this metabolite in the organelles.
S-adenosylmethionine (SAM), the second most widely used enzyme substrate after ATP (Cantoni, 1975), is the methyl group donor for almost all biological methylation reactions. It is also used as a source of methylene groups (in the synthesis of cyclopropyl fatty acids), amino groups (in the synthesis of 7,8-diaminoperlagonic acid), ribosyl groups (in the synthesis of epoxyqueuosine), and aminopropyl groups (in the synthesis of ethylene and polyamines; Fontecave et al., 2004).
Both in chloroplasts and mitochondria, SAM is needed for the methylation of DNA, RNA, and proteins (Montasser-Kouhsari et al., 1978; Black et al., 1987; Kobayashi et al., 1990; Block et al., 2002) and in chloroplasts for the methylation of Rubisco and for the activity of the methyltransferases involved in tocopherol and plastoquinone synthesis (d'Harlingue and Camara, 1985; Grimm et al., 1997; Ying et al., 1999; Cheng et al., 2003). Moreover, SAM is required as an essential cofactor in mitochondria for the activity of biotin synthase and lipoic acid synthase (Miller et al., 2000; Picciocchi et al., 2001, 2003; Yasuno and Wada, 2002), and in plastids for the activity of lipoic acid synthase and Thr synthase (Wallsgrove et al., 1983; Curien et al., 1996; Laber et al., 1999; Yasuno and Wada, 2002).
In plants, the genes and the enzymes involved in the metabolism of SAM have been characterized (Ravanel et al., 1998; Hanson and Roje, 2001; Hesse and Hoefgen, 2003); however, their subcellular distribution requires a network connection between cytosol, mitochondria, and chloroplasts. Because in Arabidopsis (Arabidopsis thaliana) SAM is synthesized from ATP and Met by four synthetases that are widely accepted to be localized exclusively in the cytosol (Schröder et al., 1997; Ravanel et al., 1998; Hanson and Roje, 2001), the SAM required in the chloroplasts and mitochondria must be imported from the cytosol. Furthermore, S-adenosylhomocysteine (SAHC) produced from SAM during methylation reactions, is a potent competitive inhibitor of methyltransferases (Weretilnyk et al., 2001) and, in plants, as shown in yeast (Saccharomyces cerevisiae) SAHC hydrolase is most probably present exclusively in the cytosol (Ravanel et al., 1998; Hanson et al., 2000; Hanson and Roje, 2001). Therefore, it was proposed that chloroplasts and mitochondria have to export SAHC to the cytosol to maintain the SAM-to-SAHC ratio that regulates methyltransferase activity (Ravanel et al., 2004). Recently, in our laboratory, a mitochondrial carrier, which is able to import SAM and export SAHC into and from the mitochondria in yeast and man (Marobbio et al., 2003; Agrimi et al., 2004), has been cloned and characterized; however, in plant mitochondria or chloroplasts, the one or more proteins responsible for this transport activity have not been hitherto isolated or identified at the genetic level.
In this study, we provide evidence that the gene products of At4g39460 and At1g34065, named SAMC1 and SAMC2, respectively, are two isoforms of a SAM transporter in Arabidopsis. These proteins are 325 and 321 amino acids long, respectively, possess the characteristic sequence features of the mitochondrial carrier family (Millar and Heazlewood, 2003; Fernie et al., 2004; Palmieri, 2004; Picault et al., 2004) and display a high degree (75%) of homology. The SAMC1 protein was expressed in Escherichia coli, purified, reconstituted in phospholipid vesicles, and identified from its transport properties as a carrier for SAM. SAMC1 mRNA is expressed at higher levels than SAMC2 RNA in autotrophic and heterotrophic tissues. The green fluorescent protein (GFP) fused to SAMC1 was found to be targeted to mitochondria. However, a proteomic study demonstrated previously that SAMC1 is a component of the chloroplast envelope (Ferro et al., 2002). These findings are consistent with the subcellular localizations predicted by targeting prediction programs and suggest that SAMC1 could be dual targeted.
RESULTS
Isolation and Characterization of SAMC1 and SAMC2
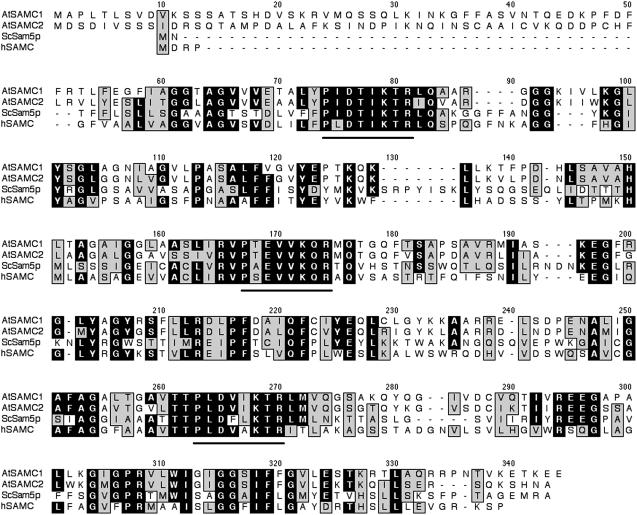
By screening the Arabidopsis genome (http://www.arabidopsis.org) with the sequences of yeast Sam5p (Marobbio et al., 2003) and human SAMC (Agrimi et al., 2004), two homologs have been identified: At4g39460 (GenBank AAK76726) and At1g34065 (GenBank NP_680566), hereafter named SAMC1 and SAMC2, respectively. The SAMC1 and SAMC2 coding sequences were amplified by PCR from an Arabidopsis cDNA library (Minet et al., 1992). They encoded proteins of 325 and 321 amino acids, respectively, which are 64% identical to one another. The amplified SAMC2 differs from the sequence NP_680566 derived from conceptual translation in that it lacks residues 91 to 96 (NFSGWW) of the previously annotated sequence. The two novel Arabidopsis proteins belong to the mitochondrial carrier family because their amino acid sequences are composed of three tandem repeats of about 100 amino acids, each containing two transmembrane α-helices, linked by an extensive loop, and a conserved signature motif (see Fig. 1). However, unlike many members of this family, including Sam5p and SAMC, SAMC1 and SAMC2 appear to have a presequence of nearly 50 amino acids. They share 28% to 29% identical amino acids with Sam5p and SAMC, a percentage that increases to 31% to 34% based on the sequences of SAMC1 and SAMC2 without the first 49 residues. In The Arabidopsis Information Resource database, 28 expressed sequence tags (ESTs) were found for SAMC1, whereas no ESTs were found for SAMC2. During the preparation of this article, an EST has been released in the National Center for Biotechnology Information database (GenBank BP837715) that corresponds to 1 to 377 bp of the SAMC2 coding sequence, except for a few base discrepancies most likely due to the different ecotypes sequenced (Columbia [Col] versus Landsberg). Taken together, these data indicate that SAMC1 is expressed at a significantly higher level than SAMC2, although both genes are transcriptionally active.
Figure 1.
Alignment of the Arabidopsis SAMC1 and SAMC2 with the yeast Sam5p and the human SAMC. Black regions denote identical amino acids and gray regions conserved amino acids. Solid lines were placed below the conserved motif that is characteristic of the mitochondrial carrier protein family. AtSAMC1, Arabidopsis SAMC1; AtSAMC2, Arabidopsis SAMC2; ScSam5p, yeast Sam5p; hSAMC, human SAMC.
Expression Patterns of SAMC1 and SAMC2 in Arabidopsis
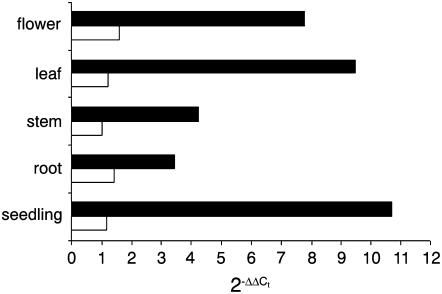
In a preliminary northern-blot analysis, SAMC1 expression was found in flowers, leaves, stems, and seedlings, whereas no SAMC2 expression was detected (data not shown). We then determined gene expression levels by real-time reverse transcription (RT)-PCR using the housekeeping elongation factor (EF) 1α gene as an internal control (Fig. 2). In all organs analyzed, SAMC1 was expressed at higher levels than SAMC2, consistent with digital and conventional northern analyses. The SAMC1 mRNA was expressed most strongly in seedlings. It was also expressed in leaves and flowers and, to a lesser extent, in stems and roots. SAMC2 mRNA was expressed at comparable low levels in all organs analyzed. These results are in close agreement with the data housed in publicly available microarray data collections (Zimmermann et al., 2004), except for stems, where we found a lower level of expression for SAMC1 than that predicted by Affimetrix and ATH1 gene chip arrays. To investigate organ specificity in more detail, the SAMC1 promoter region was fused transcriptionally to the β-glucuronidase (GUS) gene. During the first stage of vegetative growth, strong expression was observed in cotyledons of postgerminating seedlings in the base of the primary root and in the root tips (Fig. 3A, a). In young seedlings containing two to four true leaves (Fig. 3A, b–d), significant expression was found along the entire primary root, in the base and tips of the lateral roots (but not in the root hair), in the first two true leaves, and, to a lesser extent, in the cotyledons. Progressively reduced staining was observed in young and adult leaves on aging (Figs. 3A, c and e) and virtually no expression was found in senescent leaves, unless they were wounded or damaged (Fig. 3B), or stems (Fig. 3A, f). Expression was, however, also observed in sepals during the early stage of flower development and in mature flowers (Figs. 3A, f–h). No expression was found in petals, pollen grains, and the central septum of pollen tubes, whereas expression was clearly seen at the stigma of the pollen tubes (Figs. 3A, g–h). Furthermore, GUS expression was found in the silique vasculature (Fig. 3A, i). Similar studies performed with SAMC2 did not lead to any significant staining, confirming that SAMC2 is very weakly expressed.
Figure 2.
Expression of SAMC1 and SAMC2 in various organs. Real-time PCR experiments were conducted on cDNAs prepared by RT of total RNAs from various Arabidopsis organs, using gene-specific primers. Three independent preparations of total RNA (100 ng) from each organ were assayed in triplicate (ses were less than 10%). The relative quantification of SAMC1 (black bars) and SAMC2 (white bars) was performed according to the comparative method (2−ΔΔCt). EF1α was employed as a reference gene. For the calibrator (SAMC2 stem), ΔΔCt = 0 and 2−ΔΔCt = 1. For the remaining organs, the value of 2−ΔΔCt indicates the fold change in gene expression relative to the calibrator.
Figure 3.
GUS staining of Arabidopsis plants transformed with the SAMC1 promoter-GUS fusion. A, Histochemical analysis of promoter activity in seedlings (a–d), adult leaf (e), inflorescence stems (f), flowers (g and h), and siliques (i). Plants were harvested 8 (a), 11 (b), 13 (c), and 38 (e–i) d after germination. Details of the roots from the seedling in c are shown in d. B, GUS staining of wounded leaves from adult (a) and senescent (b and c) plants.
Bacterial Expression of SAMC Proteins
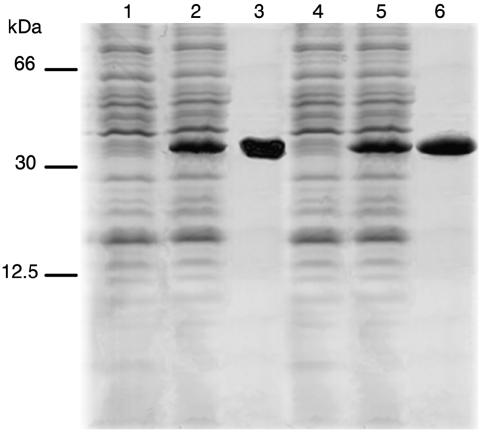
SAMC1 and SAMC2 proteins were expressed at high levels in E.coli BL21(DE3) (Fig. 4, lanes 2 and 5). They accumulated as inclusion bodies and were purified by centrifugation and washing (Fig. 4, lanes 3 and 6). The apparent molecular masses of the recombinant proteins were about 35 kD (the calculated values with initiator Met were 34,844 and 34,407 D). The identity of the purified proteins was confirmed by N-terminal sequence analysis. About 100 mg of each purified protein were obtained per liter of culture. The proteins were not detected in cells harvested after induction of expression but lacking the coding sequence in the vector (Fig. 4, lanes 1 and 4) nor in bacteria harvested immediately before induction (data not shown).
Figure 4.
Expression in E. coli and purification of SAMC1 and SAMC2. Proteins were separated by SDS-PAGE and stained with Coomassie Blue dye. The positions of the markers (bovine serum albumin, carbonic anhydrase, and cytochrome c) are shown on the left. Lanes 1 and 2, E. coli BL21(DE3) containing the expression vector with (lane 2) and without (lane 1) the coding sequence of SAMC1; lanes 4 and 5, E. coli BL21(DE3) containing the expression vector with (lane 5) and without (lane 4) the coding sequence of SAMC2. Samples were taken 5 h after induction. The same number of bacteria was analyzed in each sample. Lanes 3 and 6, Purified SAMC1 and SAMC2 (about 15 μg) proteins originated from bacteria shown in lanes 2 and 5, respectively.
Functional Characterization of SAMC1
The recombinant SAMC1 protein was reconstituted into liposomes and its transport activities for a variety of potential substrates were tested in homoexchange experiments (i.e. with the same substrate inside and outside). Using external and internal substrate concentrations of 1 and 10 mm, respectively, the recombinant and reconstituted protein catalyzed an active [3H]SAM/SAM exchange, which was inhibited by a mixture of pyridoxal-5′-P and bathophenanthroline. It did not catalyze homoexchanges for phosphate, carnitine, Glu, Asp, Met, Orn, malate, AMP, and ADP. Of importance, no [3H]SAM/SAM exchange activity was detected if SAMC1 had been boiled before incorporation into liposomes or if sarkosyl-solubilized material was reconstituted from bacterial cells either lacking the expression vector for SAMC1 or harvested immediately before induction of expression. In contrast to SAMC1, recombinant and reconstituted SAMC2 showed no activity under any of the experimental conditions tested, which include variation of the parameters that influence solubilization of the inclusion bodies and reconstitution of the protein into liposomes.
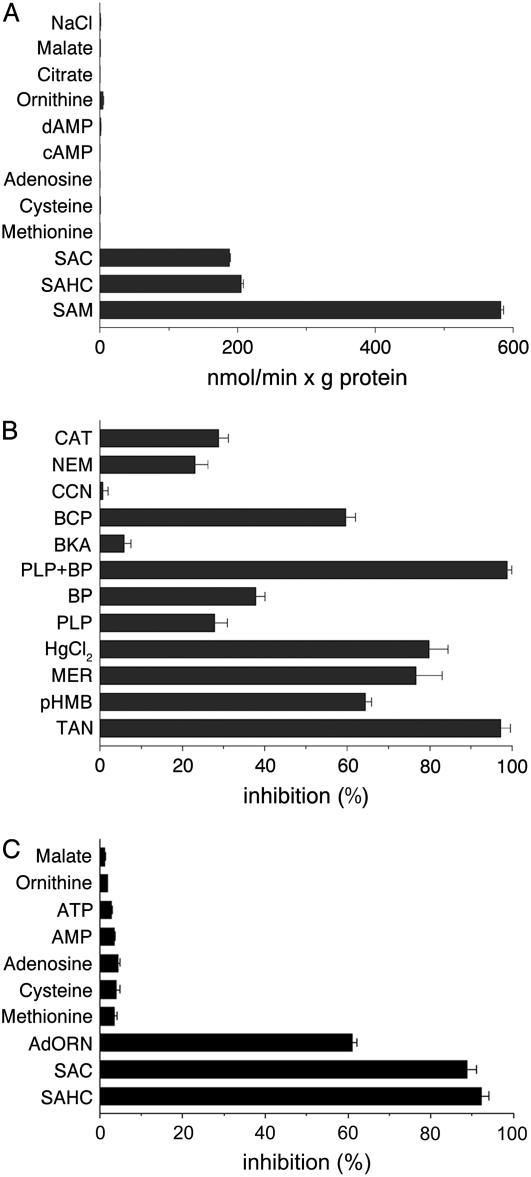
The substrate specificity of reconstituted SAMC1 was further investigated by measuring the rate of [3H]SAM uptake into proteoliposomes that had been preloaded with various potential substrates (Fig. 5A). The highest activity was observed in the presence of internal SAM. [3H]SAM was also efficiently taken up by proteoliposomes containing SAHC and S-adenosylcysteine. Negligible or zero activity was found in the absence of internal substrate (NaCl present) or in the presence of internal Met, Cys, adenosine, cAMP, dAMP, Orn, citrate, and malate. Alternative internal compounds 5′-deoxyadenosine, phosphate, pyruvate, oxoglutarate, Glu, Gln, thiamine, thiamine pyrophosphate, ADP, and NAD+ were also incapable of supporting [3H]SAM uptake (data not shown). Therefore, reconstituted SAMC1 displays a very narrow substrate specificity confined to SAM and its closest analogs.
Figure 5.
Substrate specificity and inhibitor sensitivity of SAMC1. Liposomes were reconstituted with SAMC1 and preloaded internally with various substrates (concentration, 10 mm) in A and with 10 mm SAM in B and C. Transport was started by the addition of 0.1 mm [3H]SAM and terminated after 30 s. A, Dependence of SAMC1 activity on internal substrate. The values are the means ± sd of at least three independent experiments. B, Effect of inhibitors on the [3H]SAM/SAM exchange. The final concentrations of the inhibitors, which were added 3 min before the labeled substrate, were 10 μm for carboxyatractyloside (CAT) and bongkrekic acid (BKA), 0.1 mm for p-hydroxymercuribenzoate (pHMB), mersalyl (MER), HgCl2, and bromocresol purple (BCP), 2 mm for pyridoxal-5′-phosphate (PLP), bathophenanthroline (BP), and N-ethylmaleimide (NEM), 1 mm for α-cyano-4-hydroxycinnamate (CCN), and 0.1% for tannic acid (TAN). Where indicated, PLP and BP were added together at concentrations of 40 mm and 16 mm, respectively. The extent of inhibition (%) is reported as the means ± sd of at least three independent experiments. C, Inhibition of the [3H]SAM/SAM exchange by external substrates. External substrates (concentration, 2 mm) were added together with [3H]SAM. The extent of inhibition (%) is reported as the means ± sd of at least three independent experiments. AdORN, Adenosylornithine; SAC, S-adenosylcysteine; SAHC, S-adenosylhomocysteine.
The [3H]SAM/SAM exchange reaction catalyzed by reconstituted SAMC1 was inhibited strongly by tannic acid, bromocresol purple, and the sulfydryl reagents mercuric chloride, mersalyl, and p-hydroxymercuribenzoate (Fig. 5B). It was also inhibited, to a lesser extent, by 2 mm N-ethylmaleimide, 2 mm bathophenanthroline, and 2 mm pyridoxal-5′-P. The latter two inhibitors displayed a synergistic effect, causing complete inhibition of transport activity when added together at higher concentrations. Carboxyatractyloside and bongkrekic acid, which are specific and powerful inhibitors of the mitochondrial ADP/ATP carrier (Klingenberg, 1989; Fiore et al., 1998), exhibited different behavior: carboxyatractyloside inhibited the activity of SAMC1 partially, whereas bongkrekic acid had little effect at a concentration (10 μm) that completely inhibits the ADP/ATP carrier. No inhibition was observed with the pyruvate carrier-specific inhibitor α-cyano-4-hydroxycinnamate (Halestrap, 1975), whereas [3H]SAM uptake was inhibited strongly by external addition of SAHC, S-adenosylcysteine, and adenosylornithine (sinefungin; Fig. 5C). In contrast, no significant inhibition was observed with external Met, Cys, adenosine, AMP, ATP, Orn, malate (Fig. 5C), or pyruvate, phosphate, oxoglutarate, and Asp (data not shown), when used at a concentration 20 times that of the labeled substrate.
Kinetic Characteristics of Recombinant SAMC1
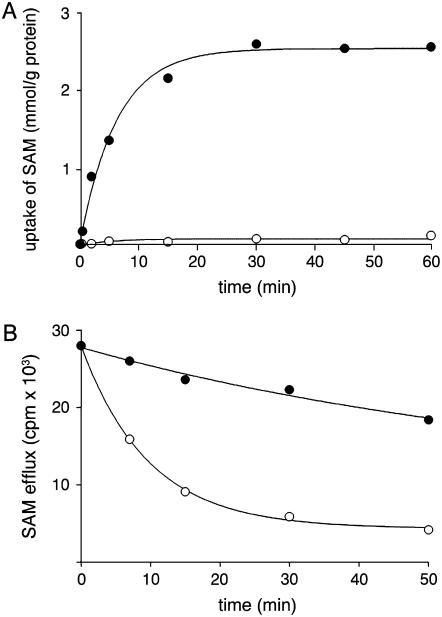
The uptake of [3H]SAM into proteoliposomes by exchange (in the presence of 10 mm internal SAM) followed first-order kinetics, isotopic equilibrium being approached exponentially (Fig. 6A). The rate constant and the initial rate of SAM exchange deduced from the time course (Palmieri et al., 1995) were 0.16 min−1 and 0.4 mmol min−1 g−1 protein, respectively. In contrast, the uniport uptake of SAM was very low in proteoliposomes preloaded with NaCl in the absence of internal substrate. The uniport mode of transport was further investigated by measuring the efflux of [3H]SAM from prelabeled active proteoliposomes because this provides a more convenient assay for unidirectional transport (Palmieri et al., 1995). In these experiments, little, yet significant, efflux of [3H]SAM from prelabeled proteoliposomes was observed in the absence of external substrate (Fig. 6B). The addition of 5 mm SAM induced a greater efflux of SAM (Fig. 6B), and the efflux either with or without added substrate was prevented by inhibitors of SAM transport (data not shown). Therefore, SAMC1 is able to catalyze both uniport and exchange reactions.
Figure 6.
Kinetics of [3H]SAM transport in proteoliposomes reconstituted with SAMC1. A, Uptake of SAM. 1 mm [3H]SAM was added to proteoliposomes containing 10 mm SAM (exchange, •) or 5 mm NaCl and no substrate (uniport, ○). Data are means ± sd for three independent experiments. B, Efflux of [3H]SAM from proteoliposomes reconstituted in the presence of 1 mm SAM. The internal substrate pool was labeled with [3H]SAM by carrier-mediated exchange equilibration. Then the proteoliposomes were passed through Sephadex G-75. The efflux of [3H]SAM was started by adding buffer A alone (•) or 5 mm SAM in buffer A (○). Data are means ± sd for three independent experiments.
The kinetic constants of the recombinant SAMC1 were determined by measuring the initial transport rate at various external [3H]SAM concentrations in the presence of a constant saturating internal concentration of 10 mm SAM. The transport affinity (Km) and specific activity (Vmax) values for SAM/SAM exchange at 25°C were 95 ± 19 μm and 1.2 ± 0.2 mmol min−1 g−1 protein, respectively, in 18 experiments. Externally added SAHC, S-adenosylcysteine, and adenosylornithine were competitive inhibitors of [3H]SAM uptake because they increased the apparent Km without changing the Vmax (data not shown). The inhibition constants (Ki) of SAHC, S-adenosylcysteine, and adenosylornithine were 20.8 ± 3.51 μm, 0.20 ± 0.08 mm, and 1.48 ± 0.33 mm, respectively, in at least three experiments for each inhibitor.
Subcellular Localization of SAMC1
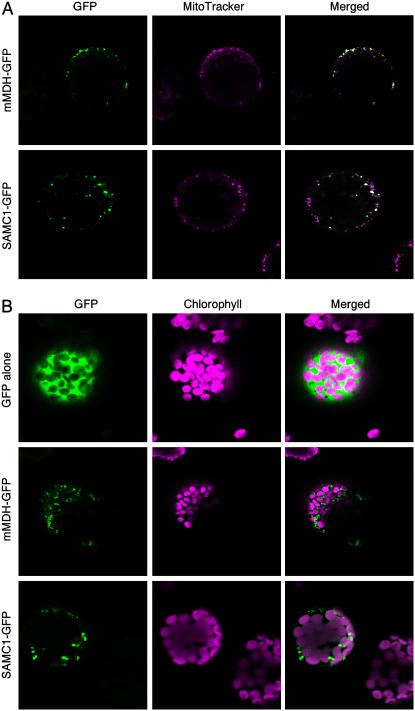
Some members of the mitochondrial carrier family are localized in nonmitochondrial membranes (Palmieri et al., 2001b; Bedhomme et al., 2005; Leroch et al., 2005; Weber et al., 2005). Using different computer prediction programs, the subcellular localization of SAMC1 was not unequivocal. For example, the ChloroP program predicted plastidial localization with high probability (score 0.85), whereas the iPSort program predicted mitochondrial localization also with high probability (score 0.75). Furthermore, the scores for the chloroplastic and mitochondrial localization obtained by the TargetP and Predotar programs were similar to those found for other proteins that are targeted to both organelles in Arabidopsis (Duchene et al., 2005). We therefore investigated the intracellular localization of ectopically expressed SAMC1 using a GFP-tagged protein (Fig. 7). Cells expressing SAMC1-GFP showed a punctate pattern of green fluorescence typical of the mitochondrial network that overlapped with the fluorescence displayed by the mitochondrial-specific dye, MitoTracker Red (Fig. 7A). This situation was similar to that obtained with GFP fused with the functionally characterized mitochondrial malate dehydrogenase (mMDH; Nunes-Nesi et al., 2005; Fig. 7A). In contrast, the green fluorescence of either SAMC1-GFP or mMDH-GFP did not colocalize with the red autofluorescence of chlorophyll (Fig. 7B). These data show that SAMC1 is efficiently targeted to mitochondria even though it does not exhibit a canonical presequence; they do not, however, refute earlier evidence (Ferro et al., 2002), suggesting that the transporter is also present in the chloroplast envelope.
Figure 7.
Subcellular localization of SAMC1. Transient expression in Arabidopsis protoplasts of GFP fused in frame with the full-length SAMC1 or mMDH. GFP alone and chlorophyll were used as controls for targeting to cytosol and plastids, respectively; GFP fused to mMDH and MitoTracker Red for targeting to mitochondria. Merged, Merged image of GFP fluorescence with MitoTracker Red fluorescence (A) and of GFP fluorescence and chlorophyll autofluorescence (B). The bars indicated in the figures represent 8 μm (GFP alone and SAMC1-GFP) and 20 μm (mMDH-GFP).
DISCUSSION
In this work, SAMC1 was shown, by direct transport assays, to be capable of transporting SAM and SAHC upon expression in E. coli and reconstitution into liposomes. This approach has been used for the identification of mitochondrial carriers from yeast (Palmieri et al., 2006a) and mammals (Palmieri, 2004), and has previously allowed the mitochondrial carriers for dicarboxylate/tricarboxylates and basic amino acids to be identified in Arabidopsis (Picault et al., 2002; Hoyos et al., 2003; Palmieri et al., 2006b). For the closely related sequence of SAMC2, which was also cloned and expressed in E. coli in this study, no biochemical data are available because we were incapable of renaturing and/or reconstituting it functionally. However, because of the high degree of homology between SAMC1 and SAMC2 (75% homology and 64% identical amino acids), which is similar to that found for other mitochondrial carrier isoforms (Fiermonte et al., 2002, 2003, 2004), and because of their close phylogenetic relationship (they form a unique clade among members of the mitochondrial carrier family in Arabidopsis; Picault et al., 2004), it is likely that SAMC2 is an isoform of the SAM transporter in Arabidopsis.
Similar to the yeast Sam5p and the human SAMC, SAMC1 transports SAM, SAHC, and, to a lesser extent, the nonphysiological structurally related compounds SAC and adenosylornitine, but none of the many other compounds tested. However, SAMC1 differs from yeast and human orthologs in several respects. SAMC1 protein catalyzes both the uniport and the exchange modes of transport like Sam5p, but differently from SAMC. The specific activity of SAMC1 is 1 order of magnitude higher than that of Sam5p and more than 2-fold higher than that of SAMC. The affinity of SAMC1 for SAM is similar to that of Sam5p, but 4.1-fold lower than that of SAMC. When comparing the effects of known mitochondrial carrier inhibitors on the three orthologs, the Arabidopsis SAM carrier exhibits a sensitivity to tannic acid and bromocresol purple much stronger than the yeast ortholog, but similar to that displayed by the human carrier. Carboxyatractyloside and bathophenantroline inhibit SAMC1 more than Sam5p and SAMC, whereas sensitivity to p-hydroxymercuribenzoate, mersalyl, and mercuric chloride is about the same for all three transporters. In addition, like its yeast and human counterparts, the Arabidopsis carrier is not inhibited by bongkrekic acid, α-cyano-4-hydroxycinnamate, and many compounds structurally related to SAM, such as amino acids, nucleosides, and nucleotides.
The green fluorescence of the GFP-tagged SAMC1 completely overlapped with the fluorescence mitochondrion-selective dye MitoTracker Red, demonstrating that the Arabidopsis SAMC1 transporter has a mitochondrial localization. However, the data available suggest that SAMC1 may have a dual mitochondrial and chloroplastic localization, as demonstrated for several plant proteins (for reviews, see Peeters and Small, 2001; Karniely and Pines, 2005; Mackenzie, 2005). First, using an elegant strategy based on the use of highly purified and characterized membrane fractions, extraction of hydrophobic proteins with organic solvents, SDS-PAGE separation, and tandem mass spectrometry analysis, Ferro et al. (2002) identified SAMC1 as a component of the chloroplast envelope. Second, the results reported in this article based on the use of GFP fused proteins show that this protein is colocalized with a mitochondrial marker. Whereas we were unable to detect fluorescence in the plastid, this may merely reflect a lack of sensitivity in the GFP assay. It has been shown that, in the presence of an uneven dual distribution, the large amount in one localization makes difficult the detection of the small amount in the other (Duchene et al., 2005; Regev-Rudzki et al., 2005). It is also possible that the GFP promoter fusion missed essential cis-elements that aid its targeting to the plastid, but are not necessary for mitochondrial targeting (Christensen et al., 2005; Kabeya and Sato, 2005). Two further lines of evidence support the dual localization of SAMC1. First, it was recently shown that, in isolated spinach (Spinacia oleracea) leaf chloroplasts, SAM is taken up by a transport system (Ravanel et al., 2004) that exhibits properties very similar to those displayed by the recombinant SAMC1. Second, bioinformatic analysis of the protein sequence revealed a long N-terminal extension of SAMC1, indicative of joint plastid and mitochondrial targeting.
Because mitochondrial and plastidic demands for this metabolite must be met by transport of cytosolically derived SAM (Schröder et al., 1997; Ravanel et al., 1998; Hanson and Roje, 2001), our data strongly suggest that a primary physiological role of SAMC1 in autotrophic and heterotrophic tissues is probably to catalyze the uptake of SAM in exchange for SAHC, which is produced from SAM in the mitochondrial matrix and plastidial stroma by methyltransferase activities. By fulfilling this task, SAMC1 allows regeneration of the methyl group of SAM in the cytosol, provides the organelles with methyl groups, and regulates methyltransferase activities in the organelles by removing the potent competitive inhibitor SAHC.
In addition, SAM is also a substrate for many other enzyme-catalyzed reactions in plant organelles. For example, in plants, lipoic acid synthase is localized both in chloroplasts and mitochondria (Yasuno and Wada, 2002), biotin synthase only in mitochondria (Picciocchi et al., 2001, 2003), and Thr synthase only in plastids (Wallsgrove et al., 1983; Curien et al., 1996; Laber et al., 1999). Because all of these enzymes require SAM as an essential cofactor, a further important role of SAMC1 is probably to catalyze the uniport of SAM into mitochondria and possibly also into chloroplasts. Whereas further experimentation is required to confirm the putative plastidial localization, it should be noted that the other metabolites produced from SAM within the mitochondria or plastids by the action of biotin synthetase, lipoate synthetase, and Thr synthase (i.e. 5′-deoxyadenosine and Met) cannot be exported by SAMC1 because they are not substrates of this carrier, implying that the presence of further transporters is responsible for their removal. Identification of these proteins should be the focus of further studies aimed at understanding the cellular network of SAM metabolism.
By activating Thr synthase, SAM modulates carbon flux partition between members of the Asp family of amino acids and regulates its own biosynthesis (Hesse and Hoefgen, 2003). Because de novo Met synthesis is exclusively chloroplastic, whereas SAM synthetase is cytosolic, SAMC1 may play a key role in the interaction between Met synthesis and SAM metabolism. Transgenic plants with reduced SAMC1 activity might enable us to determine the physiological influence of SAMC1 on de novo Met biosynthesis. First investigations of SAMC1 Arabidopsis knockout plants revealed that homozygous seeds of these knockout lines are unable to germinate (data not shown). Interestingly, it has been shown that de novo synthesis of Met plays an important role in seed germination and seedling growth (Gallardo et al., 2002). The strong up-regulation of Met synthase and SAM synthetase observed during seed germination and seedling establishment by Gallardo et al. (2002) parallels the high levels of SAMC1 expression in the early stages of plant development. SAM metabolism could affect plant growth and development in several ways. For example, SAM serves as a substrate for SAM decarboxylase, the key regulatory enzyme responsible for the production of the polyamines spermidine and spermine, which are believed to be involved in plant development (Alcazar et al., 2005; Cona et al., 2006) and stress response (Panicot et al., 2002; Perez-Amador et al., 2002; Zapata et al., 2004). Such studies would be expected to complement the findings of Moffatt and coworkers, who demonstrated that a reduction in adenosine kinase activity by antisense inhibition resulted in decreased cellular levels of SAM and developmental abnormalities (Moffatt et al., 2002). In addition, they would likely provide more direct proof as to whether these effects are directly ascribable to altered SAM metabolism. The importance of SAM in ethylene biosynthesis and the observation that proteins involved in SAM metabolism are up-regulated on wounding and herbivory (this study; Arimura et al., 2002) suggest that it will also be interesting to analyze how the plant prioritizes the gene regulatory and biosynthetic functions of SAM in response to both biotic and abiotic stresses.
MATERIALS AND METHODS
Materials
[3H]SAM was purchased from NEN Life Science Products. Cardiolipin and sarkosyl (N-lauroylsarcosine) were supplied by Sigma. Egg-yolk phospholipids (egg lecithin) were obtained from Fluka and Amberlite XAD-2 from Supelco. All other reagents were analytical grade.
Sequence Search and Analysis
The genome of Arabidopsis (Arabidopsis thaliana) was screened with the sequences of the yeast (Saccharomyces cerevisiae) Sam5p (Marobbio et al., 2003) and the human SAMC (Agrimi et al., 2004) using BLASTP and TBLASTN. The amino acid sequences were aligned with the ClustalW multiple sequence alignment program (version 1.8; http://www.clustalw.genome.ad.jp). The SAMC1 (accession no. AM260490) and SAMC2 (accession no. AM260491) sequences, showing the lowest e-values, were selected for further study.
RNA Isolation and Northern-Blot Analysis
Total RNA was isolated from frozen organs using TRIzol (Gibco BRL) according to the manufacturer's instructions. RNA concentration was measured and its integrity was checked on a 1.5% agarose gel (w/v). Hybridization was performed as described in Sambrook et al. (1989), using specific probes of 237 bp (from −110 to +127 nucleotides of the SAMC1 cDNA) and 176 bp (from +1 to +176 nucleotides of the SAMC2 cDNA) for SAMC1 and SAMC2, respectively. The probes were labeled with a [32P]dCTP by random priming, using the random prime labeling system (Amersham Bioscience). RNA loading was checked with ethidium bromide. Membranes were autoradiographed at −70°C for several days with an intensifying screen.
Expression Analysis by Real-Time RT-PCR
Total RNAs from different organs were reverse transcribed using the GeneAmp RNA PCR core kit (Applied Biosystems) with random hexamers as primers. For real-time PCRs, primers based on the cDNA sequences of SAMC1 and SAMC2 were designed with Primer Express (Applied Biosystems). The forward and reverse primers corresponded to nucleotides 318 to 340 and 398 to 418 for SAMC1, nucleotides 386 to 406 and 406 to 436 for SAMC2, and nucleotides 618 to 639 and 698 to 718 for EF1α. Real-time PCR was performed in an optical 96-well plate using the automated ABI Prism 7000 sequence detection system (Applied Biosystems). Fifty microliters of reaction volume contained 2 μL of template (reverse transcribed first-strand cDNA), 25 μL SYBR Green PCR master mix (Applied Biosystems), and 300 nm each primer. The specificity of PCR amplification was checked with the heat dissociation protocol following the final cycle of PCR. Separation of real-time PCR products on 4% (w/v) agarose gel revealed single bands of the expected size whose identity was confirmed by direct sequencing. To correct for differences in the amount of starting first-strand cDNAs, the Arabidopsis EF1α gene was amplified in parallel as a reference gene. The relative quantification of SAMC1 and SAMC2 in various organs was performed according to the comparative method (2−ΔΔCt; Bustin, 2000; Fiermonte et al., 2002; user bulletin no. 2 [P/N 4303859]; Applied Biosystems), with the stem ΔCt for SAMC2 as calibrator. 2−ΔΔCt = 2−(ΔCt sample−ΔCt calibrator), where ΔCt sample is Ct sample − Ct reference gene and Ct is the threshold cycle (i.e. the PCR cycle number at which emitted fluorescence exceeds 10 times the sd of baseline emissions).
Promoter-GUS Fusion Experiments
For promoter-GUS fusion experiments, the promoter regions of SAMC1 (from −1,146 bp to +6 bp) and SAMC2 (from −1,396 bp to +12 bp) were amplified by PCR from Arabidopsis Col-0 genomic DNA using the following primers: forward, 5′-CACCGGGAGATAATTGAAAGC-3′ and reverse, 5′-AGCCATGAGAAACGCCTCTGACCTAAT-3′ for SAMC1; and forward, 5′-CACCTGAGAGAAAAAGAAAGAAGAAAAAGAAGAGA-3′ and reverse, 5′-GTCACTATCCATCTAAAACCATT-3′ for SAMC2. The PCR products were first cloned into the shuttle vector pENTR/d-TOPO (Invitrogen) and then transferred into the binary Gateway vector pKGWFS7 in frame with the GUS gene (Karimi et al., 2002). The resulting constructs were introduced into Arabidopsis Col-0 plants by Agrobacterium-mediated transformation according to the floral-dip method (Clough and Bent, 1998). To select transgenic plants, seeds were germinated on Murashige and Skoog plates (Murashige and Skoog, 1962) containing 1% Suc and supplemented with kanamycin in a growth chamber (250 μmol photons m−2 s−1; 22°C) under a 16-h light/8-h dark regime before transfer to soil in a climate-controlled chamber under the same photoperiod. Samples for GUS activity analysis were harvested 8, 11, 13, or 38 d after germination and stained overnight according to standard protocols (Clough and Bent, 1998).
Construction of Expression Plasmids
PCR primers for SAMC1 (forward, 5′-GGATCCATGGCTCCTCTTACTCTCTCCGT-3′; reverse, 5′-AAGCTTTTATTCTTCTTTGGTTTCTTTAACCGT-3′) and for SAMC2 (forward, 5′-CATAAGCTTATGGATAGTGACATTGTTTCCAGTAGCATA-3′; reverse, 5′-GAATTCTTAAGCATTGTGACTCTTTTGGCTTCT-3′) carrying BamHI and HindIII and HindIII and EcoRI sites, respectively, were used to amplify the predicted open reading frames from an Arabidopsis cDNA library (Minet et al., 1992). The resulting products were cloned into the pRUN vector (derived from pKN172; Fiermonte et al., 1993) for expression in Escherichia coli.
For subcellular localization of SAMC1 and mMDH, the SAMC1-GFP and the mMDH-GFP fusion constructs were prepared. The coding sequences of SAMC1 and of mMDH without the terminal codon were amplified by PCR using the following primers: forward, 5′-CACCATGGCTCCTCTTACTCTCTCGT-3′ and reverse, 5′-TTCTTCTTTGGTTTCTTTAACCGTATT-3′ for SAMC1; and forward, 5′-CACCATGAGGACCTCCATGTTG-3′ and reverse, 5′-GTTTTCTTTGGCAAACTTGATTC-3′ for tomato (Lycopersicon esculentum) mMDH (encoded by AY725474). The resulting products were cloned into the entry vector pENTR/d-TOPO (Invitrogen) using the Gateway recombination system. Subsequently, the SAMC1 or the mMDH coding sequence was recombined into the destination vector pK7FWG2 encoding a C-terminal enhanced GFP under the cauliflower mosaic virus 35S promoter (Karimi et al., 2002; CLONTECH). The inserts were sequenced to confirm identity prior to plant transformation.
In Vivo Targeting of GFP Fusion Constructs
Protoplasts were prepared from Arabidopsis Col-0 plants (Jin et al., 2001) grown as described above in the absence of kanamycin. The pK7FWG2 vector containing the coding sequence of GFP, SAMC1-GFP, or mMDH-GFP was introduced into Arabidopsis protoplasts by the polyethylene glycol-mediated transformation method (Kang et al., 1998). After 36 to 48 h of incubation in the dark at 22°C, protoplasts were imaged using a laser-scanning confocal microscope (Leica DM IRBE microscope; TCS SPII confocal scanner). In some experiments, the transformed protoplasts were incubated in the presence of 400 nm MitoTracker Red CMXRos (Molecular Probes) at 25°C for 30 min and then extensively washed before imaging following the protocol of Studart-Guimaraes et al. (2005).
Bacterial Expression and Purification of SAMC1 and SAMC2
The overproduction of SAMC1 and SAMC2 as inclusion bodies in the cytosol of E. coli BL21(DE3) was accomplished as described (Fiermonte et al., 1993). Control cultures with empty vector were processed in parallel. Inclusion bodies were purified on a Suc density gradient (Fiermonte et al., 1993), washed at 4°C with Tris-EDTA buffer (10 mm Tris/HCl, 1 mm EDTA, pH 7.0), then twice with a buffer containing Triton X-114 (3%, w/v), 1 mm EDTA, and 10 mm PIPES/NaOH, pH 7.0, and once again with Tris-EDTA buffer (Palmieri et al., 2001a; Picault et al., 2002; Hoyos et al., 2003). The SAMC1 and SAMC2 proteins were solubilized in 1.6% sarkosyl (w/v). Small residues were removed by centrifugation (258,000g for 20 min at 4°C).
Reconstitution of Recombinant SAMC1 and SAMC2 into Liposomes
The recombinant proteins were diluted 7-fold with buffer containing 0.6% Triton X-114, 0.2 mm EDTA, 10 mm PIPES, pH 7.0, and then reconstituted by cyclic removal of detergent (Palmieri et al., 1995). The reconstitution mixture consisted of protein solution (0.12 mg), 10% Triton X-114, 10% phospholipids as sonicated liposomes, 10 mm SAM (except where otherwise indicated), cardiolipin (0.7 mg), 20 mm PIPES, pH 7.0, and water (final volume 700 μL). The mixture was recycled 13 times through an Amberlite column (3.2 cm × 0.5 cm) preequilibrated with buffer containing 10 mm PIPES, pH 7.0, and substrate at the same concentration as in the reconstitution mixture. All operations were performed at 4°C, except the passages through Amberlite, which were carried out at room temperature.
Transport Measurements
External substrate was removed from the proteoliposomes on Sephadex G-75 columns preequilibrated with a buffer containing 50 mm NaCl and 10 mm PIPES, pH 7.0 (buffer A). Transport at 25°C was started by adding [3H]SAM to substrate-loaded proteoliposomes (exchange) or to empty proteoliposomes (uniport) and terminated, after the desired time, by the addition of 40 mm pyridoxal-5′-P and 16 mm bathophenantroline (the inhibitor stop method; Palmieri et al., 1995). In controls, inhibitors were added together with the labeled substrate. Finally, the external radioactivity was removed on Sephadex G-75 and the radioactivity in the liposomes was measured (Palmieri et al., 1995). The experimental values were corrected by subtracting control values. The initial transport rate was calculated from the radioactivity taken up by proteoliposomes in the initial linear range of substrate uptake (Palmieri et al., 1995). For efflux measurements, proteoliposomes containing 1 mm SAM were labeled with 5 μm [3H]SAM by carrier-mediated exchange equilibration (Palmieri et al., 1995). After 60 min, external radioactivity was removed by passing the proteoliposomes through Sephadex G-75. Efflux was started by adding unlabeled external substrate or buffer A alone and terminated by adding the inhibitors indicated above.
Other Methods
Proteins were separated by SDS-PAGE and stained with Coomassie Blue. The N termini were sequenced and the amount of purified proteins was estimated by laser densitometry of stained samples using carbonic anhydrase as a protein standard (Fiermonte et al., 1998). The amount of protein incorporated into liposomes was measured as described previously (Fiermonte et al., 1998). In all cases, it was about 25% of the protein added to the reconstitution mixture.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AM260490 (SAMC1) and AM260491 (SAMC2).
This work was supported by grants from Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR), Consiglio Nazionale delle Ricerche-MIUR project “Functional genomics,” the Centro di Eccellenza Geni in campo Biosanitario e Agroalimentare, and the Consorzio interuniversitario per le biotecnologie.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ferdinando Palmieri (fpalm@farmbiol.uniba.it).
References
- Agrimi G, Di Noia MA, Marobbio CMT, Fiermonte G, Lasorsa FM, Palmieri F (2004) Identification of the human mitochondrial S-adenosylmethionine transporter: bacterial expression, reconstitution, functional characterization and tissue distribution. Biochem J 379: 183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcazar R, Garcia-Martinez JL, Cuevas JC, Tiburcio AF, Altabella T (2005) Overexpression of ADC2 in Arabidopsis induces dwarfism and late-flowering through GA deficiency. Plant J 43: 425–436 [DOI] [PubMed] [Google Scholar]
- Arimura G, Ozawa R, Nishioka T, Boland W, Koch T, Kühnemann F, Takabayashi J (2002) Herbivore-induced volatiles induce the emission of ethylene in neighboring lima bean plants. Plant J 29: 87–98 [DOI] [PubMed] [Google Scholar]
- Bedhomme M, Hoffmann M, McCarthy EA, Gambonnet B, Moran RG, Rébbeillé F, Ravanel S (2005) Folate metabolism in plants: an Arabidopsis homolog of the mammalian mitochondrial folate transporter mediates folate import into chloroplasts. J Biol Chem 280: 34823–34831 [DOI] [PubMed] [Google Scholar]
- Black MT, Meyer D, Widger WR, Cramer WA (1987) Light-regulated methylation of chloroplast proteins. J Biol Chem 262: 9803–9807 [PubMed] [Google Scholar]
- Block MA, Tewari AK, Albrieux C, Marechal E, Joyard J (2002) The plant S-adenosyl-L-methionine: Mg-protoporphyrin IX methyltransferase is located in both envelope and thylakoid chloroplast membranes. Eur J Biochem 269: 240–248 [DOI] [PubMed] [Google Scholar]
- Bustin SA (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25: 169–193 [DOI] [PubMed] [Google Scholar]
- Cantoni GL (1975) Biological methylation: selected aspects. Annu Rev Biochem 44: 435–451 [DOI] [PubMed] [Google Scholar]
- Cheng Z, Sattler S, Maeda H, Sakuragi Y, Bryant DA, DellaPenna D (2003) Highly divergent methyltransferases catalyze a conserved reaction in tocopherol and plastoquinone synthesis in cyanobacteria and photosynthetic eukaryotes. Plant Cell 15: 2343–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AC, Lyznik A, Mohammed S, Elowsky CG, Elo A, Yule R, Mackenzie SA (2005) Dual-domain, dual-targeting organellar protein presequences in Arabidopsis can use non-AUG start codons. Plant Cell 17: 2805–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cona A, Rea G, Angelini R, Federico R, Tavladoraki P (2006) Functions of amine oxidases in plant development and defence. Trends Plant Sci 11: 80–88 [DOI] [PubMed] [Google Scholar]
- Curien G, Dumas R, Ravanel S, Douce R (1996) Characterization of an Arabidopsis thaliana cDNA encoding an S-adenosylmethionine-sensitive threonine synthase: threonine synthase from higher plants. FEBS Lett 390: 85–90 [DOI] [PubMed] [Google Scholar]
- d'Harlingue A, Camara B (1985) Plastid enzymes of terpenoid biosynthesis: purification and characterization of γ-tocopherol methyltransferase from Capsicum chromoplasts. J Biol Chem 260: 15200–15203 [PubMed] [Google Scholar]
- Duchene AM, Giritch A, Hoffmann B, Cognat V, Lancelin D, Peeters NM, Zaepfel M, Marechàl-Drouard L, Small ID (2005) Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc Natl Acad Sci USA 102: 16484–16489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Carrari F, Sweetlove LJ (2004) Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr Opin Plant Biol 7: 254–261 [DOI] [PubMed] [Google Scholar]
- Ferro M, Salvi D, Riviere-Rolland H, Vermat T, Seigneurin-Berny D, Grunwald D, Garin J, Joyard J, Rolland N (2002) Integral membrane proteins of the chloroplast envelope: identification and subcellular localization of new transporters. Proc Natl Acad Sci USA 99: 11487–11492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiermonte G, De Leonardis F, Todisco S, Palmieri L, Lasorsa FM, Palmieri F (2004) Identification of the mitochondrial ATP-Mg/Pi transporter: bacterial expression, reconstitution, functional characterization, and tissue distribution. J Biol Chem 279: 30722–30730 [DOI] [PubMed] [Google Scholar]
- Fiermonte G, Dolce V, David L, Santorelli FM, Dionisi-Vici C, Palmieri F, Walker JE (2003) The mitochondrial ornithine transporter: bacterial expression, reconstitution, functional characterization, and tissue distribution of two human isoforms. J Biol Chem 278: 32778–32783 [DOI] [PubMed] [Google Scholar]
- Fiermonte G, Dolce V, Palmieri F (1998) Expression in Escherichia coli, functional characterization and tissue distribution of isoforms A and B of the phosphate carrier from bovine mitochondria. J Biol Chem 273: 22782–22787 [DOI] [PubMed] [Google Scholar]
- Fiermonte G, Palmieri L, Todisco S, Agrimi G, Palmieri F, Walker JE (2002) Identification of the mitochondrial glutamate transporter: bacterial expression, reconstitution, functional characterization, and tissue distribution of two human isoforms. J Biol Chem 277: 19289–19294 [DOI] [PubMed] [Google Scholar]
- Fiermonte G, Walker JE, Palmieri F (1993) Abundant bacterial expression and reconstitution of an intrinsic membrane transport protein from bovine mitochondria. Biochem J 294: 293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore C, Trezeguet V, Le Saux A, Roux P, Schwimmer C, Dianoux AC, Noel F, Lauquin GJ, Brandolin G, Vignais PV (1998) The mitochondrial ADP/ATP carrier: structural, physiological and pathological aspects. Biochimie 80: 137–150 [DOI] [PubMed] [Google Scholar]
- Fontecave M, Atta M, Mulliez E (2004) S-adenosylmethionine: nothing goes to waste. Trends Biochem Sci 29: 243–249 [DOI] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SP, Puype M, Demol H, Vandekerckhove J, Job D (2002) Importance of methionine biosynthesis for Arabidopsis seed germination and seedling growth. Physiol Plant 116: 238–247 [DOI] [PubMed] [Google Scholar]
- Grimm R, Grimm M, Eckerskorn C, Pohlmeyer K, Rohl T, Soll J (1997) Postimport methylation of the small subunit of ribulose-1,5-biphosphate carboxylase in chloroplasts. FEBS Lett 408: 350–354 [DOI] [PubMed] [Google Scholar]
- Halestrap AP (1975) The mitochondrial pyruvate carrier: kinetics and specificity for substrates and inhibitors. Biochem J 148: 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AD, Gage DA, Shachar-Hill Y (2000) Plant one-carbon metabolism and its engineering. Trends Plant Sci 5: 206–213 [DOI] [PubMed] [Google Scholar]
- Hanson AD, Roje S (2001) One-carbon metabolism in higher plants. Annu Rev Plant Physiol Plant Mol Biol 52: 119–137 [DOI] [PubMed] [Google Scholar]
- Hesse H, Hoefgen R (2003) Molecular aspects of methionine biosynthesis. Trends Plant Sci 8: 259–262 [DOI] [PubMed] [Google Scholar]
- Hoyos ME, Palmieri L, Wertin T, Arrigoni R, Polacco JC, Palmieri F (2003) Identification of a mitochondrial transporter for basic amino acids in Arabidopsis thaliana by functional reconstitution into liposomes and complementation in yeast. Plant J 33: 1027–1035 [DOI] [PubMed] [Google Scholar]
- Jin JB, Kim YA, Kim SJ, Lee SH, Kim DH, Cheong GW, Hwang I (2001) A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13: 1511–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Sato N (2005) Unique translation initiation at the second AUG codon determines mitochondrial localization of the phage-type RNA polymerases in the moss Physcomitrella patens. Plant Physiol 138: 369–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SG, Jin JB, Piao HL, Pih KT, Jang HJ, Lim JH, Hwang I (1998) Molecular cloning of an Arabidopsis cDNA encoding a dynamin-like protein that is localized to plastids. Plant Mol Biol 38: 437–447 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Karniely S, Pines O (2005) Single translation—dual destination: mechanisms of dual protein targeting in eukaryotes. EMBO Rep 6: 420–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg M (1989) Molecular aspects of the adenine nucleotide carrier from mitochondria. Arch Biochem Biophys 270: 1–14 [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Ngernprasirtsiri J, Akazawa T (1990) Transcriptional regulation and DNA methylation in plastids during transitional conversion of chloroplasts to chromoplasts. EMBO J 9: 307–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laber B, Maurer W, Hanke C, Grafe S, Ehlert S, Messerschmidt A, Clausen T (1999) Characterization of recombinant Arabidopsis thaliana threonine synthase. Eur J Biochem 263: 212–221 [DOI] [PubMed] [Google Scholar]
- Leroch M, Kirchberger S, Haferkamp I, Wahl M, Neuhaus HE, Tjaden J (2005) Identification and characterization of a novel plastidic adenine nucleotide uniporter from Solanum tuberosum. J Biol Chem 280: 17992–18000 [DOI] [PubMed] [Google Scholar]
- Mackenzie SA (2005) Plant organellar protein targeting: a traffic plan still under construction. Trends Cell Biol 15: 548–554 [DOI] [PubMed] [Google Scholar]
- Marobbio CM, Agrimi G, Lasorsa FM, Palmieri F (2003) Identification and functional reconstitution of yeast mitochondrial carrier of S-adenosylmethionine. EMBO J 22: 5975–5982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Heazlewood JL (2003) Genomic and proteomic analysis of mitochondrial carrier proteins in Arabidopsis. Plant Physiol 131: 443–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JR, Busby RW, Jordan SW, Cheek J, Henshaw TF, Ashley GW, Broderick JB, Cronan JE, Marletta MA (2000) Escherichia coli LipA is a lipoyl synthase: in vitro biosynthesis of lipoylated pyruvate dehydrogenase complex from octanoyl-acyl carrier protein. Biochemistry 39: 15166–15178 [DOI] [PubMed] [Google Scholar]
- Minet M, Dufour ME, Lacroute F (1992) Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J 2: 417–422 [DOI] [PubMed] [Google Scholar]
- Moffatt BA, Stevens YY, Allen MS, Snider JD, Pereira LA, Todorova MI, Summers PS, Weretilnyk EA, Martin-McCaffrey L, Wagner C (2002) Adenosine kinase deficiency is associated with developmental abnormalities and reduced transmethylation. Plant Physiol 128: 812–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montasser Kouhsari S, Keith G, Weil GH (1978) Methylation of yeast tRNAPhe by enzymes from cytoplasm, chloroplasts and mitochondria of Phaseolus vulgaris. Biochim Biophys Acta 521: 576–583 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Plant Physiol 15: 473–497 [Google Scholar]
- Nunes-Nesi A, Carrari F, Lytovchenko A, Smith AM, Loureiro ME, Ratcliffe RG, Sweetlove LJ, Fernie AR (2005) Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol 137: 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri F (2004) The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Arch 447: 689–709 [DOI] [PubMed] [Google Scholar]
- Palmieri F, Agrimi G, Blanco E, Castegna A, Di Noia MA, Iacobazzi V, Lasorsa FM, Marobbio CMT, Palmieri L, Scarcia P, et al (2006. a) Identification of mitochondrial carriers in Saccharomyces cerevisiae by transport assay of reconstituted recombinant proteins. Biochim Biophys Acta doi/10.1016/j.bbabio.2006.05.023 [DOI] [PubMed]
- Palmieri F, Indiveri C, Bisaccia F, Iacobazzi V (1995) Mitochondrial metabolite carrier proteins: purification, reconstitution, and transport studies. Methods Enzymol 260: 349–369 [DOI] [PubMed] [Google Scholar]
- Palmieri L, Pardo B, Lasorsa FM, Del Arco A, Kobayashi K, Iijima M, Runswick MJ, Walker JE, Saheki T, Satrustegui J, et al (2001. a) Citrin and Aralar1 are Ca2+-stimulated aspartate/glutamate transporters in mitochondria. EMBO J 20: 5060–5069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri L, Rottensteiner H, Girzalsky W, Scarcia P, Palmieri F, Erdmann R (2001. b) Identification and functional reconstitution of the yeast peroxisomal adenine nucleotide transporter. EMBO J 20: 5049–5059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri L, Todd CD, Arrigoni R, Hoyos ME, Santoro A, Polacco CJ, Palmieri F (2006. b) Arabidopsis mitochondria have two basic amino acid transporters with partially overlapping specificities and differential expression in seedling development. Biochim Biophys Acta doi/10.1016/j.bbabio.2006.03.025 [DOI] [PubMed]
- Panicot M, Minguet EG, Ferrando A, Alcazar R, Blazquez MA, Carbonell J, Altabella T, Koncz C, Tiburcio AF (2002) A polyamine metabolon involving aminopropyl transferase complexes in Arabidopsis. Plant Cell 14: 2539–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters N, Small I (2001) Dual targeting to mitochondria and chloroplasts. Biochim Biophys Acta 1541: 54–63 [DOI] [PubMed] [Google Scholar]
- Perez-Amador MA, Leon J, Green PJ, Carbonell J (2002) Induction of the arginine decarboxylase ADC2 gene provides evidence for the involvement of polyamines in the wound response in Arabidopsis. Plant Physiol 130: 1454–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picault N, Hodges M, Palmieri L, Palmieri F (2004) The growing family of mitochondrial carrier in Arabidopsis. Trends Plant Sci 9: 138–146 [DOI] [PubMed] [Google Scholar]
- Picault N, Palmieri L, Pisano I, Hodges M, Palmieri F (2002) Identification of a novel transporter for dicarboxylates and tricarboxylates in plant mitochondria: bacterial expression, reconstitution, functional characterization and tissue distribution. J Biol Chem 277: 24204–24211 [DOI] [PubMed] [Google Scholar]
- Picciocchi A, Douce R, Alban C (2001) Biochemical characterization of the Arabidopsis biotin synthase reaction: the importance of mitochondria in biotin synthesis. Plant Physiol 127: 1224–1233 [PMC free article] [PubMed] [Google Scholar]
- Picciocchi A, Douce R, Alban C (2003) The plant biotin synthase reaction: identification and characterization of essential mitochondrial accessory protein components. J Biol Chem 278: 24966–24975 [DOI] [PubMed] [Google Scholar]
- Ravanel S, Block MA, Rippert P, Jabrin S, Curien G, Rébeillé F, Douce R (2004) Methionine metabolism in plants: chloroplasts are autonomous for de novo methionine synthesis and can import S-adenosylmethionine from the cytosol. J Biol Chem 279: 22548–22557 [DOI] [PubMed] [Google Scholar]
- Ravanel S, Gakiere B, Job D, Douce R (1998) The specific features of methionine biosynthesis and metabolism in plants. Proc Natl Acad Sci USA 95: 7805–7812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev-Rudzki N, Karniely S, Ben-Haim NN, Pines O (2005) Yeast aconitase in two location and two metabolic pathways: seeing small amount is believing. Mol Biol Cell 16: 4163–4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schröder G, Eichel J, Breinig S, Schroder J (1997) Three differentially expressed S-adenosylmethionine synthetases from Catharanthus roseus: molecular and functional characterization. Plant Mol Biol 33: 211–222 [DOI] [PubMed] [Google Scholar]
- Studart-Guimeraes C, Gibon Y, Frankel N, Wood CC, Zanor MI, Fernie AR, Carrari F (2005) Identification and characterisation of the alfa and beta subunits of the succinyl CoA ligase of tomato. Plant Mol Biol 59: 781–791 [DOI] [PubMed] [Google Scholar]
- Wallsgrove RM, Lea PJ, Miflin BJ (1983) Intracellular localization of aspartate kinase and the enzyme of threonine and methionine biosynthesis in green leaves. Plant Physiol 71: 780–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber AP, Schwacke R, Flugge UI (2005) Solute transporters of the plastid envelope membrane. Annu Rev Plant Biol 56: 133–164 [DOI] [PubMed] [Google Scholar]
- Weretilnyk EA, Alexander KJ, Drebenstedt M, Snider JD, Summers PS, Moffatt BA (2001) Maintaining methylation activities during salt stress: the involvement of adenosine kinase. Plant Physiol 125: 856–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuno R, Wada H (2002) The biosynthetic pathway for lipoic acid is present in plastids and mitochondria in Arabidopsis thaliana. FEBS Lett 517: 110–114 [DOI] [PubMed] [Google Scholar]
- Ying Z, Mulligan RM, Janney N, Houtz RL (1999) Rubisco small and large subunit N-methyltransferases: bi- and mono-functional methyltransferases that methylate the small and large subunits of Rubisco. J Biol Chem 274: 36750–36756 [DOI] [PubMed] [Google Scholar]
- Zapata PJ, Serrano M, Pretel MT, Amoros A, Botella MA (2004) Polyamines and ethylene changes during germination of different plant species under salinity. Plant Sci 167: 781–788 [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]